Abstract
In Arabidopsis (Arabidopsis thaliana), the NRT2.1 gene codes for the main component of the root nitrate (NO3−) high-affinity transport system (HATS). Due to the strong correlation generally found between high-affinity root NO3− influx and NRT2.1 mRNA level, it has been postulated that transcriptional regulation of NRT2.1 is a key mechanism for modulation of the HATS activity. However, this hypothesis has never been demonstrated, and is challenged by studies suggesting the occurrence of posttranscriptional regulation at the NRT2.1 protein level. To unambiguously clarify the respective roles of transcriptional and posttranscriptional regulations of NRT2.1, we generated transgenic lines expressing a functional 35S::NRT2.1 transgene in an atnrt2.1 mutant background. Despite a high and constitutive NRT2.1 transcript accumulation in the roots, the HATS activity was still down-regulated in the 35S::NRT2.1 transformants in response to repressive nitrogen or dark treatments that strongly reduce NRT2.1 transcription and NO3− HATS activity in the wild type. In some treatments, this was associated with a decline of NRT2.1 protein abundance, indicating posttranscriptional regulation of NRT2.1. However, in other instances, NRT2.1 protein level remained constant. Changes in abundance of NAR2.1, a partner protein of NRT2.1, closely followed those of NRT2.1, and thus could not explain the close-to-normal regulation of the HATS in the 35S::NRT2.1 transformants. Even if in certain conditions the transcriptional regulation of NRT2.1 contributes to a limited extent to the control of the HATS, we conclude from this study that posttranscriptional regulation of NRT2.1 and/or NAR2.1 plays a predominant role in the control of the NO3− HATS in Arabidopsis.
The uptake of nitrate (NO3−) by roots cells from the soil solution is the main process of nitrogen (N) acquisition in most herbaceous plant species (Marschner, 1995). It relies on the combined activity of both high- and low-affinity transport systems (HATS and LATS, respectively) that together ensure an efficient intake over a wide range of external NO3− concentrations (Crawford and Glass, 1998; Miller et al., 2007). However, with the exception of well-fertilized agricultural systems, NO3− concentrations in the soil are often below the millimolar range, making the HATS particularly important for nutrition of the plant (Marschner, 1995; Malagoli et al., 2004). To date, the membrane carriers associated with root uptake of NO3− belong to either NRT1 or NRT2 transporter families (Miller et al., 2007; Tsay et al., 2007). In general, NRT1 proteins are low-affinity transports, whereas NRT2 proteins are believed to be active in high-affinity transport only (Miller et al., 2007; Tsay et al., 2007).
In Arabidopsis (Arabidopsis thaliana), at least three transporters (NRT1.1, NRT2.1, and NRT2.2) have been proposed to contribute to the NO3− HATS (Li et al., 2007; Tsay et al., 2007), but it is now clear that the HATS activity, in most environmental conditions, is predominantly dependent on the NRT2.1 protein. Null mutants for NRT2.1 have lost up to 75% of the HATS activity (Cerezo et al., 2001; Filleur et al., 2001; Li et al., 2007), and consequently cannot grow normally with NO3− as sole nitrogen (N) source when provided at a low concentration (e.g. <1 mm; Orsel et al., 2004). However, as other NRT2 proteins identified in Chlamydomonas reinhardtii and barley (Hordeum vulgare; Quesada et al., 1994; Tong et al., 2005), the Arabidopsis NRT2.1 protein needs to interact with a NAR2-like partner protein (NAR2.1/NRT3.1) to be functional in NO3− transport (Okamoto et al., 2006; Orsel et al., 2006). Both gene products need to be coexpressed to yield NO3− uptake in Xenopus oocytes (Orsel et al., 2006), and nar2.1 null mutants lack the NRT2.1 protein at the plasma membrane (Wirth et al., 2007), and are strongly deficient in NO3− HATS (Okamoto et al., 2006; Orsel et al., 2006). The precise function of NAR2.1 remains unclear, but it has been recently proposed that the active form of the transporter is in fact a NRT2.1/NAR2.1 heterooligomer (Yong et al., 2010).
A key feature of the NO3− HATS activity is that it can be rapidly and markedly modulated, to allow a fine coordination of the high-affinity root NO3− uptake with the N demand of the whole plant as well as its response to signaling mechanisms integrating N and carbon metabolisms (Crawford and Glass, 1998; Forde, 2002; Glass et al., 2002; Miller et al., 2007; Gojon et al., 2009; Nunes-Nesi et al., 2010). Furthermore, many studies have shown that regulation of the HATS is always highly correlated with changes in NRT2.1 transcript accumulation in the roots. In more detail, both NO3− HATS activity and NRT2.1 expression are similarly induced by NO3− (Filleur and Daniel-Vedele, 1999; Lejay et al., 1999; Zhuo et al., 1999), repressed by high N supply to the plant (Lejay et al., 1999; Zhuo et al., 1999; Cerezo et al., 2001; Gansel et al., 2001), repressed by darkness (Lejay et al., 1999), and stimulated by sugar supply to the roots (Lejay et al., 2003, 2008). All these changes in NRT2.1 expression (mRNA accumulation) were found to be associated with parallel changes in the activity of the NRT2.1 promoter (Girin et al., 2007, 2010). Thus, it has often been hypothesized that the transcriptional control of NRT2.1 is a major mechanism for regulation of the NO3− HATS in Arabidopsis (Forde, 2002; Miller et al., 2007; Gojon et al., 2009; Girin et al., 2010). As a consequence, many studies have focused on the identification of regulatory proteins governing NRT2.1 gene expression (Muños et al., 2004; Krouk et al., 2006; Castaings et al., 2009; Ho et al., 2009; Wang et al., 2009; Girin et al., 2010; Widiez et al., 2011).
