Abstract
The serine/threonine protein kinase 3-phosphoinositide-dependent protein kinase 1 (PDK1) is a highly conserved eukaryotic kinase that is a central regulator of many AGC kinase subfamily members. Through its regulation of AGC kinases, PDK1 controls many basic cellular processes, from translation to cell survival. While many of these PDK1-regulated processes are conserved across kingdoms, it is not well understood how PDK1 may have evolved within kingdoms. In order to better understand PDK1 evolution within plants, we have isolated and characterized the PDK1 gene from the moss Physcomitrella patens (PpPDK1), a nonvascular representative of early land plants. PpPDK1 is similar to other plant PDK1s in that it can functionally complement a yeast PDK1 knockout line. However, unlike PDK1 from other plants, the P. patens PDK1 protein does not bind phospholipids due to a lack of the lipid-binding pleckstrin homology domain, which is used for lipid-mediated regulation of PDK1 activity. Sequence analysis of several PDK1 proteins suggests that lipid regulation of PDK1 may not commonly occur in algae and nonvascular land plants. PpPDK1 can phosphorylate AGC kinase substrates from tomato (Solanum lycopersicum) and P. patens at the predicted PDK1 phosphorylation site, indicating that the PpPDK1 substrate phosphorylation site is conserved with higher plants. We have also identified residues within the PpPDK1 kinase domain that affect kinase activity and show that a mutant with highly reduced kinase activity can still confer cell viability in both yeast and P. patens. These studies lay the foundation for further analysis of the evolution of PDK1 within plants.
The kinase subfamily known as the AGC kinases is responsible for regulating many basic cellular functions, and these kinases are found in diverse species including yeast, mammals, and plants (Manning et al., 2002; Bögre et al., 2003; Pearce et al., 2010). This group of kinases is named for three of its representatives, cAMP-dependent protein kinase 1 (PKA), cGMP-dependent protein kinase (PKG), and protein kinase C (PKC). Given that the AGC kinase subfamily has been maintained across vast evolutionary distances and divergent life strategies, it appears that it is of ancient origin.
Protein kinases within the AGC group are regulated by two main conserved phosphorylation sites. The first site is a Ser or Thr located in a hydrophobic motif typically found at the C terminus of the protein (Frödin et al., 2002). The second phosphorylation site is a Ser or Thr located in a region of variable length referred to as the activation loop or T-loop (Bögre et al., 2003). In many AGC kinases from both mammals (Bayascas, 2008, 2010) and plants (Bögre et al., 2003; Anthony et al., 2004, 2006; Devarenne et al., 2006; Zegzouti et al., 2006a, 2006b), this activation loop site is phosphorylated by 3-phosphoinositide-dependent protein kinase 1 (PDK1). Activation loop phosphorylation by PDK1 typically occurs after docking of the phosphorylated hydrophobic motif of the AGC kinase substrate within a pocket located in the small lobe of the PDK1 kinase domain (Biondi et al., 2002). Accordingly, the hydrophobic motif of AGC kinases is also known as the PDK1-interacting fragment (PIF), and the pocket with which it interacts is called the PIF-binding pocket. Many substrate AGC kinases also contain a PIF-binding pocket for binding their own PIF, which stabilizes the kinase-active conformation generated from PDK1 phosphorylation (Frödin et al., 2002; Biondi, 2004).
PDK1 is itself an AGC kinase and was initially identified by chromatographic fractionation of muscle and brain protein extracts as the upstream kinase responsible for activation loop phosphorylation of protein kinase B (PKB [also known as Akt]; Alessi et al., 1997; Stokoe et al., 1997). In the years since its identification, PDK1 has been demonstrated to phosphorylate at least 23 of the 60 human and 12 of the 39 Arabidopsis (Arabidopsis thaliana) AGC kinases (Zegzouti et al., 2006b; Pearce et al., 2010), thereby coordinating numerous and diverse cellular processes, including cell survival and apoptosis, cell growth and division, hormone responses, and protein synthesis (Pearce et al., 2010). Evidently, the ability of PDK1 to activate a large array of AGC kinases is a crucial regulatory mechanism for many important signaling pathways.
The phosphorylation of PKB by PDK1 requires production of the signaling lipid phosphatidylinositol-3,4,5-trisphosphate [PtdIns(3,4,5)P3], which directs the colocalization of PDK1 and PKB at the plasma membrane to promote their interaction (Borgatti et al., 2003; Calleja et al., 2007). Subsequently, several other AGC kinases have been shown to be phosphorylated by PDK1 in response to PtdIns(3,4,5)P3 production (Bayascas, 2010). A domain within PDK1 known as the pleckstrin homology (PH) domain directly binds PtdIns(3,4,5)P3 (Bögre et al., 2003). Even though plants do not produce PtdIns(3,4,5)P3 (Bögre et al., 2003), the PH domain has been shown to bind several phosphoinositides (Deak et al., 1999), but phosphatidic acid appears to be the phospholipid that induces PDK1 activity toward substrates (Anthony et al., 2004, 2006).
Although not present in prokaryotes, PDK1-like sequences have been identified in almost all eukaryotes studied so far, including yeast, multicellular fungi, nematodes, insects, gastropods, mammals, and plants (Silber et al., 2004). As might be expected because of its role as a key regulator of AGC kinase signaling, loss of PDK1 is lethal in yeast (Casamayor et al., 1999; Niederberger and Schweingruber, 1999), Drosophila melanogaster (Rintelen et al., 2001), mouse (Lawlor et al., 2002), and tomato (Solanum lycopersicum; Devarenne et al., 2006). Interestingly, in contrast to the other studied PDK1 genes, Arabidopsis PDK1 (AtPDK1) appears to be nonessential for plant survival (Camehl et al., 2011). PDK1s from both human and Arabidopsis have been shown to complement loss of the Saccharomyces cerevisiae PDK1 homologs PKH1 and PKH2 (Casamayor et al., 1999; Deak et al., 1999), suggesting that while some aspects of PDK1 function are common to yeast, mammals, and plants, additional organism-specific AGC kinase-mediated cellular regimes await further exploration.
In recent years, the moss Physcomitrella patens has emerged as an exciting new model organism due to several attractive advantages over other plant models, including a dominant haploid gametophyte stage (Cove, 2005) and highly efficient homologous recombination (Schaefer, 2002). This makes targeted gene disruption and replacement possible for the first time in a plant. Additionally, as a member of the bryophytes, P. patens is an ideal model for plant evolutionary studies. Bryophytes, consisting of mosses, liverworts, and hornworts, are the closest extant relatives of early land plants, which first appear in the fossil record approximately 440 to 460 million years ago (Wellman et al., 2003; Wellman, 2010). Almost all other plant models are angiosperms, a much more recent lineage that emerged approximately 140 to 180 million years ago (Soltis et al., 2008). Therefore, studies performed in P. patens can be used to uniquely illuminate the evolutionary changes that have occurred since the ancestors of modern plants first emerged from aquatic habitats to colonize the land.
In this study, we identify and characterize a homolog of PDK1 from P. patens (PpPDK1) and analyze PDK1 sequences from divergent plant species to examine the protein features of PDK1 in an evolutionary context. These experiments present a starting point from which additional aspects of PDK1 function and evolution may be investigated.
RESULTS
Identification of a Putative PDK1 from P. patens
The P. patens genome database at Phytozome (www.phytozome.net) was searched using the AtPDK1 sequence (GenBank accession no. AF132742.1; Deak et al., 1999). The two top hits, Pp1s217_11V6.2 and Pp1s217_11V6.1 (Supplemental Fig. S1A), are alternate transcripts produced from the same genomic locus, Pp1s217_11V6, which spans 3,941 nucleotides of genomic DNA and contains a 2,803-nucleotide coding region of nine exons/eight introns (Fig. 1A; Supplemental Fig. S2A). Pp1s217_11V6.2 is listed as the primary transcript and thus will be the main focus of this study. Its coding sequence comprises 1,044 nucleotides, which produces a protein of 347 amino acids (Fig. 1; Supplemental Fig. S2, B and C). Pp1s217_11V6.1 is a splice variant with a 1,041-nucleotide coding sequence generated by including the final three nucleotides of exon 5 with intron 5. This produces a protein lacking a single Glu that would originate from the junction between exons 5 and 6 (data not shown). Pp1s217_11V6 locus transcripts are annotated as PDK1 by the Kyoto Encyclopedia of Genes and Genomes. The third and fourth hits (Pp1s118_230V6 and Pp1s4_386V6.1) are both annotated as putative homologs of ribosomal S6 kinase (S6K) by Eukaryotic Orthologous Groups and have coding sequences of 1,488 and 1,518 nucleotides, respectively (Supplemental Fig. S1A). Pp1s217_11V6.2, Pp1s118_230V6, and Pp1s4_386V6.1 were all amplified from total moss mRNA, cloned into Escherichia coli and yeast expression vectors, and sequenced. Hereafter, the Pp1s217_11V6.2, Pp1s118_230V6, and Pp1s4_386V6.1 genes are referred to as PpPDK1, Pp188412, and Pp174181, respectively (Supplemental Table S1).
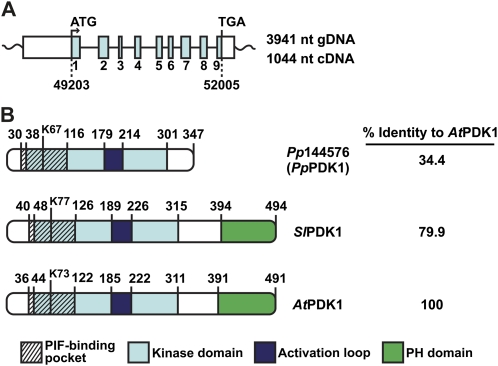
Figure 1.
Features of P. patens PDK1 (PpPDK1) and comparison with PDK1 from tomato (SlPDK1) and Arabidopsis (AtPDK1). A, Diagram of locus Pp1s217_11V6, which is located on P. patens genomic scaffold 217. The Pp1s217_11V6 locus spans 3,941 nucleotides (nt) of genomic DNA consisting of a 901-nucleotide 5′ untranslated region, a 2,803-nucleotide coding region of nine exons/eight introns, and a 237-nucleotide 3′ untranslated region. Expression of this gene produces a transcript with 1,044 nucleotides of coding sequence. Numbers below the dashed lines indicate the start and end locations of the PpPDK1 coding sequence on genomic scaffold 217. B, Features of PpPDK1 protein compared with SlPDK1-1 and AtPDK1-1, as determined by ClustalW multiple alignment of protein sequences. The conserved Lys that is required for Mg2+ coordination in all kinases is K67 in PpPDK1, K77 in SlPDK1, and K73 in AtPDK1. The percentage identity of each protein to AtPDK1 was determined using the EMBOSS Needle pairwise sequence alignment program. [See online article for color version of this figure.]
