Abstract
The rhizosphere is teemed with organisms that coordinate their symbioses using chemical signals traversing between the host root and symbionts. Chemical signals also mediate interactions between roots of different plants, perhaps the most obvious being those between parasitic Orobanchaceae and their plant hosts. Parasitic plants use specific molecules provided by host roots to initiate the development of haustoria, invasive structures critical for plant parasitism. We took a transcriptomics approach to identify parasitic plant genes associated with host factor recognition and haustorium signaling and previously identified a gene, TvPirin, which is transcriptionally up-regulated in roots of the parasitic plant Triphysaria versicolor after being exposed to the haustorium-inducing molecule 2,6-dimethoxybenzoquinone (DMBQ). Because TvPirin shares homology with proteins associated with environmental signaling in some plants, we hypothesized that TvPirin may function in host factor recognition in parasitic plants. We tested the function of TvPirin in T. versicolor roots using hairpin-mediated RNA interference. Reducing TvPirin transcripts in T. versicolor roots resulted in significantly less haustoria development in response to DMBQ exposure. We determined the transcript levels of other root expressed transcripts and found that several had reduced basal levels of gene expression but were similarly regulated by quinone exposure. Phylogenic investigations showed that TvPirin homologs are present in most flowering plants, and we found no evidence of parasite-specific gene duplication or expansion. We propose that TvPirin is a generalized transcription factor associated with the expression of a number of genes, some of which are involved in haustorium development.
The Orobanchaceae comprises roughly 90 genera of facultative and obligate parasitic plants that directly invade host roots to rob them of water and nutrients (Kuijt, 1969; Musselman, 1980; Nickrent, 2011). The Orobanchaceae invade their hosts via haustoria, parasitic organs that attach to and penetrate host tissues. Haustoria later develop physiological connections between host and parasite vasculature systems that provide the conduit for the transfer of metabolites, proteins, nucleic acids, and viruses between host and parasite (Westwood et al., 2009). Root parasitism can be debilitating to host plants, and some of the world’s most devastating agricultural pests are weedy Orobanchaceae (Parker and Riches, 1993).
Haustoria develop on parasite roots in response to contact with host roots or host recognition molecules termed xenognosins (Lynn et al., 1981). Haustorium ontogeny can be observed under a microscope by exposing aseptic parasite roots in vitro to host exudates or purified xenognosins (William, 1961; Atsatt et al., 1978; Riopel and Timko, 1995). Within minutes of xenognosin exposure, the parasite root stops elongating and its growth is redirected toward a radial expansion of the root tip. After a few hours, the root tips noticeably swell and epidermal hairs begin to elongate over the swollen root tissue. Depending on the species, the swelling and hair proliferation are localized at the apex of the emerging radical or at lateral positions just behind the root cap. In the presence of a host, the haustorial hairs attach the parasite to the host root and provide the foundation for invasive cells internal to the haustorium to penetrate host epidermal and cortical cell layers (Baird and Riopel, 1984). Upon contact with the host stele, the haustorium develops a vascular bridge connecting host and parasite vascular systems.
Several phenols and quinones have been identified that induce haustorium development in vitro (Steffens et al., 1982; Chang and Lynn, 1986; Albrecht et al., 1999). Xenognosin activity is redox dependent, and phenols need to be oxidized to their sister quinones to be active haustorium inducers. In Striga, this reaction is driven by hydrogen peroxide-dependent peroxidases acting on monolignol molecules in the host cell walls (Smith et al., 1990). The correlation between quinone redox potential and inducing activity suggests that a key feature of xenognosins is their participation in oxidoreduction reactions (Smith et al., 1996). The involvement of radical semiquinones in haustorium signaling was suggested by the inhibitory effect of cyclopropyl-p-benzoquinone, a spin trap whose single electron reduction generates a reactive electrophile at the cyclopropyl ring that is predicted to irreversibly bind the xenognosin receptor (Zeng et al., 1996; Keyes et al., 2001). The requirement for a semiquinone was shown genetically by RNA interference (RNAi) silencing of quinone oxidoreductases in roots of the hemiparasite Triphysaria versicolor (Bandaranayake et al., 2010). Two xenognosin-regulated quinone oxidoreductases, one catalyzing single electron reductions and the other bivalent reductions, were silenced in transgenic T. versicolor roots. Silencing the single electron-reducing quinone oxidoreductase reduced haustorium development, while silencing the bivalent reducing enzyme had no effect. These results suggest that the first step in haustorium signaling is the redox activation of xenognosins to their semiquinone states (Bandaranayake et al., 2010).
We took a transcriptomic approach to identify parasite genes associated with xenognosin signaling (Matvienko et al., 2001a; Torres et al., 2005; Westwood et al., 2010). One interesting gene candidate for haustorium signaling is TvPirin (Matvienko et al., 2001a). Pirin was first described as a human protein that interacts with the transcription factor NF I in yeast two-hybrid systems (Wendler et al., 1997). In another two-hybrid screen, Pirin was identified as binding the oncoprotein Bcl3, a modulator of NF-KB transcription factor activity (Dechend et al., 1999). The Pirin protein is more abundant in C-Jun and rat sarcoma-transformed rat fibroblasts than in normal cells (Bergman et al., 1999). In Serratia marcescens, Pirin modifies pyruvate catabolism by interacting with pyruvate dehydrogenase (Soo et al., 2007). Pirins can also function catalytically as quercetin dioxygenases (Adams and Jia, 2005). The crystalline structure of human Pirin indicates two similar β-barrel domains arranged face to face with an iron cofactor within a cavity formed at the N-terminal domain, suggesting possible involvement in redox reactions (Pang et al., 2004).
