Abstract
Immune cells respond to chemotactic signals by means of G protein-coupled receptors. Attempts to elucidate the function of specific G protein family members in these responses is complicated by redundancy among the different G protein isoforms. We have used lentiviral-based RNA interference to eliminate expression of specific G protein subunits selectively in J774A.1 mouse macrophages. The chemotactic response to the complement factors C5a and C3a is ablated in cells lacking Gβ2 but is unaffected in cells lacking Gβ1, Gαi2, or Gαi3. Similarly, the C5a-mediated calcium response of single cells is either absent or significantly delayed and weakened by Gβ2 knockdown. Assessment of Akt1 phosphorylation levels in response to C5a shows rapid and sustained phosphorylation in both wild-type cells and cells lacking Gβ1. Cells lacking Gβ2 retain the rapid response but cannot sustain phospho-Akt1 levels. The phenotype of cells lacking Gβ2 can be reversed by overexpression of either human Gβ2 or mouse Gβ1. These data demonstrate the usefulness of lentiviral-based RNA interference in the systematic analysis of a signaling pathway, and they suggest that in J774A.1 cells, Gβ2-derived Gβγ is the most effective mediator of chemotaxis to C5a.
The specificity of G protein-coupled receptors derives both from ligand binding and from the activation of intracellular signaling pathways. Biochemical and molecular genetic approaches have been used to map the intracellular interactions that underlie information processing. This task is complex because it involves resolving interactions among homologous components with similar biochemical activity. Thus, for example, G protein-coupled receptors that control chemotaxis in immune cells (e.g., chemokine or C5a receptors), when activated by specific ligand binding at the cell surface, catalyze the disassociation of the intracellular heterotrimeric G proteins, releasing the GTP-bound Gαi subunit and the cognate Gβγ dimer (1–3). The free βγ dimer is required for the initiation of cellular chemotaxis because treatment of the cells with pertussis toxin (PTx), which interferes specifically with the disassociation of Gβγ from members of the Gαi family, inhibits chemotaxis (4, 5). There may be some residual role also for the Gαi subunit (5). Although the inhibitor studies provide some clarity, they do not distinguish readily between the Gαi or βγ homologs. There are 3 genes that encode different Gαi subunits, 5 genes that encode β subunits, and 12 genes that encode γ subunits (6). Only subsets of these genes are expressed in immune cells; however, it is difficult to determine the precise roles of the isoforms that are expressed because they have very similar biochemical activity (7). With respect to chemotaxis, Gβγ has been shown to interact with and to activate a variety of proteins, including phospholipase C (PLC)-β2, PLC-β3, PLC-ε, p21-activated kinase 1, phosphatidylinositol 3-kinase, and isoforms of adenylyl cyclase (8–11). In addition, βγ has been shown to modulate the activity of a various ion channels (12, 13). There is evidence that, in some contexts, different βγ combinations may show specificity with respect to the effectors with which they interact (14). However, βγ dimers of different composition have also been found to replace each other equally well in other circumstances (15).
It has been difficult to apply genetic techniques to the question of specific isoform function in mammalian cells. However, the recent development of gene silencing with double-stranded small interfering RNA (siRNA) potentially may provide the level of resolution that is necessary to eliminate specific isoforms so that the effects of their loss on cellular function can be observed (16, 17). siRNA sequences have been shown to ablate gene activity in a sequence-specific fashion (18). Furthermore, lentiviral vectors have been shown to act as an efficient means for generating siRNA complexes (19). By inserting into the cellular genome, they can be used to generate cell lines that express a specific siRNA as a hairpin structure, which is processed subsequently into siRNA. The lentiviral vectors can be used for extensive studies in tissue culture. In addition, these constructs infect stem cells, and they can be transplanted into irradiated hosts to reconstitute the immune system for physiological studies (20). In this article, we demonstrate the systematic application of gene elimination by siRNA to the analysis of the function of individual components of a G protein-coupled receptor signaling pathway. We used the analysis of the C5a-mediated signaling in the J774A.1 macrophage cell line as a model system to explore this approach.
Materials and Methods
Vector Construction. A mouse H1 RNA polymerase III promoter was amplified from mouse genomic DNA and cloned at EcoRI–BamHI sites of pBS-SKII plasmid, which was obtained from Stratagene (21). To construct the 19–21 nucleotide hairpin siRNA cassettes, two complementary DNA oligonucleotides were chemically synthesized, annealed, and inserted between the BamHI and XhoI sites immediately downstream of the H1 promoter: 5′-GATCCCC19–21TTCAAGAGA19–21TTTTTC-3′ and 3′-GGG19–21AAGTTCTCT19–21AAAAAGAGCT-5′. We picked more than four siRNA target sequences for each gene and tested the expression efficiency by cotransfecting into human embryonic kidney (HEK)-293 cells with a GFP gene-fusion target gene. After expression for 48 h, the levels of residual GFP fusion proteins were assessed by Western blotting with anti-GFP serum (Santa Cruz Biotechnology). The target sequences for each of the genes were as follows: Gβ1, 5′-CATTATCTGTGGTATCACATC-3′, corresponding to nucleotide positions 804–824; Gβ2, 5′-CATCTGCTCCATCTATAGTC-3′, corresponding to positions 357–376; Gβ4, 5′-CATCTGCTCCATATACAAC-3′, corresponding to positions 357–375; Gαi2, 5′-GCACAGAGTGACTACATCC-3′, corresponding to positions 490–508; and Gαi3, 5′-GGAGTGCTGAAGAAGGAGT-3′, corresponding to positions 335–353.