Based on ClustalW protein sequence alignment with AtPDK1 and tomato PDK1 (SlPDK1), PpPDK1 appears to possess characteristics of a typical PDK1: a pocket for binding the PIF motif of substrate AGC kinases, a kinase domain containing the conserved Lys residue (Lys-67) required for ATP coordination by all protein kinases, and an activation loop within the kinase domain (Fig. 1B). However, PpPDK1 lacks a C-terminal lipid-binding PH domain and thus shares only 34.4% amino acid identity with AtPDK1, whereas SlPDK1 and AtPDK1 are 79.9% identical (Fig. 1B). Like PpPDK1, yeast Pkh1 and Pkh2 also lack PH domains, but both proteins are much larger than PpPDK1 (Pkh1, 766 amino acids; Pkh2, 1,061 amino acids), and their overall sequence identity to other PDK1s is low (Casamayor et al., 1999).
PpPDK1 Is a Functional Homolog of S. cerevisiae Pkh1/2
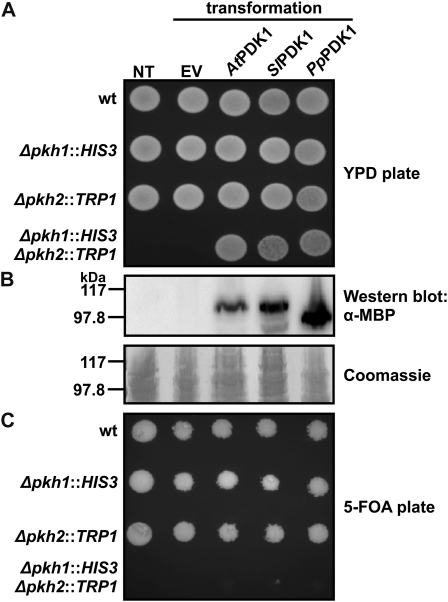
AtPDK1 was initially identified as a functional homolog of PDK1 by its ability to complement a deletion strain of the S. cerevisiae PDK1 homologs PKH1 and PKH2 (Deak et al., 1999). The previously characterized diploid yeast strain AC306 is heterozygous for the deletion of both PKH1 and PKH2 (Casamayor et al., 1999). After sporulation of this strain, haploid cells lacking both PKH1 and PKH2 (pkh1Δ::HIS3/pkh2Δ::TRP1) are inviable, but expression of either human PDK1 or AtPDK1 in these cells rescues the lethal phenotype (Casamayor et al., 1999; Deak et al., 1999). A similar complementation experiment was performed to test whether PpPDK1 is a functional homolog of Pkh1/2. AC306 yeast transformed with empty vector, MBP-AtPDK1-6His, MBP-SlPDK1-6His, or MBP-PpPDK1-6His were sporulated, and haploid spores were grown on nonselective yeast peptone dextrose (YPD) medium to enable the growth of all viable spores. Analysis of at least 30 tetrads from each yeast culture recovered no viable pkh1Δ::HIS3/pkh2Δ::TRP1 spores from untransformed yeast or from yeast transformed with empty vector (Fig. 2A). However, after transformation with MBP-AtPDK1-6His, MBP-SlPDK1-6His, or MBP-PpPDK1-6His, viable pkh1Δ::HIS3/pkh2Δ::TRP1 spores were recovered, indicating that each PDK1 tested was able to rescue the lethal phenotype of the PKH1/2 deletion (Fig. 2A). Western blotting with α-MBP confirmed expression of the PDK1 proteins (Fig. 2B). The ability of each PDK1 to confer pkh1Δ::HIS3/pkh2Δ::TRP1 spore viability was confirmed by growing spores on medium with 5-fluoroorotic acid (5-FOA), which induces loss of the URA3-marked plasmids containing the PDK1 constructs. No viable pkh1Δ::HIS3/pkh2Δ::TRP1 spores were recovered from 5-FOA plates (Fig. 2C), suggesting that, like AtPDK1 and SlPDK1, PpPDK1 is a functional homolog of Pkh1/2.
Figure 2.
PpPDK1 rescues lethality caused by the deletion of both S. cerevisiae PDK1 homologs, PKH1 and PKH2. Yeast strain AC306 was transformed with p416GPD containing the indicated PDK1 cDNAs. NT, Untransformed AC306; EV, empty vector-transformed AC306. Sporulation was induced, and the four haploid cells produced by each diploid cell were grown on rich medium (YPD) and medium containing 5-FOA. Spore phenotypes are indicated on the left and were determined by replica plating on medium lacking His, Trp, or uracil. A, On YPD plates, pkh1Δ::HIS3/pkh2Δ::TRP1 spores are only viable when AtPDK1, SlPDK1, or PpPDK1 is expressed. B, α-MBP western blot showing protein expression. C, On 5-FOA plates, pkh1Δ::HIS3/pkh2Δ::TRP1 spores are not viable due to loss of the URA3-marked p416GPD plasmid. wt, Wild type.
The Pp174181 and Pp188412 kinase genes identified in our search of P. patens for PDK1-like sequences appear to be more closely related to the AGC kinase S6K than to PDK1, based on protein BLAST analysis (Supplemental Fig. S1A). To verify that P. patens possesses a single functional homolog of PDK1, a second complementation experiment was performed with PpPDK1, Pp174181, and Pp188412. The previously characterized haploid S. cerevisiae strain INA106-3B lacks PKH2 and possesses a point mutation in PKH1 (D398G; pkh2Δ::LEU2/pkh1D398G) that confers a temperature-sensitive phenotype; INA106-3B yeast is able to grow at the permissive temperature of 25°C but not at the restrictive temperature of 35°C (Inagaki et al., 1999). INA106-3B yeast were transformed with empty vector, MBP-PpPDK1-6His, MBP-Pp174181, or MBP-Pp188412. Transformed yeast cultures were grown in liquid medium lacking uracil at 25°C and spotted onto plates that were incubated at either 25°C or 35°C. Only yeast transformed with MBP-PpPDK1-6His were able to complement pkh2Δ::LEU2/pkh1D398G as assessed by growth at 35°C (Supplemental Fig. S1B). Western blotting with α-MBP confirmed the expression of all proteins (Supplemental Fig. S1B). Taken together, these results suggest that, like rice (Oryza sativa; Matsui et al., 2010) and in contrast to Arabidopsis (Bögre et al., 2003; Camehl et al., 2011) and tomato (Devarenne et al., 2006), the P. patens genome contains a single PDK1 gene.
Lipid-Binding Ability of PpPDK1
Because of its lack of a PH domain (Fig. 1A), the ability of PpPDK1 to bind lipids was analyzed. PpPDK1 did not appear to strongly bind phospholipids, although very weak binding to several phospholipids was detected using protein-lipid overlay assays (Supplemental Fig. S3A). This result is in contrast to AtPDK1 and SlPDK1, which both strongly interact with a number of phospholipids, including multiple phosphorylated phosphatidylinositols and phosphatidic acid (Supplemental Fig. S3A; Deak et al., 1999). PpPDK1 also appeared not to bind sphingolipids (Supplemental Fig. S3B), contrary to human PDK1, which is activated by sphingosine (King et al., 2000), and Pkh1/2, which are activated by sphingoid bases (Friant et al., 2001). These results raise the possibility that, unlike the other plant PDK1s analyzed so far (Deak et al., 1999; Bögre et al., 2003; Anthony et al., 2004, 2006; Zegzouti et al., 2006a), PpPDK1 activity and signaling may not be lipid regulated. However, a more detailed study is required to confirm or deny lipid control of PpPDK1.
Characterization of PpPDK1 Activity
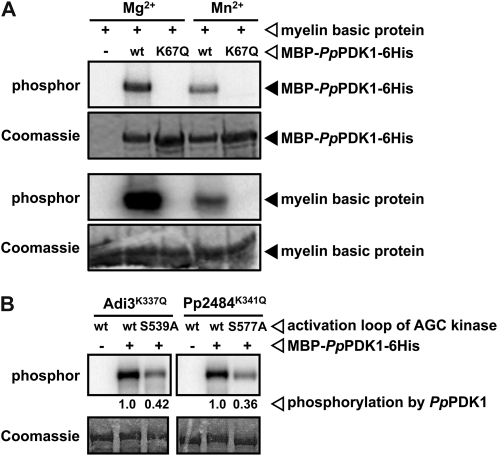
The kinase activity (autophosphorylation and transphosphorylation) of PpPDK1 was tested by in vitro kinase assays. MBP-PpPDK1-6His used either Mg2+ or Mn2+ as a divalent cation for autophosphorylation and phosphorylation of the artificial kinase substrate myelin basic protein (Fig. 3A). As expected, since Lys-67 coordinates ATP, mutation of Lys-67 to Gln (K67Q) abolished both PpPDK1 autophosphorylation and phosphorylation of myelin basic protein (Fig. 3A). To test whether PpPDK1 can activate AGC kinases, PpPDK1 was incubated with either a known PDK1 AGC kinase substrate, Adi3 from tomato (Devarenne et al., 2006), or a novel putative AGC kinase isolated from P. patens, Pp2484, which was identified by searching the P. patens genome database at Phytozome using the Adi3 sequence (accession no. AY849914). Based on ClustalW protein sequence alignment with Adi3, Pp2484 appears to possess characteristics of a typical AGC kinase: a kinase domain containing the invariant ATP-coordinating Lys (Lys-341), an activation loop within the kinase domain, and a C-terminal PIF motif for interaction with PDK1 (Supplemental Fig. S4A). Pp2484 was further verified as a functional kinase using in vitro kinase assays to confirm the contribution of conserved residues to its activity. As expected, mutation of the conserved ATP-coordinating Lys-341 to Gln (K341Q) completely abolished MBP-Pp2484 autophosphorylation and phosphorylation of myelin basic protein (Supplemental Fig. S4B). Mutation of the conserved activation loop Ser to the phosphomimetic amino acid Asp (S577D) increased MBP-Pp2484 kinase activity compared with wild-type protein (Supplemental Fig. S4B). Next, the ability of PpPDK1 to phosphorylate these AGC kinases was tested. MBP-PpPDK1-6His phosphorylated kinase-inactive MBP-Adi3K337Q and MBP-Pp2484K341Q (Fig. 3B). As has been seen for SlPDK1 phosphorylation of Adi3 (Devarenne et al., 2006), mutation of the conserved PDK1 phosphorylation site in the activation loop of MBP-Adi3K337Q and MBP-Pp2484K341Q (Ser-539 in Adi3 and Ser-577 in Pp2484) to Ala decreased the total phosphorylation of both proteins by approximately 60% (Fig. 3B). These results provide evidence that, like other PDK1s, PpPDK1 is able to phosphorylate AGC kinases in vitro, including a P. patens AGC kinase that is a potential substrate in vivo. Taken with the yeast complementation results, it seems likely that PpPDK1 functions as the only PDK1 used by P. patens to regulate the activity of its AGC kinases.
Figure 3.
Characterization of PpPDK1 kinase activity on myelin basic protein and AGC kinases from tomato and P. patens. A, Wild-type (wt) MBP-PpPDK1-6His autophosphorylates and phosphorylates myelin basic protein in the presence of either Mg2+ or Mn2+, but the K67Q mutation abolishes all kinase activity. The top two panels show PpPDK1 autophosphorylation, and the bottom two panels show PpPDK1 phosphorylation of myelin basic protein. B, PpPDK1 phosphorylates AGC kinases Adi3 (tomato) and Pp2484 (P. patens) at the conserved PDK1 phosphorylation site in the activation loop of both kinases. Mutation of the conserved activation loop Ser to Ala (S539A in Adi3 and S577A in Pp2484) results in reduced phosphorylation of Adi3 and Pp2484 by PpPDK1.