However, several reports have pointed out that NRT2 transporters may also be strongly regulated at the posttranscriptional level. First, overexpression of the NpNRT2.1 gene in Nicotiana plumbaginifolia succeeded in stimulating the HATS activity when plants were supplied with NO3− as sole N source, but not when plants were fed with a mixed ammonium nitrate (NH4NO3) N source (Fraisier et al., 2000), indicating that NRT2.1 transcript accumulation was not the limiting factor in the latter case. More recently, the abundance of the Arabidopsis NRT2.1 protein in the root plasma membrane was reported to be little affected by treatments that strongly regulate both NRT2.1 transcript accumulation and NO3− HATS activity (Wirth et al., 2007). In barley too, changes in HvNRT2.1 protein were not correlated with those of HvNRT2.1 transcript accumulation during induction by NO3− (Ishikawa et al., 2009). Finally, studies on NRT2 homologs in lower eukaryotes, e.g. NrtA in Aspergillus nidulans (Wang et al., 2007) and YNT1 in Hansenula polymorpha (Navarro et al., 2008) have also evidenced the occurrence of posttranscriptional regulatory mechanisms for these proteins. Altogether, these data question the role of NRT2.1 transcription as a major process for the regulation of root NO3− uptake, and suggest that posttranscriptional events affecting abundance and/or activity of the NRT2.1 protein may actually have a stronger impact on functionality of the HATS.
The aim of our work was to clarify the respective importance of transcriptional and posttranscriptional regulations of NRT2.1 expression in the modulation of the NO3− HATS activity in Arabidopsis. Therefore, our strategy was to suppress the transcriptional level of control by expressing a 35S::NRT2.1 transgene in a nrt2.1 null-mutant background, and to determine whether this prevented, or not, regulation of the NO3− HATS in response to N or light treatments known to affect both NRT2.1 transcription and root NO3− acquisition in wild-type plants. The outcome of this study is that, although the transcriptional regulation of NRT2.1 may account for part of the control, posttranscriptional regulatory mechanisms clearly play a predominant role in the response of the HATS to environmental cues.
RESULTS
Functional Complementation of the atnrt2.1-2 Mutant by a 35S::NRT2.1 Transgene
To prevent transcriptional regulation of NRT2.1, we transformed the atnrt2.1-2 mutant (Little et al., 2005), with a 35S::NRT2.1 construct containing the genomic sequence of NRT2.1, fused to the constitutive cauliflower mosaic virus 35S promoter (see “Materials and Methods”). After selection of the transformants, we obtained three independent, homozygous, single-insertion transgenic lines (L5, L6, and L10) that we selected for further investigation on the basis of high NRT2.1 mRNA level in the roots.
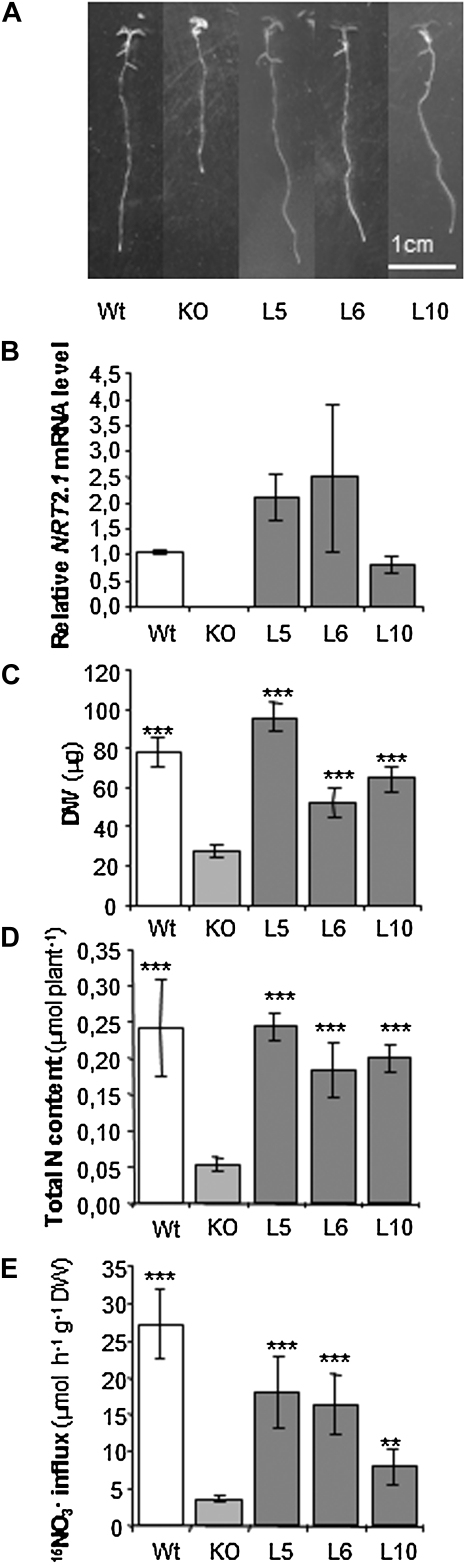
Because of its crucial role in the NO3− HATS (Cerezo et al., 2001; Filleur et al., 2001; Li et al., 2007), disruption of NRT2.1 results in a dramatic growth reduction when plants are cultivated on media with low NO3− (<1 mm) concentration (Orsel et al., 2004; Li et al., 2007). Indeed, when grown for 8 d in vertical petri agar plates containing 0.2 mm NO3− as a N source, atnrt2.1-2 plants show severely impaired primary root growth (Fig. 1A), and an average of 3.1- and 4.6-fold decrease in total dry biomass accumulation and total N content as compared to wild-type plants, respectively (Fig. 1, C and D). Root 15NO3− influx assays by short-term labeling at 0.2 mm external 15NO3− concentration confirmed that under these conditions, the mutant has lost up to 85% to 90% of the HATS activity as compared to the wild type (Fig. 1E). Therefore, we used these conditions to determine whether the 35S::NRT2.1 transgene was actually expressed in the transformants, and could functionally complement the atnrt2.1-2 mutant phenotype in restoring both root NO3− acquisition and growth at the level of wild-type plants. When assayed at day 8, the three transgenic lines (L5, L6, and L10) displayed a similar or higher level of NRT2.1 mRNA accumulation in the roots as compared to the wild type (Fig. 1B). The 35S::NRT2.1 construct was able to rescue, either totally (L5) or partially (L6 and L10), defects in primary root growth (Fig. 1A), dry biomass accumulation (Fig. 1C), and total N content of the seedlings (Fig. 1D). This shows that the transformants performed an efficient high-affinity NO3− uptake, resulting from the expression of a functional HATS under these conditions. Accordingly, high-affinity root 15NO3− influx in all three transformants lines was strongly enhanced as compared to the mutant, although it remained lower than that in the wild type (Fig. 1E).
Figure 1.