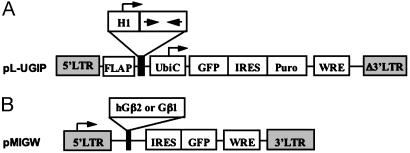
The lentiviral vector pL-UGIP was derived from FG12 (22). Internal ribosome entry site and puromycin-resistance gene sequences were inserted downstream of the GFP gene. The expression of both of the genes was driven by the ubiquitin C (UbiC) promoter. To construct the siRNA-expressing lentiviral vectors, the siRNA cassette containing H1 promoter and hairpin sequences was subcloned into the XbaI and XhoI sites upstream of the UbiC promoter (Fig. 1A). The cDNAs corresponding to the Gβ subunits were obtained by PCR from a mouse brain cDNA library and cloned into the SalI and BamHI sites of pEGFP-C1 (Clontech).
Fig. 1.
Construction of virus-based vectors. (A) Schematic diagram of the siRNA-expressing lentiviral vector pL-UGIP. The hairpin form of siRNA is expressed under the control of a mouse H1RNA polymerase III promoter. The vector also contains the enhanced GFP marker gene and the puromycin-resistance gene (Puro) regulated by a UbiC promoter and separated by an internal ribosome entry site (IRES) sequence. (B) Schematic diagram of Gβ subunit expressing the retroviral vector pMIGW. Expression of the Gβ cDNA and the enhanced GFP gene is controlled by an LTR region having promoter activity. FLAP, HIV-1 FLAP element; WRE, woodchuck hepatitis B virus RNA regulatory element.
Human cDNA of Gβ2 was isolated from HEK-293 cells by RT-PCR. We used a modified form of pMIG that contained woodchuck hepatitis B virus RNA regulatory element (WRE) sequences to generate constructs of the full-length human Gβ2 or mouse Gβ1 cDNA in a retroviral vector containing a GFP gene as a marker (23). The retroviruses were generated in HEK-293T cells cotransfected with the expression vector pMIGW-hGβ2 and the accessory vector pCMV/gag-pol (Fig. 1B). All resulting plasmids were confirmed by restriction enzyme analysis and DNA sequencing.
Lentivirus Generation. Lentivirus production was done as described (22). Briefly, HEK-293T cells were cotransfected with appropriate amounts of vector plasmid, the HIV-1 lentiviral packaging constructs pRSVEV and pMDLg/pRRE (24), and the VSV-G expression plasmid pHCMVG (25) by using the calcium phosphate transfection method. The viruses were collected from the culture supernatants after 28 h and concentrated 100-fold by ultracentrifugation (25). The concentrated virus stocks were titered on HEK-293 cells based on GFP expression. Titers for the siRNA expression constructs were ≈5 × 108 plaque-forming units per ml.
Cell Culture and Lentivirus Transduction. J774A.1 cells (obtained from the American Type Culture Collection) were maintained in DMEM containing 10% FCS, 100 units/ml penicillin, and 100 μg/ml streptomycin. The cells were transduced with concentrated virus at a multiplicity of infection of 10–20 in the presence of 8 μg/ml Polybrene (Sigma). Supernatant was removed after 8 h and replaced with growth medium. Cells were incubated 4 days after infection with 2 μg/ml puromycin for 1 day to select infected cells. GFP expression in selected cells was confirmed by fluorescence-activated cell sorting.
Western Blotting. All cells were lysed with radioimmunoprecipitation assay buffer (150 mM NaCl/50 mM NaF/20 mM Tris, pH 7.5/1% Triton X-100/0.5% sodium deoxycholate), supplemented with complete protease inhibitor tablets (Roche Diagnostics). Protein concentration was measured by using protein assay buffer (Bio-Rad). Equal amounts of protein (30 μg) were loaded per lane and separated on an SDS/4–20% PAGE gel (Invitrogen). Protein was transferred to a nitrocellulose membrane and incubated with specific antibody at the appropriate concentration and goat anti-rabbit or anti-mouse antibody conjugated with horseradish peroxidase (1:5,000). Anti-Gαi2 antibody was provided by Susanne Mumby (University of Texas Southwestern Medical Center, Dallas). Phospho-specific antibodies were purchased from Cell Signaling Technology (Beverly, MA), and the other antibodies were purchased from Santa Cruz Biotechnology. The blot was developed with enhanced chemiluminescence reagent (Amersham Biosciences).
Chemotaxis. J774A.1 cells were eluted with PBS containing 2 mM EDTA, washed twice, and suspended at a concentration of 1 × 106 cells per ml in migration assay buffer (DMEM containing 2 mg/ml BSA). Migration was measured by using 96-well chemotaxis chambers (Neuroprobe, Cabin John, MD) with polycarbonate filter (5 μm pore size). The lower wells were filled with 29 μl of assay buffer containing various concentrations of chemoattractants. After addition of 25 μl of cell suspension to the upper part of each well, the chamber was incubated at 37°C for 2 h. The filter was removed from between the chambers, and cells adhering to upper surface were washed off with PBS and stained with Diff-Quick (Baxter Diagnostics, McGaw Park, IL). Migrated cells were counted at five randomly selected fields under a microscope (×1,000 magnification). All experiments were performed in duplicated wells.
