Abstract
3-methylcrotonyl-CoA carboxylase (MCC), a member of the biotin-dependent carboxylase superfamily, is essential for the metabolism of leucine, and deficient mutations in this enzyme are linked to methylcrotonylglycinuria (MCG) and other serious diseases in humans 1–8. MCC has strong sequence conservation with propionyl-CoA carboxylase (PCC), and their holoenzymes are both 750 kD α6β6 dodecamers. Therefore the architecture of the MCC holoenzyme is expected to be highly similar to that of PCC 9. Here we report the crystal structures of the Pseudomonas aeruginosa MCC (PaMCC) holoenzyme, alone and in complex with coenzyme A. Surprisingly, the structures show that the architecture and overall shape of PaMCC are strikingly different when compared to PCC. The α subunits display trimeric association in the PaMCC holoenzyme while they have no contacts with each other in PCC. Moreover, the positions of the two domains in the β subunit in PaMCC are swapped relative to those in PCC. The structural information establishes a foundation for understanding the disease-causing mutations of MCC and provides new insights into the catalytic mechanism and evolution of biotin-dependent carboxylases. The large structural differences between MCC and PCC also have general implications for the relationship between sequence conservation and structural similarity.
Keywords: fatty acid metabolism, amino acid metabolism, propionic acidemia, 3-methylcrotonylglycinuria, protein complex, biotin-dependent carboxylase, acetyl-CoA carboxylase, propionyl-CoA carboxylase
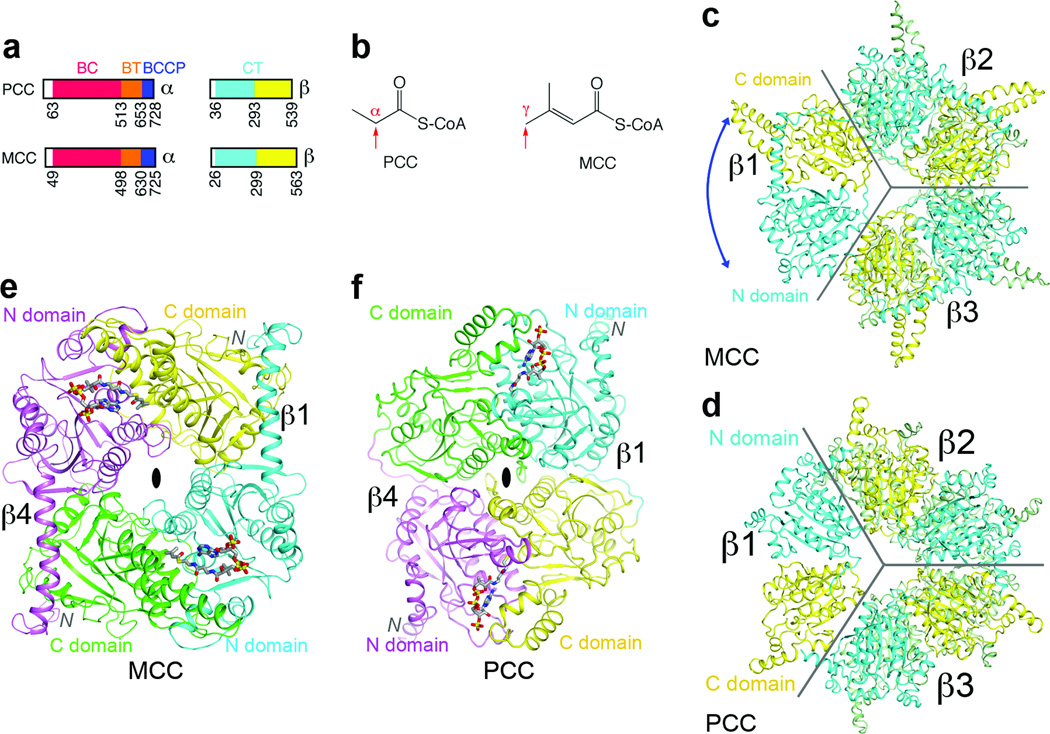
The α and β subunits of human MCC have 42% and 34% sequence identity with those of human PCC (Supplementary Figs. 1, 2). The α subunit contains the biotin carboxylase (BC) and biotin carboxyl carrier protein (BCCP) domains (Fig. 1a), and a domain that mediates BC-CT interactions (BT domain) 9. BC catalyzes the MgATP-dependent carboxylation of biotin, and then a carboxyltransferase (CT) activity, supplied by the β subunit, catalyzes the transfer of the carboxyl group to the acceptor. The β subunit contains two domains (Fig. 1a), N and C domains, with the same backbone fold, and its active site is located at the interface of a dimer.
