Abstract
Oncogenic activation of KRAS occurs commonly in non-small cell lung cancer (NSCLC), but strategies to therapeutically target this pathway have been challenging to develop. Information about downstream effectors of KRAS remains incomplete and tractable targets are yet to be defined. In this study we investigated the role of Protein Kinase C delta (PKCδ) in KRAS dependent lung tumorigenesis using a mouse carcinogen model and human NSCLC cells. The incidence of urethane-induced lung tumors was decreased by 69% in PKCδ deficient (δKO) mice compared to wild type (δWT) mice. δKO tumors are smaller and showed reduced proliferation. DNA sequencing indicated that all δWT tumors had activating mutations in KRAS, whereas only 69% of δKO tumors did, suggesting that PKCδ acts as a tumor promoter downstream of oncogenic KRAS, while acting as a tumor suppressor in other oncogenic contexts. Similar results were obtained in a panel of NSCLC cell lines with oncogenic KRAS, but which differ in their dependence on KRAS for survival. RNAi-mediated attenuation of PKCδ inhibited anchorage-independent growth, invasion, migration and tumorigenesis in KRAS-dependent cells. These effects were associated with suppression of MAPK pathway activation. In contrast, PKCδ attenuation enhanced anchorage-independent growth, invasion and migration in NSCLC cells that were either KRAS-independent or that had wild-type KRAS. Unexpectedly, our studies indicate that the function of PKCδ in tumor cells depends on a specific oncogenic context, as loss of PKCδ in NSCLC cells suppressed transformed growth only in cells dependent upon oncogenic KRAS for proliferation and survival.
Keywords: PKC delta, K-Ras, lung cancer, transformation
Introduction
Non-small cell lung cancers (NSCLC) account for the majority of lung cancers, with adenocarcinomas being the predominant subtype (1). Mutation of KRAS occurs in about 30% of lung adenocarcinomas, resulting in its constitutive activation (2). To identify potential downstream effectors of oncogenic K-Ras, we have explored the contribution of Protein Kinase C delta (PKCδ) to K-Ras dependent lung tumorigenesis. The PKC family of serine/threonine protein kinases consists of 11 isoforms that regulate a wide variety of biological functions (3). Studies in PKCδ knock-out (δKO) mice have confirmed a role for this kinase in cell proliferation and apoptosis (3–10). In the δKO mouse, cell death in response to irradiation is suppressed, and smooth muscle and epithelial cells cultured from these mice are resistant to multiple apoptotic stimuli (5, 7, 11). PKCδ may regulate apoptosis via p53, as p53 transcriptional activation in response to genotoxins and oxidative stress requires PKCδ (12–15). In the context of proliferation, PKCδ has been shown to be a downstream effector of the EGFR (6, 16–18) and collaborates with the Hedgehog pathway to regulate ERK signaling (19). PKCδ has also been shown to both positively and negatively regulate cell cycle progression (19, 20).
The demonstration of a pro-apoptotic role for PKCδ has lead to the suggestion that it may function as a tumor suppressor (3, 21). In support of this, PKCδ protein expression is reduced in human squamous cell and bladder carcinomas (22, 23), and decreases with increasing tumor grade in human endometrial carcinomas (24). In contrast, PKCδ expression is elevated in pancreatic cancer, myelogenous leukemia and hepatocellular carcinoma (25–27). To address these potentially diverse functions of PKCδ, we have used a chemically induced mouse model in which lung tumorigenesis is associated with activating mutations in K-Ras, and a panel of human NSCLC cells that harbor oncogenic KRAS. Our studies show that PKCδ functions as a tumor promoter in transformed cells that are dependent upon K-Ras for proliferation and survival, while in tumor cells that do not rely on K-Ras, PKCδ may function as a tumor suppressor.
Methods
Animal models
FVB δKO mice were generated by backcrossing C57/Bl6 δKO mice (28) with FVB mice for >10 generations. Nude mice were purchased from Jackson laboratories (Bar Harbor, ME). Animals were maintained at the University of Colorado Denver Anschutz Medical Campus in accordance with Laboratory Animal Care guidelines and protocols, and with approval of the University of Colorado Denver Institutional Animal Use and Care Committee. Male FVB δWT and δKO mice (6 to 8 weeks of age) were injected with 1 mg/kg urethane in sterile water and sacrificed at 10 or 20 weeks post injection. Mice were perfused with 10 ml of sterile-filtered heparinized-PBS (500 U/ml) and the lungs were inflated with 10% formalin. Fixed lungs were processed and paraffin-embedded, or stored for DNA extraction at −80°C.
Tumor analysis
Tumor sections (5 μm) were stained with hematoxylin and eosin (H&E) and anti-Ki67 as previously described (5). Quantification of tumors at 20 weeks was done by counting macroscopic tumors in dissected lungs. Tumor diameter was measured using digital micro-calipers; due to the spherical shape of the tumors, tumor volume was calculated using the formula 4/3 π × r3. For quantification of microscopic tumors at 10 weeks, paraffin embedded tumors were cut into sequential 4 μm thick sections and stained with H+E. Tumor number was determined by analysis of sections representing the entire lung by light microscopy. Tumor volume was determined using the formula 4/3 × π × r3 where tumor diameter equaled 4 μm × the number of sections the tumor spanned.