RT-PCR and Quantitative RT-PCR. Total RNA was prepared from J774A.1 by using TRIzol (Invitrogen). cDNA library was synthesized with SuperScript II reverse transcriptase (Invitrogen) and screened for the mRNA of each G protein by PCR with specific primers that covered the ORF. Then, PCR products were subcloned into pBluescript SK vector and confirmed by sequencing.
mRNA expression levels of each gene were quantitated by TaqMan real-time RT-PCR (Applied Biosystems, Foster City, CA) using gene-specific primers and normalized with GAPDH mRNA. The primers of genes are as follows: Gβ1, 5′-TGGACACGACAACCGAGTCA-3′ (sense) and 5′-GCCATGCCATCATCAGTCAC-3′ (antisense); Gβ2, 5′-GGATTCCATGTGCCGACAG-3′ (sense) and 5′-GCATTGATGTCCGACTCGTG-3′ (antisense); Gαi2, 5′-TGCCGTGGTCTACAGCAACA-3′ (sense) and 5′-CGCTTCACGATGGCCAGTATA-3′ (antisense); Gαi3, 5′-TGGCTCTCAGTGATTACGACCTT-3′ (sense) and 5′-GGTTCATTTCCTCATCCTCAGC-3′ (antisense); Gγ2, 5′-CTGGTAGAACAGCTGAAGATGGAA-3′ (sense) and 5′-GGACACCTTTATCCTGTCGATGT-3′ (antisense); Gγ5, 5′-CATGACCCTCTGCTGACTGGA-3′ (sense) and 5′-GGGTCTGAAGGGATTCGTACTTG-3′ (antisense); and GAPDH, 5′-AGTATGACTCCACTCACGGCAA-3′ (sense) and 5′-CCATTCTCGGCCTTGACTGT-3′ (antisense).
Measurement of Intracellular Ca2+ Concentration ([Ca2+]i). Cells were grown in glass-bottom dishes (MatTek, Ashland, MA) and loaded with fura 2 acetoxymethyl ester (2 μM; Molecular Probes) in assay buffer (Hanks' balanced salt solution/10 mM Hepes, pH 7.2) for 30 min at room temperature. After washing, the dish was placed on the stage of an IX71 inverted fluorescence microscope (Olympus, Melville, NY) to which a digital imaging system with electronically controlled excitation filter positions and an associated software system (slidebook, Version 4.0; Intelligent Imaging Innovations, Denver) was attached. Measurements of [Ca2+]i were performed on individual cells at excitation wavelengths of 340 and 380 nm and an emission wavelength of 510 nm at 1.5-s intervals.
Results
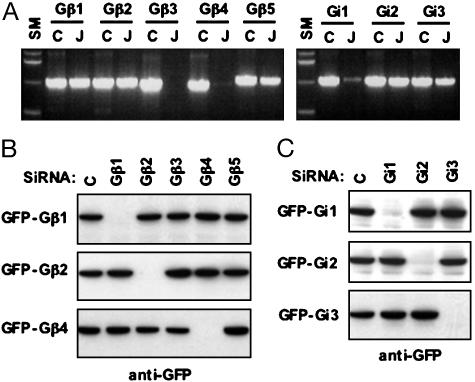
Suppression of G Proteins by Subtype-Specific siRNAs. The mouse macrophage cell line J774A.1 contained mRNA corresponding to the Gβ1, Gβ2, and Gβ5 genes and had low levels of mRNA for Gβ4; Gβ3 was not detected. Specific transcripts were detected also for the Gαi subunits, with relatively low levels for Gαi1 (Fig. 2A). Proteins corresponding to Gβ4, Gβ5, or Gαi1 were not observed in Western blotting with isotype-specific antibodies (data not shown), suggesting that the expression of these proteins was absent or too low to be detected.
Fig. 2.
Expression of G protein mRNAs in J774A.1 and silencing of G proteins by subtype-specific siRNAs. (A) RT-PCR was performed by using total RNA isolated from J774A.1. After PCR (30 cycles) with subtype-specific primers, one-fifth of the product was loaded on a 1% agarose gel. For controls (C), cDNA corresponding to the Gβ or Gα subunit was used. Alternatively, the RT product derived from the J774A.1 cells (J) was used. SM, size markers. (B and C) siRNA constructs contained in pSK vectors were transfected into HEK-293 cells with plasmid, maintaining the cDNA corresponding to the enhanced GFP fusion form of each G protein. After 48 h, Western blotting was performed with an anti-GFP antibody.
To obtain an effective siRNA sequence, we synthesized more than four different siRNA hairpins specific for mRNA corresponding to each mouse G protein subunit. Initial experiments were performed in HEK-293 cells to test the efficiency of inhibition of expression elicited after transfecting the cells with plasmids expressing GFP-tagged forms of the G protein subunit. We chose the siRNA that showed >95% inhibition of the expression of the target proteins. Then, the specificity of inhibition of the target gene was examined by cotransfecting siRNA constructs with constructs expressing each isotype of the G protein subunit. The target protein of each siRNA almost disappeared, whereas the expression of the other isotypes was unchanged (Fig. 2 B and C).