PIF-Binding Pocket Mutations That Affect PpPDK1 Activity
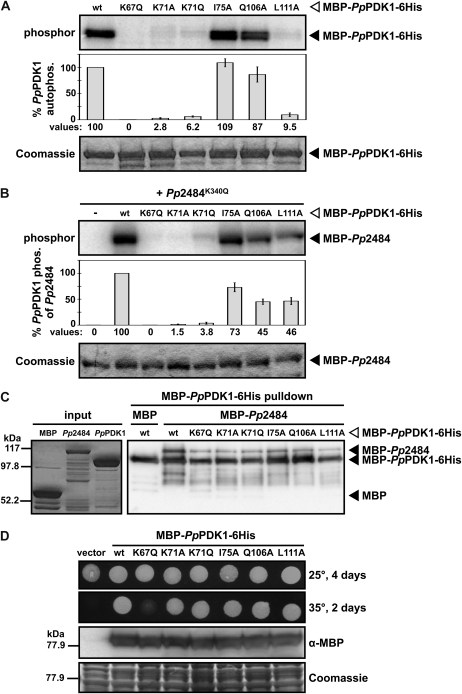
Interaction between many AGC kinase substrates and PDK1 is mediated by hydrophobic interactions between the PIF motif at the C terminus of the substrate and the PIF-binding pocket of PDK1 (Bögre et al., 2003). Mutating any of several important residues in the PIF-binding pocket decreases PDK1 interaction with substrates (Frödin et al., 2002). In human PDK1, the PIF-binding pocket residues that interact with the PIF motif of PKA include Lys-115, Ile-119, Gln-150, or Leu-155, and each residue is required for efficient PDK1-PIF interaction (Biondi et al., 2000). These residues correspond to Lys-341, Ala-345, Glu-376, and Leu-383 of Adi3, which also has a PIF-binding pocket that interacts with its own PIF motif, and mutation of Lys-341, Gln-376, or Leu-383 reduces Adi3 autophosphorylation (Devarenne et al., 2006). Based on ClustalW protein sequence alignment with human PDK1 and Adi3, the corresponding PIF-binding pocket residues of PpPDK1 are Lys-71, Ile-75, Gln-106, and Leu-111 (Supplemental Fig. S5A). Each of these residues was individually mutated to test the effects on PpPDK1 activity and interaction with Pp2484.
First, the autophosphorylation activity of PpPDK1 PIF-binding pocket mutants was tested by in vitro kinase assays. As seen in Figure 3A, autophosphorylation was completely abolished in the kinase-inactive MBP-PpPDK1K67Q-6His (Fig. 4A). The K71A, K71Q, and L111A mutations all drastically reduced MBP-PpPDK1-6His autophosphorylation compared with wild-type PpPDK1 (Fig. 4A). In contrast, the I75A and Q106A mutations produced similar autophosphorylation levels compared with wild-type PpPDK1 (Fig. 4A). These results are somewhat unexpected, because PDK1 proteins lack a PIF motif, and autophosphorylation is not known to require PIF-binding pocket residues. It is possible that in PpPDK1, Lys-71 and Leu-111 are required for the general maintenance of PpPDK1 in a kinase-active state, possibly by ensuring proper conformation of the small lobe of the kinase domain where the PIF-binding pocket is found.
Figure 4.
Functional analysis of conserved PpPDK1 PIF-binding pocket residues. A, Mutation of PpPDK1 PIF-binding pocket residues reduces autophosphorylation. Values are reported as percentages of wild-type (wt) PpPDK1 autophosphorylation and are means of three independent experiments. Error bars indicate se. B, Mutation of PpPDK1 PIF-binding pocket residues reduces the phosphorylation of Pp2484. Values are reported as percentages of wild-type PpPDK1 phosphorylation of Pp2484 and are means of four independent experiments. Error bars indicate se. C, Mutation of PpPDK1 PIF-binding pocket residues reduces, but does not abolish, interaction with Pp2484. The left panel shows protein inputs analyzed by SDS-PAGE and staining with Coomassie blue, and the right panel shows Ni+2 resin pulldowns of MBP-PpPDK1-6His analyzed by α-MBP western blot. D, All PpPDK1 PIF-binding pocket mutants that possess kinase activity are able to fully complement a temperature-sensitive allele of S. cerevisiae PKH1. Haploid strain pkh2Δ/pkh1D398G temperature-sensitive yeast was transformed with p416GPD containing the indicated PpPDK1 constructs. Transformed yeast were grown in liquid medium lacking uracil, spotted on plates lacking uracil, and grown at the indicated temperatures and times. Total protein was extracted from cultures grown in liquid medium at 25°C and analyzed by α-MBP western blot to verify the expression of each PpPDK1.
Next, the PpPDK1 PIF-binding pocket mutants were incubated with kinase-inactive Pp2484K341Q in in vitro kinase assays to test the activity of the PpPDK1 mutants on a potential substrate. MBP-PpPDK1K67Q-6His displayed no detectable activity on MBP-Pp2484K341Q (Fig. 4B). The activity of the K71A and K71Q MBP-PpPDK1-6His mutants toward MBP-Pp2484K341Q was drastically reduced compared with wild-type PpPDK1 (Fig. 4B). The activity of MBP-PpPDK1L111A-6His on MBP-Pp2484K341Q was also reduced compared with wild-type PpPDK1 (Fig. 4B) but was not as dramatic as the decrease in autophosphorylation shown in Figure 4A. Interestingly, although the autophosphorylation of MBP-PpPDK1I75A-6His and MBP-PpPDK1Q106A-6His was comparable to wild-type PpPDK1 (Fig. 4A), these mutants displayed reduced activity on MBP-Pp2484K341Q compared with wild-type PpPDK1 (Fig. 4B). In kinase assays containing the PpPDK1 PIF-binding pocket mutants and kinase-inactive Adi3K337Q, similar results were obtained for the K67Q, K71A, K71Q, and L111A mutants (Supplemental Fig. S5B). However, in contrast to the kinase assays with Pp2484, the activity of MBP-PpPDK1-6HisI75A and MBP-PpPDK1Q106A-6His on MBP-Adi3K337Q was comparable to that of wild-type PpPDK1 (Supplemental Fig. S5B).
To further investigate the contributions of Lys-71, Ile-75, Gln-106, and Leu-111 to substrate binding, in vitro pulldown experiments were performed between PpPDK1 and Pp2484. Wild-type and PIF-binding pocket mutants of MBP-PpPDK1-6His were preincubated with Ni2+ resin, then either MBP or MBP-Pp2484 was added, and the interactions were assessed by western blotting with α-MBP. No individual point mutation was able to abolish the interaction between PpPDK1 and Pp2484, although several PpPDK1 mutants (K67Q, K71A, K71Q, and L111A) were able to pull down less Pp2484 than wild-type PpPDK1 (Fig. 4C). Taken together with the kinase assays, these data may indicate that the analyzed PpPDK1 PIF-binding pocket residues are required for proper conformation of an active kinase as well as for participation in substrate binding.
Finally, the ability of PpPDK1 PIF-binding pocket mutants to complement PKH1/2 was tested with the same INA106-3B yeast used to test Pp174181 and Pp188412 complementation (Supplemental Fig. S1B). INA106-3B yeast were transformed with empty vector, wild-type MBP-PpPDK1-6His, or an MBP-PpPDK1-6His PIF-binding pocket mutant and analyzed as in Supplemental Figure S1B. Unexpectedly, only yeast transformed with MBP-PpPDK1K67Q-6His, which completely lacks kinase activity (Fig. 4, A and B), was unable to fully complement PKH1/2, as assessed by growth at 35°C (Fig. 4D). Yeast transformed with MBP-PpPDK1K71A-6His, which is capable of minimal autophosphorylation (Fig. 4A) and transphosphorylation (Fig. 4B), displayed comparable growth at 35°C to yeast transformed with wild-type MBP-PpPDK1-6His (Fig. 4D). Western blotting with α-MBP confirmed the expression of all MBP-PpPDK1-6His proteins (Fig. 4D). These results suggest that, while kinase-active PpPDK1 is required to perform tasks related to cell survival and growth, very low levels of activity are sufficient to enable PpPDK1 to fulfill these vital functions.
Analysis of pdk1 Knockout and PpPDK1-6His and PpPDK1K71A-6His Transformed Moss
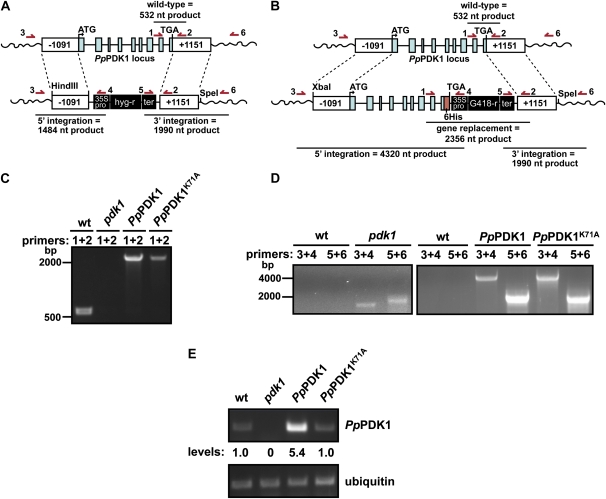
In every organism studied so far except Arabidopsis (Camehl et al., 2011), deletion of PDK1 is lethal (Casamayor et al., 1999; Niederberger and Schweingruber, 1999; Rintelen et al., 2001; Lawlor et al., 2002; Devarenne et al., 2006). To test whether deletion of PpPDK1 is lethal, we attempted to knock out PpPDK1 via homologous recombination using a construct containing the hygromycin resistance marker between 5′ and 3′ PpPDK1-targeting fragments (Fig. 5A). At the same time, we took an alternative approach in case the pdk1 knockout line was lethal. Because PpPDK1K71A had the largest reduction in kinase activity (Fig. 4, A and B) but still enabled yeast viability (Fig. 4D), we used gene targeting to replace the endogenous PpPDK1 with either PpPDK1-6His or PpPDK1K71A-6His (Fig. 5B). After transformation and two rounds of antibiotic selection, the surviving moss colonies were PCR genotyped using primers 1 and 2 shown in Figure 5, A and B.
Figure 5.