Sequencing of KRAS in urethane-induced tumors
DNA was extracted using the DNeasy Blood and Tissue kit and the Qiacube Automated Extraction System (Qiagen, Valencia, CA). Exons 2 and 3 of the mouse KRAS gene were sequenced using the Applied Biosystems Incorporated (Foster City, CA) Big Dye Cycle Sequencing Kit and ABI 3730 Sequencer. The following primer sets were used:
| Exon 2 Forward External | 5′ TTTACACACAAAGGTGAGTGT 3′ |
| Exon 2 Forward Internal | TGTGTGAGACATGTTCTAATTTAGTTG |
| Exon 2 Reverse | GCACGCAGACTGTAGAGCAG |
| Exon 3 Forward | CCAGACTGTGTTTCTCCCTTC |
| Exon 3 Reverse External | TGCAGGCATAACAATTAGCAA |
| Exon 3 Reverse Internal | TCACATGCCAACTTTCTTATTC |
NSCLC cell culture and depletion of PKCδ
NSCLC cell lines, A549, NCI-H2009, NCI-H441, NCI-H727, NCI-H460, SW1-573, Colo699 and NCI-H226, were acquired from the University of Colorado Denver lung SPORE cell bank. Cell line profiling for authentication was done at the DNA sequencing Core at University of Colorado Anschutz Medical Campus using the ABI profiler plus and ABI Identifiler profiling kits. Cells were maintained in RPMI-1640 with 2 mM L-glutamine and 10% fetal bovine serum. Transient depletion of PKCδ by siRNA was done using ON-TARGETplus SMARTpool siRNA for human PKCδ (Dharmacon, Thermo Fisher, Lafayette, CO) consisting of the following sequences: 5′CCAUGUAUCCUGAGUGGAA, 5′CCAAGGUGUUGAUGUCUGU, 5′AAAGAACGCUUCAACAUCG, and 5′CCGCACCGCUUCAAGGUUC, and the ON-TARGETplus non-targeting Pool. Stable depletion of PKCδ was performed using lentiviral constructs containing shRNA to human PKCδ (Open Biosystems, pLKO-TRC00010193 or pLKO-TRC00010203) or an shRNA control (pLKO-scrambled). Lentiviral particle containing media was prepared as previously described (29). NSCLC cell lines were infected with 1 ml of lentiviral particle containing media plus 500 μg/ml polybrene for 1 hour, followed by the addition of complete media. Cell lines were maintained in selection media with 2 μg/ml puromycin and used at low passage (<8). Stable depletion of K-Ras was performed using lentiviral constructs as described (36).
Cell proliferation and anchorage-independent growth
Cells were plated at low density and harvested for determination of cell number at days 1 through 6 post-plating. For analysis of cell proliferation by BrdU incorporation, 0.5×104 cells/well were grown in 96 - well plates and transiently depleted of PKCδ using pooled siRNA as described above. After 72 hours, proliferating cells were labeled with 10 mM 5-bromo-2′–deoxyuridine for 2 hours. BrdU incorporation was detected using the Cell Proliferation Elisa kit (Roche Applied Science, Indianapolis, IN). A WST-1 proliferation kit was used according to the manufacturer’s specification for analysis of cell proliferation in cells depleted of K-Ras (Roche Applied Science, Indianapolis IN). To assay anchorage-independent growth, NSCLC lung cancer cell lines were suspended (0.5×104–5×104 cells per well) in a top layer of 0.36% noble agar (USB, Cleveland, Ohio) in RPMI-1640 + 10% FBS and a bottom layer of 0.4% noble agar in RPMI-1640 + 10% FBS in 6 - well multiplate tissue culture dishes. After 1–6 weeks, colonies were stained with NBT (Sigma, St. Louis, MO), pictures were taken with a S600 Canon camera and colonies were quantified using MetaMorph software (Molecular Devices, Downingtown, PA).
Invasion and migration
Boyden chambers (BD Biosciences, San Jose, CA) with Matrigel coating were used for invasion assays; chambers without Matrigel were used for migration assays. Cells (5 × 104) were plated in serum free media in the top chamber, while media containing 10% serum was added to the bottom chamber. After 22 hours, cells were fixed in 10% normal buffered formalin and stained with Crystal Violet. Cells that did not migrate or invade through the filters were then scraped off, and the remaining migrated/invaded cells were counted. Five fields (200X magnification) were counted for each filter; each experimental condition was assayed on triplicate filters.
Ras pull-down assay
Cell lysates were prepared using the Ras Activation assay kit from Millipore (Billerica, MA) and pre-cleared with 50 μL of packed glutathione sepharose 4B (GE Healthcare, Piscataway, NJ) per 500 μL of lysate. For controls, lysates plus GTPγS (positive) or GDP (negative) were used. For the Ras pull-down, 350 μL of pre-cleared lysate was transferred to a tube containing 10 μg of PBD-Pak1 agarose and incubated for 1 hour at 4°C. Pellets were washed, resuspended in 2x Laemmli sample buffer and separated by SDS-PAGE. Proteins were detected by immunoblotting for Ras as described below.
Adenovirus infection
NSCLC cells were plated at 1×106 cells per 60mm dish. The following day cells were infected with either the Ad-LacZ control or Ad-PKCδKD adenovirus at an MOI=250 as previously described (10). Cells were harvested for protein 24 hours post-transduction and ran on a 10% SDS-PAGE. Proteins were immobilized on PVDF membrane and then immunoblotted for the indicated proteins.
Immunoblot analysis
Immunoblotting was done as previously described (5). Antibodies to PKCδ (sc-937) and actin (sc-1616) were purchased from Santa Cruz Biotech (Santa Cruz, CA). The following antibodies were purchased from Cell Signaling Technologies (Danvers, MA): phospho-Akt (pAKT; #4060); Akt (#9272); phospho-ERK1/2 (pERK1/2; #9101); ERK1/2 (#4695); phospho-MEK1/2 (pMEK; #9121); MEK1/2 (#9122); phospho-PDK1 (pPDK1; #3061); PDK1 (#3062). Anti-Ras was purchased from Millipore (Billerica, MA) and antibodies to RSK1 and phospho-RSK1 (pRSK90; #AF889) were purchased from R&D Systems (Minneapolis, MN).
Growth of H441 cells as xenografts
Nude mice were injected with control (scr; n=10) H441 cells in the left flank or H441 cells expressing either δ193 (n=5) or δ203 (n=5) in the right flank. Tumors were measured using digital micro-calipers every second day starting at day 9 and tumor volume was calculated using the formula π/6(L × 0.5L2). Mice were sacrificed at day 27 and tumor lysates were immunoblotted for pERK and stripped and re-probed for total ERK or immunoblotted for PKCδ and stripped and re-probed for actin.