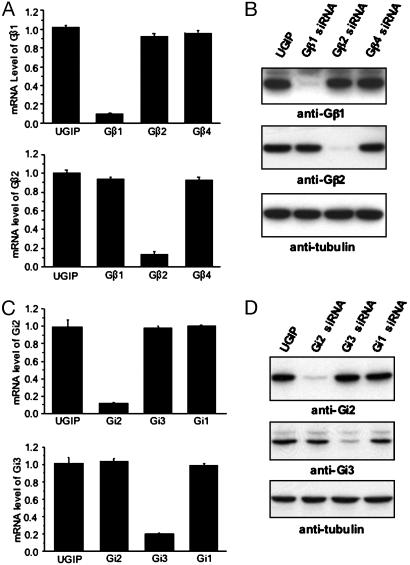
Suppression of Endogenous G Proteins by Lentivirus Carrying siRNA. Electroporation or lipid-mediated transfection methods developed to date give low gene delivery in several types of cells, including J774A.1. To increase the efficiency, we used lentivirus-mediated gene transfer (22). The lentiviral vector expresses a human UbiC-driven GFP gene and a puromycin-resistance gene, which provide a visual marker and a selection for tracking or enriching for transduced cells, respectively (Fig. 1 A). After virus infection and selection with puromycin, >98% of the cells showed a GFP-positive signal when tested by fluorescence-activated cell sorter analysis. To confirm the specific gene silencing effect of siRNA in the J774A.1 cells, we used quantitative RT-PCR with specific primers and immunoblotting against endogenous proteins with isotype-specific antibodies. The relative mRNA level, normalized with GAPDH mRNA, was decreased by ≈90% in cells transduced by virus carrying each isotype-specific siRNA (Fig. 3 A and C). Consistent with quantitative RT-PCR data, significant down-regulation of target gene expression was observed by Western blot analysis, whereas expression of the other isotypes was not affected by the siRNA (Fig. 3 B and C).
Fig. 3.
siRNA-mediated inhibition of endogenous G protein expression. (A and C) After puromycin selection, mRNA levels of G proteins in lentivirus-transduced cells were analyzed by TaqMan RT-PCR. The mRNA level of each G protein was reduced specifically in the cells containing its siRNA. mRNA levels were normalized to GAPDH mRNA. (B and D) Western blotting using subtype-specific antibodies. G proteins were significantly down-regulated in the cells containing the corresponding siRNA. Protein expression was normalized to tubulin levels in total cell extracts.
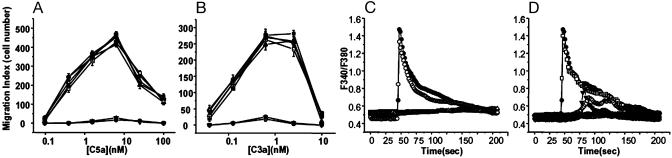
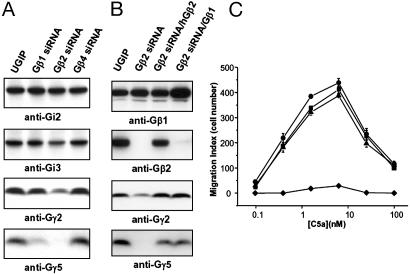
The Cells Lacking Gβ2 Did Not Migrate to Chemotactic Factors. To study the biological relevance of the suppression of endogenous G protein expression by siRNA, we investigated cellular responses to extracellular stimulation. J774A.1 cells are known to express classical types of the C5a and C3a chemokine receptors (26). By using chemotaxis assays to various chemokines, we also found activity corresponding to the CXCR4 and the SDF1α receptor; expression levels, however, were very low (data not shown). In the present study, we used C5a and C3a as agonists. As shown in Fig. 4 A and B, J774A.1 cells migrated across polycarbonate filters toward a source of chemokines. The concentration–response curves were bell-shaped, peaking at 1–10 nM for the C5a ligand and at 0.6–2.5 nM for the C3a ligand. Pretreatment of cells with PTx (100 ng/ml) for 12 h completely inhibited the ability of the cells to generate chemotactic responses to C5a and C3a (Fig. 4 A and B), consistent with previous results, which suggested that Gαi subfamily-mediated signaling pathways were involved in cell migration in the chemoattractant gradient (27, 28). Given this result, we examined migration of the cells containing siRNA directed against Gαi2 or Gαi3. The migration-response curves of these cells were similar to those of wild-type or empty virus-transduced cells, suggesting that the presence of only one subtype of Gαi family protein expressed in the cell was sufficient to compensate for the absence of the other subtype. The cells containing Gβ2 siRNA did not migrate toward C5a and C3a, whereas the cells containing Gβ1 and Gβ4 siRNA showed migration efficiency similar to that of control cells. Loss of chemotaxis in the cells treated with Gβ2 siRNA was similar to the effect of PTx and was not compensated by the endogenous residual Gβ subtypes.
Fig. 4.