Characterization of Arabidopsis transgenic lines expressing NRT2.1 under the control of the 35S promoter. Wild type (WT), nrt2.1-2 knockout mutant (KO), and three transgenic lines (L5, L6, and L10) were grown in vitro during 8 d on 0.2 mm KNO3 as N source. A, Primary root growth after 8 d. B, Root NRT2.1 expression quantified by quantitative (Q)-PCR. Values are means of two biological replicates ± sd. C, Dry weight (DW) of 8-d-old plants. Values are means of six replicates ± sd. D, Total N contents quantified after 8 d of growth. Values are means of six replicates ± sd. E, Root NO3− influx measured at the external concentration of 0.2 mm 15NO3−. Values are means of 12 replicates ± sd. Differences between WT/transformants and the KO mutant are significant at *P < 0.05, **P < 0.01, ***P < 0.001 (Student’s t test).
Constitutive Expression of NRT2.1 Does Not Suppress Feedback Repression of the HATS by High N Supply
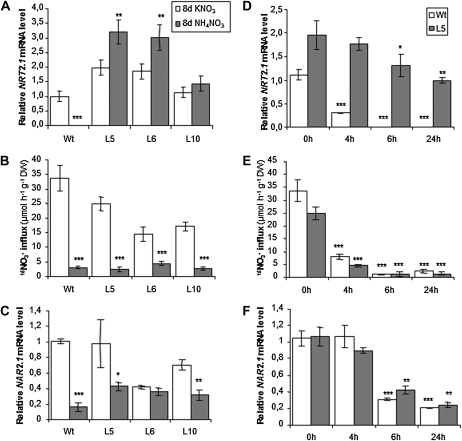
To investigate the role of the transcriptional regulation of NRT2.1 in the control of the HATS activity, we first explored the response of both root NRT2.1 mRNA accumulation and root 15NO3− influx in the transformants to the repressive action of high N provision in the medium. Furthermore, root NAR2.1 mRNA accumulation was also measured, to determine whether putative changes of the HATS activity in the transformants could be accounted for by regulation of NAR2.1 expression. Since supply of ammonium (NH4+) in addition to NO3− was previously proposed to trigger posttranscriptional regulation of the HATS in N. plumbaginifolia (Fraisier et al., 2000), our first series of experiments aimed at comparing L5, L6, and L10 transgenic lines with wild-type plants after 8 d of growth either with 0.2 mm KNO3 or 10 mm NH4NO3 as a N source. As shown in Figure 2A, NRT2.1 expression was almost totally suppressed in wild-type plants grown on 10 mm NH4NO3, while the NRT2.1 mRNA level in the roots of the 35S::NRT2.1 lines remained constitutively high under both N regimes. On 10 mm NH4NO3, due to the strong repression in wild-type plants, this level was 530-, 500-, and 240-fold higher in L5, L6, and L10 than in Columbia-0 (Col-0), respectively. Nevertheless, despite this tremendous difference in NRT2.1 mRNA accumulation, root 15NO3− influx was markedly inhibited on 10 mm NH4NO3 as compared with 0.2 mm KNO3 in all genotypes (Fig. 2B). NAR2.1 mRNA accumulation in roots was also down-regulated by high N supply (Fig. 2C), but to a much lower extent than root 15NO3− influx, especially in the transgenic plants. These data demonstrate that transcriptional regulation of NRT2.1 is not strictly required for feedback repression of HATS activity by high NH4NO3 provision, and that posttranscriptional mechanisms, possibly associated with altered NAR2.1 expression, are sufficient to inhibit high-affinity NO3− uptake under these conditions.
Figure 2.
NO3− influx, NRT2.1, and NAR2.1 mRNA level in roots of wild-type (WT) and transgenic plants in response to 10 mm NH4NO3. A to C, Plants were grown in vitro during 8 d on 0.2 mm KNO3 (white bars) or 10 mm NH4NO3 (gray bars) as N source. D to F, WT plants (white bars) and the transgenic line L5 (gray bars) were grown in vitro on 0.2 mm KNO3 and treated during 4, 6, and 24 h by the addition of 2 mL of a 250 mm NH4NO3 solution to reach a final concentration of 10 mm NH4NO3. A and D, Root NRT2.1 expression quantified by Q-PCR. Values are means of two biological replicates ± sd. B and E, Root NO3− influx measured at the external concentration of 0.2 mm 15NO3−. Values are means of 12 replicates ± sd. C and F, Root NAR2.1 expression quantified by Q-PCR. Values are means of two biological replicates ± sd. Differences between plants on KNO3 and NH4NO3 are significant at *P < 0.05, **P < 0.01, ***P < 0.001 (Student’s t test).
In the above experiments, we analyzed the HATS regulation in plants subjected to long-term (8 d) repressive treatment on 10 mm NH4NO3. This prevented unraveling any putative delay in the HATS repression that could have been caused by constitutive expression of NRT2.1. To rule out this hypothesis, we performed time-course experiments with L5 and wild-type plants supplied with 10 mm NH4NO3 for up to 24 h. As compared to control plants left on 0.2 mm KNO3, expression of NRT2.1 in the wild type was already repressed 3.6-fold as soon as 4 h following addition of 10 mm NH4NO3 in the medium, and decreased further to become barely detectable after 6 and 24 h (Fig. 2D). In L5 plants, the level of NRT2.1 mRNA remained very high after 10 mm NH4NO3 supply, although it was slightly reduced after 6 and 24 h of repressive conditions (Fig. 2D). Despite the high and relatively stable level of NRT2.1 mRNA in L5 plants, root 15NO3− influx was rapidly and similarly down-regulated in wild-type and L5 plants when compared to the controls on 0.2 mm KNO3 (Fig. 2E). Unlike NRT2.1 mRNA in wild-type plants, NAR2.1 transcript accumulation in both Col-0 and L5 plants started to decrease only after 6 h of NH4NO3 supply, and remained significant (approximately 20% of the controls) even after 24 h of treatment (Fig. 2F). These conclusions were confirmed with the L10 transgenic line (Supplemental Fig. S1). Thus, these data show that down-regulation of HATS activity by high NH4NO3 supply occurs as fast in the transgenic lines as in the wild type, and occurs before any visible change in NAR2.1 mRNA accumulation.