Results
Reduced urethane-induced lung tumorigenesis in the δKO mouse
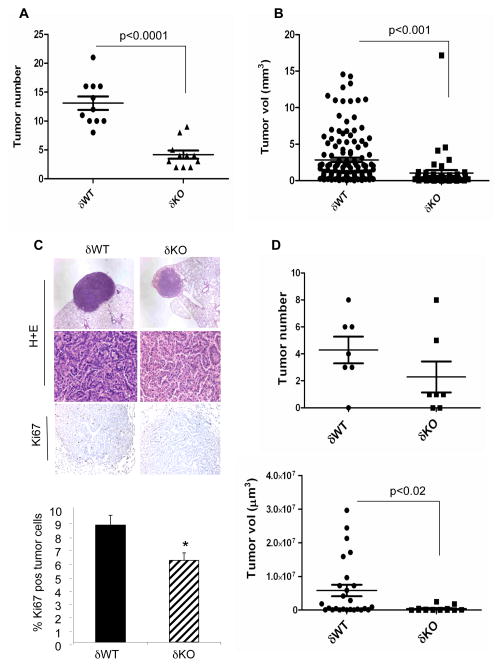
To ask if PKCδ contributes to lung tumorigenesis, we have analyzed the development of urethane-induced tumors in mice in which the PKCδ gene has been disrupted (δKO) and their wild type (δWT) littermates. As shown in Figure 1A, lung tumors in δKO mice treated with urethane for 20 weeks were reduced by 69% compared to δWT mice, with an average of 13.1 +/− 1.1 tumors per δWT mouse and 4.1 +/− 0.7 tumors per δKO mouse (p<0.0001). Tumors from both genotypes resembled well-differentiated adenomas; however tumors from δKO mice were significantly smaller than those from δWT mice (Figure 1B and Figure 1C, top). Expression of the proliferation marker Ki67 was reduced by 30% in δKO tumors as compared to δWT tumors (Figure 1C, bottom). Cells positive for cleaved caspase-3 were only very rarely observed in any tumors (data not shown), suggesting that aberrant regulation of apoptosis is unlikely to explain the reduced lung tumorigenesis in δKO mice.
Figure 1. Urethane-induced lung tumorigenesis is suppressed in δKO mice.
FVB δWT or δKO mice were injected with 1 mg/kg urethane in sterile water and sacrificed as described in Materials and Methods. (A) Lung tumors/mouse at 20 weeks (n=11 each genotype) post-urethane injection. (B) Quantification of δWT and δKO tumor size at 20 weeks post-urethane injection. (C) Top, H+E staining (40X and 400X) and Ki67 immunohistochemistry (200X) of δWT and δKO tumors from 20 week treated mice; bottom, quantification of Ki67 expression. Data represents the average number of Ki67 positive cells/total tumor cells +/− SEM (p<0.001). Greater than 1000 cells were quantified for each tumor; n=17 tumors derived from 4 δWT mice and 14 tumors derived from 4 δKO mice. (D) Top, lung tumors per mouse at 10 weeks (n=7 each genotype) post-urethane injection; bottom, quantification of δWT and δKO tumor size at 10 weeks post-urethane injection. Significance for all experiments was determined using a 2-tailed Students t test.
The dramatic reduction in tumor number in δKO mice could result from delayed tumor growth or possibly tumor regression. To address this further, we analyzed tumor number and size in δKO and δWT mice at 10 weeks following urethane treatment (Figure 1D). Microscopic and macroscopic tumors were apparent in the lungs of both genotypes; 6/7 δWT mice had 3 or more tumors, while only 2/7 δKO mice had a similar tumor burden. This suggests a trend toward reduced tumor number in δKO mice; however tumor number per mouse was not statistically different between the two groups (Figure 1D, top). In contrast, a statistically significant reduction in tumor size was seen in 10 week urethane-treated δKO mice (Figure 1D, bottom). In addition, while tumors >2.5 × 106μm3 comprised 40% of the tumors found in δWT mice; this population was absent from the δKO mice. This suggests that in the absence of PKCδ, urethane-induced tumors either do not progress or progress more slowly.
To determine the KRAS mutational status of δWT and δKO tumors, we sequenced exons two and three of KRAS, which contain the codons (12, 13 and 61) most frequently mutated in lung cancer. All 20 tumors analyzed from δWT mice had oncogenic mutations in codon 61 of KRAS. No mutations in codons 12 or 13 were found. The reduction in urethane-induced tumors in δKO mice suggests that in the context of oncogenic KRAS, PKCδ functions as a tumor promoter. However, only 22 of 32 tumors sequenced from δKO mice had K-Ras codon 61 mutations (p<0.008; Table 1). Hence, over 30% of the δKO tumors arise via a K-Ras-independent mechanism. Furthermore, as wild type KRAS tumors are only found in δKO mice, in this subset of tumors loss of PKCδ appears to be permissive for tumorigenesis, suggesting that in some oncogenic contexts PKCδ may function as a tumor suppressor.
Table 1. KRAS mutations in lung adenomas from δWT and δKO urethane treated mice.
Number of lung tumors from δWT and δKO mice with KRAS codon 61 mutations. KRAS codons 12, 13 and 61 were sequenced in 32 δKO and 20 δWT tumors; no codon 12 or 13 mutations were found in either tumor group.
| Genotype | wt KRAS | mutant KRAS | Q61R | Q61L |
|---|---|---|---|---|
|
| ||||
| δWT | 0 | 20 | 19 | 1 |
| δKO | 10* | 22 | 19 | 3 |
Significantly different from δWT (p<0.008) by Fishers Exact 2-tailed test.
PKCδ is required for transformed growth of human K-Ras dependent NSCLC cells
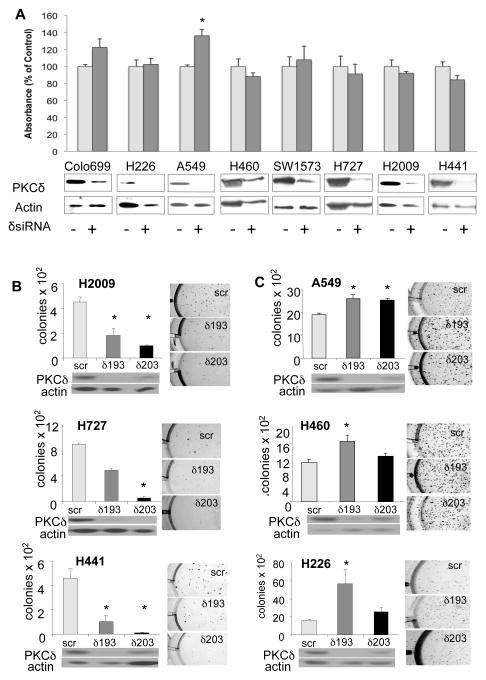
While many NSCLC cell lines harbor oncogenic KRAS, recent studies from Settleman and colleagues show that only a subset of these cells continue to require activated K-Ras for proliferation and survival (36). To determine the role of PKCδ in transformed growth, we used a panel of NSCLC cell lines that are either dependent on activated K-Ras for survival (H2009, H441, H727), that are independent of K-Ras for survival (A549, H460 and SW1573), or that express wild type K-Ras protein (Colo669 and H226). The K-Ras dependency of these cell lines for growth was verified in cells depleted of K-Ras. As reported previously, H2009 and H441 cells were highly dependent on K-Ras for growth, while H727 cells were only slightly dependent (Figure S1) (36). PKCδ expression in these eight NSCLC cell lines and a human cell line derived from normal bronchial epithelial cells (HBEC) does not correlate per se with the presence of, or dependence on, activated K-Ras (Figure S2A).