Effects of G protein silencing on cellular responses to chemokines. (A and B) Chemotaxis in J774A.1 cells toward C5a (A) and C3a (B). Chemotaxis was analyzed by using transwell migration, as described in Materials and Methods. Cells expressing siRNA corresponding to each G protein were incubated with or without 100 ng/ml PTx for 12 h before the chemotaxis assays. The data are expressed as the mean ± SE of three independent experiments. •, pL-UGIP (cells infected with lentivirus lacking insert, see Fig. 1); ○, PTx-treated pL-UGIP; ▴, Gβ1 siRNA; ▾, Gβ2 siRNA; ▪, Gβ4 siRNA; □, Gi2 siRNA; ▵, Gi3 siRNA. (C) C5a-induced calcium transient was PLC-β-dependent. Cells loaded with fura 2 acetoxymethyl ester were stimulated with C5a, and changes in fluorescence in single cells were monitored. Similar traces were obtained in 20 recordings of different cells. U73122 (▵), a PLC inhibitor, or thapsigargin (▪), an endoplasmic reticular Ca2+-ATPase inhibitor, blocked C5a-induced [Ca2+]i increase. After a 10-min treatment with 10 μM U73122 or 1 μM thapsigargin, C5a was added. C5a-induced [Ca2+]i increase was unchanged in calcium-free buffer containing 200 μM EGTA (○). •, No pretreatment. (D) Inhibition of [Ca2+]i increase by C5a in the cells containing Gβ2 siRNA. In control cells and Gβ1 siRNA-containing cells, C5a induced [Ca2+]i increases, which were blocked by PTx pretreatment for 12 h. In contrast, Gβ2 siRNA-containing cells showed no or delayed calcium responses to C5a. The arrow indicates C5a time of addition. •, pL-UGIP; ○, PTx-treated pL-UGIP; □, Gβ1 siRNA; ♦, Gβ2 siRNA (no response); ▾, Gβ2 siRNA (delayed weak response); ⋄, Gβ2 siRNA (delayed weak biphasic response).
Gβ2 siRNA Inhibited [Ca2+]i Transients Induced by C5a. Gβγ subunits released from the GTP-bound form of Gαi increase cytoplasmic Ca2+ concentration by activating PLC-β (29, 30). Therefore, we measured cytoplasmic Ca2+ increase in response to C5a stimulation by using fluorescence microscopy. To determine the source of Ca2+ influx, C5a-induced [Ca2+]i responses were examined in the presence of Ca2+-free assay buffer containing 200 μM EGTA. In this condition, [Ca2+]i responses were similar to the responses obtained in calcium-containing buffer. Next, we examined changes of [Ca2+]i responses by inhibiting PLC activity with U73122 and depleting internal calcium stores with thapsigargin. When cells are pretreated with 10 μM U73122 or 1 μM thapsigargin for 10 min, the responses to C5a were completely inhibited (Fig. 4C). Treatment of control cells and Gβ1 siRNA-containing cells showed maximum Ca2+ increase at 20 nM C5a (Fig. 4D). PTx pretreatment completely inhibited the Ca2+ increase, confirming that the Ca2+ increase was derived from a Gαi-mediated pathway. The responses of Gβ2 siRNA-containing single cells to C5a were heterogeneous: approximately half of the tested cells showed little or no response, and the remaining half showed delayed weak response or a delayed weak biphasic response. Some of the heterogeneity in the low responding cells may result from residual levels of Gβ2 protein. We used puromycin selection, and the resulting population may include cells with different numbers or positions of viral insertion leading to variable degrees of Gβ2 suppression. Consistent with previous work, these data indicate that C5a stimulates calcium mobilization from inositol trisphosphate-dependent internal calcium stores and that Gβ2 is involved in robust PLC-β activation.
Expression of Gγ Subunits Was Down-Regulated in the Cells Lacking Gβ2. Some protein interaction is necessary for the stability of the members of signaling-protein complexes. Thus, for example, ablation of Gγ subunit expression destabilizes uncomplexed Gβ (31). Furthermore, RGS proteins containing the G protein γ-like domain (RGS6, RGS7, RGS9, and RGS11) are Gβ5 binding partners; they are destabilized and degraded in mice lacking Gβ5, and reciprocally, Gβ5 protein is degraded in the absence of a suitable interacting subunit (32, 33). To examine the stability of Gα and Gγ subunits as a function of Gβ suppression, we performed quantitative RT-PCR and Western blotting. The steady-state mRNA levels corresponding to Gαi2, Gαi3, Gγ2, and Gγ5 were not affected in the cells lacking Gβ subunits, compared with control cells (data not shown). Protein levels of Gαi2 and Gαi3 were not changed significantly, whereas Gγ2 was considerably down-regulated in the cells lacking Gβ2 but not Gβ1 or Gβ4. Gγ5 was down-regulated in the cells lacking Gβ1 and was not detected in the cells lacking Gβ2 (Fig. 5A).
Fig. 5.
Changes in G protein expression by silencing or expression of Gβ cDNA. (A) Western blotting for GαiorGγ subunits. Protein levels of Gαi2 and Gαi3 were not changed compared with control cells transfected with pL-UGIP (empty viral vector). Gγ2 expression was reduced significantly in Gβ2-silenced cells. Gγ5 expression was reduced in Gβ1-silenced cells and was not detected in Gβ2-silenced cells. (B) Western blotting for Gβ and Gγ subunits. Gβ1 expression was enhanced by transduction of retrovirus carrying Gβ1 gene (top blot, lane four). Gβ2 protein was detected in mouse Gβ2-silenced cells transduced with retrovirus carrying human Gβ2 gene (second blot from the top, lane three). Expressions of Gγ2 and Gγ5 were restored by exogenous expression of human Gβ2 or Gβ1.(C) Expression of exogenous Gβ subunits restored cellular responses to C5a. Migration efficiency of Gβ2-silenced cells was restored by exogenous expressions of human Gβ2 (▴) or Gβ1 (▪). The data are expressed as the mean ± SE of three independent experiments. •, pL-UGIP; ♦, Gβ2 siRNA.