Production of a pdk1 knockout line and replacement of endogenous PpPDK1 with PpPDK1-6His. A and B, The constructs used to replace the endogenous PpPDK1 locus with a hygromycin marker to create a pdk1 knockout line (A) and to replace the endogenous PpPDK1 locus with PpPDK1-6His or PpPDK1K71A-6His (B). The 6× His tag (maroon box) is not drawn to scale for ease of visibility. Dashed lines indicate regions of homologous recombination. The locations of genotyping primers used in C and D are depicted with arrows above the diagrams. nt, Nucleotides. C, PCR-based genotyping with primers 1 + 2 shows that the indicated P. patens transformants do not contain a wild-type (wt) PpPDK1 gene. D, PCR-based genotyping of the indicated P. patens transformants showing the proper integration of constructs into the endogenous PpPDK1 locus. Wild-type P. patens genomic DNA was used as a negative control for integration of the constructs. E, RT-PCR analysis of gene expression for the indicated genes. Analysis of ubiquitin gene expression was used as an internal control. [See online article for color version of this figure.]
Initial genotyping showed that the majority of the pdk1 knockout transformants did not lack the endogenous PpPDK1 gene, whereas the majority of gene replacement transformants yielded genotyping products for both the endogenous PpPDK1 (532 nucleotides) and PpPDK1-6His (2,356 nucleotides; data not shown), suggesting that these colonies might have been unstably transformed. Further analysis identified one pdk1 knockout line, one PpPDK1-6His transformant, and one PpPDK1K71A-6His transformant. A representative genotyping PCR is shown in Figure 5C, indicating that the pdk1 knockout line lacked the endogenous PpPDK1 gene and that the gene replacement lines lacked the endogenous copy of PpPDK1 and were most likely stable transformants.
Additional genotyping was used to verify 5′ and 3′ integration of all constructs into the endogenous PpPDK1 locus of the P. patens genome. Using the primer combinations 3 + 4 and 5 + 6 shown in Figure 5, A and B, PCR products of the correct size were amplified from genomic DNA extracted from the pdk1 knockout line and the gene replacement lines, but not from wild-type P. patens (Fig. 5D). This indicated that all transformants had integrated the exogenous DNA into the PpPDK1 locus. These PCR products were cloned and sequenced to confirm that proper 5′ and 3′ integration had occurred.
Finally, Southern-blot hybridization was performed to verify that PpPDK1-6His and PpPDK1K71A-6His were present in the correct location in the P. patens genome. Southern-blot analysis was not carried out on the pdk1 knockout line due to a lack of large amounts of viable tissue (see below). A probe based in the 35S promoter of the G418 resistance cassette (Supplemental Fig. S6A) hybridized to a predicted DNA fragment of approximately 5 kb in NdeI/SmaI/XbaI-digested genomic DNA from PpPDK1-6His and PpPDK1K71A-6His strains, but not from wild-type P. patens (Supplemental Fig. S6B), confirming that the PpPDK1-6His constructs had integrated into the endogenous PpPDK1 locus. The PpPDK1-6His transformant produced four additional hybridizing DNA fragments, whereas the PpPDK1K71A-6His transformant produced one additional hybridizing fragment, suggesting that both strains probably contain multiple insertions of the PpPDK1-6His constructs into the PpPDK1 locus, which is a common occurrence in transformed lines of P. patens (Schaefer and Zrÿd, 1997; Schaefer, 2002). Overall, these results indicate that the transformed moss strains lack the endogenous PpPDK1 gene and contain at least one copy of PpPDK1-6His or PpPDK1K71A-6His DNA that have been integrated into the PpPDK1 locus by homologous recombination.
In order to confirm the loss of PpPDK1 expression in the pdk1 knockout line and the expression of the PpPDK1-6His and PpPDK1K71A-6His constructs, reverse transcription (RT)-PCR expression analysis was performed on all transformants as described previously using ubiquitin as a control (Harries et al., 2005). PpPDK1 mRNA was absent from the pdk1 knockout line and was present at similar levels in wild-type and PpPDK1K71A-6His moss, whereas PpPDK1-6His moss contained higher levels of PpPDK1 mRNA (Fig. 5E). These results indicate that the pdk1 knockout line lacks the endogenous PpPDK1 and the gene replacement lines express either PpPDK1-6His or PpPDK1K71A-6His. Attempts to pull down PpPDK1-6His and PpPDK1K71A-6His proteins from moss using Ni2+ resin were unsuccessful (data not shown), so future efforts to purify PpPDK1 from moss may require the use of additional tags, such as a tandem affinity purification tag, or robust overexpression of PpPDK1.
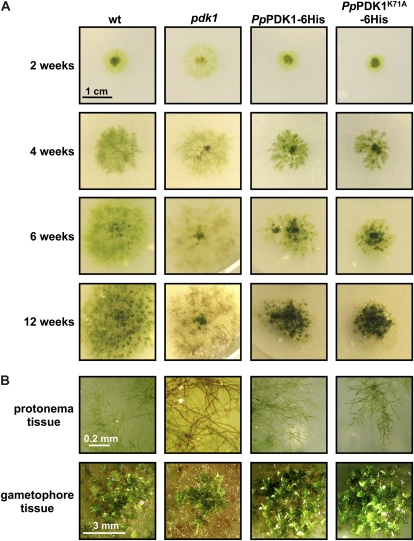
To assess macroscopic effects from the loss of PDK1 and the K71A mutation on P. patens growth and development, wild-type, pdk1 knockout, PpPDK1-6His, and PpPDK1K71A-6His strains were grown and allowed to develop protonema and leafy gametophore tissue over a 12-week time course. The pdk1 knockout line and wild-type moss grew similarly for the first 2 weeks (Fig. 6A). However, at later time points, the pdk1 knockout line produced fewer protonema, some of which began to turn brown and die (Supplemental Fig. S7). By 6 weeks, significant browning of the pdk1 knockout protonema was seen, and at 12 weeks, much of the protonemal tissue was brown, although some green filaments were still visible (Fig. 6). A small amount of apparently healthy gametophore tissue formed in the center of the pdk1 knockout colony (Fig. 6), but the majority of the tissue appeared to be dead. For the gene replacement lines, the colony viability and the sizes of both transformed strains appeared to be similar to wild-type moss for the first 4 weeks, after which expansion of the transformed moss colonies through protonemal growth was reduced compared with wild-type moss (Fig. 6A). At the same time, the production of gametophore tissue in the gene replacement lines appeared to form earlier than in the wild-type moss, but it was not apparently different in morphology from the wild-type moss (Fig. 6). This result is in agreement with yeast complementation results (Fig. 4D) and suggests that the minimally active PpPDK1K71A is able to carry out essential functions related to moss growth and viability. It should be noted that the reduced colony size of the gene replacement moss lines could be due to improper RNA processing, since the vector used for gene replacement lacks a terminator sequence for the introduced PpPDK1 gene. However, this vector has been used previously for gene replacement without noticeable differences from the wild type (Shakirov et al., 2010; Spinner et al., 2010). Additionally, from the pdk1 knockout line, it appears that the loss of PpPDK1 is not completely lethal but does not produce healthy moss tissue. In comparison, this would suggest that the PpPDK1-6His and PpPDK1K71A-6His constructs in the gene replacement lines are functional, since this moss tissue was fully viable and did not show browning like the pdk1 knockout line.
Figure 6.
pdk1 knockout, PpPDK1-6His, and PpPDK1K71A-6His moss macroscopic phenotypes compared with wild-type (wt) P. patens. A, The indicated moss lines were plated and photographed over a 12-week period. B, Images of protonema and gametophore tissue for the indicated moss lines taken at 12 weeks. [See online article for color version of this figure.]
As an additional test of the functionality of the PpPDK1-6His and PpPDK1K71A-6His transformants and the impaired growth of the pdk1 knockout line, we tested whether these moss lines were hypersensitive to heat (30°C) and osmotic stress (0.9 m mannitol). For heat stress, incubation at 30°C for 14 d gave surviving tissue in all moss lines except for the pdk1 knockout line, which appeared to be completely dead (Supplemental Fig. S8A). After 14 d of recovery at 25°C, all moss lines except the pdk1 knockout line were able to recover and grow healthy, viable tissue that resembled the unstressed control (Supplemental Fig. S8A). Similarly, the pdk1 knockout line was most severely affected by 30 d of growth on 0.9 m mannitol, and 14 d of recovery without mannitol restored the growth of all lines except the pdk1 knockout line (Supplemental Fig. S8B). These data suggest that the PpPDK1-6His and PpPDK1K71A-6His constructs in the gene replacement lines are able to function comparably to the wild-type PpPDK1, but the pdk1 knockout line is compromised in both normal growth and in response to heat and osmotic stresses.
Features of PDK1s from Algae, Primitive Land Plants, and Angiosperms
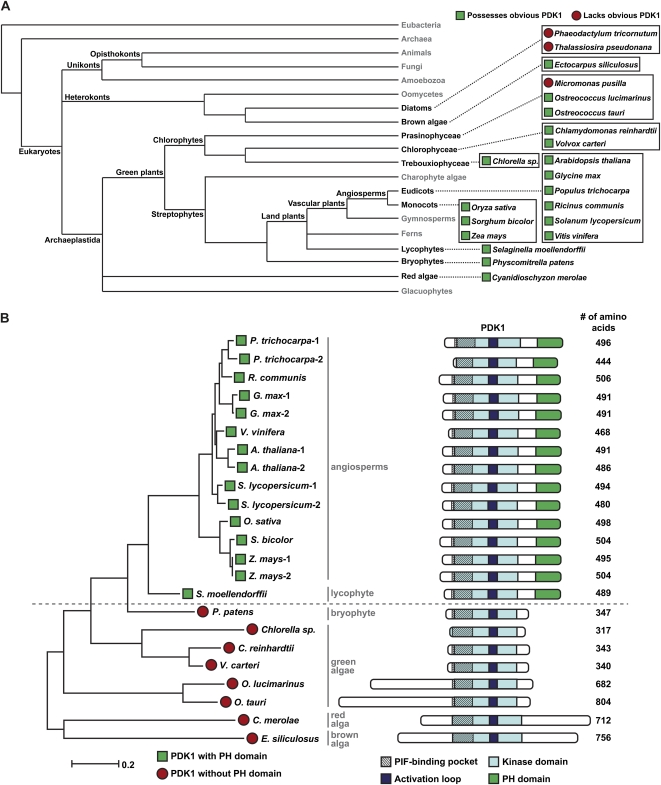
Given the position of P. patens as a representative of early land plants, we sought to compare the PpPDK1 sequence with those of other plants to gain insights into the evolution of plant PDK1 protein domain composition and functions. With the genome sequences from many different higher and lower plants currently available, it is now possible to do such an analysis based on a comparison of PDK1 protein domain features. We obtained 23 PDK1 sequences from 10 higher plants (nine angiosperms, one lycophyte) and eight lower plants (one bryophyte, five green algae, one red alga, one brown alga) for a comparative analysis (Fig. 7A). Interestingly, a search of the genomes of two diatoms (Thalassiosira pseudonana, http://genome.jgi-psf.org/Thaps3/Thaps3.home.html;Phaeodactylum tricornutum, http://genome.jgi-psf.org/Phatr2/Phatr2.home.html) and one additional green alga (Micromonas pusilla, http://genome.jgi-psf.org/MicpuC2/MicpuC2.home.html) did not reveal obvious PDK1 sequence homologs (Fig. 7A), raising the possibility that these organisms either lack PDK1 altogether or possess PDK1 genes that are very different from known sequences, which could make them difficult to identify by sequence homology. It is also possible that their genome sequences require further annotation to identify PDK1 sequences.