To determine the effect of PKCδ depletion on growth of NSCLC cells as monolayers, PKCδ was transiently depleted by transfection of pooled targeted siRNA’s. Assay of BrdU incorporation 72 hours after the addition of siRNA indicated that the proliferation of A549 cells was slightly, but significantly, increased by depletion of PKCδ, however PKCδ depletion had no effect on the proliferation of any of the other NSCLC cell lines (Figure 2A). When PKCδ was depleted with shRNA and cell growth assayed by counting cells on successive days, depletion of PKCδ had no effect on cell proliferation (Figures S2B, S2C and S2D). Anchorage-independent growth of cancer cells is a marker of transformed growth in vitro and correlates with tumor aggressiveness and metastatic potential in vivo (31). To assay the effect of PKCδ depletion on anchorage-independent growth, we depleted PKCδ in NSCLC cells using a lentiviral delivery of shRNA directed to PKCδ (δ193 and δ203), or a scrambled shRNA control (scr). δ193 expression resulted in nearly complete depletion of PKCδ in all cell lines, while δ203 resulted in partial depletion of PKCδ protein (Figure 2B, 2C and S2B). Neither shRNA altered the expression of other PKC isoforms (data not shown). As shown in Figure 2B, depletion of PKCδ suppressed colony formation in soft agar by 60–80% in three K-Ras dependent NSCLC cell lines (H2009, H441 and H727). A fourth K-Ras dependent cell line, H358, showed a 55% decrease in anchorage-independent growth in cells expressing δ193 shRNA as compared to scr shRNA (data not shown). In contrast, the K-Ras independent cell lines (A549 and H460) and the K-Ras WT cells (H226) showed a small but significant increase in colony formation when PKCδ was depleted, suggesting that PKCδ suppresses anchorage-independent cell growth in this subset of NSCLC cell lines (Figure 2B).
Figure 2. PKCδ is required for anchorage-independent growth of NSCLC cells that are K-Ras dependent for survival.
(A) BrdU incorporation in eight NSCLC cells in which PKCδ was transiently depleted using siRNA oligos as indicated. Control cells (δsiRNA minus) were treated with scrambled siRNA oligos. Immunoblots for PKCδ to show protein depletion are shown below; blots were stripped and probed for actin. For all panels, data from a representative experiment is shown which was done in triplicate +/− SEM. (B) and (C): Control NSCLC cells (scr), and cells expressing δ193 or δ203, were suspended in soft agar and colony number determined as described in Materials and Methods. (B) K-Ras dependent NSCLC cell lines H2009, H727 and H441. (C) K-Ras independent NSCLC cell lines A549 and H460, and H226 cells that have wild type K-Ras. Graphs show triplicate measurements in one representative experiment +/− SEM; pictures of representative soft agar colonies are included. * = significantly different from control (p<0.05) by 2-tailed Students t test. Immunoblots showing expression of PKCδ and actin for each cell line are shown below the graphs. Each experiment was repeated 2 to 6 times.
PKCδ regulates MAPK signaling in NSCLC cells
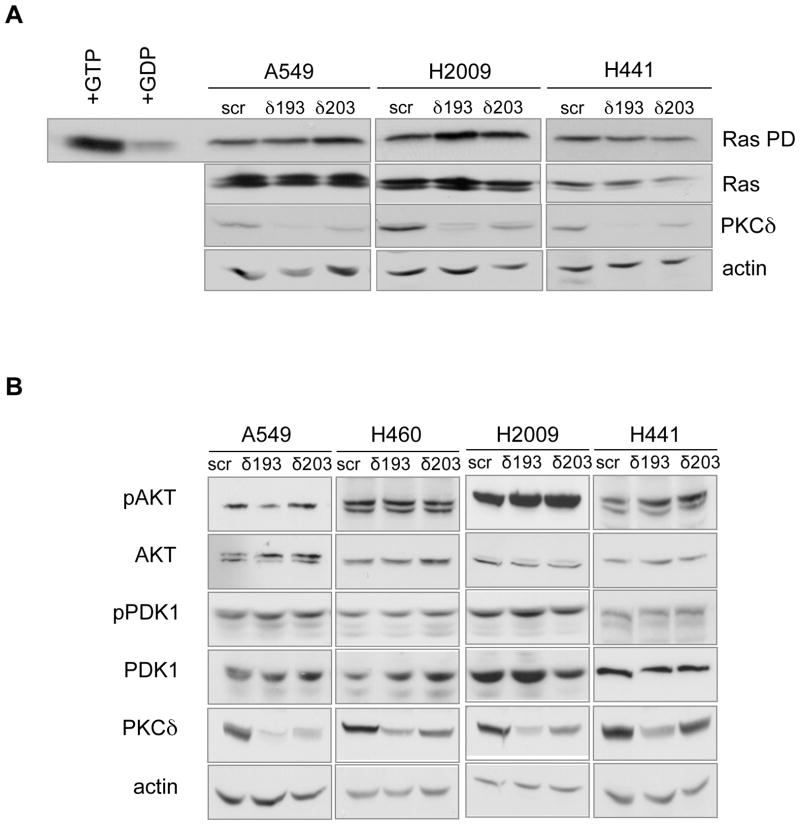
To probe the mechanism by which PKCδ regulates transformed growth of K-Ras dependent NSCLC cells, we investigated activation of the MAPK and AKT pathways, both of which are known to drive proliferation downstream of oncogenic K-Ras. No significant differences were found in the amount of activated Ras between the control cell lines and cell lines in which PKCδ had been depleted (Figure 3A). As PKCδ has previously been implicated in regulation of PDK1 (32), an upstream activator of Akt, we next investigated whether PDK1 and/or AKT activation was regulated by PKCδ. As shown in Figure 3B, although AKT activation varies between the NSCLC cell lines, neither PDK1 nor AKT were regulated by depletion of PKCδ.
Figure 3. PKCδ is not required for activation of Ras or PDK1/AKT in NSCLC cells.
(A) Ras activation in control (scr), δ193, and δ203 expressing A549, H2009 and H441 cells. Top, pull-down of GTP-bound Ras; far left, GTPγS or GDP was added to cell lysates prior to pull-down. Second, third and fourth rows show immunoblots for total Ras, PKCδ and actin. (B) Cell lysates prepared from control (scr), δ193, and δ203 expressing K-Ras independent (A549 and H460) and K-Ras dependent (H2009 and H441) NSCLC cells were immunoblotted for phospho (S241) PDK-1, phospho (S473) AKT, phospho (S217/221) as indicated. Blots were stripped and probed for total PDK-1, AKT, PKCδ and actin, as indicated. Experiments were repeated a minimum of 4 times.