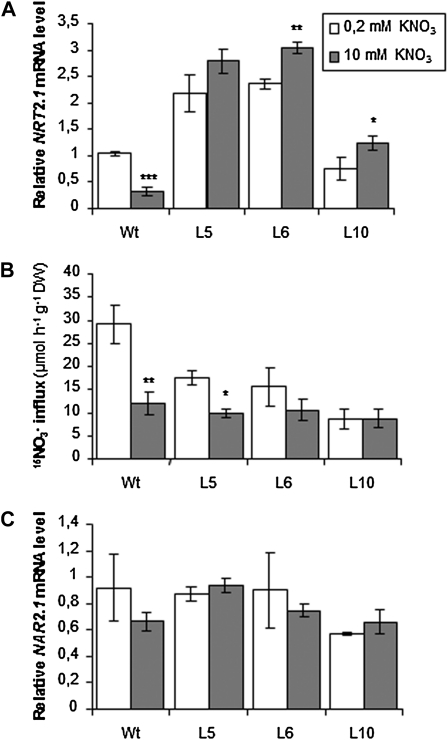
To determine if the repression of HATS activity in 35S::NRT2.1 transformants was specifically due to the presence of NH4+ in the external medium (as suggested by Fraisier et al., 2000), or was the consequence of high N provision, plants were grown for 8 d on either 0.2 or 10 mm KNO3 as the sole N source. As compared to 0.2 mm, supply of 10 mm KNO3 significantly repressed NRT2.1 expression in the roots of wild-type plants, although to a lesser extent than 10 mm NH4NO3 (compare Fig. 3A with 2A), but had little effect on NRT2.1 mRNA accumulation in transgenic lines (Fig. 3A). As a consequence, the transgenic lines overexpressed NRT2.1 mRNA 3- to 9-fold when compared to the wild type on 10 mm NO3−. Interestingly, high NO3− supply also markedly reduced HATS-mediated 15NO3− influx in wild-type plants (by 60%), but not so much in the transgenic lines where root 15NO3− influx was lowered at most by 30% to 40% as compared to the controls (Fig. 3B). Unlike with 10 mm NH4NO3, NAR2.1 mRNA levels were not significantly affected by 10 mm instead of 0.2 mm KNO3 supply, in any of the genotypes (Fig. 3C). Altogether, these data suggest that down-regulation of the HATS activity by high NO3− (10 mm) supply (1) cannot be suppressed, but can possibly be attenuated by constitutive expression of NRT2.1, and (2) cannot be explained by concurrent repression of NAR2.1 expression.
Figure 3.
NO3− influx, NRT2.1, and NAR2.1 mRNA level in roots of wild-type and transgenic plants in response to 10 mm KNO3. Plants were grown in vitro during 8 d on 0.2 mm KNO3 (white bars) or 10 mm KNO3 (gray bars). A, Root NRT2.1 expression quantified by Q-PCR. Values are means of two biological replicates ± sd. B, Root NO3− influx measured at the external concentration of 0.2 mm 15NO3−. Values are means of 12 replicates ± sd. C, Root NAR2.1 expression quantified by Q-PCR. Values are means of two biological replicates ± sd. Differences between plants grown on 0.2 mm KNO3 and 10 mm KNO3 are significant at *P < 0.05, **P < 0.01, ***P < 0.001 (Student’s t test).
Constitutive Expression of NRT2.1 Does Not Suppress Feedback Repression of the HATS by Darkness
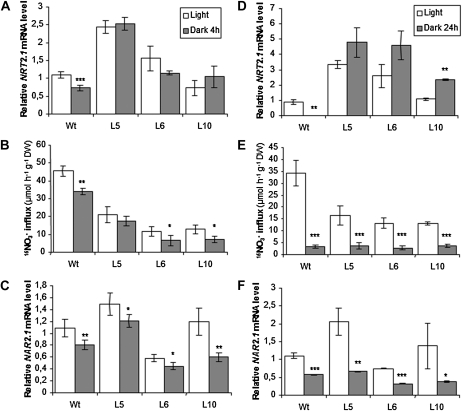
Sugar production through photosynthesis in the light is another main regulatory factor stimulating NRT2.1 expression and NO3− HATS activity (Lejay et al., 2003, 2008). As a consequence, NRT2.1 mRNA accumulation and HATS activity undergo marked diurnal rhythms, with a gradual repression following light/dark transition (Lejay et al., 1999). To determine the functional impact of constitutive NRT2.1 expression on the response of the HATS to photosynthesis, 8-d-old plants grown in vitro on a medium containing 0.2 mm KNO3 were transferred to the dark for 4 or 24 h following a period of 10 or 8 h in the light, respectively. In the wild type, as expected, NRT2.1 expression decreased by 34% and 98% after 4 and 24 h in darkness, respectively (Fig. 4, A and D). Accordingly, these treatments resulted in a 24% and 90% decrease in root 15NO3− influx, respectively (Fig. 4, B and E). In the transgenic lines, light/dark transition had no impact on the NRT2.1 mRNA levels, which remained high even after 4 or 24 h in the dark. After 4 h in darkness, root 15NO3− influx was only slightly reduced in the L6 and L10 lines, and not affected in the L5 line, as compared to the control plants kept in the light (Fig. 4B), whereas after 24 h in the dark, strong down-regulation of root 15NO3− influx was observed in all transgenic lines, in spite of the fact that they dramatically overaccumulated NRT2.1 mRNA as compared to the wild type. The response of NAR2.1 expression to dark treatments in both wild-type and transgenic lines mirrored that of NRT2.1 in the wild type (Fig. 4, C and F), however with a much reduced amplitude since NAR2.1 mRNA level was only decreased by approximately 50% to 70% after 24 h of darkness (against 98% for NRT2.1 mRNA in wild-type plants). Thus, as it was the case for repression by high N supply, repression of the HATS activity by darkness (1) cannot be suppressed by constitutive expression of NRT2.1, and (2) is much stronger than down-regulation of NAR2.1 expression.
Figure 4.
NO3− influx, NRT2.1, and NAR2.1 mRNA level in roots of wild-type and transgenic plants in response to darkness. A to C, Plants were grown in vitro for 8 d on 0.2 mm KNO3 as N source and harvested after 14 h of light (white bars) or 10 h of light + 4 h of darkness (dark bars). D to F, Plants were grown in vitro for 7 d on 0.2 mm KNO3 as N source and transferred, after 8 h of light, either in continuous dark (dark bars) or in continuous light (white bars) for 24 h. A and D, Root NRT2.1 expression quantified by Q-PCR. Values are means of two biological replicates ± sd. B and E, Root NO3− influx measured at the external concentration of 0.2 mm 15NO3−. Values are means of 12 replicates ± sd. C and F, Root NAR2.1 expression quantified by Q-PCR. Values are means of two biological replicates ± sd. Differences between plants treated in the light and in the dark are significant at *P < 0.05, **P < 0.01, ***P < 0.001 (Student’s t test).