Figure 7.
Phylogenetic relationship and protein domains of PDK1 proteins from plants. A, Cladogram showing the evolutionary relationship of plants used in B. B, Comparison of PDK1 protein sequences. On the left side, the indicated proteins were aligned and a maximum-likelihood phylogenetic tree was produced using MEGA5. On the right side, the indicated domains were identified in each protein and aligned based on the activation loop domain. The horizontal dashed line indicates the division between vascular (above the line) and nonvascular (below the line) plants. [See online article for color version of this figure.]
Next, we analyzed all 23 identified PDK1 protein sequences for conserved features, including kinase domain, PIF-binding pocket, activation loop, and PH domain. Multiple sequence alignment revealed that the kinase domain of every PDK1 analyzed possesses conserved PIF-binding pocket residues but that algal and P. patens PDK1s differ from vascular plant PDK1s in the lack of a PH domain (Fig. 7B; Supplemental Fig. S9). As might be expected, a maximum-likelihood tree constructed from all 23 plant PDK1 sequences closely resembled the known phylogenetic placement of these plants, with higher plant PDK1s much more closely related to each other than to primitive land plant or algal PDK1s (Fig. 7B). Our sequence analysis suggests that, while many residues in the PDK1 catalytic domain and PIF-binding pocket have been maintained in highly divergent plant taxa throughout hundreds of millions of years of evolution, the lipid-binding PH domain may only be required in higher plant PDK1s and is not a characteristic feature of PDK1s from nonvascular land plants and algae.
DISCUSSION
Regulation of many basic processes in eukaryotic cells occurs through the phosphorylation of several members of the AGC kinase subfamily by PDK1 (Bögre et al., 2003; Bayascas, 2010). Thus, PDK1 function appears to be highly conserved among many different organisms, suggesting that PDK1 is an evolutionarily ancient gene. While several PDK1 genes have been reported from higher plants (Deak et al., 1999; Devarenne et al., 2006; Matsui et al., 2010), PDK1 from organisms representative of more ancient plant systems, such as lower, nonvascular plants, has not been reported. Here, were identified and characterized such a PDK1 gene from the moss P. patens, and these studies offer insights into deciphering how the PDK1 gene may have evolved within plants.
Is PpPDK1 a True PDK1?
From our studies, it may be questioned whether PpPDK1 is truly a PDK1, since it lacks a PH domain. However, in the 14 years since the human PDK1 was identified, the definition of what constitutes a PDK1 protein has emerged. Containing a PH domain does not appear to be a requirement to catalog a protein as a PDK1, since a PH domain it is not required for the function of yeast Pkh1/2 (Casamayor et al., 1999). These proteins were identified as PDK1s by the similarity of the kinase domains to known PDK1s, the ability of Pkh1/2 to phosphorylate known AGC kinase substrates, and the ability of human PDK1 to rescue yeast lacking PKH1/2 (Casamayor et al., 1999). Thus, the conserved characteristics that define a PDK1 protein include the presence of a PIF-binding pocket and the ability to phosphorylate AGC kinase substrates at the conserved activation loop phosphorylation site (Bögre et al., 2003). Some PDK1 proteins have also been shown to phosphorylate the activation loop site on AGC kinases from a different species. For example, AtPDK1 can phosphorylate and activate endogenous AGC kinases (Anthony et al., 2004, 2006; Zegzouti et al., 2006a, 2006b) as well as the mammalian AGC kinase PKB (Deak et al., 1999). PDK1 proteins from other species have also been identified by the ability of the associated gene to complement the lethality of the yeast pkh1/2 knockout (Deak et al., 1999).
The PpPDK1 identified here meets all of these definitions of PDK1s and thus should be considered a true PDK1. PpPDK1 contains a PIF-binding pocket that mediates the interaction with substrates (Fig. 4D), phosphorylates P. patens and tomato AGC kinases at the conserved activation loop site (Fig. 3B), and can complement the yeast pkh1/2 knockout (Figs. 2 and 4D). These complementation assays indicate that PpPDK1 can regulate some of the known PDK1 functions in a heterologous system and suggest that PpPDK1 may carry out the same functions within P. patens. These studies would also suggest that the mechanism for activation of the substrates by PpPDK1 is conserved with what has been discovered for other PDK1s.
The PpPDK1 Knockout Is Developmentally Compromised
Unlike in other organisms, deletion of PpPDK1 was not lethal, even though much of the pdk1 knockout moss colony appeared dead after 12 weeks of growth. Subsequent replating can restore the growth of green protonema, but the same browning, nonviable phenotype appears as the moss ages. This would suggest that the pdk1 knockout line has severe developmental abnormalities. The only other report of a nonlethal pdk1 knockout is for Arabidopsis (Camehl et al., 2011). However, plants lacking both AtPDK1-1 and AtPDK1-2 are stunted and less fertile than the wild type (Camehl et al., 2011), suggesting that a loss of PDK1 in plants leads to growth retardation. The only other plant that was analyzed for the loss of PDK1 was tomato, where loss of PDK1 by virus-induced gene silencing was lethal (Devarenne et al., 2006). Confirmation of lethality in tomato would require the production of a PDK1 knockout, which is currently not possible in tomato. These contradictory results regarding the effects of PDK1 loss in photosynthetic organisms suggest that divergent roles for PDK1 may have evolved within plants. Further analysis of PDK1 from additional plants will be required to clarify this situation.
It is also interesting that the minimally kinase-active PpPDK1K71A protein could confer viability and does not produce the browning, nonviable phenotype seen with the pdk1 knockout line (Fig. 6). The PpPDK1K71A protein was also capable of supporting viability in yeast (Fig. 4D). There are several possible explanations for why PpPDK1 proteins with drastically reduced kinase activity are still able to confer cell viability. First, the PpPDK1 mutations did not completely eliminate interaction with a P. patens AGC kinase (Fig. 4C); thus, any phosphorylation taking place could be sufficient for cell viability. Second, some functions of PDK1 may be independent of interaction with AGC kinases. For example, several non-AGC kinase proteins have been identified in mammals that are required for PDK1 function (Nakamura et al., 2008; Sephton et al., 2009). Finally, interaction of PDK1 with some substrates is independent of PIF binding (Collins et al., 2003). For example, in mammalian cells, PKB, which also has a PH domain for lipid binding, appears to associate with PDK1 through the lipid interaction of their PH domains (Collins et al., 2003). This would suggest that lipid binding is sufficient to activate some PDK1 substrates. However, PpPDK1 does not have a PH domain (Fig. 1B) and does not strongly bind lipids (Supplemental Fig. S3). Thus, it will be of interest to determine if novel mechanisms exist in P. patens for PDK1 interaction with substrates. Additionally, more studies with the P. patens PDK1K71A line generated here will be needed to determine if there is altered interaction with and activation of endogenous AGC kinase substrates.
Comparison of PpPDK1 with Other PDK1 Genes
Our phylogenetic analysis shows that there is a clear difference between higher (vascular) and lower (nonvascular) plants; the PDK1 proteins from these classes of plants can be distinguished by the presence or absence of the PH domain, respectively (Fig. 7B). Thus, it is possible that the PH domain for lipid binding is an aspect of PDK1 function that developed at the time of vascular system formation. Additionally, the sequences for the green algae Osterococcus lucimarinus and Osterococcus tauri, the red alga Cyanidioschyzon merolae, and the brown alga Ectocarpus siliculosus contain extensive amino acid sequences on the N-terminal and C-terminal sides of the kinase domain (Fig. 7B). The significance of these extra sequences toward function remains to be determined, since the activity of these proteins has not been reported.
The possible lack of PDK1 sequences from diatoms raises some interesting questions. Diatoms can be considered more recently evolved algae in relation to green and red algae (Sims et al., 2006; Moustafa et al., 2009). Thus, the diatom eukaryotic nonphotosynthetic ancestor may not have contained a PDK1-like sequence(s), they may have lost their PDK1 gene(s) since the main endosymbiotic event, or they may have PDK1-like sequences divergent enough from conserved PDK1s that they are not recognizable in typical comparison searches. Further analysis of diatom genomes for PDK1-like sequences will help to clarify this situation.
Our analysis of plant PDK1 proteins is limited by the number of PDK1 sequences from lower plants and nonflowering vascular plants. With the addition of genome sequences from additional lower plants, such as the bryophyte liverwort Marchantia polymorpha (currently, the M. polymorpha EST sequences contain 4,024 contigs but not an obvious PDK1 sequence homolog; http://www.genome.jp/kegg-bin/show_organism?org=empm), more green, red, and brown algae, and vascular plants such as ferns, this comparison will become more robust and allow for a more concrete assessment of the domain evolution of PDK1 proteins.
MATERIALS AND METHODS
Cloning and Site-Directed Mutagenesis
Physcomitrella patens PDK1 (Pp1s217_11V6.2 at Phytozome; GenBank accession no. N049607), Pp188412 (Pp1s118_230V6 at Phytozome; GenBank accession no. JN049610), and Pp174181 (Pp1s4_386V6.1 at Phytozome; GenBank accession no. JN049609) coding sequences were identified by searching Phytozome using the Arabidopsis (Arabidopsis thaliana) AtPDK1 sequence (GenBank accession no. AF132742.1; Deak et al., 1999). Pp2484 (Pp1s224_73V6.1 at Phytozome; GenBank accession no. JN049608) was identified by searching Phytozome using the tomato (Solanum lycopersicum) Adi3 sequence (GenBank accession no. AY849914; Devarenne et al., 2006).
Total RNA was extracted from P. patens and Arabidopsis tissue using TRIzol reagent (Invitrogen), and RT was performed using SuperScript III reverse transcriptase (Invitrogen). Full-length cDNAs for the following genes were amplified from first-strand cDNA using forward primers beginning at the ATG start codon and reverse primers beginning at the stop codon or last amino acid: PpPDK1, forward primer 5′-ATGGCCATGGATGGGACC-3′, reverse primer 5′-TACGTCGTATACAAATGCATCCAAACC-3′; Pp188412, forward primer 5′-ATGGCGGCAACAAATCGCATTAAC-3′, reverse primer 5′-CTATGATGAGCCCAGCCACG-3′; Pp174181, forward primer 5′-ATGACACTTGCTACCACTTTCAGG-3′, reverse primer 5′-TTATGAGGATTCCCCACTCTTGAG-3′; AtPDK1, forward primer 5′-ATGTTGGCAATGGAG-3′, reverse primer 5′-GCGGTTGTGAAGAGTC-3′. The annotated full-length 2,328-nucleotide Pp2484 cDNA could not be amplified from first-strand cDNA. Instead, a 1,560-nucleotide cDNA for Pp2484 was amplified from first-strand cDNA using forward primer 5′-ATGAGTGGAAAGTTGAGCATGAG-3′ and reverse primer 5′-TCAAAAAAAGTCAAAATCCACGTATGAC-3′. The amplified 1,560-nucleotide Pp2484 cDNA lacks the first 768 nucleotides of exon 1, but the encoded protein contains all conserved AGC kinase elements. Cloning of SlPDK1 and Adi3 was previously reported (Devarenne et al., 2006). PpPDK1, AtPDK1, and SlPDK1 lacking the endogenous stop codon were cloned into pET22-b(+) (Novagen) to incorporate a C-terminal 6× His tag. PpPDK1-6His, AtPDK1-6His, and SlPDK1-6His were amplified from pET22-b(+) and cloned into pMAL-c2x (New England Biolabs) for N-terminal MBP translational fusions. Pp188412, Pp147181, and Pp2484 without a 6× His tag were also cloned into pMAL-c2x.