The Arabidopsis (Arabidopsis thaliana) protein AtPirin1 binds the Gα-subunit (GPA1) of the heterotrimeric guanine nucleotide binding protein (G protein) in yeast two-hybrid and in vitro binding assays (Lapik and Kaufman, 2003). The Pirin/GPA1 complex acts in concert with the NF-Y transcription factor to regulate the abscisic acid and blue light induction of the light-harvesting chlorophyll a/b-binding gene in Arabidopsis (Warpeha et al., 2007). Plants homozygous for atpirin1 have delayed germination, inhibited seedling development, and earlier flowering times than the wild type (Lapik and Kaufman, 2003). The tomato (Solanum lycopersicum) pirin Le-Pirin is transcriptionally induced during camptothecin-induced programmed cell death (Orzaez et al., 2001). Because Pirin is transcriptionally regulated during haustorium development and is associated with signal transduction in plants, we investigated its function in parasitic plants.
RESULTS
TvPirin Gene Structure and Phylogeny
TvPirin was originally identified from a cDNA library enriched for transcripts up-regulated in T. versicolor root tips exposed to 2,6-dimethoxybenzoquinone (DMBQ; Matvienko et al., 2001a). The original TvPirin cDNA (BE574904.1) was predicted to be missing about 30 nucleotides of 5′ coding sequences, so RACE reactions and sequence assemblies were carried out to obtain the complete coding sequence. The transcript is predicted to encode a 322-amino acid protein with about 70% amino acid identity to the human Pirin (NP_003653.1; Wendler et al., 1997). There was no evidence of a nuclear localization signal using the protein localizations predictor WoLF PSORT (Horton et al., 2007).
Primers were designed from each end of the cDNA and used to amplify and sequence T. versicolor genomic DNA. Comparison of the genomic and cDNA sequences indicated that TvPirin has six introns. Two Arabidopsis Pirin homologs, At3g59260 and At2g43120, also have six introns located at analogous positions as in TvPirin. The Arabidopsis AtPirin1 characterized as a Gα binding protein (At3G59220) has slightly less homology to TvPirin and has five introns at distinct positions from those in TvPirin. Therefore, it is unlikely that the characterized Arabidopsis gene AtPirin1 is the ortholog of TvPirin.
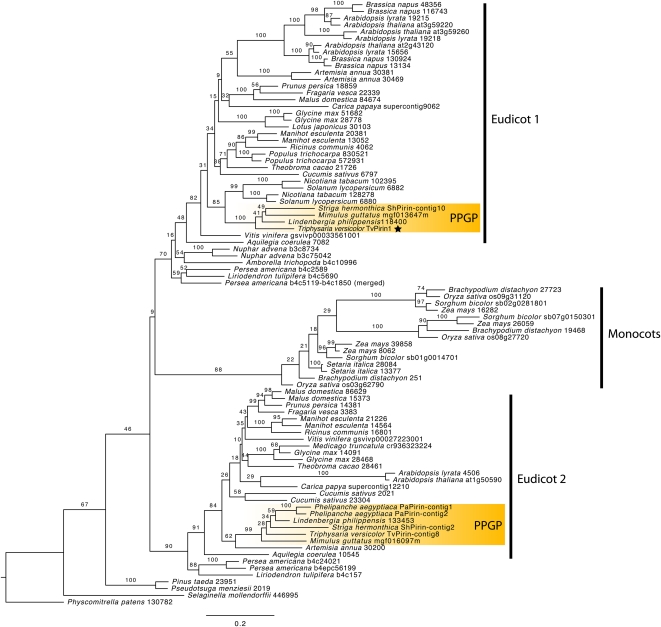
We searched the Parasitic Plant Genome Project (PPGP) EST database using BLASTN to determine how many homologs of TvPirin are present in Orobanchaceae. At the time of this analysis, the database reflected >1.3 billion reads derived from Sanger, 454, and Illumina cDNA sequencing of various tissues and growth stages of T. versicolor, Striga hermonthica, Phelipanche aegyptiaca, and Lindenbergia philippensis (Westwood et al., 2010). We also searched for homologous sequences in public databases containing fully sequenced plant genomes to develop a phylogeny of plant Pirins (Fig. 1). This analysis indicated two clades of Pirins in eudicots, both represented in the Orobanchaceae. The TvPirin in this study (starred in Fig. 1) is part of a well-supported clade that includes the parasite S. hermonthica and the nonparasitic plants Mimulus guttatus and L. philippensis. Surprisingly, we did not detect a P. aegyptiaca homolog in this clade, but there was a P. aegyptiaca homolog in clade 2. There is 77% amino acid identity between the clade 1 protein (TvPirin1) and clade 2 protein (TvPirin-contig 8). We cannot say if the clade 2 transcript is silenced by the hairpin constructions because the primers used in the quantitative reverse transcription (qRT)-PCR analyses were not designed to amplify TvPirin-contig8 transcripts. There was no evidence of parasite-specific duplications or expansions of Pirin as might be expected if it evolved novel functions in the parasite clade.
Figure 1.
Pirin phylogeny. The phylogeny of the Pirin gene family was derived from sequence data of 10 fully sequenced plant genomes together with sequences from the PPGP (shaded and labeled PPGP). The TvPirin gene described in this manuscript is marked with a star. [See online article for color version of this figure.]