Figure 1. The domains of MCCβ are swapped compared to PCCβ.
(a). Domain organization of human MCC and PCC. (b). Distinct carboxylation targets of MCC and PCC, indicated by the red arrow. (c). Crystal structure of the β6 hexamer of PaMCC. The subunit beneath β1 is omitted for clarity, and the other two subunits in the bottom layer are colored in green. The blue arrow indicates the swapping of the positions of the N and C domains relative to PCCβ. Gray lines mark the boundaries of the subunits. (d). Structure of Roseobacter denitrificans PCCβ 9. (e). Structure of the β2 dimer of PaMCC. The N and C domains of the subunit in the bottom layer (β4) are colored in magenta and green, respectively. (f). Structure of the β2 dimer of PCC. All the structure figures were produced with PyMOL (www.pymol.org) unless stated otherwise.
Our initial interest in MCC stemmed from its distinct site of carboxylation in the substrate (Fig. 1b). To understand the molecular basis for this activity, we produced crystals of the MCCβ hexamer from Pseudomonas aeruginosa (PaMCCβ) that diffracted to 1.5 Å resolution (Supplementary Table 1). This bacterial enzyme is highly homologous to human MCC (HsMCC), with sequence identities of 47% and 65% for the α and β subunits, respectively. To facilitate comparisons between these highly conserved enzymes, we have numbered the residues in PaMCC according to their equivalents in HsMCC. In Pseudomonas organisms, MCC is also involved in terpenoids metabolism 10,11.
We solved the structure of PaMCCβ using PCCβ as the model 9. However, subsequent crystallographic analysis revealed that the positions of the N and C domains in PaMCCβ (Fig. 1c) are swapped relative to those in PCCβ (Fig. 1d), even though the overall shapes of the two β6 hexamers are similar. This distinct domain organization of PaMCCβ is primarily due to a different connectivity between its N and C domains, rather than a swap of these two domains in the primary sequence (Supplementary Fig. 3, Supplementary text). This also leads to a large difference in the organization of the PaMCCβ dimer compared to PCCβ (Figs. 1e, 1f). The closest structural homolog of PaMCCβ is the α subunit of glutaconyl-CoA decarboxylase (GCDα) 12,13 (Supplementary Fig. 4). However, the sequence conservation between PaMCCβ and GCDα (27% identity) is actually lower than that between PaMCCβ and PCCβ (34% identity). The CoA binding sites, located in the N domain, are swapped between MCCβ and PCCβ as well (Figs. 1e, 1f). This may be related to the activity of MCC on the γ carbon of the substrate (Fig. 1b), as the activity of GCDα is also on the γ carbon (Supplementary text).
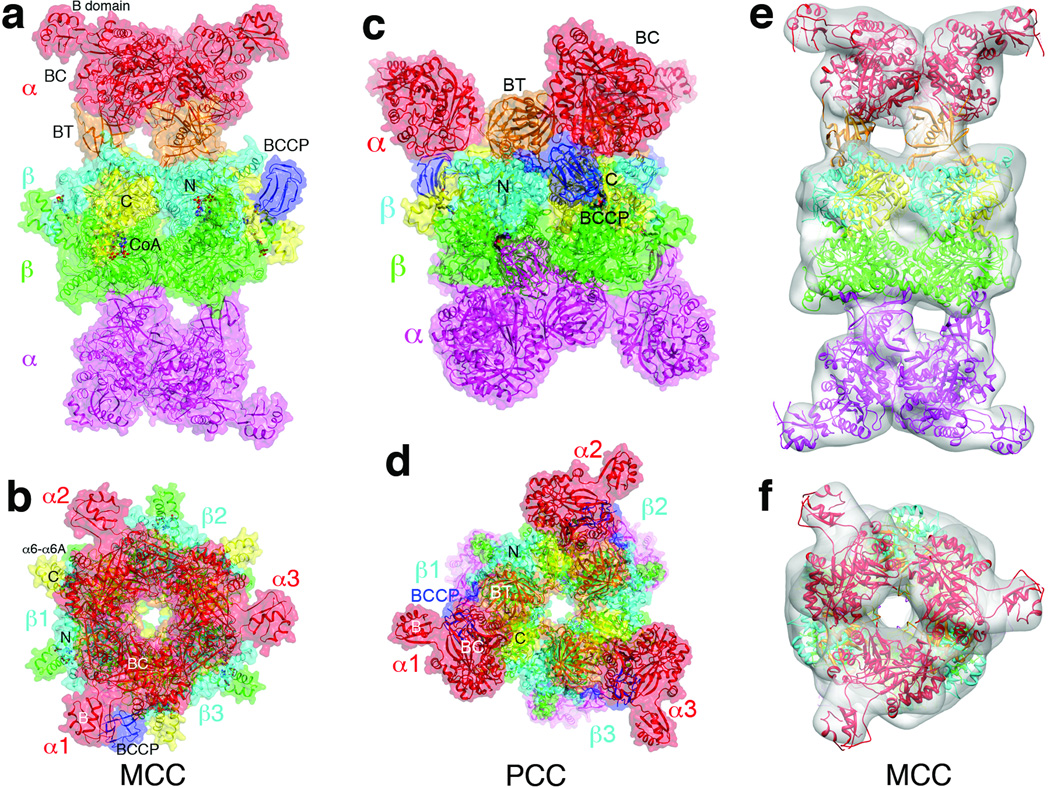
Most importantly, the change in domain organization of MCCβ suggests that the overall architecture of the MCC holoenzyme may be different as well (Supplementary text). Therefore, we next determined the structures of PaMCC free enzyme and CoA complex at 2.9 and 3.5 Å resolution, respectively (Fig. 2a, Supplementary Table 1). Like PCC, the holoenzyme of PaMCC contains a central β6 cylindrical core, with 3 α subunits at each end (Fig. 2b). The overall structures of the free enzyme and CoA complex of PaMCC are similar, although there are also recognizable differences (Supplementary Fig. 5, Supplementary text).