Site-directed mutagenesis was performed on genes cloned into pMAL-c2x using Pfu Turbo (Stratagene) according to the manufacturer’s instructions. Finally, MBP-PpPDK1-6His, MBP-AtPDK1-6His, MBP-SlPDK1-6His, MBP-Pp188412, MBP-Pp174181, and MBP-PpPDK1-6His point mutants were cloned into the plasmid p416GPD (Mumberg et al., 1995) for yeast complementation studies. p416GPD contains a URA3 marker and a constitutive GPD promoter to drive the expression of the gene of interest.
Yeast Strains and Complementation of PKH1 and PKH2
Two previously described strains of Saccharomyces cerevisiae were used in complementation experiments: strain AC306 (Casamayor et al., 1999) was used for tetrad analysis of PKH1/2 complementation, and strain INA106-3B (Inagaki et al., 1999) was used for temperature sensitivity PKH1/2 complementation. AC306 is a diploid strain that is heterozygous for PKH1 and PKH2 deletion (genotype MATa/MATα PKH1/pkh1Δ::HIS3 PKH2/pkh2Δ::TRP1 ade2-1 can1-100 his3-11,15 leu2-13,112 trp1-1 ura3-1). INA106-3B is a haploid strain that lacks PKH2 and contains a point mutation in PKH1 (genotype MATa pkh1D398G pkh2Δ::LEU2 ade1 his3-2 trp1 ura3). Both strains were transformed with the PDK1 constructs in p416GPD.
After transformation, AC306 yeast transformed with the desired plasmid was grown at 30°C on plates lacking uracil. Sporulation was induced by incubating transformed yeast in 1% potassium acetate at 25°C for 7 d. After sporulation, tetrad dissections were performed on at least 30 tetrads of each sample using standard techniques. After dissection, spore genotype and viability were analyzed by replica plating on minimal medium and on medium lacking His, Trp, or uracil. After spore analysis, representative cultures of each haploid spore were grown at 30°C in liquid YPD medium and spotted on both YPD plates and plates containing 5-FOA (Research Products International), which selects for loss of the URA3-marked p416GPD plasmid.
For temperature-sensitive PKH1/2 complementation, liquid cultures of INA106-3B yeast transformed with the desired plasmid were grown at 25°C in liquid medium lacking uracil and spotted on two identical plates lacking uracil. One plate was incubated at 25°C for 4 d and the other plate was incubated at 35°C for 2 d, then the plates were viewed and photographs were taken.
To analyze the expression of all genes cloned into p416GPD, total protein was extracted as described previously (Yaffe and Schatz, 1984) from yeast grown at 30°C in liquid YPD medium (AC306 yeast) or at 25°C in liquid medium lacking uracil (INA106-3B yeast). MBP fusion proteins were then detected by α-MBP (1:10,000; New England Biolabs) western blot.
In Vitro Kinase Assays
PpPDK1-6His, Pp2411, AtPDK1-6His, SlPDK1-6His, Pp2411, and Adi3 were expressed as MBP fusion proteins using pMAL-c2 in Escherichia coli BL21(DE3) and purified with amylose resin (New England Biolabs) according to the manufacturer’s instructions. In vitro kinase assays were performed by combining the purified proteins in a 30-μL final volume of kinase buffer containing 10 mm Tris, pH 7.5, 10 mm MgCl2 or 10 mm MnCl2, and 1 mm dithiothreitol. The reactions were started by the addition of 1 μCi of [γ-32P]ATP and nonradiolabeled ATP to a final concentration of 20 μm followed by incubation at room temperature for 15 min. Reactions were stopped by the addition of 4× SDS-PAGE sample buffer. Protein phosphorylation was visualized by a phosphorimager (Bio-Rad Molecular Imager) after separation by SDS-PAGE, and signals were quantified using Quantity One software (Bio-Rad). All kinase assays were performed a minimum of three times, with representative images shown in the figures.
In Vitro Pulldown Assays
MBP, MBP-Pp2411, and MBP-PpPDK1-6His were purified with amylose resin as described above. Five micrograms of purified MBP-PpPDK1-6His was added to 10 μL of Ni2+ resin (Novagen) in 200 μL of binding buffer (20 mm Tris-HCl, pH 7.9, 500 mm NaCl, 5 mm imidazole, and 0.1% Triton-X-100) and mixed at 4°C for 1 h to allow MBP-PpPDK1-6His to bind the resin. Next, 5 μg of purified MBP or MBP-Pp2411 was added to the sample and mixed at 4°C for 1 h to allow the proteins to interact with MBP-PpPDK1-6His. After the incubation period, the samples were centrifuged at 400g for 1 min, the supernatant was removed, and the samples were washed three times with 1 mL of binding buffer followed by the addition of 100 μL of 4× SDS-PAGE sample buffer. Ten microliters of each sample was then separated by SDS-PAGE, and MBP fusion proteins were detected by α-MBP western blot. Protein inputs were analyzed by performing SDS-PAGE on 5 μg of purified MBP, MBP-Pp2411, and MBP-PpPDK1-6His and staining with Coomassie blue.
Protein-Lipid Overlays
Binding of MBP, MBP-AtPDK1-6His, MBP-SlPDK1-6His, and MBP-PpPDK1-6His to several common phospholipids and sphingolipids was tested by protein-lipid overlay using PIP Strips and Sphingo Strips (Echelon Bioscience) according to the manufacturer’s instructions and as described previously (Zegzouti et al., 2006b). Briefly, lipid strips were blocked with 3% fatty acid-free bovine serum albumin in 1× phosphate-buffered saline for 16 h, 5 μg of each purified protein was incubated with a lipid strip for 1.5 h at room temperature, the lipid strips were washed three times with 1× phosphate-buffered with 0.1% Tween 20, and proteins bound to lipids were detected using α-MBP as with typical western blots.
Tissue Culture of P. patens and Generation of pdk1 Knockout and PpPDK1-6His Constructs
Wild-type P. patens cv Gransden (Ashton and Cove, 1977) was routinely grown on BCD plates (Roberts et al., 2011) overlaid with cellophane discs (AA Packaging) at 25°C in continuous light following standard protocols (Roberts et al., 2011). Seven days before transformation, wild-type P. patens was passaged to BCD plates containing 5 mm diammonium tartrate. For each moss stress treatment, all four moss lines (wild type, pdk1 knockout, PpPDK1-6His, and PpPDK1K71A-6His) were grown on the same plate to ensure that results were not due to variation between plates. For heat stress, moss lines were moved to BCD plates and incubated at 30°C for 14 d, then allowed to recover on the same plate at 25°C for 14 d. For osmotic stress, moss lines were moved to BCD plates containing 0.9 m mannitol for 14 d, then moved to new BCD plates lacking mannitol and allowed to recover for 14 d.
For both knockout and gene replacement generation, pBHRF and pBNRF vectors were used, respectively. In short, pBNRF and pBHRF contain 35S:NptII (G418/kanamycin resistance) or 35S:Hph (hygromycin resistance) EcoRI fragments from pHP23b and pGL2, respectively, cloned into the EcoRI site of pBilox, a derivative of pMCS5 (MoBiTec), with two direct repeats of the loxP sites cloned into the XhoI/KpnI and BglII/SpeI sites (Schaefer et al., 2010). The construct used to generate the pdk1 knockout line was made by amplifying a 1,091-nucleotide 5′ targeting fragment from genomic DNA with primers 5′-GACAAGCTTCTCAGAAGTGCAAAGGCTTTCATTC-3′ (HindIII restriction site in boldface) and 5′-GACCTCGAGTCCTGTAATGTTGGTGCC-3′ (XhoI restriction site in boldface) and cloned into the HindIII and XhoI sites of the vector pBHRF (Schaefer et al., 2010) upstream of Hph. Similarly, a 1,151-nucleotide 3′ targeting fragment was amplified from genomic DNA with primers 5′-GACGCGGCCGCATTATCAGTAGACTCACATG-3′ (NotI restriction site in boldface) and 5′-GACAGATCTGATATGTAACACCAACCTAG-3′ (BglII restriction site in boldface) and cloned into the NotI and BglII sites of pBHRF, 3′ to Hph. A diagram of the complete construct can be found in Figure 5A.
The construct used to replace the endogenous PpPDK1 locus with PpPDK1-6His or PpPDK1K71A-6His was made by amplifying a 1,091-nucleotide 5′ targeting fragment plus the entire PpPDK1 coding region from genomic DNA with primers 5′-AAGTGCAAAGGCTTTCAATC-3′ and 5′-TACGTCGTATACAAATGCATCC-3′ and cloning into pCR2.1-TOPO (Invitrogen). A C-terminal 6× His tag, a 5′ XbaI site, and a 3′ XhoI site were added to this PpPDK1 by PCR and cloned into the XbaI and XhoI sites of pBNRF upstream of NptII. Similarly, a 1,151-nucleotide 3′ targeting fragment was amplified from genomic DNA with primers 5′-GACGCGGCCGCATTATCAGTAGACTCACATG-3′ (NotI restriction site in boldface) and 5′-GACAGATCTGATATGTAACACCAACCTAG-3′ (BglII restriction site in boldface) and cloned into the NotI and BglII sites of pBNRF, 3′ to NptII. A diagram of the complete construct can be found in Figure 5B. The K71A mutation was then introduced by site-directed mutagenesis using Pfu Turbo according to the manufacturer’s instructions.
To transform P. patens, 50 μg of each construct was linearized by simultaneous digestion with 80 units of HindIII and SpeI for pdk1 knockout generation or XbaI and SpeI for the PpPDK1 gene replacement. Linearized DNA was introduced into wild-type P. patens cv Gransden protoplasts using a previously described polyethylene glycol transformation protocol (Roberts et al., 2011). Transformants were initially selected by 7 d of growth on plates containing 20 μg mL−1 hygromycin or 25 μg mL−1 G418 followed by 10 d of growth on nonselective plates and an additional 7 d of growth on hygromycin or G418 plates. Surviving colonies were analyzed by genotyping PCR and Southern blotting (see below) for integration of the PpPDK1-6His constructs into the correct location in the genome.