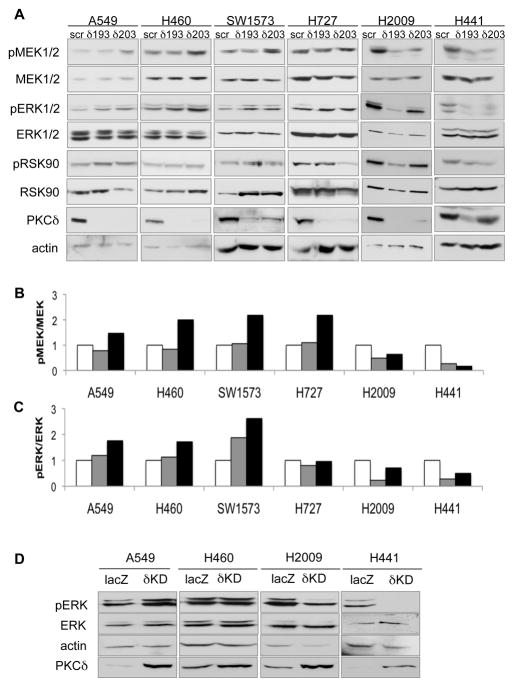
Analysis of the MAPK pathway revealed that depletion of PKCδ suppresses proliferative signaling via this pathway dramatically in K-Ras dependent H2009 and H441 cells (Figure 4A). Phosphorylation of MEK was suppressed in H2009 and H441 cells depleted of PKCδ, especially in cells expressing the δ193 shRNA, as was phosphorylation of its downstream target, ERK, (quantified in Figure 4B and 4C). Depletion of PKCδ in the H727 cells had only a slight effect on ERK activation, and appeared to increased pMEK, perhaps reflecting the observation by our lab and Singh et al. that these cells are intermediate between K-Ras dependent and K-Ras independent NSCLC cells (Fig S1A) (36). Phosphorylation of RSK90, a downstream target of activated ERK, was also slightly suppressed in PKCδ depleted H727, H2009 and H441 cells, although this was some what variable, perhaps reflecting the fact that multiple pathways can regulate RSK90. In contrast to K-Ras dependent cells, depletion of PKCδ in K-Ras independent A549, H460 and SW1573 cells resulted in an increase in phospho-MEK, which correlated with a more robust activation of ERK (Figures 4A, 4B and 4C). Regulation of ERK activation by PKCδ was verified by transducing A549, H460, H2009 and H441 cells with an adenovirus encoding a kinase dead (δKD) PKCδ which functions as a dominant negative (10). A decrease in pERK was found only in the K-Ras dependent H2009 and H441 cells, while a small increase in pERK was observed in the K-Ras independent A549 and H460 cells. These studies indicate that PKCδ positively regulates proliferative signaling through the MEK/ERK pathway in K-Ras dependent NSCLC cells, while in K-Ras independent cells PKCδ may suppress this proliferative pathway.
Figure 4. PKCδ is required for activation of MEK/ERK in NSCLC cells that are K-Ras dependent for survival.
(A) Cell lysates prepared from control (scr), δ193, and δ203 expressing K-Ras mutated NSCLC cells were immunoblotted for phospho (S217/221) MEK1/2, phospho (T202/Y204) ERK1/2, and phospho (S380) RSK90. Blots were stripped and probed for total MEK1/2, ERK1/2, RSK90, PKCδ and actin, as indicated. Representative experiments are shown; experiments were repeated a minimum of 4 times. (B) pMEK1/2 levels from (A) were quantified by densitometry, normalized to total MEK1/2, and plotted relative to control (scr). (C) pERK1/2 levels from (A) were quantified by densitometry, normalized to total ERK1/2 and plotted relative to control (scr). For (B) and (C), scr=white bars, δ193=gray bars, δ203=black bars. (D) A549, H460, H2009 and H441 cells were infected with adenovirus encoding either control (Ad-lacZ) or kinase dead (Ad-δKD). Cell lysates were prepared and immunoblotted for phospho ERK1/2, ERK, actin and PKCδ. Increased PKCδ protein in δKD transduced cells indicates expression of δKD. This experiment was repeated three times; a representative blot is shown.
Depletion of PKCδ suppresses invasion, migration and tumor growth in nude mice
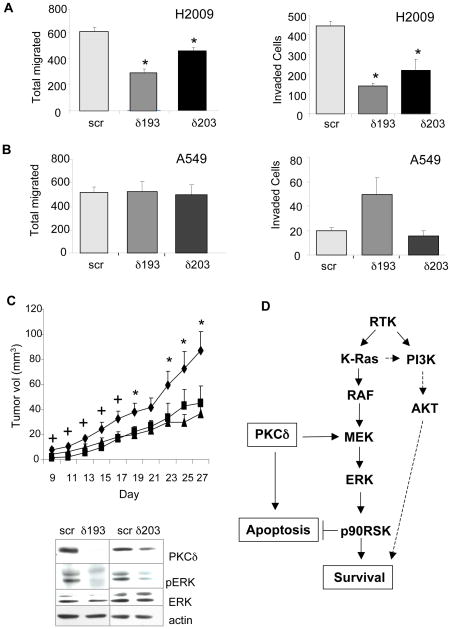
As PKCδ has been shown to regulate motility and invasion of breast carcinoma cells through regulation of the ERK pathway (33), we next sought to determine if depletion of PKCδ suppresses the ability of A549 or H2009 cells to migrate in response to serum, or to invade through a Matrigel cushion. As shown in Figure 5A, depletion of PKCδ significantly suppressed cell migration (up to 50%) and invasion (up to 70%) in H2009 cells as compared to cells expressing a scrambled shRNA. In contrast, no inhibition of migration or invasion was seen in PKCδ depleted A549 cells (Figure 5B). This indicates that similar to anchorage-independent growth, PKCδ regulates invasion and migration of NSCLC cells only in the context of dependency on activated K-Ras.
Figure 5. Depletion of PKCδ suppresses invasion, migration and tumor growth in nude mice.