Figure 2. The MCC holoenzyme has a strikingly different architecture compared to PCC.
(a). Structure of the CoA complex of PaMCC holoenzyme, side view. Domains in the α and β subunits in the top half of the structure are colored as in Fig. 1a. The α and β subunits in the bottom half are colored in magenta and green, respectively. The molecular surface is shown in a semi-transparent rendering. (b). Structure of the PaMCC holoenzyme, top view. (c). Structure of the PCC holoenzyme 9, side view. (d). Structure of the PCC holoenzyme, top view. (e). EM reconstruction of PaMCC at 12 Å resolution, side view. The crystal structure of the PaMCC free enzyme can be readily fit into the EM density. (f). EM reconstruction of PaMCC, top view. Panels e and f were produced with Chimera 22.
Strikingly, the positions of the α subunits in PaMCC, especially their BC domains, are entirely different from those in PCC (Fig. 2c). Rather than being splayed far apart from each other like in PCC (Fig. 2d), the three BC domains on either end of MCC are in direct contact with each other (Fig. 2b), burying 640 Å2 of the surface area of each BC domain. Therefore, the BC domain shows trimeric association in the MCC holoenzyme, although this trimer is probably unstable on its own due to the relatively small buried surface area. Overall, the MCC holoenzyme has the shape of a cylinder, approximately 100 Å in diameter and 200 Å tall, and this shape is remarkably different from that of PCC (Supplementary Fig. 6) 9.
As an independent verification for the crystal structure of PaMCC, we carried out electron microscopy (EM) studies on this holoenzyme and produced a reconstruction at 12 Å resolution (Supplementary text, Supplementary Figs. 7, 8). The crystal structure and the EM density are in excellent agreement with each other (Figs. 2e, 2f), confirming the large structural differences between the MCC and PCC holoenzymes.
Interactions between the α and β subunits in PaMCC are mediated predominantly by the BT domain, and by the BCCP domain when it is in the active site of the β subunit for catalysis (see below). Approximately 3,000 Å2 of the surface area of each α subunit is buried in the interface of the holoenzyme. The BT domain buries 1,500 Å2 in the interface with the β subunit, as well as 200 Å2 in a contact with the BC domain of a neighboring α subunit (see below). The BCCP domain in the active site of the β subunit contributes 700 Å2 to the surface area burial, primarily through residues around the biotinylated Lys681 residue (Supplementary Fig. 1).