Hairpin-Mediated Silencing of TvPirin Reduces Haustorium Development
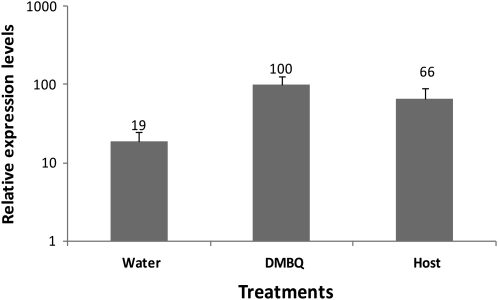
Prior investigations showed that TvPirin is transcriptionally up-regulated in parasite roots by DMBQ (Matvienko et al., 2001a). In this study, we quantified the degree of TvPirin regulation in response to host root contact by qRT-PCR. As seen in Figure 2, contact with Medicago truncatula roots resulted in about a 3- to 4-fold increase in TvPirin transcripts in T. versicolor root tips, about the same as obtained with DMBQ.
Figure 2.
Transcriptional regulation of TvPirin. RNA was extracted from T. versicolor seedlings after exposure to water, DMBQ, or M. truncatula (host) roots for 2 h. Steady-state transcript levels of TvPirin were determined by qRT-PCR and normalized to the constitutively expressed gene TvQN8 for each sample. Data are means ± sd of three technical replicates of three biological replicates (n = 9). Each biological replicate was a square petri dish with 20 to 50 seedlings. Expression levels of roots treated with DMBQ was set to 100%. Note the relative expression values are written on top of each bar.
A hairpin RNAi vector targeted against TvPirin sequences, called pHpPRN, and the parent vector, pHG8-YFP, were transformed into T. versicolor hypocotyls via Agrobacterium rhizogenes and transgenic roots identified by their yellow fluorescent protein (YFP) fluorescence. Based on YFP fluorescence, 25% to 30% of the seedlings had at least one transgenic root. Roots transgenic for pHpPRN or pHG8-YFP had similar growth rates and morphologies (Table I).
Table I.
Efficiency and phenotypes of transgenic rootsa
| Phenotype | pHG8-YFP | pHpPRN | t-Value |
| Transformation efficiencyb | 28 ± 2.2 | 26 ± 2.8 | 0.61 |
| n = 1271 | n = 1539 | ||
| YFP-positive roots per plantc | 1.8 ± 0.4 | 1.7 ± 0.4 | 0.88 |
| n = 352 | n = 403 | ||
| YFP-positive roots per plantd | 2.0 ± 0.9 | 2.4 ± 0.7 | 0.05 |
| n = 49 | n = 57 | ||
| Root length (cm)d | 3.5 ± 1.8 | 3.1 ± 1.0 | 0.74 |
| n = 49 | n = 57 | ||
| Lateral roots per YFP rootd | 0.1 ± 0.3 | 0.1 ± 0.4 | 0.20 |
| n = 49 | n = 57 | ||
| Root growth no DMBQ (mm/24 h)d | 2.6 ± 0.4 | 2.5 ± 0.6 | 0.85 |
| n = 40 | n = 40 | ||
| Root growth with DMBQ (mm/24 h)d | 2.7 ± 0.5 | 2.6 ± 0.7 | 0.12 |
| n = 40 | n = 40 |
Values are mean ± sd with n = total number of plants or roots in all replicates. Means are the averages of three independent transformation experiments per construction. Data were analyzed using SAS statistical software (pooled t test/equal/unequal variance).
Percentage of plants with at least one yellow root.
Twenty-one days after transformation.
Thirty-three days after transformation.
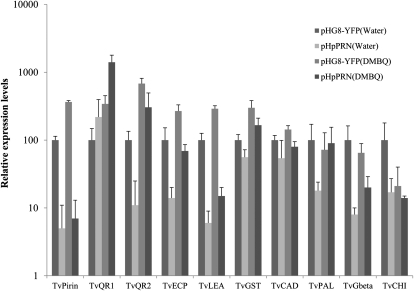
RNA was harvested from pHG8-YFP and pHpPRN roots with and without DMBQ exposure and TvPirin transcripts quantified. Basal levels of TvPirin were about 20-fold lower in pHpPRN roots than pHG8-YFP roots (Fig. 3). Similar reductions were obtained in previous experiments targeting TvQR1 and TvQR2 by RNAi (Bandaranayake et al., 2010). When pHG8-YFP roots were exposed to DMBQ, there was a 3- to 4-fold induction of TvPirin compared to the water treatment. When pHpPRN roots were similarly treated with DMBQ, we detected no up-regulation of TvPirin (Fig. 3).
Figure 3.
Gene expression in pHpPRN roots. Steady-state transcript levels of 10 parasite genes in TvPirin-silenced roots were determined by quantitative PCR and normalized to the constitutively expressed gene TvQN8 for each sample. Data are means ± sd of three technical replicates of three to four plants (n = 9–12). Expression levels in pHG8-YFP transgenic roots treated with water was set to 100% and considered as the basal level of expression of the interested gene. Note the log scale y axis.
The ability of transgenic roots to develop haustoria was investigated by exposing transgenic roots to host roots, host root exudates, DMBQ, or water and assaying haustorium development 24 h later (Table II). Nontransgenic seedlings and control transgenic roots formed haustoria in approximately 70% to 85% of the roots after treatment with host root, root exudates, or DMBQ. In contrast, only about 20% to 50% of roots transformed with pHpPRN developed haustoria. No haustoria were seen to develop after the 24-h assay period.
Table II.