Migration and invasion of control (scr), δ193, or δ203 expressing H2009 (A) and A549 (B) cells was assayed as described in Materials and Methods. Five fields were counted for each filter and each experimental condition was assayed on triplicate filters. Each experiment was repeated three or more times. (C) Top, nude mice were injected with control H441 cells (diamonds; n=10) in the left flank or H441 cells expressing either δ193 (triangles; n=5) or δ203 (squares; n=5) in the right flank. Tumors were measured starting on day nine. Graphs show average of measurements +/− SEM. * = significantly different from δ203 (p<0.05); + = significantly different from δ193 (p<0.05). (C), Bottom, tumor lysates were immunoblotted for pERK and stripped and re-probed for total ERK, or immunoblotted for PKCδ and stripped and re-probed for actin. Representative images are shown. (D), Model for PKCδ regulation of apoptosis and survival pathways in K-Ras-dependent NSCLC. Ras can also activate proliferation via the PI3K/AKT pathway, although our studies indicate this is not dependent on PKCδ. See text for more details. RTK, receptor tyrosine kinase; PI3K, phosphotidylinositol 3′ kinase.
To investigate whether depletion of PKCδ alters the tumorigenicity of NSCLC cells in vivo, K-Ras dependent H441 cells expressing either scr shRNA, δ193, or δ203 were injected in contralateral flanks of nude mice and tumor size was measured beginning at day nine post-injection. Tumors from H441 cells expressing the δ193 construct were smaller than those expressing the scr control at all time points (40–80%) but this difference was not significant until day 19. Tumors derived from H441 cells expressing the δ203 construct also showed reduced growth compared to the scr shRNA control (40–60%), however in this case the difference was only significant until day 17 (Figure 5C, top). To verify that PKCδ depletion is sustained in the tumors, tumor lysates were prepared when mice were sacrificed at day 27 and probed for PKCδ expression (Figure 5C, bottom). Analysis of pERK in these tumor lysates indicated that the reduction in ERK activation observed in vitro was also maintained during tumor growth in vivo (Figure 5C, bottom), suggesting that reduced tumor growth is coupled to reduced proliferative signaling via this pathway.
Discussion
The development of new therapeutics to target the K-Ras pathway depends on the identification of specific mediators of K-Ras dependent tumorigenesis. While a number of laboratories have defined oncogenic KRAS gene signatures (34–36), the molecular effectors of K-Ras that drive transformation and proliferation in NSCLC are still largely unknown. Our studies suggest that PKCδ functions as a tumor promoter in the context of oncogenic KRAS, as it is required for urethane-induced lung tumorigenesis and for transformed growth of a subset of NSCLC cell lines that are dependent upon activated K-Ras for survival. Conversely, PKCδ may suppress tumorigenesis through other oncogenic pathways, as urethane-induced tumors with WT KRAS arise in the absence of PKCδ, and transformed growth of K-Ras independent NSCLC cells is enhanced when PKCδ is depleted.
Our studies suggest that in the urethane-induced tumor model, PKCδ is required for the progression of K-Ras driven lung tumors and hence functions as a tumor promoter. Likewise, depletion of PKCδ suppresses growth of K-Ras dependent NSCLC cells in soft agar, and inhibits migration and invasion, confirming that this kinase is functionally important for the malignant phenotype. Based on our observation that urethane-induced tumors are decreased dramatically in δKO mice, together with the finding that 31% of the tumors that arise do not have KRAS mutations, the probability of oncogenic KRAS driving tumorigenesis in δKO mice appears to be reduced nearly 5-fold. This is consistent with a recent study by Mauro et al. which shows that overexpression of PKCδ can promote tumorigenesis in a mouse model of human pancreatic cancer, the majority of which harbor oncogenic KRAS (37). Studies from Xia et al. likewise show that PKCδ is required for survival signaling of 3T3 cells expressing activated K-Ras, albeit in these cells PKCδ appears to regulate AKT signaling downstream of activated K-Ras, while our studies suggest PKCδ regulation of MEK/ERK may be more critical for NSCLC cells (32, 38). Of note, Fields and colleagues have shown that atypical PKCι is an oncogene in human NSCLCs. Interestingly, amplification of PKCι appears to occur primarily in squamous cell carcinomas, a subtype of NCSLC that do not typically have activated K-Ras (39).
A particularly novel outcome of our studies is the finding that 31% of the urethane tumors in δKO mice do not have activating mutations in KRAS. We propose that in this group of tumors PKCδ functions as a tumor suppressor. Similarly, our studies suggest that in K-Ras independent NSCLC cell lines, loss of PKCδ enhances transformed growth and proliferative signaling, again suggesting a role as a tumor suppressor. Taken together, this suggests that alternative proliferative pathways that are normally suppressed by PKCδ may sustain growth in K-Ras independent NSCLC cell lines, and in the subset of urethane tumors that do not have mutated KRAS. In this regard, preliminary data from our laboratory indicates that about 10% of this later group of tumors has mutations in either codon 9 or codon 20 of PI3KCA (data not shown). Future studies are needed to characterize these mutations and to understand why they are only seen in the context of the δKO mouse. Activating mutations in components of the PTEN/AKT pathway have recently been reported in a small subset of human lung cancers (40). Curiously, in some cells these mutations occur in conjunction with mutation of KRAS, suggesting a mechanism by which K-Ras dependent cancer cells may become K-Ras independent for proliferation and survival. Singh et al. report that in pancreatic cancer cell lines with oncogenic KRAS, K-Ras independence correlates with increased activation of AKT, however this correlation does not hold true for the subset of NSCLC cell lines examined in this study (36). Instead, in the K-Ras independent NSCLC cell lines, depletion of PKCδ increases activation of MEK/ERK. Several mechanisms have been suggested for PKCδ regulation for MEK/ERK including regulation of the ERK phosphatase, MKP3, or interaction of PKCδ with the adaptor protein, Sprouty 2 (41, 42).
In addition to its well-established role in apoptosis, our current studies support a role for PKCδ in cell survival and transformation, and begin to define the molecular context for these seemingly disparate functions of PKCδ. An important question is what “switches” PKCδ from its pro-apoptotic role to its pro-survival/transformation role. We propose that in K-Ras dependent cells PKCδ function to integrate cell proliferation and survival signals through regulation of MEK/ERK signaling (Figure 5D). In other cells, particularly in non-transformed cells, and in K-Ras independent NSCLC cells, PKCδ may functions primarily to regulate apoptosis and suppress proliferation. The plasticity of PKCδ signaling in lung cancer cells illustrates the need to understand the genetic context of specific cancer subtypes so as to more accurately target therapies.