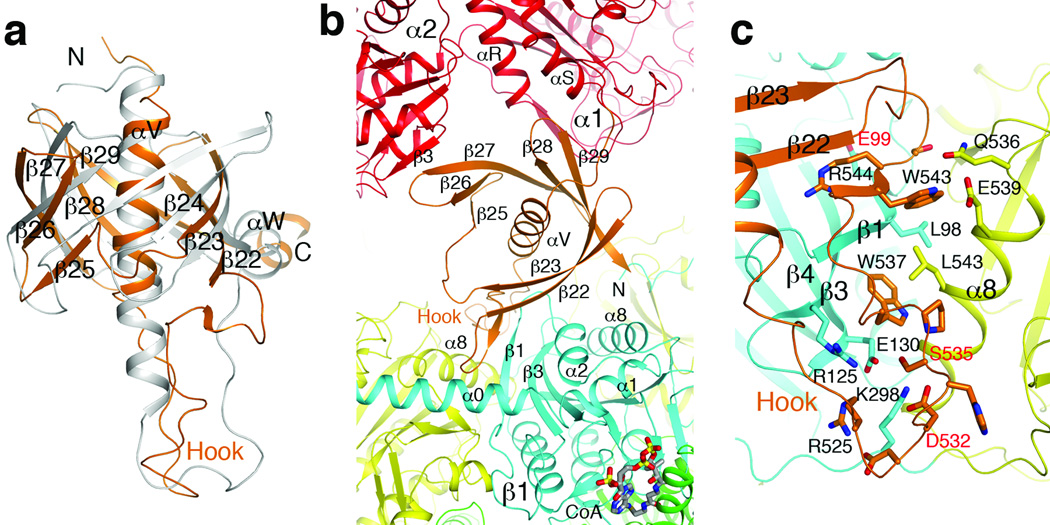
The BT domain in PaMCC contains a central α-helix surrounded by a seven-stranded, highly-twisted anti-parallel β-sheet (Fig. 3a, Supplementary Fig. 9). The overall structure of this domain is similar to that in PCC, with an rms distance of 1.8 Å for their equivalent Cα atoms. In addition, sequence comparisons suggest that the BT domain of HsMCC and most other MCCs may have an eight-stranded β-barrel, which would be equivalent to that in PCC (Fig. 3a, Supplementary text).
Figure 3. The BT domain mediates interactions in the MCC holoenzyme.
(a). Overlay of the structure of PaMCC BT domain (in orange) with that of PCC (gray). A large conformational difference for the hook is visible. The exact positions of many of the β-strands are different as well. (b). The BT domain (orange) contacts a β subunit (β1, N domain in cyan, C domain yellow) as well as a neighboring α subunit (α2, red) in the PaMCC holoenzyme. (c). Detailed interactions between the hook of the BT domain and the β subunit in PaMCC. Three disease-causing mutation sites near this interface are labeled in red. For stereo version of the panels, please see Supplementary Figs. 9, 10.
However, the position of the BT domain relative to the β subunit and its interactions in the PaMCC holoenzyme are different compared to PCC (Fig. 3b, Supplementary Figs. 10, 11). The hook of the BT domain 9, connecting the central helix to the first β-strand, is crucial for interactions with the β subunit but has a significantly different conformation in PaMCC (Fig. 3a). The hook interacts with both the N and C domains of the β subunit in PaMCC, and residues 542–544 in the hook form a parallel β-sheet with strand β1 (residues 95–100) in the N domain of the β subunit (Fig. 3c, Supplementary Fig. 10, Supplementary text). Consistent with these observations, we found that structure-based mutations in the hook could destabilize the PaMCC holoenzyme (Supplementary Fig. 12).
In addition to the hook, there is direct contact between the BT domain of one α subunit and the BC domain of a neighboring α subunit, where residues 604–608 (strand β27 of the BT domain) form an anti-parallel β-sheet with strand β3 of the BC domain (Fig. 3b, Supplementary text).
Overall, the unique interactions for the BT domain and the trimeric association of the BC domain in the MCC holoenzyme suggest that PCC cannot form a similar architecture, and therefore it is unlikely that the observed structural differences between MCC and PCC represent different stages of catalysis for these enzymes.
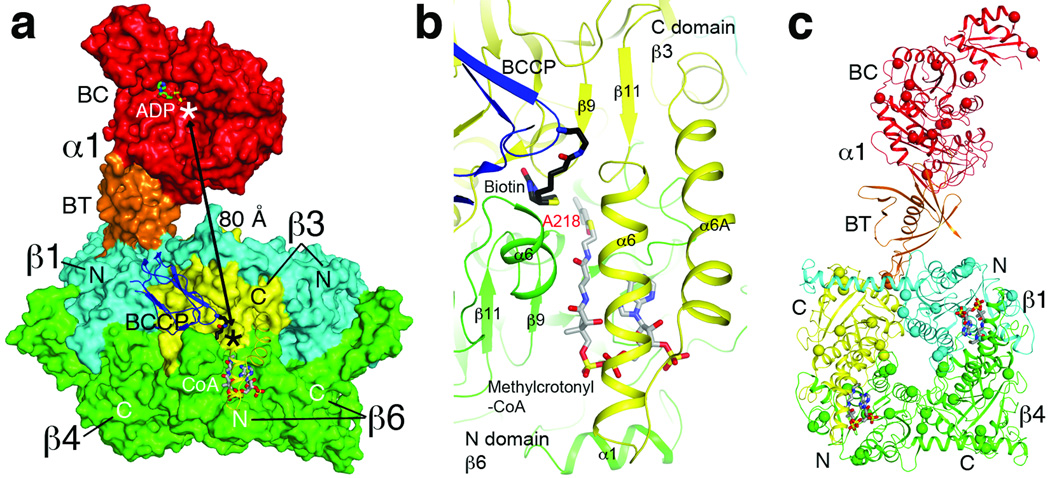
The distance between the active sites of BC and CT is approximately 80 Å in MCC (Fig. 4a). Therefore, the entire BCCP domain must translocate during MCC catalysis, as is the case with PCC 9 and pyruvate carboxylase (PC) 14–17. On the other hand, MCC is distinct from PCC in that while the BT domain of an α subunit contacts its closest β subunit, its BCCP domain is actually located in the active site of a neighboring β subunit (Fig. 4a), another consequence of the swapping of the N and C domains in the β subunit of MCC.