Genomic DNA Extraction
Genomic DNA was extracted from each moss colony surviving two rounds of hygromycin or G418 selection by grinding the tissue (approximately 10–25 mm2 per colony) in a microcentrifuge tube with 200 μL of extraction buffer (2% hexadecyl-trimethyl ammonium bromide, 100 mm Tris, pH 8.0, 20 mm EDTA, and 1.4 m NaCl) followed by incubation at 65°C for 30 min. An equal volume (200 μL) of phenol/chloroform was added, and the mixture was vortexed vigorously for 15 s, followed by centrifugation at 15,000g for 15 min. The upper aqueous phase was added to a new microcentrifuge tube containing 170 μL of isopropanol and 10 μL of 3 m sodium acetate, pH 5.2. Samples were incubated at −20°C for 1 h followed by centrifugation at 15,000g for 20 min. The supernatant was aspirated, and the pellets were washed with 200 μL of 70% ethanol followed by centrifugation at 15,000g for 20 min. The pellets were dried for 1 h at room temperature and resuspended at 37°C in 20 μL of autoclaved deionized water containing 10 μg of RNase A.
Genotyping
Genomic DNA extracted as described above was subjected to genotyping PCR. Initial genotyping was performed with primers that amplify a 532-nucleotide product if the endogenous PpPDK1 is present, a 2,356-nucleotide product if a PpPDK1-6His construct is present, and no product if the endogenous PpPDK1 is not present: primer 1, 5′-GAGAGACTGGGAGTTCAAGGCTATG-3′, and primer 2, 5′-CTCCAACTGGTAATTAATGCTGCAATGG-3′. A diagram of the locations of these genotyping primers can be found in Figure 5, A and B.
Additional genotyping PCR was used to test for proper 5′ and 3′ integration of the pdk1 knockout and PpPDK1-6His constructs. The first set of primers tests for 5′ integration. These primers are based outside the 5′ targeting fragment (primer 3) and in the 35S promoter (primer 4) and amplify the entire 5′ targeting fragment for the pdk1 knockout construct and the entire 5′ targeting fragment plus the PpPDK1-6His coding sequence for the gene replacement constructs (for primer locations, see Fig. 5, A and B): primer 3, 5′-GGTAGGTGGTATTTCTAACACTCAATGATG-3′, and primer 4, 5′-CGTGCTCCACCATGTTGACGAAG-3′. The second set of primers tests for 3′ integration. These primers are based in the NptII gene (primer 5) and outside the 3′ targeting fragment (primer 6) and amplify the entire 3′ targeting fragment for all constructs (for primer locations, see Fig. 5, A and B): primer 5, 5′-GCTGAAATCACCAGTCTCTCTCTAC-3′, and primer 6, 5′-GGCAATGGTTCAAAAACCTCTTATAAGTCC-3′.
Southern Blot
Southern-blot analysis was performed essentially as described previously (Nelson et al., 2011) to verify that PpPDK1-6His or PpPDK1K71A-6His was integrated into the correct location in the P. patens genome and to assess the number of integration events. Forty micrograms of genomic DNA extracted from wild-type, PpPDK1-6His, or PpPDK1K71A-6His moss was simultaneously digested with 120 units each of NdeI, SalI, and XbaI and separated on a 1% agarose gel, which was then transferred to a nylon membrane (GE Healthcare). The membrane was hybridized at 65°C with a High Prime (Roche) internally labeled DNA probe composed of 234 nucleotides of the 35S promoter used to drive NptII expression in pBNRF. This probe was amplified from the pBNRF vector using primers 5′-CGTCAACATGGTGGAGCACG-3′ and 5′-GCAGAGGCATCTTCAACGATGG-3′. A diagram of the location of the probe can be found in Supplemental Figure S6A.
Expression Analysis of PpPDK1 in Wild-Type, pdk1 Knockout, PpPDK1-6His, and PpPDK1K71A-6His Moss
Total RNA was extracted from P. patens tissue using TRIzol reagent (Invitrogen), and RT was performed using qScript cDNA SuperMix (Quanta Biosciences) according to the manufacturer’s instructions. RT-PCR was performed as described previously (Harries et al., 2005). As an expression control, ubiquitin was amplified with forward primer 5′-ACTACCCTGAAGTTGTATAGTTCGG-3′ and reverse primer 5′-CAAGTCACATTACTTCGCTGTCTAG-3′. PpPDK1 was amplified with forward primer 5′-TTCAAAGCTGCGACAGAATATTTGAC-3′ and reverse primer 5′-CTTGCCATTTTTCTTCTTCATCCAAAC-3′. Each gene was amplified by 30 cycles of PCR, utilizing a 30-s denaturing step at 94°C, a 30-s annealing step at 57°C, and a 30-s extension step at 72°C.
Phylogenetic Analysis of PDK1 Proteins
The following PDK1 protein sequences were obtained from the National Center for Biotechnology Information, Phytozome, and individual species genome databases using a BLAST search with either the Osterococcus tauri PDK1 sequence (accession no. XP_003078129.1 at GenBank) or the Arabidopsis PDK1-1 sequence (accession nos. AED90755.1 at GenBank and AT5G04510 at Phytozome): Arabidopsis PDK1-2 (accession nos. NP_187665.2 at GenBank and AT3G10540 at Phytozome), Cyanidioschyzon merolae PDK1 (CDS no. 2387, locus CMO090C from genome Web site http://merolae.biol.s.u-tokyo.ac.jp/), Chlamydomonas reinhardtii PDK1 (accession nos. XP_001701378.1 at GenBank and Cre18.g749950 at Phytozome), Chlorella variabilis NC64A PDK1 (protein identifier 26267 from genome Web site http://genome.jgi-psf.org/ChlNC64A_1/ChlNC64A_1.home.html), Ectocarpus siliculosus PDK1 (accession nos. CBJ33478 at GenBank and Esi0491_0016 at http://bioinformatics.psb.ugent.be/webtools/bogas/overview/Ectsi), Glycine max PDK1-1 (accession nos. XP_002262670.1 at GenBank and Glyma10g34430 at Phytozome), G. max PDK1-2 (accession no. Glyma20g33140 at Phytozome), Osterococcus lucimarinus PDK1 (protein identifier 14571 from genome Web site http://genome.jgi-psf.org/Ost9901_3/Ost9901_3.home.html), Oryza sativa PDK1 (accession nos. BAF06862 at GenBank and LOC_Os01g65230 at Phytozome), Physcomitrella patens PDK1 (accession nos. JN049607 at GenBank and Pp1s217_11V6 at Phytozome), Populus trichocarpa PDK1-1 (accession nos. XP_002315349.1 at GenBank and POPTR_0010s23910.1 at Phytozome), P. trichocarpa PDK1-2 (accession no. POPTR_0008s02890.1 at Phytozome), Ricinus communis PDK1 (accession nos. XP_002533941.1 at GenBank and 27428.t000007 at Phytozome), Sorghum bicolor PDK1 (accession nos. XP_002458841.1 at GenBank and Sb03g041290 at Phytozome), tomato PDK1-1 (accession no. AAW38936 at GenBank), tomato PDK1-2 (gene model SL1.00sc03032_9.1.1 at genome Web site http://solgenomics.net), Selaginella moellendorffii PDK1 (accession nos. XP_002960408.1 at GenBank and 164063 at Phytozome), Volvox carteri PDK1 (accession nos. XP_002957069.1 at GenBank and 83913 at Phytozome), Vitis vinifera PDK1 (accession nos. CBI40191.3 at GenBank and GSVIVG01034673001 at Phytozome), Zea mays PDK1-1 (accession no. ABB71956.1 at GenBank), and Z. mays PDK1-2 (accession nos. ACG46841.1 at GenBank and GRMZM2G097821 at Phytozome).
A multiple alignment of all 23 PDK1 protein sequences was created using MUSCLE. A maximum-likelihood phylogenetic tree was then created from aligned PDK1 sequences using MEGA version 5 (Tamura et al., 2011), and the resulting phylogram was labeled in Adobe Illustrator. The cladogram showing phylogenetic relationships between plants whose PDK1 sequences were analyzed (Fig. 7A) was created in Adobe Illustrator based on published literature (Bhattacharya and Medlin, 1998; Merchant et al., 2007; Cock et al., 2010; Banks et al., 2011).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers N049607, JN049610, JN049609, JN049608, and JN049606.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Evidence for one PDK1 in the P. patens genome.
Supplemental Figure S2. PpPDK1 genomic DNA, cDNA, and protein sequences.
Supplemental Figure S3. PpPDK1 does not strongly bind phospholipids or sphingolipids.
Supplemental Figure S4. Characterization of the kinase activity for the P. patens AGC kinase Pp2484.
Supplemental Figure S5. Phosphorylation of Adi3 by PpPDK1 PIF-binding pocket mutants.
Supplemental Figure S6. Southern blot analysis of PpPDK1-His transformed moss.
Supplemental Figure S7. Protonema growth of wild-type and the pdk1 moss.
Supplemental Figure S8. Stress treatment of moss lines.
Supplemental Figure S9. Alignment of PDK1 proteins used for phylogenetic analysis.
Supplemental Table S1. Names of all genes used in this study.
Supplementary Material
Acknowledgments
We thank the other members of the Devarenne laboratory for comments and constructive discussions; Eugene Shakirov (Timothy Hall laboratory at Texas A&M University) and Pierre-Francois Perroud (Ralph Quatrano laboratory at Washington University) for advice with P. patens transformation; Mary Bryk (Texas A&M University) and Rachel Jordan (Mary Bryk laboratory) for help with tetrad analysis; Jeremy Thorner (University of California, Berkeley) for donation of the AC306 Δpkh1/Δpkh2 heterozygous yeast strain; Ted Powers (University of California, Davis) for donation of the haploid INA106-3B pkh2Δ/pkh1D398G temperature-sensitive yeast strain; and Andrew Nelson (Dorothy Shippen laboratory at Texas A&M University) for help with Southern-blot analysis.