Supplementary Material
Acknowledgments
This publication was supported by NIH/NCRR Colorado CTSI Grant Number TL1 RR025778 (JMS), the Colorado SPORE in Lung Cancer, NIH NCI p50-CA58187 (WAF and MER), and the Colorado Cancer League (MER). Its contents are the authors’ sole responsibility and do not necessarily represent official NIH views.
References
- 1.Schiller JH. Current standards of care in small-cell and non-small-cell lung cancer. Oncology. 2001;61(Suppl1):3–13. doi: 10.1159/000055386. [DOI] [PubMed] [Google Scholar]
- 2.Beau-Faller M, Legrain M, Voegeli AC, Guerin E, Lavaux T, Ruppert AM, et al. Detection of K-Ras mutations in tumour samples of patients with non-small cell lung cancer using PNA-mediated PCR clamping. Br J Cancer. 2009;100:985–92. doi: 10.1038/sj.bjc.6604925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reyland ME. Protein kinase C isoforms: Multi-functional regulators of cell life and death. Front Biosci. 2009;14:2386–99. doi: 10.2741/3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeVries TA, Kalkofen RL, Matassa AA, Reyland ME. Protein kinase Cdelta regulates apoptosis via activation of STAT1. J Biol Chem. 2004;279:45603–12. doi: 10.1074/jbc.M407448200. [DOI] [PubMed] [Google Scholar]
- 5.Humphries MJ, Limesand KH, Schneider JC, Nakayama KI, Anderson SM, Reyland ME. Suppression of apoptosis in the protein kinase Cdelta null mouse in vivo. J Biol Chem. 2006;281:9728–37. doi: 10.1074/jbc.M507851200. [DOI] [PubMed] [Google Scholar]
- 6.Kharait S, Dhir R, Lauffenburger D, Wells A. Protein kinase Cdelta signaling downstream of the EGF receptor mediates migration and invasiveness of prostate cancer cells. Biochem Biophys Res Commun. 2006;343:848–56. doi: 10.1016/j.bbrc.2006.03.044. [DOI] [PubMed] [Google Scholar]
- 7.Leitges M, Mayr M, Braun U, Mayr U, Li C, Pfister G, et al. Exacerbated vein graft arteriosclerosis in protein kinase Cdelta-null mice. J Clin Invest. 2001;108:1505–12. doi: 10.1172/JCI12902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Majumder PK, Mishra NC, Sun X, Bharti A, Kharbanda S, Saxena S, et al. Targeting of protein kinase C delta to mitochondria in the oxidative stress response. Cell Growth Differ. 2001;12:465–70. [PubMed] [Google Scholar]
- 9.Majumder PK, Pandey P, Sun X, Cheng K, Datta R, Saxena S, et al. Mitochondrial translocation of protein kinase C delta in phorbol ester-induced cytochrome c release and apoptosis. J Biol Chem. 2000;275:21793–6. doi: 10.1074/jbc.C000048200. [DOI] [PubMed] [Google Scholar]
- 10.Matassa AA, Carpenter L, Biden TJ, Humphries MJ, Reyland ME. PKCdelta is required for mitochondrial-dependent apoptosis in salivary epithelial cells. J Biol Chem. 2001;276:29719–28. doi: 10.1074/jbc.M100273200. [DOI] [PubMed] [Google Scholar]
- 11.Allen-Petersen BL, Miller M, Neville M, Anderson S, Nakayama K, Reyland ME. Loss of protein kinase C delta alters mammary gland development and apoptosis. Cell Death Dis. 2010;1:e17. doi: 10.1038/cddis.2009.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ryer EJ, Sakakibara K, Wang C, Sarkar D, Fisher PB, Faries PL, et al. Protein kinase C delta induces apoptosis of vascular smooth muscle cells through induction of the tumor suppressor p53 by both p38-dependent and p38-independent mechanisms. J Biol Chem. 2005;280:35310–7. doi: 10.1074/jbc.M507187200. [DOI] [PubMed] [Google Scholar]
- 13.Yamaguchi T, Miki Y, Yoshida K. Protein kinase C delta activates IkappaB-kinase alpha to induce the p53 tumor suppressor in response to oxidative stress. Cell Signal. 2007;19:2088–97. doi: 10.1016/j.cellsig.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 14.Liu H, Lu ZG, Miki Y, Yoshida K. Protein kinase C delta induces transcription of the TP53 tumor suppressor gene by controlling death-promoting factor Btf in the apoptotic response to DNA damage. Mol Cell Biol. 2007;27:8480–91. doi: 10.1128/MCB.01126-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson CL, Lu D, Huang J, Basu A. Regulation of p53 stabilization by DNA damage and protein kinase C. Mol Cancer Ther. 2002;1:861–7. [PubMed] [Google Scholar]
- 16.Paugh BS, Paugh SW, Bryan L, Kapitonov D, Wilczynska KM, Gopalan SM, et al. EGF regulates plasminogen activator inhibitor-1 (PAI-1) by a pathway involving c-Src, PKCdelta, and sphingosine kinase 1 in glioblastoma cells. Faseb J. 2008;22:455–65. doi: 10.1096/fj.07-8276com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao Y, He D, Saatian B, Watkins T, Spannhake EW, Pyne NJ, et al. Regulation of lysophosphatidic acid-induced epidermal growth factor receptor transactivation and interleukin-8 secretion in human bronchial epithelial cells by protein kinase Cdelta, Lyn kinase, and matrix metalloproteinases. J Biol Chem. 2006;281:19501–11. doi: 10.1074/jbc.M511224200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iwabu A, Smith K, Allen FD, Lauffenburger DA, Wells A. Epidermal growth factor induces fibroblast contractility and motility via a protein kinase C delta-dependent pathway. J Biol Chem. 2004;279:14551–60. doi: 10.1074/jbc.M311981200. [DOI] [PubMed] [Google Scholar]
- 19.Riobo NA, Haines GM, Emerson CP., Jr Protein kinase C-delta and mitogen-activated protein/extracellular signal-regulated kinase-1 control GLI activation in hedgehog signaling. Cancer Res. 2006;66:839–45. doi: 10.1158/0008-5472.CAN-05-2539. [DOI] [PubMed] [Google Scholar]
- 20.Santiago-Walker AE, Fikaris AJ, Kao GD, Brown EJ, Kazanietz MG, Meinkoth JL. Protein kinase C delta stimulates apoptosis by initiating G1 phase cell cycle progression and S phase arrest. J Biol Chem. 2005;280:32107–14. doi: 10.1074/jbc.M504432200. [DOI] [PubMed] [Google Scholar]