Haustorium development in transgenic roots silenced for TvPIRIN
| Treatmenta | Percentage of Roots with Haustoria | ||
| Seedlingsb | pHG8YFP | pHpPRN | |
| 30 μM DMBQ | 75 ± 11 | 71 ± 12* | 38 ± 10** |
| n = 565 | n = 532 | n = 505 | |
| Host root exudates | 85 ± 3 | 73 ± 9* | 18 ± 5** |
| n = 214 | n = 66 | n = 27 | |
| Host roots | 87 ± 8 | 84 ± 9* | 47 ± 10** |
| n = 90 | n = 113 | n = 23 | |
| Water | 0 ± 0 | 0 ± 0 | 0 ± 0 |
| n = 186 | n = 104 | n = 97 | |
T. versicolor roots were exposed to DMBQ or host exudates and assayed 24 h later or exposed to Arabidopsis roots and assayed after 5 d. Data are averages ± sd of three to seven plates with four to six independently transformed plants in each plate. Experiment with DMBQ treatment was repeated three times with plants from three independent transformation experiments. Each plant had 1 to 30 roots depending on the age of the plant (n = total number of roots in all replicates). Each treatment was applied to similarly aged plants from the same transformation experiment. Pairwise comparisons of treatments within a row labeled with a different number of asterisks are significantly different at α = 0.05 (t test).
Seedling data were not included in statistical analysis because they were differently aged.
Hairpin-Mediated Silencing and Regulation of Additional Genes by TvPirin
We determined steady state levels of 10 transcripts (including TvPirin) in T. versicolor roots transformed with either the empty parent vector (pHG8-YFP) or the hairpin construct (pHpPRN) by quantitative real-time PCR (Fig. 3). Five genes were up-regulated by DMBQ: TvPirin, TvQR1, TvQR2, TvGST, TvLEA, and TvECP. The genes TvQR1 and TvQR2 encode quinone oxidoreductases and previously were reported as up-regulated by DMBQ (Matvienko et al., 2001b; Bandaranayake et al., 2010). TvGST encodes a putative glutathione transferase, a ubiquitous class of enzymes involved in detoxification by conjugation (Hayes et al., 2005). TvLEA is homologous to genes encoding Late Embryogenesis Abundant (LEA) proteins in Arabidopsis. Both glutathione S-transferase (GST) and LEA transcripts are differentially regulated in nonparasitic plants by various biotic and abiotic stresses (Mowla et al., 2006; Sappl et al., 2009). TvECP has weak homology to several putative plant cytochrome P450 monooxygenases. We also examined the expression of four genes not up-regulated by DMBQ: TvCAD, TvPAL, TvCHI, and TvGbeta. TvPAL is predicted to encode Phe ammonia-lyase and TvCHI chalcone isomerase, two genes on the phenylpropanoid biosynthesis pathway (Dixon and Paiva, 1995). TvCAD is a putative cinnamyl-alcohol dehydrogenase that catalyzes monolignol biosynthesis, a key step in lignin biosynthesis (Raes et al., 2003). TvGbeta is predicted to encode the β-subunit of the G protein complex (Assmann, 2002).
The basal expression level of each gene was taken as the transcript abundance in control pHG8-YFP roots treated with water. Seven genes, TvPirin, TvQR2, TvECP, TvLEA, TvPAL, TvGbeta, and TvCHI, were significantly reduced in pHpPRN compared to control roots (Fig. 3). Because there is no apparent homology between pHpPRN and the other transcripts (except TvPirin), the reduction in gene expression is likely a secondary effect of TvPirin silencing. Basal expression of TvQR1, TvGST, and TvCAD was not significantly affected in TvPirin-silenced roots.
The relative expression levels after DMBQ exposure were then determined (Fig. 3). All of the transcripts, except TvPirin, that were induced by DMBQ in control roots were similarly induced in pHpPRN roots. These results suggest that TvPirin functions in establishing the basal transcript levels of a set of genes in parasite roots. However, TvPirin does not affect the responsiveness of genes to DMBQ.
DISCUSSION
The chemical influence of some plants on the growth and development of others has been appreciated for centuries (Willis, 1985). While exploiting the potential of allelopathy to improve crop performance is often discussed, little is known about the mechanisms by which plants detect and process chemical signals from other plants. Haustorium development in response to xenognosins provides a useful model for investigating chemical signaling between plant roots in general. Both xenognosin response and allelopathy are mediated by similar molecules: quinones and oxidized phenols. Depending on its concentration, DMBQ can be either a developmental stimulant that induces haustorium development or a plant toxin (Tomilov et al., 2006). Parasite quinone oxidoreductases use both xenognosins and allelopathic phytotoxins as biochemical substrates (Wrobel et al., 2002; Bandaranayake et al., 2010). Furthermore, both allelopathy and haustorium induction are dependent upon the redox state of the chemical agent. Juglone, the active allelopathic agent from black walnut trees, is synthesized in the inactive hydrojuglone state that is then activated to the toxic state upon exposure to oxygen (Lee and Campbell, 1969). As discussed previously, haustorium development is also dependent on the redox state of the inducer. Similarities between quinone associated allelopathy and haustorium initiation led to the hypothesis that the processes may share common molecular mechanisms (Tomilov et al., 2006).