Figure 4. Molecular basis for catalysis and disease-causing mutations in the MCC holoenzyme.
(a). The BC and CT active sites (indicated with the asterisks) are separated by 80 Å in the PaMCC holoenzyme. Molecular surfaces of one α subunit and two β2 dimers are shown. The position of ADP bound to the BC subunit of E. coli acetyl-CoA carboxylase 23 indicates the BC active site. Helices α6-α6A are shown as a ribbon in order to make CoA visible. (b). Schematic drawing of the active site of the β subunit. Biotin (black) and the modeled conformation of methylcrotonyl-CoA (gray) are shown as stick models. Residue Ala218 is the site of a disease-causing mutation. For stereo version of this panel, please see Supplementary Fig. 14. (c). Disease-causing mutation sites are shown as spheres in the PaMCC structure. The mutations are distributed throughout the holoenzyme.
We observed the binding of both CoA and BCCP-biotin to the active site of one of the β subunits (Fig. 4a, Supplementary Fig. 13), and built a model for the bound conformation of methylcrotonyl-CoA (Fig. 4b, Supplementary Fig. 14, Supplementary text). The binding modes are consistent with the expected kinetic mechanism of the CT reaction 18. The N1’ atom of biotin is ~6 Å from the reactive γ carbon of methylcrotonyl-CoA in this model (Supplementary Fig. 14). There is a large conformational change for two helices in the active site upon CoA binding (Supplementary Fig. 15, Supplementary text).
The structure of PaMCC provides a foundation for understanding the molecular basis of its disease-causing mutations in HsMCC, which represent one of the most frequently observed inborn errors of metabolism 1–4,19–21. The missense mutations are distributed throughout the holoenzyme (Fig. 4c), but their effects can be interpreted based on the structure (Supplementary text; Supplementary Table 2). Many of the mutations are located in or near the BC or CT active site (for example R385S in the α subunit, A218T and V375F in the β subunit; Fig. 4b, Supplementary Figs. 14, 16, 17). The V375F mutation may block the binding of BCCP to the active site of the β subunit (Supplementary Fig. 17). A group of mutations are located in or near the subunit interface in the holoenzyme, such as S535F in the hook of the BT domain (Fig. 3c). Additional mutations are located in the hydrophobic core of the structure, and may disrupt folding and/or stability of the enzyme.
Our structure of the PaMCC holoenzyme also has implications for the evolution of biotin-dependent carboxylases. The structural differences between MCC and PCC suggest that there may be two separate lineages of such enzymes that carboxylate CoA esters of organic acids. One lineage includes PCC and acetyl-CoA carboxylase (ACC), which carboxylates the α carbon of the acid. The other lineage targets the γ carbon of an α-β unsaturated acid and includes MCC, GCDα, and possibly also geranyl-CoA carboxylase (GCC, Supplementary Fig. 18) 10,11.
More importantly, the structures of MCC and PCC show that their strong sequence conservation only ensures that the backbone folds of the domains in the two enzymes are the same. On the other hand, the organization of these domains in the individual subunits and especially the architecture of the subunits in the holoenzymes are remarkably different. These observations may also have wide-ranging implications for the relationship between sequence conservation and structural similarity in general.
Methods summary
Crystallography
The α and β subunits of PaMCC were co-expressed in E. coli, with a His-tag on the β subunit. The PaMCC holoenzyme was purified by nickel affinity and gel filtration chromatography. Crystals were obtained by the microbatch method under oil, and the structures were determined by the molecular replacement method.
Electron microscopy
PaMCC sample was stained with uranyl acetate and electron micrographs were recorded at 70,000× magnification in a 200 kV electron microscope. A 12 Å resolution 3D reconstruction was obtained from ~12,000 particle images.