Gβ subunits, especially Gβ2, affected the stability of their Gγ subunit binding partners, suggesting that Gβ2 may be the primary molecule binding to Gγ2 and Gγ5. There may be other γ subunits expressed as well; however, we were not able to obtain reagents to test for the presence of all of the γ subunits.
Expression of Exogenous Gβ Subunits Restored Cellular Responses to C5a. To examine the specificity of the response to the absence of Gβ2 further, we studied the effect of reexpression of Gβ subunits on cellular responses. We generated retroviruses (Fig. 1) expressing either human Gβ2 or Gβ1. Although the amino acid sequences of the human and mouse Gβ proteins are the same, the nucleic acid sequences in third codon positions are different. Gβ2 siRNA target sequences corresponding to the mouse gene were coincident with that of the human gene except for two nucleotides. Mouse Gβ2 siRNA did not block expression of human Gβ2 (Fig. 5B). Exogenous expression of human Gβ2 restored expression of Gγ2 and Gγ5. However, Gγ subunits were restored also by overexpression of Gβ1 in the absence of Gβ2 (Fig. 5B). Moreover, in Gβ2 siRNA-containing cells transduced with retroviruses carrying either the human Gβ2 or Gβ1 gene, chemotactic efficiency was similar to that of wild-type cells (Fig. 5C). This result suggests that overproduction of Gβ1 can compensate for the decreased level of Gβ2. These data are consistent with results that showed no difference in activity between Gβ1 and Gβ2 in in vitro experiments and with overexpression experiments used to define the roles of each of the Gβ subunits (34, 35).
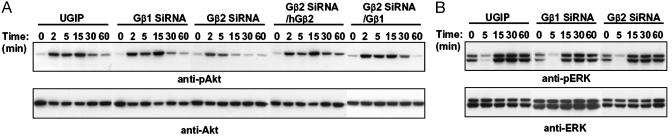
Gβ2 siRNA Treatment Modified the Kinetics of Akt Phosphorylation. Phosphatidylinositol 3-kinase γ (PI3Kγ) plays a major role in cell migration toward chemokines by interacting with Gβγ subunits (9, 36). To follow PI3Kγ activity, we examined Akt phosphorylation, an event reflective of PI3K activation. On stimulation with C5a, the Akt phosphorylation level on Ser-473 was increased at 2 min and sustained for 30 min in control cells. This sustained phosphorylation was found also in cells containing Gβ1 siRNA. In Gβ2 siRNA-containing cells, phosphorylation occurred at 2 min but vanished by 15 min, implying that interaction of Gβ2 with a downstream element (possibly PI3Kγ) extended Akt phosphorylation (Fig. 6A). Exogenous expression of human Gβ2 or Gβ1 in Gβ2 siRNA-containing cells restored prolonged phosphorylation to the level seen in control cells, consistent with the restoration of migration efficiency observed in these cells. In contrast to Akt phosphorylation, ERK phosphorylation after C5a addition did not change in cells lacking Gβ2, compared with control cells and Gβ1 siRNA-containing cells (Fig. 6B). Thus, in contrast to Akt phosphorylation, the signaling pathways for ERK phosphorylation were not affected by the lack of one of the Gβ subunits.
Fig. 6.
C5a-stimulated phosphorylation of Akt and extracellular signal-regulated kinase (ERK). After incubation with media containing 1% FBS for 12 h, cells were stimulated with C5a for the indicated times. Cell lysates were subjected to immunoblotting with specific antibodies against phosphorylated forms of Akt (A) or ERK (B). The immunoblots were stripped and reprobed with anti-Akt or anti-ERK antibodies. (A) Akt phosphorylation by C5a was sustained >30 min in control cells and Gβ1-silenced cells, whereas phosphorylation was ended by 15 min after treatment in Gβ2-silenced cells. Extended phosphorylation was restored by expression of human Gβ2 or Gβ1. (B) In contrast to the results shown in A, C5a stimulated ERK phosphorylation by the same degree in control cells and Gβ-silenced cells.
Discussion
RNA interference delivered by lentiviral vectors was used to probe the relative contribution of components of the C5a receptor-mediated chemotaxis pathway in the J774A.1 macrophage cell line. Cells were infected with virus carrying a variety of different hairpin sequences that were processed into double-stranded RNA, ablating target transcripts and blocking specific protein expression. We found that the loss of either of the two major members of the Gi family of Gα subunits in these cells, Gαi2 or Gαi3, had little or no effect on chemotaxis. Furthermore, the absence of the Gβ1 subunit also did not affect chemotaxis. However, the loss of the Gβ2 subunit blocked chemotaxis completely. The interpretation of these data is complicated because the presence of the Gγ subunit depends on the stability of the Gβ subunit, thus its expression is affected also by specific Gβ RNA interference. Much of the evidence suggests that Gβ and Gγ act together in the dimer (37) to activate downstream effectors and that the function of the Gγ subunit is integral to the activity of the Gβ subunit. Furthermore, both Gβ1 and Gβ2 can bind various Gγ subunits. Biochemical studies (38, 39) suggest also that different Gβγ subunit combinations may have very similar functions. It was surprising, therefore, that we saw specificity for the loss of the Gβ2 subunit. To determine whether this specificity resided in the sequence of the Gβ2 subunit or in the level of Gβ subunit, we replaced the mouse Gβ subunits with their human Gβ subunit counterparts, which have the same amino acid sequence but different transcripts, so that the human protein could be expressed in the presence of mouse specific siRNA. We found that replacement of mouse Gβ2 by human Gβ2 or by overexpression of Gβ1 restored chemotaxis, suggesting that a critical level of Gβγ subunit is required but that either Gβ1 or Gβ2 could fulfill the downstream functions required for chemotaxis. One model would suggest that the kinetics of accumulation of free βγ and its localization in the cell is critical to its signaling function. Free βγ accumulation depends on many factors. These factors include the catalytic activity of the receptor, the lifetime of the cognate Gαi–GTP complex, and the interaction of Gβγ with various effectors and small molecules that can lead to its sequestration and translocation. Thus, although both β1 and β2 can function, a variety of kinetic factors could make the β2 dimers more effective sources for signal transmission. On the other hand, Gβ2 may have some specific affinity for a signaling complex downstream of the C5a receptor. In its absence, endogenous Gβ1 is not able to function because it is sequestered. However, exogenous overexpression of human Gβ1 does compensate. We were able to show in these experiments that at least two downstream effectors were activated. One of these effectors was PLC-β, which leads to the rapid release of calcium, and the other was the phosphorylation of Akt, which was extended and stabilized by the presence of free Gβγ.