Haustorium development is a multistep process that requires the coordinated expression of a number of genes and pathways. Most of the processes associated with haustorium development have almost certainly been derived from autotrophic plant processes. In this study, we found that the TvPirin gene associated with haustorium development has homologs in autotrophic plants that must provide functions unrelated to parasitism. Other genes associated with haustorium development similarly have nonparasitic functions in autotrophic plants. Expansins, nonenzymatic proteins that promote cell wall loosening, are used in both parasitic and nonparasitic plant processes (O’Malley and Lynn, 2000; Wrobel and Yoder, 2001). ζ Crystallin quinone oxidoreductases catalyze similar biochemical reactions in many organisms but have specific functions in triggering haustorium development in parasites (Wrobel et al., 2002; Bandaranayake et al., 2010). TvPirin provides both parasite specific and nonparasitic functions without apparent gene duplication.
Our results are consistent with TvPirin functioning in transcription regulation because its inhibition results in several transcripts being down-regulated. The basal expression levels of seven of 10 genes investigated were significantly lower in pHpPRN than pHG8-YFP roots. These included both DMBQ-induced and -noninduced genes, so there was no apparent specificity for quinone-responsive transcripts. However, DMBQ up-regulated genes maintained the same level of responsiveness in TvPirin-silenced roots as in control transgenics, suggesting the role of TvPirin is restricted to establishing basal levels of gene expression rather than xenognosin responsiveness. We propose that TvPirin is a generalized transcription factor associated protein that functions in the transcription of several different genes, some of which are needed for haustorium development.
MATERIALS AND METHODS
Chemicals, Plants, and Genes
Seeds of the outcrossing species Triphysaria versicolor were collected from an open pollinated population near Napa, CA. DMBQ was purchased from Pfaltz & Bauer, dissolved in water at 30 μM, filter sterilized, and stored at 4°C until used. PCR primers were designed using Primer 3 software (Rozen and Skaletsky, 2000) and synthesized by Integrated DNA Technologies. Primer sequences are shown in Supplemental Table S1.
Phylogenic Analyses
The Pirin gene family was identified from PlantTribes 2.0 (http://fgp.bio.psu.edu/tribedb/10_genomes/index.pl), an objective classification of plant proteins based on a cluster analysis of the inferred proteins of 10 fully sequenced plant genomes (Wall et al., 2008). Using the PlantTribes annotations, a single Tribe (roughly analogous to a gene family) was identified that contained four Arabidopsis (Arabidopsis thaliana) Pirin protein sequences. All embryophyte protein sequences that comprise the Pirin Tribe were extracted from PlantTribes and aligned using MAFFT v6.717b (Katoh et al., 2002; Katoh and Toh, 2008). The alignment was used to make a profile hidden Markov model with which publicly available EST sequences (http://www.plantgdb.org/) and gene models from genome sequences (http://www.phytozome.net/; Schnable et al., 2009; Schmutz et al., 2010; Velasco et al., 2010; Vogel et al., 2010; Shulaev et al., 2011) were searched using HMMER version 3.0 to identify putative homologs to the Pirin gene family genome sequences (http://hmmer.janelia.org/). All ESTs that were searched against the HMM were first translated with GeneWise from the Wise2 package (Birney et al., 2004) using the Vitis vinifera sequence as a reference. Additional parasite sequences were generated through the PPGP, a National Science Foundation-funded program aimed at obtaining deep transcriptome data sets from several species of Orobanchaceae (Westwood et al., 2010). Putative Pirin homologs from all PPGP assemblies (individual tissues and combo builds) were reassembled for each species using CAP3 (Huang and Madan, 1999) to identify a nonredundant set of transcripts (i.e. consolidate contigs that remained unassembled due to minor polymorphisms or sequencing errors or insufficient depth for automated assembly). Contigs and singletons from these assemblies as well as from all additional genome and transcriptome sequences were aligned using MAFFT, and a preliminary phylogenetic analysis was carried out with RAxML v.7.0.4 (Stamatakis, 2006) to identify and remove any contaminating host sequences, as well as nearly identical, unassembled sequences (likely due to sequencing error or putative alleles). The nonredundant alignment was then realigned using SATé (Liu et al., 2009) and adjusted manually. The phylogenetic tree was produced with RAxML, with 1000 fast bootstrap replicates.
Plasmid Constructions
Hairpin constructions were made using the Gateway-compatible vector pHellsgate 8 that contained a YFP reporter gene for selection in T. versicolor roots (Helliwell et al., 2002; Subramanian et al., 2006; Bandaranayake et al., 2010). A 462-nucleotide region of the TvPirin open reading frame was amplified using PCR primers flanked by attB recombinase sites (Supplemental Table S1). The gel-purified PCR products were recombined into the Gateway donor vector pDONR211 following the manufacturer’s protocol for BP recombination (Invitrogen). After the pDONR construct was confirmed by sequencing, Gateway LR recombination was performed with pHG8-YFP generating the hairpin vector pHpPRN. The vector was confirmed by restriction digestions.
T. versicolor Root Transformation
T. versicolor roots were transformed using an Agrobacterium rhizogenes-based protocol as described (Bandaranayake et al., 2010). The plasmid constructions were transformed into A. rhizogenes MSU440 by electroporation (Sonti et al., 1995). Roots were excised from aseptic T. versicolor seedlings and the cut ends inoculated with MSU440 bearing the appropriate constructions. Seedlings were maintained on agar plates at 25°C for about 3 weeks, at which time transgenic roots were identified by visualization of YFP fluorescence with a Zeiss Stemi SV11 dissecting microscope equipped with an YFP filter set. Transformation efficiencies were calculated as the number of plants with at least one yellow root.
Haustorium Assay
Root exudates were collected from Medicago truncatula plants grown in Magenta boxes containing 20 mL of 0.5% agar. After 10 to 14 d, plants were removed and the agar was centrifuged at 20,000 rpm for 30 min. The supernatant was filter sterilized and stored at 4°C.