Mutagenesis and kinetic studies
Site-specific mutants were designed based on the structural information, and their effects on the formation of the holoenzyme were assessed by nickel affinity chromatography. The catalytic activity of PaMCC was determined by a coupled enzyme assay, monitoring the hydrolysis of ATP.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Methods
Protein expression and purification
Full-length PaMCCβ was subcloned into the pET28a vector (Novagen). The expression construct contained an N-terminal hexa-histidine tag, which was not removed for crystallization. The native protein was over-expressed overnight in E. coli BL21 Rosetta (DE3) cells (Novagen) at 20°C in the presence of 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) (Gold Biotechnology, Inc). The soluble protein was eluted from nickel affinity beads (Qiagen) and was further purified by gel filtration chromatography with a running buffer of 25 mM Tris (pH 7.4), 250 mM NaCl, and 2 mM DTT. The purified protein was concentrated to 20 mg/ml, supplemented with 5 % (v/v) glycerol, flash-frozen with liquid nitrogen and then stored at −80 °C.
The PaMCC holoenzyme was over-expressed using a bi-cistronic plasmid, with PaMCCα (untagged) placed downstream of PaMCCβ in the pET28a vector, similar to the strategy used for the co-expression of the PCC holoenzyme 24. The holoenzyme was purified following the same protocol as that for PaMCCβ.
Protein crystallization
Crystals were obtained with the microbatch under-oil (Paraffin oil, Hampton Research) method at 20°C. The protein was at 20 mg/ml concentration. For PaMCCβ, the precipitant solution contained 100 mM Tris (pH 8.5), 0.2 M (NH4)2HPO4, and 30% (v/v) PEG 200.
For the PaMCC holoenzyme, the protein was first incubated with 2 mM methylcrotonyl-CoA for 30 min on ice. The precipitant solution contained 20% (w/v) PEG 3350 and 0.2 M sodium malonate (pH 7.0) or 0.2 M Na2SO4. Crystals with the shape of thin plates appeared after 10–14 days and belonged to space group P21. Most of these crystals diffracted X-rays poorly. CoA was observed in the active site of the β subunit based on the subsequent crystallographic analysis.
Under the same conditions, crystals with the shape of rhomboid blocks were observed after 6–8 weeks, after significant evaporation of the crystallization drops. These crystals belonged to space group R32, and showed better X-ray diffraction. They contained the free enzyme of PaMCC.
The crystals were flash-frozen in liquid nitrogen for diffraction analysis and data collection at 100 K.
Data collection and structure determination
X-ray diffraction data sets were collected on an ADSC Q315 CCD at the X29A beamline of the National Synchrontron Light Source (NSLS) at Brookhaven National Laboratory. The diffraction images were processed using the HKL package 25. The data processing and refinement statistics are summarized in Supplementary Table 1.
Crystals of PaMCCβ contained one β subunit in the asymmetric unit. The structure was determined by the molecular replacement method with the program Phaser 26, using the structure of PCCβ as the model 24. Structure refinement was carried out with the programs CNS 27 and Refmac 28, and programs O 29 and Coot 30 were used for manual model rebuilding. Water molecules were located automatically with the program CNS.
Crystals of the CoA complex of PaMCC holoenzyme contained one dodecamer in the asymmetric unit. The structure was determined by the molecular replacement method with the program Phaser, using the structure of PaMCCβ hexamer and the BC domain of PCCα 24 as the models. One copy of the BCCP domain was also located, using the BCCP domain of PCC as the model. However, molecular replacement calculations with the BT domain of PCC were not successful, and the BT domain model was built based on the (omit) electron density after structure refinement.
Crystals of the free enzyme of PaMCC contained one αβ protomer in the asymmetric unit. The structure was determined by the molecular replacement method, using the structure of one αβ protomer of PaMCC CoA complex as the model. No water molecules were included in the atomic models of the holoenzyme due to the limited resolution.
Electron microscopy
PaMCC sample was diluted to appropriate concentrations for electron microscopy (EM) with a buffer containing 25 mM Tris (pH 7.4) and 250 mM NaCl. An aliquot of 2.5 µl of each of the diluted samples was placed onto a carbon film-coated, glow-discharged 300-mesh copper grid. Excess sample was blotted away by filter paper after one minute. The sample was immediately stained with 2% uranyl acetate solution and air dried. Electron micrographs were recorded in an FEI TF20 electron microscope (operated at 200 kV) on a TVIPS 16-megapixel CCD camera at 70,000× magnification. The sample with optimal particle concentration as shown in Supplementary Fig. 6 was used for extended imaging and in-depth 3D reconstruction analysis.