Regulation of NRT2.1 and NAR2.1 Protein Abundance Does Not Always Explain Repression of HATS by High N Supply or Darkness
According to the above results, changes in NAR2.1 expression only poorly correlate with those of the HATS activity. However, changes in NAR2.1 mRNA may not reflect those of NAR2.1 protein. Therefore, a specific polyclonal anti-NAR2.1 antibody was raised in rabbit, against a peptidic sequence within the N terminus of the protein, to further investigate if changes in NAR2.1 protein level could explain the regulation of the HATS activity. The affinity-purified anti-NAR2.1 antibody was tested on western blots with total microsomal membranes purified from roots of hydroponically grown wild-type plants or nrt2.1-2 and nar2.1-1 mutants (Supplemental Fig. S2). In the wild-type and nrt2.1-2 mutant, the anti-NAR2.1 antibody revealed one band at approximately 25 kD that corresponds to the theoretical Mr of NAR2.1 at 23.4 kD. This band was specific for NAR2.1 since it was absent in the microsomal membranes from nar2.1-1 mutant. It is interesting to note that the level of NAR2.1 protein was much lower in the nrt2.1-2 mutant compared to the wild type, suggesting that the lack of NRT2.1 has an impact on NAR2.1 accumulation. This link between the level of NRT2.1 and NAR2.1 proteins has already been observed in NAR2.1 KO mutants, in which the lack of NAR2.1 prevents accumulation of the NRT2.1 protein at the plasma membrane (Wirth et al., 2007; Yong et al., 2010).
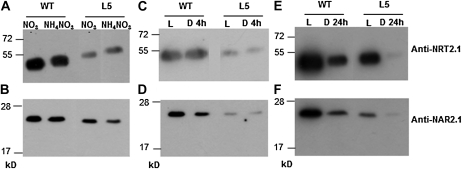
In previous studies the level of NRT2.1 protein has been shown to be very stable compared to NRT2.1 mRNA level and HATS activity in response to repressive environmental conditions such as high N or darkness (Girin et al., 2007; Wirth et al., 2007). NRT2.1 protein level was only decreased after long-term treatments with 10 mm NH4NO3. To determine if the level of NAR2.1 protein could explain the repression of HATS activity in conditions where NRT2.1 level is stable, NAR2.1 and NRT2.1 protein levels were measured in response to 4 h of 10 mm NH4NO3 provision, and to 4 or 24 h of darkness in the wild-type and in L5 plants (Fig. 5). Western blots, using the anti-NAR2.1 antibody described above and the antibody against NRT2.1 previously described by Wirth et al. (2007), revealed two different situations for both NRT2.1 and NAR2.1. In response to treatments of 4 h with NH4NO3 or 4 h of darkness the level of NRT2.1 and NAR2.1 proteins were not affected in both wild-type and L5 roots (Fig. 5, A–D), whereas after 24 h of darkness the level of NAR2.1 protein was reduced along with the level of NRT2.1 protein (Fig. 5, E and F). This last result was surprising for NRT2.1 since Wirth et al. (2007) found that the accumulation of NRT2.1 was not affected after 24 h of darkness in wild-type plants. However, the growth conditions strongly differed between our present study (8 d of growth in vitro) and that of Wirth et al. (2007; 6 weeks of growth in hydroponics). When we performed western blots on wild-type plants grown for 6 weeks in hydroponics, like in Wirth et al. (2007), we also found that in the wild type the levels of both NRT2.1 and NAR2.1 proteins were stable after 20 h of darkness (Supplemental Fig. S3). It suggests that, when plants were grown in vitro during 8 d, the levels of NRT2.1 and NAR2.1 proteins respond much faster to darkness than when plants were older and grown in hydroponics. Furthermore, as observed in the nrt2.1-2 mutant (Supplemental Fig. S2), the level of NAR2.1 was always lower in L5 plants compared to the wild type along with the level of NRT2.1 protein. It confirms that the 35S::NRT2.1 construction does not fully complement the nrt2.1-2 mutant and further illustrates the apparent strong link between the level of NRT2.1 and NAR2.1 proteins.
Figure 5.
Immunoblot for NRT2.1 (A, C, and E) and NAR2.1 (B, D, and F) using microsomes from roots of wild type (WT) and the transgenic line L5. A and B, Plants were grown in vitro during 8 d on 0.2 mm KNO3 and treated during 4 h by the addition of 2 mL of a 250 mm NH4NO3 solution to reach a final concentration of 10 mm NH4NO3. C and D, Plants were grown in vitro for 8 d on 0.2 mm KNO3 as N source and harvested after 14 h of light or 10 h of light + 4 h of darkness. E and F, Plants were grown in vitro for 7 d on 0.2 mm KNO3 as N source and transferred, after 8 h of light, either in continuous dark or in continuous light for 24 h. Samples were separated on a 11% SDS-PAGE gel (12 μg protein/lane).
Altogether the results show that, for short treatments of 4 h of 10 mm NH4NO3 or darkness, the regulation of NRT2.1 and NAR2.1 protein levels cannot explain the repression of HATS activity. However, for long treatments with high N or darkness, posttranscriptional mechanisms are involved to decrease the synthesis or increase the degradation of NRT2.1, and possibly NAR2.1, proteins, and thus participate in the repression of HATS activity.
The Quantitative Importance of the Transcriptional Control of NRT2.1 and Other High-Affinity NO3− Transporters in the HATS Regulation
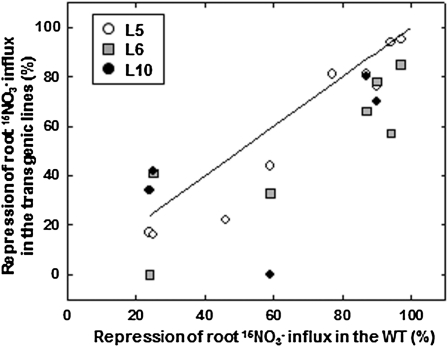
Altogether, the data presented above clearly indicate that constitutive expression of NRT2.1 does not eliminate the NO3− HATS repression by either high N supply or darkness. However, this may not mean that changes in NRT2.1 transcription play no role at all in this regulation. Indeed, in some instances (see Figs. 3 and 4), it appeared that down-regulation of the HATS activity was somehow less pronounced in the transgenic plants as compared to the wild type. To precisely quantify this apparent difference in a more general way, we plotted together all individual values obtained for repression of root 15NO3− influx in the various genotypes and in response to the various treatments (Fig. 6). This unraveled that in most cases, down-regulation of HATS activity by repressive treatments was attenuated in transgenic plants as compared to the wild type. Thus, although certainly not decisive, transcriptional regulation of NRT2.1 does seem to contribute to the response of the HATS to the N and light signals investigated here.