References
- Alessi DR, James SR, Downes CP, Holmes AB, Gaffney PR, Reese CB, Cohen P. (1997) Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Bα. Curr Biol 7: 261–269 [DOI] [PubMed] [Google Scholar]
- Anthony RG, Henriques R, Helfer A, Mészáros T, Rios G, Testerink C, Munnik T, Deák M, Koncz C, Bögre L. (2004) A protein kinase target of a PDK1 signalling pathway is involved in root hair growth in Arabidopsis. EMBO J 23: 572–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony RG, Khan S, Costa J, Pais MS, Bögre L. (2006) The Arabidopsis protein kinase PTI1-2 is activated by convergent phosphatidic acid and oxidative stress signaling pathways downstream of PDK1 and OXI1. J Biol Chem 281: 37536–37546 [DOI] [PubMed] [Google Scholar]
- Ashton NW, Cove DJ. (1977) Isolation and preliminary characterisation of auxotrophic and analogue resistant mutants of the moss, Physcomitrella patens. Mol Gen Genet 154: 87–95 [Google Scholar]
- Banks JA, Nishiyama T, Hasebe M, Bowman JL, Gribskov M, dePamphilis C, Albert VA, Aono N, Aoyama T, Ambrose BA, et al. (2011) The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science 332: 960–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayascas JR. (2008) Dissecting the role of the 3-phosphoinositide-dependent protein kinase-1 (PDK1) signalling pathways. Cell Cycle 7: 2978–2982 [DOI] [PubMed] [Google Scholar]
- Bayascas JR. (2010) PDK1: the major transducer of PI 3-kinase actions. Curr Top Microbiol Immunol 346: 9–29 [DOI] [PubMed] [Google Scholar]
- Bhattacharya D, Medlin L. (1998) Algal phylogeny and the origin of land plants. Plant Physiol 116: 9–15 [Google Scholar]
- Biondi RM. (2004) Phosphoinositide-dependent protein kinase 1, a sensor of protein conformation. Trends Biochem Sci 29: 136–142 [DOI] [PubMed] [Google Scholar]
- Biondi RM, Cheung PC, Casamayor A, Deak M, Currie RA, Alessi DR. (2000) Identification of a pocket in the PDK1 kinase domain that interacts with PIF and the C-terminal residues of PKA. EMBO J 19: 979–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biondi RM, Komander D, Thomas CC, Lizcano JM, Deak M, Alessi DR, van Aalten DM. (2002) High resolution crystal structure of the human PDK1 catalytic domain defines the regulatory phosphopeptide docking site. EMBO J 21: 4219–4228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bögre L, Okrész L, Henriques R, Anthony RG. (2003) Growth signalling pathways in Arabidopsis and the AGC protein kinases. Trends Plant Sci 8: 424–431 [DOI] [PubMed] [Google Scholar]
- Borgatti P, Martelli AM, Tabellini G, Bellacosa A, Capitani S, Neri LM. (2003) Threonine 308 phosphorylated form of Akt translocates to the nucleus of PC12 cells under nerve growth factor stimulation and associates with the nuclear matrix protein nucleolin. J Cell Physiol 196: 79–88 [DOI] [PubMed] [Google Scholar]
- Calleja V, Alcor D, Laguerre M, Park J, Vojnovic B, Hemmings BA, Downward J, Parker PJ, Larijani B. (2007) Intramolecular and intermolecular interactions of protein kinase B define its activation in vivo. PLoS Biol 5: e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camehl I, Drzewiecki C, Vadassery J, Shahollari B, Sherameti I, Forzani C, Munnik T, Hirt H, Oelmüller R. (2011) The OXI1 kinase pathway mediates Piriformospora indica-induced growth promotion in Arabidopsis. PLoS Pathog 7: e1002051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casamayor A, Torrance PD, Kobayashi T, Thorner J, Alessi DR. (1999) Functional counterparts of mammalian protein kinases PDK1 and SGK in budding yeast. Curr Biol 9: 186–197 [DOI] [PubMed] [Google Scholar]
- Cock JM, Sterck L, Rouzé P, Scornet D, Allen AE, Amoutzias G, Anthouard V, Artiguenave F, Aury JM, Badger JH, et al. (2010) The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature 465: 617–621 [DOI] [PubMed] [Google Scholar]
- Collins BJ, Deak M, Arthur JS, Armit LJ, Alessi DR. (2003) In vivo role of the PIF-binding docking site of PDK1 defined by knock-in mutation. EMBO J 22: 4202–4211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cove D. (2005) The moss Physcomitrella patens. Annu Rev Genet 39: 339–358 [DOI] [PubMed] [Google Scholar]
- Deak M, Casamayor A, Currie RA, Downes CP, Alessi DR. (1999) Characterisation of a plant 3-phosphoinositide-dependent protein kinase-1 homologue which contains a pleckstrin homology domain. FEBS Lett 451: 220–226 [DOI] [PubMed] [Google Scholar]
- Devarenne TP, Ekengren SK, Pedley KF, Martin GB. (2006) Adi3 is a Pdk1-interacting AGC kinase that negatively regulates plant cell death. EMBO J 25: 255–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friant S, Lombardi R, Schmelzle T, Hall MN, Riezman H. (2001) Sphingoid base signaling via Pkh kinases is required for endocytosis in yeast. EMBO J 20: 6783–6792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frödin M, Antal TL, Dümmler BA, Jensen CJ, Deak M, Gammeltoft S, Biondi RM. (2002) A phosphoserine/threonine-binding pocket in AGC kinases and PDK1 mediates activation by hydrophobic motif phosphorylation. EMBO J 21: 5396–5407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harries PA, Pan A, Quatrano RS. (2005) Actin-related protein2/3 complex component ARPC1 is required for proper cell morphogenesis and polarized cell growth in Physcomitrella patens. Plant Cell 17: 2327–2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki M, Schmelzle T, Yamaguchi K, Irie K, Hall MN, Matsumoto K. (1999) PDK1 homologs activate the Pkc1-mitogen-activated protein kinase pathway in yeast. Mol Cell Biol 19: 8344–8352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King CC, Zenke FT, Dawson PE, Dutil EM, Newton AC, Hemmings BA, Bokoch GM. (2000) Sphingosine is a novel activator of 3-phosphoinositide-dependent kinase 1. J Biol Chem 275: 18108–18113 [DOI] [PubMed] [Google Scholar]
- Lawlor MA, Mora A, Ashby PR, Williams MR, Murray-Tait V, Malone L, Prescott AR, Lucocq JM, Alessi DR. (2002) Essential role of PDK1 in regulating cell size and development in mice. EMBO J 21: 3728–3738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning G, Plowman GD, Hunter T, Sudarsanam S. (2002) Evolution of protein kinase signaling from yeast to man. Trends Biochem Sci 27: 514–520 [DOI] [PubMed] [Google Scholar]
- Matsui H, Miyao A, Takahashi A, Hirochika H. (2010) Pdk1 kinase regulates basal disease resistance through the OsOxi1-OsPti1a phosphorylation cascade in rice. Plant Cell Physiol 51: 2082–2091 [DOI] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L, et al. (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustafa A, Beszteri B, Maier UG, Bowler C, Valentin K, Bhattacharya D. (2009) Genomic footprints of a cryptic plastid endosymbiosis in diatoms. Science 324: 1724–1726 [DOI] [PubMed] [Google Scholar]
- Mumberg D, Müller R, Funk M. (1995) Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156: 119–122 [DOI] [PubMed] [Google Scholar]
- Nakamura A, Naito M, Tsuruo T, Fujita N. (2008) Freud-1/Aki1, a novel PDK1-interacting protein, functions as a scaffold to activate the PDK1/Akt pathway in epidermal growth factor signaling. Mol Cell Biol 28: 5996–6009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson AD, Lamb JC, Kobrossly PS, Shippen DE. (2011) Parameters affecting telomere-mediated chromosomal truncation in Arabidopsis. Plant Cell 23: 2263–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederberger C, Schweingruber ME. (1999) A Schizosaccharomyces pombe gene, ksg1, that shows structural homology to the human phosphoinositide-dependent protein kinase PDK1, is essential for growth, mating and sporulation. Mol Gen Genet 261: 177–183 [DOI] [PubMed] [Google Scholar]
- Pearce LR, Komander D, Alessi DR. (2010) The nuts and bolts of AGC protein kinases. Nat Rev Mol Cell Biol 11: 9–22 [DOI] [PubMed] [Google Scholar]
- Rintelen F, Stocker H, Thomas G, Hafen E. (2001) PDK1 regulates growth through Akt and S6K in Drosophila. Proc Natl Acad Sci USA 98: 15020–15025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AW, Dimos CS, Budziszek MJ, Jr, Goss CA, Lai V. (2011) Knocking out the wall: protocols for gene targeting in Physcomitrella patens. Methods Mol Biol 715: 273–290 [DOI] [PubMed] [Google Scholar]
- Schaefer DG. (2002) A new moss genetics: targeted mutagenesis in Physcomitrella patens. Annu Rev Plant Biol 53: 477–501 [DOI] [PubMed] [Google Scholar]
- Schaefer DG, Delacote F, Charlot F, Vrielynck N, Guyon-Debast A, Le Guin S, Neuhaus JM, Doutriaux MP, Nogué F. (2010) RAD51 loss of function abolishes gene targeting and de-represses illegitimate integration in the moss Physcomitrella patens. DNA Repair (Amst) 9: 526–533 [DOI] [PubMed] [Google Scholar]
- Schaefer DG, Zrÿd JP. (1997) Efficient gene targeting in the moss Physcomitrella patens. Plant J 11: 1195–1206 [DOI] [PubMed] [Google Scholar]
- Sephton CF, Zhang D, Lehmann TM, Pennington PR, Scheid MP, Mousseau DD. (2009) The nuclear localization of 3′-phosphoinositide-dependent kinase-1 is dependent on its association with the protein tyrosine phosphatase SHP-1. Cell Signal 21: 1634–1644 [DOI] [PubMed] [Google Scholar]
- Shakirov EV, Perroud PF, Nelson AD, Cannell ME, Quatrano RS, Shippen DE. (2010) Protection of Telomeres 1 is required for telomere integrity in the moss Physcomitrella patens. Plant Cell 22: 1838–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silber J, Antal TL, Gammeltoft S, Rasmussen TE. (2004) Phosphoinositide-dependent kinase-1 orthologues from five eukaryotes are activated by the hydrophobic motif in AGC kinases. Biochem Biophys Res Commun 321: 823–827 [DOI] [PubMed] [Google Scholar]
- Sims PA, Mann DG, Medlin LK. (2006) Evolution of the diatoms: insights from fossil, biological and molecular data. Phycologia 45: 361–402 [Google Scholar]
- Soltis DE, Bell CD, Kim S, Soltis PS. (2008) Origin and early evolution of angiosperms. Ann N Y Acad Sci 1133: 3–25 [DOI] [PubMed] [Google Scholar]
- Spinner L, Pastuglia M, Belcram K, Pegoraro M, Goussot M, Bouchez D, Schaefer DG. (2010) The function of TONNEAU1 in moss reveals ancient mechanisms of division plane specification and cell elongation in land plants. Development 137: 2733–2742 [DOI] [PubMed] [Google Scholar]
- Stokoe D, Stephens LR, Copeland T, Gaffney PR, Reese CB, Painter GF, Holmes AB, McCormick F, Hawkins PT. (1997) Dual role of phosphatidylinositol-3,4,5-trisphosphate in the activation of protein kinase B. Science 277: 567–570 [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellman CH. (2010) The invasion of the land by plants: when and where? New Phytol 188: 306–309 [DOI] [PubMed] [Google Scholar]
- Wellman CH, Osterloff PL, Mohiuddin U. (2003) Fragments of the earliest land plants. Nature 425: 282–285 [DOI] [PubMed] [Google Scholar]
- Yaffe MP, Schatz G. (1984) Two nuclear mutations that block mitochondrial protein import in yeast. Proc Natl Acad Sci USA 81: 4819–4823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegzouti H, Anthony RG, Jahchan N, Bögre L, Christensen SK. (2006a) Phosphorylation and activation of PINOID by the phospholipid signaling kinase 3-phosphoinositide-dependent protein kinase 1 (PDK1) in Arabidopsis. Proc Natl Acad Sci USA 103: 6404–6409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegzouti H, Li W, Lorenz TC, Xie M, Payne CT, Smith K, Glenny S, Payne GS, Christensen SK. (2006b) Structural and functional insights into the regulation of Arabidopsis AGC VIIIa kinases. J Biol Chem 281: 35520–35530 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.