- 21.Jackson DN, Foster DA. The enigmatic protein kinase Cdelta: complex roles in cell proliferation and survival. Faseb J. 2004;18:627–36. doi: 10.1096/fj.03-0979rev. [DOI] [PubMed] [Google Scholar]
- 22.D’Costa AM, Robinson JK, Maududi T, Chaturvedi V, Nickoloff BJ, Denning MF. The proapoptotic tumor suppressor protein kinase C-delta is lost in human squamous cell carcinomas. Oncogene. 2006;25:378–86. doi: 10.1038/sj.onc.1209065. [DOI] [PubMed] [Google Scholar]
- 23.Langzam L, Koren R, Gal R, Kugel V, Paz A, Farkas A, et al. Patterns of protein kinase C isoenzyme expression in transitional cell carcinoma of bladder. Relation to degree of malignancy. Am J Clin Pathol. 2001;116:377–85. doi: 10.1309/1VKK-HWH7-YVJN-7UF7. [DOI] [PubMed] [Google Scholar]
- 24.Reno EM, Haughian JM, Dimitrova IK, Jackson TA, Shroyer KR, Bradford AP. Analysis of protein kinase C delta (PKC delta) expression in endometrial tumors. Hum Pathol. 2008;39:21–9. doi: 10.1016/j.humpath.2007.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Evans JD, Cornford PA, Dodson A, Neoptolemos JP, Foster CS. Expression patterns of protein kinase C isoenzymes are characteristically modulated in chronic pancreatitis and pancreatic cancer. Am J Clin Pathol. 2003;119:392–402. doi: 10.1309/bkpc9dx98r781b87. [DOI] [PubMed] [Google Scholar]
- 26.Balasubramanian N, Advani SH, Zingde SM. Protein kinase C isoforms in normal and leukemic neutrophils: altered levels in leukemic neutrophils and changes during myeloid maturation in chronic myeloid leukemia. Leuk Res. 2002;26:67–81. doi: 10.1016/s0145-2126(01)00098-4. [DOI] [PubMed] [Google Scholar]
- 27.Tsai JH, Hsieh YS, Kuo SJ, Chen ST, Yu SY, Huang CY, et al. Alteration in the expression of protein kinase C isoforms in human hepatocellular carcinoma. Cancer Lett. 2000;161:171–5. doi: 10.1016/s0304-3835(00)00597-8. [DOI] [PubMed] [Google Scholar]
- 28.Miyamoto A, Nakayama K, Imaki H, Hirose S, Jiang Y, Abe M, et al. Increased proliferation of B cells and auto-immunity in mice lacking protein kinase Cdelta. Nature. 2002;416:865–9. doi: 10.1038/416865a. [DOI] [PubMed] [Google Scholar]
- 29.Marek L, Ware KE, Fritzsche A, Hercule P, Helton WR, Smith JE, et al. Fibroblast growth factor (FGF) and FGF receptor-mediated autocrine signaling in non-small-cell lung cancer cells. Mol Pharmacol. 2009;75:196–207. doi: 10.1124/mol.108.049544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh A, Settleman J. Oncogenic K-ras “addiction” and synthetic lethality. Cell Cycle. 2009;8:2676–7. doi: 10.4161/cc.8.17.9336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mori S, Chang JT, Andrechek ER, Matsumura N, Baba T, Yao G, et al. Anchorage-independent cell growth signature identifies tumors with metastatic potential. Oncogene. 2009;28:2796–805. doi: 10.1038/onc.2009.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xia S, Chen Z, Forman LW, Faller DV. PKCdelta survival signaling in cells containing an activated p21Ras protein requires PDK1. Cell Signal. 2009;21:502–8. doi: 10.1016/j.cellsig.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin CW, Hou WC, Shen SC, Juan SH, Ko CH, Wang LM, et al. Quercetin inhibition of tumor invasion via suppressing PKC delta/ERK/AP-1-dependent matrix metalloproteinase-9 activation in breast carcinoma cells. Carcinogenesis. 2008;29:1807–15. doi: 10.1093/carcin/bgn162. [DOI] [PubMed] [Google Scholar]
- 34.Luo J, Emanuele MJ, Li D, Creighton CJ, Schlabach MR, Westbrook TF, et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009;137:835–48. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sweet-Cordero A, Mukherjee S, Subramanian A, You H, Roix JJ, Ladd-Acosta C, et al. An oncogenic KRAS2 expression signature identified by cross-species gene-expression analysis. Nat Genet. 2005;37:48–55. doi: 10.1038/ng1490. [DOI] [PubMed] [Google Scholar]
- 36.Singh A, Greninger P, Rhodes D, Koopman L, Violette S, Bardeesy N, et al. A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell. 2009;15:489–500. doi: 10.1016/j.ccr.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mauro LV, Grossoni VC, Urtreger AJ, Yang C, Colombo LL, Morandi A, et al. PKC Delta (PKCdelta) promotes tumoral progression of human ductal pancreatic cancer. Pancreas. 2010;39:e31–41. doi: 10.1097/MPA.0b013e3181bce796. [DOI] [PubMed] [Google Scholar]
- 38.Xia S, Forman LW, Faller DV. Protein kinase C delta is required for survival of cells expressing activated p21RAS. J Biol Chem. 2007;282:13199–210. doi: 10.1074/jbc.M610225200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Regala RP, Weems C, Jamieson L, Khoor A, Edell ES, Lohse CM, et al. Atypical protein kinase C iota is an oncogene in human non-small cell lung cancer. Cancer Res. 2005;65:8905–11. doi: 10.1158/0008-5472.CAN-05-2372. [DOI] [PubMed] [Google Scholar]
- 40.Yamamoto H, Shigematsu H, Nomura M, Lockwood WW, Sato M, Okumura N, et al. PIK3CA mutations and copy number gains in human lung cancers. Cancer Res. 2008;68:6913–21. doi: 10.1158/0008-5472.CAN-07-5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lonne GK, Masoumi KC, Lennartsson J, Larsson C. Protein kinase Cdelta supports survival of MDA-MB-231 breast cancer cells by suppressing the ERK1/2 pathway. J Biol Chem. 2009;284:33456–65. doi: 10.1074/jbc.M109.036186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chow SY, Yu CY, Guy GR. Sprouty2 interacts with protein kinase C delta and disrupts phosphorylation of protein kinase D1. J Biol Chem. 2009;284:19623–36. doi: 10.1074/jbc.M109.021600. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.