First, we picked 500 particles manually from 5 CCD images with EMAN boxer program and obtained a preliminary reconstruction to 22 Å resolution with EMAN 31. Approximately 20,000 particles were subsequently picked automatically from 30 CCD images with SIGNATURE 32, using the previous 22 Å structure as reference. D3 symmetry was imposed during refinement and 3D reconstruction. Approximately 12,000 particles were selected for the final 3D reconstruction. The resolution of the final 3D reconstruction was assessed to be 12 Å based on the 0.5 cutoff in the Fourier shell correlation between 3D reconstructions from the two half sets of the whole data set (Fig. S7). The UCSF Chimera program was used to create 3D graphical representations 33. The atomic model of MCC was fitted to the density map using the fit-model-to-map module of Chimera.
Mutagenesis and kinetic studies
Site-specific mutations were introduced with the QuikChange kit (Agilent) and sequenced for confirmation. The mutant plasmids were transformed into E. coli, and the formation of the PaMCC holoenzyme was assessed by nickel affinity chromatography.
The catalytic activity of PaMCC was determined using a coupled enzyme assay, converting the hydrolysis of ATP to the disappearance of NADH 34,35. The reaction mixture contained 100 mM HEPES (pH 8.0), 0.5 mM ATP, 8 mM MgCl2, 40 mM KHCO3, 0.5 mM methylcrotonyl-CoA or crotonyl-CoA, 0.2 mM NADH, 0.5 mM phosphoenolpyruvate, 7 units of lactate dehydrogenase, 4.2 units of pyruvate kinase, and 200 mM KCl. The absorbance at 340nm was monitored for 5 min.
Supplementary Material
Acknowledgments
We thank Yang Shen for carrying out initial studies on MCC; Neil Whalen and Stuart Myers for setting up the X29A beamline at the NSLS. This research was supported in part by NIH grants DK067238 (to LT) and GM071940 (to ZHZ). CSH was also supported by an NIH training program in molecular biophysics (GM08281).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions. C.S.H. carried out protein expression, purification and crystallization experiments, mutagenesis and enzymatic assays. C.S.H. and L.T. carried out crystallographic data collection and processing, structure determination and refinement. P.G. and Z.H.Z. carried out electron microscopy experiments. All authors commented on the manuscript. L.T. supervised the project, analyzed the data and wrote the paper.
Author Information. The atomic coordinates have been deposited at the Protein Data Bank: 3U9R, 3U9S, and 3U9T.
The authors declare no competing financial interests.
References
- 1.Baumgartner MR, et al. The molecular basis of human 3-methylcrotonyl-CoA carboxylase deficiency. J. Clin. Investig. 2001;107:495–504. doi: 10.1172/JCI11948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gallardo ME, et al. The molecular basis of 3-methylcrotonylglycinuria, a disorder of leucine metabolism. Am. J. Hum. Genet. 2001;68:334–346. doi: 10.1086/318202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holzinger A, et al. Cloning of the human MCCA and MCCB genes and mutations therein reveal the molecular cause of 3-methylcrotonyl-CoA carboxylase deficiency. Human Mol. Gen. 2001;10:1299–1306. doi: 10.1093/hmg/10.12.1299. [DOI] [PubMed] [Google Scholar]
- 4.Desviat LR, et al. Functional analysis of MCCA and MCCB mutations causing methylcrotonylglycinuria. Mol. Gen. Metab. 2003;80:315–320. doi: 10.1016/S1096-7192(03)00130-6. [DOI] [PubMed] [Google Scholar]
- 5.Wakil SJ, Stoops JK, Joshi VC. Fatty acid synthesis and its regulation. Ann. Rev. Biochem. 1983;52:537–579. doi: 10.1146/annurev.bi.52.070183.002541. [DOI] [PubMed] [Google Scholar]
- 6.Tong L. Acetyl-coenzyme A carboxylase: crucial metabolic enzyme and attractive target for drug discovery. Cell. Mol. Life Sci. 2005;62:1784–1803. doi: 10.1007/s00018-005-5121-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cronan JE, Jr, Waldrop GL. Multi-subunit acetyl-CoA carboxylases. Prog. Lipid Res. 2002;41:407–435. doi: 10.1016/s0163-7827(02)00007-3. [DOI] [PubMed] [Google Scholar]
- 8.Jitrapakdee S, et al. Structure, mechanism and regulation of pyruvate carboxylase. Biochem. J. 2008;413:369–387. doi: 10.1042/BJ20080709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang CS, et al. Crystal structure of the a6b6 holoenzyme of propionylcoenzyme A carboxylase. Nature. 2010;466:1001–1005. doi: 10.1038/nature09302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forster-Fromme K, Jendrossek D. Catabolism of citronellol and related acyclic terpenoids in pseudomonads. Appl. Microbiol. Biotechnol. 2010;87:859–869. doi: 10.1007/s00253-010-2644-x. [DOI] [PubMed] [Google Scholar]