Although these initial experiments do not point unambiguously to the downstream pathway that connects the release of free βγ to the cytoskeletal changes that lead to chemotaxis, they do demonstrate the usefulness of the application of siRNA. The technique provides us with a precise tool to eliminate single genes in a specific manner and to restore gene functions in these backgrounds. The restored genes can be mutated to study their specific activity in a background lacking the initial function. Furthermore, it should be possible to apply double-stranded RNA or antisense RNA directly to the lentiviral-infected cells to generate a transient deficiency for two or three genes in a specific pathway. Finally, these cells, with their lentiviral constructs, provide a relatively stable platform that can be used for kinetic studies of signaling pathways. Thus, we can generate models for understanding the detailed functions of the signaling elements and probe the quantitative relationships among these functions in vivo.
Acknowledgments
This work was supported by National Institutes of Health Grants GM34236 (to M.I.S.) and GM 62114 (to I.D.C.F. and S.C. under the Alliance for Cellular Signaling Cooperative Agreement), and by Damon Runyon–Walter Winchell Fellowship DRG-1568 (to X.-F.Q.).
Abbreviations: HEK, human embryonic kidney; PLC, phospholipase C; siRNA, small interfering RNA; UbiC, ubiquitin C; PTx, pertussis toxin; [Ca2+]i, intracellular Ca2+ concentration; ERK, extracellular signal-regulated kinase.
References
- 1.Gerard, G. & Gerard, N. P. (1994) Annu. Rev. Immunol. 12, 775–808. [DOI] [PubMed] [Google Scholar]
- 2.Murphy, P. M. (1994) Annu. Rev. Immunol. 12, 593–633. [DOI] [PubMed] [Google Scholar]
- 3.Premack, B. A. & Schall, T. J. (1996) Nat. Med. 2, 1174–1178. [DOI] [PubMed] [Google Scholar]
- 4.Neptune, E. R. & Bourne, H. R. (1997) Proc. Natl. Acad. Sci. USA 94, 14489–14494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neptune, E. R., Iiri, T. & Bourne, H. R. (1999) J. Biol. Chem. 274, 2824–2828. [DOI] [PubMed] [Google Scholar]
- 6.Clapham, D. E. (1997) Annu. Rev. Pharmacol. Toxicol. 37, 167–203. [DOI] [PubMed] [Google Scholar]
- 7.Daaka, Y., Pitcher, J. A., Richardson, M., Stoffel, R. H., Robishaw, J. D. & Lefkowitz, R. J. (1997) Proc. Natl. Acad. Sci. USA 94, 2180–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang, H., Kuang, Y., Wu, Y., Xie, W., Simon, M. I. & Wu, D. (1997) Proc. Natl. Acad. Sci. USA 94, 7971–7975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li, Z., Jiang, H., Xie, W., Zhang, Z., Smrcka, A. V. & Wu, D. (2000) Science 287, 1046–1049. [DOI] [PubMed] [Google Scholar]
- 10.Hirsch, E., Katanaev, V. L., Garlanda, C., Azzolino, O., Pirola, L., Silengo, L., Sozzani, S., Mantovani, A., Altruda, F. & Wymann, M. P. (2000) Science 287, 1049–1053. [DOI] [PubMed] [Google Scholar]
- 11.Li, Z., Hannigan, M., Mo, Z., Liu, B., Lu, W., Wu, Y., Smrcka, A. V., Wu, G., Li, L., Liu, M., et al. (2003) Cell 114, 215–227. [DOI] [PubMed] [Google Scholar]
- 12.Krapivinsky, G., Gordon, E., Wickman, K., Velimirovic, B., Krapivinsky, L. & Clapham, D. E. (1995) Nature 374, 135–141. [DOI] [PubMed] [Google Scholar]
- 13.Clapham, D. E. (1996) Curr. Biol. 6, 814–816. [DOI] [PubMed] [Google Scholar]
- 14.Wolfe, J. T., Wang, H., Howard, J., Garrison, J. C. & Barrett, P. Q. (2003) Nature 424, 209–213. [DOI] [PubMed] [Google Scholar]
- 15.Hou, Y., Chang, V., Capper, A. B., Taussig, R. & Gautam, N. (2001) J. Biol. Chem. 276, 19982–19988. [DOI] [PubMed] [Google Scholar]