Haustorium development was assayed as previously described (Jamison and Yoder, 2001). Transgenic roots were maintained on agar plates at 25°C. Two milliliters of DMBQ (30 μM) or host root exudates were added to the roots on each plate and haustoria assayed 24 h later. Alternatively, haustoria were induced by overlaying T. versicolor roots on the surface of an agar plate with Arabidopsis roots for 5 d. In all cases, the number of YFP positive roots that made haustoria was expressed as a fraction of the total number of YFP roots treated.
Transcriptional Analyses
T. versicolor roots were exposed to M. truncatula roots or DMBQ for various times before being flash frozen in liquid nitrogen and RNA isolated using the TRIzol reagent (Invitrogen). RNA was treated with DNase1 and further purified using RNeasy Mini Spin columns (Qiagen). One microgram of RNA from each sample was converted to cDNA using the SuperScript III first-strand synthesis system for RT-PCR (Invitrogen). The reverse transcription reactions were diluted 20-fold and 2 μL used for SYBR green-based quantitative PCR assays using a LightCycler480 real-time PCR system (Roche) or ABI 7300 quantitative PCR system (Applied Biosystems). The cycle conditions were similar for both systems: 94°C for 3 min as initial denaturation followed by 35 cycles of 94°C for 15 s, 58°C for 30 s for primer annealing, and 72°C for 15 s. Melting curves of PCR products were obtained by gradually heating to 95°C, and only those producing a single melting peak were considered in the analysis. Target gene expression was measured relative to the constitutively expressed gene TvQN8. For LightCycler480 data, expression calculations used the standard curve method, taking into account the efficiencies of the PCR reactions, calculated by log-linear regression LightCycler480 analysis software (Roche). The data obtained with ABI 7300 system were analyzed with SDS Software using the ΔΔ Ct ratio ratio real-time PCR method (relative quantification), which determines the δ Ct of the gene of interest and TvQN8 and subtracts this from the analogous measurement obtained from the internal control (the δ − δ Ct).
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: TvPirin, JN606867; TvQR1, AF304461; TvQR2, AF304462; TvECP, DR173551; TvGST, DR173703; TvLEA, DR172703; and TvCAD, DR175870.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. The sequences of the primers used in these experiments.
Supplementary Material
Acknowledgments
We thank the enthusiastic support of our colleagues in the PPGP, including the contribution of materials of Striga hermonthica (Mike Timko group, University of Virginia), Phelipanche (=Orobanche) aegyptiaca (Jim Westwood group, Virginia Tech), and Lindenbergia philipensis (Paula Ralph, Penn State University). We thank the sequencing labs at University of Virginia (Yongde Bao) and Penn State University (Stephan Schuster and Lynn Tomsho) and Eric Wafula (Penn State University) for construction of the PPGP database. Gene sequences of Arabidopsis lyrata, Aquilegia coerulea, Manihot esculenta, Prunus persica, Ricinus communis, and Setaria italica were produced by the U.S. Department of Energy Joint Genome Institute (http://www.jgi.doe.gov/) in collaboration with the user community. Gene sequences of Amborella trichopoda, Liriodendron tulipifera, Nuphar advena, and Persea Americana were produced by the Ancestral Angiosperm Genome Project (http://ancangio.uga.edu).
References
- Adams M, Jia Z. (2005) Structural and biochemical analysis reveal pirins to possess quercetinase activity. J Biol Chem 280: 28675–28682 [DOI] [PubMed] [Google Scholar]
- Albrecht H, Yoder JI, Phillips DA. (1999) Flavonoids promote haustoria formation in the root parasite Triphysaria versicolor. Plant Physiol 119: 585–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assmann SM. (2002) Heterotrimeric and unconventional GTP binding proteins in plant cell signaling. Plant Cell 14(Suppl): S355–S373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atsatt PR, Hearn TF, Nelson RL, Heineman RT. (1978) Chemical induction and repression of haustoria in Orthocarpus purpurascens (Scophulariaceae). Ann Bot (Lond) 42: 1177–1184 [Google Scholar]
- Baird WV, Riopel JL. (1984) Experimental studies of haustorium initiation and early development in Agalinis purpurea (L.) Raf. (Scrophulariaceae). Am J Bot 71: 803–814 [Google Scholar]
- Bandaranayake PCG, Filappova T, Tomilov A, Tomilova NB, Jamison-McClung D, Ngo Q, Inoue K, Yoder JI. (2010) A single-electron reducing quinone oxidoreductase is necessary to induce haustorium development in the root parasitic plant Triphysaria. Plant Cell 22: 1404–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman AC, Alaiya AA, Wendler W, Binetruy B, Shoshan M, Sakaguchi K, Bergman T, Kronenwett U, Auer G, Appella E, et al. (1999) Protein kinase-dependent overexpression of the nuclear protein pirin in c-JUN and RAS transformed fibroblasts. Cell Mol Life Sci 55: 467–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Clamp M, Durbin R. (2004) GeneWise and Genomewise. Genome Res 14: 988–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M, Lynn DG. (1986) The haustorium and the chemistry of host recognition in parasitic angiosperms. J Chem Ecol 12: 561–579 [DOI] [PubMed] [Google Scholar]