- 11.Aguilar JA, et al. Substrate specificity of the 3-methylcrotonyl coenzyme A (CoA) and geranyl-CoA carboxylases from Pseudomonas aeruginosa. J. Bacteriol. 2008;190:4888–4893. doi: 10.1128/JB.00454-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wendt KS, Schall I, Huber R, Buckel W, Jacob U. Crystal structure of the carboxyltransferase subunit of the bacterial sodium ion pump glutaconyl-coenzyme A decarboxylase. EMBO J. 2003;22:3493–3502. doi: 10.1093/emboj/cdg358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kress D, et al. An asymmetric model for Na+-translocating glutaconyl-CoA decarboxylase. J. Biol. Chem. 2009;284:28401–28409. doi: 10.1074/jbc.M109.037762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.St. Maurice M, et al. Domain architecture of pyruvate carboxylase, a biotin-dependent multifunctional enzyme. Science. 2007;317:1076–1079. doi: 10.1126/science.1144504. [DOI] [PubMed] [Google Scholar]
- 15.Xiang S, Tong L. Crystal structures of human and Staphylococcus aureus pyruvate carboxylase and molecular insights into the carboxyltransfer reaction. Nat. Struct. Mol. Biol. 2008;15:295–302. doi: 10.1038/nsmb.1393. [DOI] [PubMed] [Google Scholar]
- 16.Yu LPC, et al. A symmetrical tetramer for S. aureus pyruvate carboxylase in complex with coenzyme A. Structure. 2009;17:823–832. doi: 10.1016/j.str.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lasso G, et al. Cryo-EM analysis reveals new insights into the mechanism of action of pyruvate carboxylase. Structure. 2010;18:1300–1310. doi: 10.1016/j.str.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knowles JR. The mechanism of biotin-dependent enzymes. Ann. Rev. Biochem. 1989;58:195–221. doi: 10.1146/annurev.bi.58.070189.001211. [DOI] [PubMed] [Google Scholar]
- 19.Stadler SC, et al. Newborn screening for 3-methylcrotonyl-CoA carboxylase deficiency: population heterogeneity of MCCA and MCCB mutations and impact on risk assessment. Human Mutation. 2006;27:748–759. doi: 10.1002/humu.20349. [DOI] [PubMed] [Google Scholar]
- 20.Nguyen KV, Naviaux RK, Patra S, Barshop BA, Nyhan WL. Novel mutations in the human MCCA and MCCB gene causing methylcrotonylglycinuria. Mol. Gen. Metab. 2011;102:218–221. doi: 10.1016/j.ymgme.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 21.Uematsu M, et al. Novel mutations in five Japanese patients with 3-methylcrotonyl-CoA carboxylase deficiency. J. Hum. Genet. 2007;52:1040–1043. doi: 10.1007/s10038-007-0211-9. [DOI] [PubMed] [Google Scholar]
- 22.Pettersen EF, et al. UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 23.Chou C-Y, Yu LPC, Tong L. Crystal structure of biotin carboxylase in complex with substrates and implications for its catalytic mechanism. J. Biol. Chem. 2009;284:11690–11697. doi: 10.1074/jbc.M805783200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang CS, et al. Crystal structure of the a6b6 holoenzyme of propionylcoenzyme A carboxylase. Nature. 2010;466:1001–1005. doi: 10.1038/nature09302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 26.McCoy AJ, et al. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brunger AT, et al. Crystallography & NMR System: A new software suite for macromolecular structure determination. Acta Cryst. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 28.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Cryst. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 29.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Cryst. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 30.Emsley P, Cowtan KD. Coot: model-building tools for molecular graphics. Acta Cryst. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 31.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 32.Chen JZ, Grigorieff N. SIGNATURE: a single-particle selection system for molecular electron microscopy. J. Struct. Biol. 2007;157:168–173. doi: 10.1016/j.jsb.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 33.Pettersen EF, et al. UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 34.Diacovich L, et al. Crystal structure of the b-subunit of acyl-CoA carboxylase: structure-based engineering of substrate specificity. Biochem. 2004;43:14027–14036. doi: 10.1021/bi049065v. [DOI] [PubMed] [Google Scholar]
- 35.Blanchard CZ, Lee YM, Frantom PA, Waldrop GL. Mutations at four active site residues of biotin carboxylase abolish substrate-induced synergism by biotin. Biochem. 1999;38:3393–3400. doi: 10.1021/bi982660a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.