- 16.Elbashir, S. M., Lendeckel, W., Yalcin, A., Weber, K. & Tuschl, T. (2001) Nature 411, 494–498. [DOI] [PubMed] [Google Scholar]
- 17.Harborth, J., Elbashir, S. M., Bechert, K., Tuschl, T. & Weber, K. (2001) J. Cell Sci. 114, 4557–4565. [DOI] [PubMed] [Google Scholar]
- 18.Hemann, M. T., Fridman, J. S., Zilfou, J. T., Hernando, E., Paddison, P. J., Cordon-Cardo, C., Hannon, G. J. & Lowe, S. W. (2003) Nat. Genet. 33, 396–400. [DOI] [PubMed] [Google Scholar]
- 19.Stewart, S. A., Dykxhoorn, D. M., Palliser, D., Mizuno, H., Yu, E. Y., An, D. S., Sabatini, D. M., Chen, I. S. Y., Hahn, W. C., Sharp, P. A., et al. (2003) RNA 9, 493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rubison, D. A., Dillon, C. P., Kwiatkowski, A. V., Sievers, C., Yang, L., Kopinja, J., Zhang, M., McManus, M. T., Gertler, F. B., Scott, M. L. & Parijs, L. V. (2003) Nat. Genet. 33, 401–406. [DOI] [PubMed] [Google Scholar]
- 21.Myslinski, E., Ame, J. C., Krol, A. & Carbon, P. (2001) Nucleic Acids Res. 29, 2502–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qin, X.-F., An, D. S., Chen, I. S. Y. & Baltimore, D. (2003) Proc. Natl. Acad. Sci. USA 100, 183–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Parijs, L., Refaeli, Y., Lord, J. D., Nelson, B. H., Abbas, A. K. & Baltimore, D. (1999) Immunity 11, 281–288. [DOI] [PubMed] [Google Scholar]
- 24.Dull, T., Zufferey, R., Kelly, M., Mandel, R. J., Nguyen, M., Trono, D. & Naldini, L. (1998) J. Virol. 72, 8463–8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yee, J. K., Friedmann, T. & Burns, J. C. (1994) Methods Cell Biol. 43, 99–122. [DOI] [PubMed] [Google Scholar]
- 26.Fan, Y. & McCloskey, M. A. (1994) J. Biol. Chem. 269, 31533–31543. [PubMed] [Google Scholar]
- 27.Hartmann, K., Henz, B., Kruger-Krasagakes, S., Kohl, J., Burger, R., Guhl, S., Haase, I., Lippert, U. & Zuberbier, T. (1997) Blood 89, 2863–2870. [PubMed] [Google Scholar]
- 28.Nilsson, G., Johnell, M., Hammer, C. H., Tiffany, H. L., Nilsson, K., Metcalfe, D. D., Siegbahn, A. & Murphy, P. M. (1996) J. Immunol. 157, 1693–1698. [PubMed] [Google Scholar]
- 29.Jiang, H., Kuang, Y., Wu, Y., Smrcka, A., Simon, M. I. & Wu, D. (1996) J. Biol. Chem. 271, 13430–13434. [DOI] [PubMed] [Google Scholar]
- 30.Wu, D., LaRosa, G. J. & Simon, M. I. (1993) Science 261, 101–103. [DOI] [PubMed] [Google Scholar]
- 31.Wang, Q., Mullah, B. K. & Robishaw, J. D. (1999) J. Biol. Chem. 274, 17365–17371. [DOI] [PubMed] [Google Scholar]
- 32.Chen, C.-K., Eversole-Cire, P., Zhang, H., Mancino, V., Chen, Y.-J., He, W., Wensel, T. G. & Simon, M. I. (2003) Proc. Natl. Acad. Sci. USA 100, 6604–6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Witherow, D. S., Wang, Q., Levay, K., Cabrera, J. L., Chen, J., Willars, G. B. & Slepak, V. Z. (2000) J. Biol. Chem. 275, 24872–24880. [DOI] [PubMed] [Google Scholar]
- 34.Yan, K. & Gautam, N. (1996) J. Biol. Chem. 271, 17597–17600. [DOI] [PubMed] [Google Scholar]
- 35.Ueda, N., Iniguez-Lluhi, J. I., Lee, E., Smrcka, A. V., Robishow, J. D. & Gilman, A. G. (1994) J. Biol. Chem. 269, 4388–4395. [PubMed] [Google Scholar]
- 36.Brock, C., Schaefer, M., Reusch, H. P., Czupalla, C., Michalke, M., Spicher, K., Schultz, G. & Nurnberg, B. (2003) J. Cell Biol. 160, 89–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwindinger, W. F. & Robishaw, J. D. (2001) Oncogene 20, 1653–1660. [DOI] [PubMed] [Google Scholar]
- 38.Yan, K., Kalyanaraman, V. & Gautam, N. (1996) J. Biol. Chem. 271, 7141–7146. [DOI] [PubMed] [Google Scholar]
- 39.Ueda, H., Itoh, H., Yamauchi, J., Morishita, R., Kaziro, Y., Kato, K. & Asano, T. (2000) J. Biol. Chem. 275, 2098–2102. [DOI] [PubMed] [Google Scholar]