Figure 6.
Effect of constitutive expression of NRT2.1 on the down-regulation of the HATS by repressive N or dark treatments. For all individual experiments performed (those depicted in Figs. 2–4 plus replicate experiments not shown in Figs. 2–4), the percentage of repression of root 15NO3− influx in response to repressive N or dark treatments was calculated for each genotypes as: % repression = (influx in control conditions − influx in repressive conditions)/influx in control conditions × 100. The line indicates the identity function. Thus, all data points below the line correspond to assays where repression of the NO3− HATS was lower in the 35S::NRT2.1 plants as compared to the wild type. All data points are mean of six replicates.
To rule out the possibility that a lower repression of the HATS in the transgenic lines as compared to the wild type could be due to a specific up-regulation in these lines of the other transporters participating in the HATS, root mRNA levels of NRT2.2 and NRT1.1 were measured in response to the various treatments in wild-type and L5 plants (Supplemental Fig. S4). As expected NRT2.2 mRNA was not recorded in L5 plants that have a nrt2.1-2 mutant background, described as a double mutant for both NRT2.1 and NRT2.2 (Li et al., 2007). In no instance NRT1.1 was found to be overexpressed in L5 plants as compared to wild-type plants, and its regulation was similar in both genotypes.
DISCUSSION
Transcriptional versus Posttranscriptional Regulation of NRT2.1 in the HATS Control
As outlined in numerous studies (for review, see Forde, 2002; Glass et al., 2002; Tsay et al., 2007; Gojon et al., 2009), the regulation of the NO3− HATS is of major importance for adaptation of the plants to fluctuating environmental conditions. In particular, its modulation allows a tight coordination between N acquisition and growth of the plant in response to external or internal factors, such as changes in NO3− availability in the soil or changes in photosynthesis in the shoot. It has been postulated for long that the HATS regulation is predominantly ensured by transcriptional control of NRT2.1 expression (Lejay et al., 1999; Zhuo et al., 1999; Cerezo et al., 2001; Gansel et al., 2001; Glass et al., 2002; Nazoa et al., 2003; Girin et al., 2007). However, recent studies at the NRT2 protein level have challenged this conclusion by evidencing a lack of correlation between NRT2 protein abundance on the one hand, and NRT2 gene expression and HATS activity, on the other hand (Wirth et al., 2007; Ishikawa et al., 2009). These data support the previous proposal that posttranscriptional regulatory mechanisms participate in the control of the HATS, because expression of a 35S::NpNRT2.1 or RolD::NpNRT2.1 transgene in N. plumbaginifolia did not prevent down-regulation of the HATS activity by high NH4NO3 supply (Fraisier et al., 2000). However, this work in N. plumbaginifolia could not be fully conclusive because constitutive expression of NpNRT2.1 using 35S or RolD promoters was achieved in a wild-type background, making it not possible to assess the actual functionality of the transgene product, or to rule out any putative compensation by the endogenous gene.
To unambiguously determine if posttranscriptional regulation of NRT2.1 can impact the HATS activity in Arabidopsis, we generated transgenic plants expressing only a constitutive version of the NRT2.1 gene by complementing the atnrt2.1-2 mutant, defective for both NRT2.1 and NRT2.2, with a 35S::NRT2.1 transgene. Previous work showed that the constitutive expression of NRT2.1 fused to GFP had no impact on the localization of the protein in plasma membrane of epidermis and cortex root cells compared to plants expressing NRT2.1::GFP under the control of its own promoter (Chopin et al., 2007; Wirth et al., 2007). We succeeded in isolating three independent transgenic lines (L5, L6, and L10) displaying a deregulated expression of NRT2.1. In these lines, the level of NRT2.1 mRNA in the roots was similar or higher than that in the wild type under control conditions (Figs. 1–4), and remained little affected or was even slightly increased by repressive treatments that strongly diminish NRT2.1 expression in wild-type plants (Figs. 2–4). This shows that NRT2.1 mRNA accumulation in the roots is predominantly controlled by NRT2.1 transcription. Furthermore, all three lines displayed full, or almost full, complementation of the mutant defects in growth and total N accumulation (Fig. 1), demonstrating functionality of the transgene. However, although constitutive expression of NRT2.1 led to a strong stimulation of the HATS activity as compared to the atnrt2.1-2 mutant, it never succeeded in restoring the root NO3− influx measured at day 8 in wild-type plants under control conditions (Figs. 1–4). Using NRT2.1 antibody we were able to show that this lack of full complementation in L5 plants was associated with a lower level of NRT2.1 proteins compared to the wild type (Fig. 5). The reasons why rescue of the HATS activity and of NRT2.1 protein level was only partial in 35S::NRT2.1 lines are not known, but it has been recently reported that complementation of a atnar2.1 mutant by a 35S::NAR2.1-myc construct restored only 60% to 70% of the high-affinity NO3− uptake defect of the mutant (Yong et al., 2010). To maximize the putative effect of deregulated NRT2.1 expression, we measured the HATS activity in response to high N or dark treatments that are known to dramatically repress both NO3− HATS activity and NRT2.1 mRNA level in wild-type plants (Lejay et al., 1999; Zhuo et al., 1999; Cerezo et al., 2001; Girin et al., 2007). To make sure that the response of the HATS to these treatments is predominantly due to the NRT2.1 transport system, we measured, in the same experiments, the expression, in L5 plants, of NRT2.2 and NRT1.1, two other transporters involved in high-affinity NO3− uptake (Supplemental Fig. S4). The general outcome of these series of experiments is that high-affinity root 15NO3− influx in the three transgenic lines was in most cases strongly down-regulated in response to the repressive treatments (Figs. 1–4).