- Dechend R, Hirano F, Lehmann K, Heissmeyer V, Ansieau S, Wulczyn FG, Scheidereit C, Leutz A. (1999) The Bcl-3 oncoprotein acts as a bridging factor between NF-kappaB/Rel and nuclear co-regulators. Oncogene 18: 3316–3323 [DOI] [PubMed] [Google Scholar]
- Dixon RA, Paiva NL. (1995) Stress-induced phenylpropanoid metabolism. Plant Cell 7: 1085–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JD, Flanagan JU, Jowsey IR. (2005) Glutathione transferases. Annu Rev Pharmacol Toxicol 45: 51–88 [DOI] [PubMed] [Google Scholar]
- Helliwell CA, Wesley SV, Wielopolska AJ, Waterhouse PM. (2002) High-throughput vectors for efficient gene silencing in plants. Funct Plant Biol 29: 1217–1225 [DOI] [PubMed] [Google Scholar]
- Horton P, Park K-J, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K. (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res 35(Web Server issue): W585–W587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XQ, Madan A. (1999) CAP3: a DNA sequence assembly program. Genome Res 9: 868–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamison DS, Yoder JI. (2001) Heritable variation in quinone-induced haustorium development in the parasitic plant Triphysaria. Plant Physiol 125: 1870–1879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30: 3059–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Toh H. (2008) Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform 9: 286–298 [DOI] [PubMed] [Google Scholar]
- Keyes WJ, Taylor JV, Apkarian RP, Lynn DG. (2001) Dancing together. Social controls in parasitic plant development. Plant Physiol 127: 1508–1512 [PMC free article] [PubMed] [Google Scholar]
- Kuijt J. (1969) The Biology of Parasitic Flowering Plants. University of California Press, Berkeley, CA [Google Scholar]
- Lapik YR, Kaufman LS. (2003) The Arabidopsis cupin domain protein AtPirin1 interacts with the G protein alpha-subunit GPA1 and regulates seed germination and early seedling development. Plant Cell 15: 1578–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KC, Campbell RW. (1969) Nature and occurence of juglone in Juglans nigra L. Hortic Sci 4: 297–298 [Google Scholar]
- Liu K, Raghavan S, Nelesen S, Linder CR, Warnow T. (2009) Rapid and accurate large-scale coestimation of sequence alignments and phylogenetic trees. Science 324: 1561–1564 [DOI] [PubMed] [Google Scholar]
- Lynn DG, Steffens JC, Kamat VS, Graden DW, Shabanowitz J, Riopel JL. (1981) Isolation and characterization of the first host recognition substance for parasitic angiosperms. J Am Chem Soc 103: 1868–1870 [Google Scholar]
- Matvienko M, Torres MJ, Yoder JI. (2001a) Transcriptional responses in the hemiparasitic plant Triphysaria versicolor to host plant signals. Plant Physiol 127: 272–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matvienko M, Wojtowicz A, Wrobel R, Jamison D, Goldwasser Y, Yoder JI. (2001b) Quinone oxidoreductase message levels are differentially regulated in parasitic and non-parasitic plants exposed to allelopathic quinones. Plant J 25: 375–387 [DOI] [PubMed] [Google Scholar]
- Mowla SB, Cuypers A, Driscoll SP, Kiddle G, Thomson J, Foyer CH, Theodoulou FL. (2006) Yeast complementation reveals a role for an Arabidopsis thaliana late embryogenesis abundant (LEA)-like protein in oxidative stress tolerance. Plant J 48: 743–756 [DOI] [PubMed] [Google Scholar]
- Musselman LJ. (1980) The biology of Striga, Orobanche, and other root parasitic weeds. Annu Rev Phytopathol 18: 463–489 [Google Scholar]
- Nickrent D. (2011) The Parasitic Plant Connection. Southern Illinois University at Carbondale. http://www.parasiticplants.siu.edu (September 9, 2011) [Google Scholar]
- O’Malley RC, Lynn DG. (2000) Expansin message regulation in parasitic angiosperms: marking time in development. Plant Cell 12: 1455–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orzaez D, de Jong AJ, Woltering EJ. (2001) A tomato homologue of the human protein PIRIN is induced during programmed cell death. Plant Mol Biol 46: 459–468 [DOI] [PubMed] [Google Scholar]
- Pang H, Bartlam M, Zeng Q, Miyatake H, Hisano T, Miki K, Wong LL, Gao GF, Rao Z. (2004) Crystal structure of human pirin: an iron-binding nuclear protein and transcription cofactor. J Biol Chem 279: 1491–1498 [DOI] [PubMed] [Google Scholar]
- Parker C, Riches CR. (1993) Parasitic Weeds of the World: Biology and Control. CAB International, Wallingford, UK [Google Scholar]
- Raes J, Rohde A, Christensen JH, Van de Peer Y, Boerjan W. (2003) Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol 133: 1051–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riopel JL, Timko MP. (1995) Haustorial initiation and differentiation. Press MC, Graves JD, , Parasitic Plants. Chapman and Hall, London, pp 39–79 [Google Scholar]
- Rozen S, Skaletsky H. (2000) Primer3 on the WWW for general users and for biologist programmers. Krawetz S, Misener S, , Bioinformatics Methods and Protocols. Humana Press, Totowa, NJ, pp 365–386 [DOI] [PubMed] [Google Scholar]