This provides a clear demonstration that transcriptional regulation of NRT2.1 expression is not the major mechanism for control of the HATS in our conditions. However, down-regulation of the HATS appeared in many instances less pronounced in the transgenic lines than in the wild type. For instance, when plants were grown on a media containing 10 mm NO3− as the sole N source, the NO3− HATS activity was strongly lowered in the wild type as compared to control conditions (0.2 mm NO3−), but not in the 35S::NRT2.1 plants (Fig. 3B), while in response to 10 mm NH4NO3 repression of NO3− HATS was strong in both the wild-type and the transgenic lines. These results are in accordance with those of Fraisier et al. (2000), showing that the HATS activity was less repressed by 10 mm NO3− supply (as compared to 1 mm NO3−) in the RolD::NpNRT2.1 or 35S::NpNRT2.1 transformants than in the wild type while after ammonium addition NO3− influx was markedly decreased in both the wild-type and transgenic plants. One possible explanation to the discrepancy between the effects of NO3− alone and of NH4NO3 on HATS activity, is the membrane depolarization resulting from NH4+ supply, which might have impaired the energization of NO3− uptake by root cells, despite constitutive NRT2.1 expression. However, several lines of evidence make this hypothesis very unlikely. First, membrane depolarization due to NH4+ supply is rapid (within minutes) and transient, with the membrane potential gradient spontaneously recovering to the original or to an intermediate value within 30 min (Ullrich et al., 1984; Wang et al., 1994). A more recent study on tomato (Solanum lycopersicum) roots showed that growth in presence of NH4+ even led to a hyperpolarization of the plasma membrane (Nieves-Cordones et al., 2008). Thus, membrane depolarization in response to NH4+ supply cannot explain long-term repression of NO3− HATS activity observed after several hours or 8 d on 10 mm NH4NO3 (Fig. 2, B and E). Furthermore, repression of NO3− influx by NH4+ is known to be a very specific effect that can hardly be explained by a general mechanism such as decreased energization of secondary transport systems. Indeed, if NH4+-induced membrane depolarization was responsible for NO3− influx repression, the uptake of the other proton cotransported anions should also be affected. However, this is clearly not the case, and several studies showed that a range of NH4+ concentration, which caused a marked decrease in the rate of NO3− influx, either produced no consistent pattern of effect on phosphate influx or even increased phosphate and sulfate uptake (Cox and Reisenauer, 1973; Lee and Drew, 1989). Similarly, the provision of K+, which depolarizes the plasma membrane to an extent similar to that of NH4+ does not have a negative effect on root NO3− uptake (Lee and Drew, 1989; Glass and Siddiqi, 1995; Wang et al., 1996). Finally, Krouk et al. (2006) showed that NO3− HATS activity is up-regulated (along with NRT2.1 expression) when NH4+ concentration in the external medium largely exceeds that of NO3−. Altogether, these data confirm that membrane depolarization is not the main mechanism by which NH4+ down-regulates root NO3− uptake and cannot explain why 35S::NRT2.1 plants did respond the same way to treatments with 10 mm NH4NO3 compared to the wild type.
Considering the whole set of data from all experiments (Figs. 2–4; Supplemental Fig. S1; and replicate experiments not shown in Figs. 2–4) confirmed that constitutive expression of NRT2.1 often resulted in a lower level of repression of the HATS activity as compared to the wild type (Fig. 6). This shows that, although not predominant under most situations, the transcriptional regulation of NRT2.1 has a functional impact on the control of the HATS, and may account for a small but significant part of the changes in high-affinity root NO3− uptake recorded in the wild type. This conclusion is consistent with the reports indicating that up-regulation of NRT2.1 transcript accumulation in mutants or transformants altered in N signaling or metabolism actually leads to a less-than-proportional increase in NO3− uptake by the roots (Muños et al., 2004; Good et al., 2007; Hong et al., 2009; Girin et al., 2010; Widiez et al., 2011).
Putative Posttranscriptional Mechanisms Involved in the HATS Control
In Arabidopsis, constitutive expression using a 35S promoter has already been used to investigate posttranscriptional regulation of several nutrient transporters, such as AMT1.1 (Yuan et al., 2007), IRT1 (Connolly et al., 2002), BOR1 (Takano et al., 2005), SULTR1.1, and SULTR 1.2 (Yoshimoto et al., 2007). The general conclusion from these studies is that, despite a strong regulation at the transcriptional level for all these transporters, various levels of posttranscriptional regulations are also involved in the modulation of their activity. The root-specific and iron-deficiency-inducible expression of Arabidopsis IRT1, the major transporter for high-affinity iron uptake, is controlled at both transcription and protein accumulation levels (Connolly et al., 2002). For sulfate acquisition, SULTR1.1 and SULTR1.2, two essential components of the high-affinity sulfate uptake system, are controlled both transcriptionally and posttranscriptionally at the level of protein accumulation in response to changes in environmental sulfur conditions (Yoshimoto et al., 2007). Posttranscriptional regulation of the AMT1 NH4+ transporters includes both regulation of transcript stability (Yuan et al., 2007) and posttranslational control through phosphorylation at the C terminus of the protein (Loqué et al., 2007; Neuhäuser et al., 2007; Lanquar et al., 2009). Finally it was also shown that constitutively expressed GFP-tagged BOR1 transporter proteins in Arabidopsis roots were degraded upon resupply of boron to plants by a mechanism that involved endocytosis from the plasma membrane and subsequent degradation of the transporter protein (Takano et al., 2005). From these studies it appears that posttranscriptional regulation of nutrient transporters is probably a general phenomenon in plant nutrient response, but that the mechanisms involved may be very diverse, depending on the protein.