- Sappl PG, Carroll AJ, Clifton R, Lister R, Whelan J, Harvey Millar A, Singh KB. (2009) The Arabidopsis glutathione transferase gene family displays complex stress regulation and co-silencing multiple genes results in altered metabolic sensitivity to oxidative stress. Plant J 58: 53–68 [DOI] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma JX, Mitros T, Nelson W, Hyten DL, Song QJ, Thelen JJ, Cheng JL, et al. (2010) Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183 [DOI] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei FS, Pasternak S, Liang CZ, Zhang JW, Fulton L, Graves TA, et al. (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Shulaev V, Sargent DJ, Crowhurst RN, Mockler TC, Folkerts O, Delcher AL, Jaiswal P, Mockaitis K, Liston A, Mane SP, et al. (2011) The genome of woodland strawberry (Fragaria vesca). Nat Genet 43: 109–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CE, Dudley MW, Lynn DG. (1990) Vegetative/parasitic transition: control and plasticity in striga development. Plant Physiol 93: 208–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CE, Ruttledge T, Zeng Z, O’Malley RC, Lynn DG. (1996) A mechanism for inducing plant development: the genesis of a specific inhibitor. Proc Natl Acad Sci USA 93: 6986–6991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonti RV, Chiurazzi M, Wong D, Davies CS, Harlow GR, Mount DW, Signer ER. (1995) Arabidopsis mutants deficient in T-DNA integration. Proc Natl Acad Sci USA 92: 11786–11790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soo P-C, Horng Y-T, Lai M-J, Wei J-R, Hsieh S-C, Chang Y-L, Tsai Y-H, Lai H-C. (2007) Pirin regulates pyruvate catabolism by interacting with the pyruvate dehydrogenase E1 subunit and modulating pyruvate dehydrogenase activity. J Bacteriol 189: 109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690 [DOI] [PubMed] [Google Scholar]
- Steffens JC, Lynn DG, Kamat VS, Riopel JL. (1982) Molecular specificity of haustorial induction in Agalinis purpurea (L.) Raf. (Scrophulariaceae). Ann Bot (Lond) 50: 1–7 [Google Scholar]
- Subramanian C, Woo J, Cai X, Xu XD, Servick S, Johnson CH, Nebenführ A, von Arnim AG. (2006) A suite of tools and application notes for in vivo protein interaction assays using bioluminescence resonance energy transfer (BRET). Plant J 48: 138–152 [DOI] [PubMed] [Google Scholar]
- Tomilov AA, Tomilova NB, Shin DH, Jamison D, Torres M, Reagan R, Horning T, Truong R, Nava A, Nava A, et al. (2006) Chemical signaling between plants: Mechanistic similarities between phytotoxic allelopathy and host recognition by parasitic plants. Dicke M, , Chemical Ecology: From Gene to Ecosystem. Springer, Dordrecht, The Netherlands, pp 55–69 [Google Scholar]
- Torres MJ, Tomilov AA, Tomilova N, Reagan RL, Yoder JI. (2005) Pscroph, a parasitic plant EST database enriched for parasite associated transcripts. BMC Plant Biol 5: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A, Kalyanaraman A, Fontana P, Bhatnagar SK, Troggio M, Pruss D, et al. (2010) The genome of the domesticated apple (Malus × domestica Borkh.). Nat Genet 42: 833–839 [DOI] [PubMed] [Google Scholar]
- Vogel JP, Garvin DF, Mockler TC, Schmutz J, Rokhsar D, Bevan MW, Barry K, Lucas S, Harmon-Smith M, Lail K, et al. ; International Brachypodium Initiative (2010) Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463: 763–768 [DOI] [PubMed] [Google Scholar]
- Wall PK, Leebens-Mack J, Müller KF, Field D, Altman NS, dePamphilis CW. (2008) PlantTribes: a gene and gene family resource for comparative genomics in plants. Nucleic Acids Res 36(Database issue): D970–D976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warpeha KM, Upadhyay S, Yeh J, Adamiak J, Hawkins SI, Lapik YR, Anderson MB, Kaufman LS. (2007) The GCR1, GPA1, PRN1, NF-Y signal chain mediates both blue light and abscisic acid responses in Arabidopsis. Plant Physiol 143: 1590–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendler WMF, Kremmer E, Förster R, Winnacker EL. (1997) Identification of pirin, a novel highly conserved nuclear protein. J Biol Chem 272: 8482–8489 [DOI] [PubMed] [Google Scholar]
- Westwood JH, Roney JK, Khatibi PA, Stromberg VK. (2009) RNA translocation between parasitic plants and their hosts. Pest Manag Sci 65: 533–539 [DOI] [PubMed] [Google Scholar]
- Westwood JH, Yoder JI, Timko MP, dePamphilis CW. (2010) The evolution of parasitism in plants. Trends Plant Sci 15: 227–235 [DOI] [PubMed] [Google Scholar]
- William CN. (1961) Growth and morphogenesis of Striga seedlings. Nature 189: 378–381 [Google Scholar]
- Willis RJ. (1985) The historical bases of the concept of allelopathy. J Hist Biol 18: 71–102 [Google Scholar]
- Wrobel RL, Matvienko M, Yoder JI. (2002) Heterologous expression and biochemical characterization of an NAD(P)H: quinone oxidoreductase from the hemiparasitic plant Triphysaria versicolor. Plant Physiol Biochem 40: 265–272 [Google Scholar]
- Wrobel RL, Yoder JI. (2001) Differential RNA expression of alpha-expansin gene family members in the parasitic angiosperm Triphysaria versicolor (Scrophulariaceae). Gene 266: 85–93 [DOI] [PubMed] [Google Scholar]
- Zeng ZX, Cartwright CH, Lynn DG. (1996) Cyclopropyl-p-benzoquinone: a specific organogenesis inhibitor in plants. J Am Chem Soc 118: 1233–1234 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.