Concerning NRT2.1, posttranscriptional regulation does not seem to involve mechanisms that strongly modulate the transcript stability. In all the experiments we performed, NRT2.1 transcript level in the transgenic lines was not markedly affected by the treatments, indicating that the 35S promoter was sufficient on its own to yield almost constitutive NRT2.1 mRNA accumulation. At the protein level, we previously showed that the abundance of the NRT2.1 protein in the root plasma membrane of wild-type plants is rather stable and is only affected in response to long-term treatments with high N (Girin et al., 2007; Wirth et al., 2007). Western blots performed in this study confirmed that NRT2.1 abundance is not affected by short-term N or dark treatments in roots of both the wild-type and L5 plants (Fig. 5, A and C). However, after 24 h of darkness, NRT2.1 protein level was significantly decreased in the wild-type and L5 plants when grown in vitro for 8 d, compared to plants kept in the light (Fig. 5E). Since this occurred in the absence of any decrease of NRT2.1 mRNA level in L5 plants (Fig. 4A), this shows that NRT2.1 is subject to a posttranscriptional regulatory mechanism that lowers its abundance in the membrane in response to long-term repressive treatments. The effects of this mechanism are apparently dependent on the experimental conditions. According to Wirth et al. (2007), the decrease of NRT2.1 protein level following an extended dark period was not observed when plants where grown in hydroponics for 6 weeks (see also Supplemental Fig. S3A). It suggests that NRT2.1 protein is more stable in older plants or that sugar depletion is more rapid in young plants due to a lower level of sugar stores than in older plants. Interestingly, similar data were obtained with NAR2.1 (Fig. 5F; Supplemental Fig. S3B).
A major difference between NRT2.1 and most other nutrient carriers is that NRT2.1 protein expression and transport activity requires a functional NAR2.1 protein (Orsel et al., 2006; Wirth et al., 2007), which interacts with NRT2.1 to generate a heterooligomer that may be the active form of the transporter (Yong et al., 2010). Several reports have shown that NAR2.1 expression is regulated as NRT2.1 expression, although with a much-less-pronounced amplitude in the transcript-level changes (Krouk et al., 2006; Okamoto et al., 2006; Orsel et al., 2006). Furthermore, at the protein level nothing is known concerning the regulation of NAR2.1 and its possible role in the regulation of HATS activity. Thus, constitutive expression of NRT2.1 alone may not be sufficient to prevent down-regulation of the HATS if NAR2.1 is still repressed at the mRNA or protein level. Therefore, we investigated the regulation of NAR2.1 mRNA accumulation and we designed a specific antibody for NAR2.1 to follow its regulation at the protein level. Our data show that, despite a decrease in NAR2.1 transcript level in response to the most repressive treatments (10 mm NH4NO3 or 24 h of darkness), these changes were very moderate as compared to the strong down-regulation of root 15NO3− influx in the 35S::NRT2.1 transformants (Figs. 2 and 4). Furthermore, in several instances down-regulation of the HATS could be observed in both wild-type and transgenic plants in the absence of any decay of NAR2.1 transcript level (Figs. 2 and 3). Similarly, the results obtained at the protein level do not evidence a specific response of NAR2.1 that may explain on its own the regulation of the HATS activity. Like NRT2.1, NAR2.1 protein accumulation was lowered in both the wild-type and L5 plants by long-term dark treatment (Fig. 5F) but not by short-term N or dark treatments (Fig. 5, B and D). Interestingly, this is illustrative of a more general correlation between the levels of NRT2.1 and NAR2.1 proteins. Indeed, in the nrt2.1-2 mutant lacking NRT2.1, NAR2.1 protein is present at a much lower level as compared to the wild type (Supplemental Fig. S2), although NAR2.1 mRNA accumulation is not affected (Orsel et al., 2006; data not shown). Conversely, NRT2.1 protein could not be detected in nar2.1 mutants (Wirth et al., 2007; Yong et al., 2010), despite presence of the transcript (Okamoto et al., 2006; data not shown). Finally, the only partial restoration of NRT2.1 protein expression in L5 plants was associated with a lowered level of NAR2.1 in the membranes as compared to wild-type plants (Fig. 5), although NAR2.1 mRNA accumulation was similar in both genotypes (Figs. 2–4). These data strongly suggest a tight coregulation of NRT2.1 and NAR2.1 expression at the posttranscriptional level.
Altogether, the above results indicate that the regulation of NO3− HATS activity, in response to high N and darkness, is a complex mix between three levels of regulation. Both transcriptional regulation of NRT2.1 and NAR2.1 and posttranscriptional control of protein abundance may explain long-term regulation (after one or several days) of the root NO3− HATS. However, the results also collectively show that such mechanisms cannot explain why root 15NO3− influx remains normally repressed in the 35S::NRT2.1 lines in response to short-term N or dark treatments that do not modify NRT2.1 and NAR2.1 protein levels. This suggests that posttranslational modifications of NRT2.1 and/or NAR2.1 proteins also probably play an important role in the control of the HATS activity. Given the complexity of such a two-component system, many hypotheses can be envisaged, such as association/dissociation of the NRT2.1/NAR2.1 heterooligomer (Yong et al., 2010), partial proteolysis of NRT2.1 (Wirth et al., 2007), or phosphorylation events in NRT2.1 and/or NAR2.1 (Forde, 2000). Furthermore, despite that it is now quite well documented that NRT2.1 is part of a high-Mr complex in the plasma membrane (Wirth et al., 2007; Yong et al., 2010), no evidence is available yet that this complex comprises only NRT2.1 and NAR2.1, leaving the possibility that other unknown regulatory protein(s) may be involved. Whatever the posttranslational mechanisms responsible for down-regulation of the HATS in our experiments, it is quite clear that they are activated by a variety of treatments (high N supply with or without NH4+, transfer to the dark), making them likely to be of general occurrence.
MATERIALS AND METHODS
Plant Material and Culture Conditions
Arabidopsis (Arabidopsis thaliana) genotypes used in this study were the wild-type Col-0 ecotype, the atnrt2.1-2 mutant, obtained from the Salk Institute (Salk_035429; Little et al., 2005), and the nar2.1-1 mutant (Orsel et al., 2006).
For all experiments except those presented in Supplemental Figures S2 and S3, plants were grown in sterile conditions in vertical agar plates (12 × 12 cm) on solid medium (0.8% [w/v] agar type A; Sigma, product A4550) containing 0.5 mm CaSO4, 0.5 mm MgCl2, 1 mm KH2PO4, 2.5 mm MES, 50 μm NaFe EDTA, 50 μm H3BO3, 12 μm MnCl2, 1 μm CuCl2, 1 μm ZnCl2, and 0.03 μm NH4Mo. The pH was adjusted to 5.7 with KOH. This medium was supplemented with either KNO3 or NH4NO3 as described in the text. After storing for 2 d at 4°C in the dark, plates were transferred in a growth chamber with 16/8 h day/night cycle at 21°C/18°C and 70% relative humidity. Light intensity during the light period was 125 μmol photons m−2 s−1. All the plants were harvested after 8 d of growth. For NH4NO3, NO3−and long dark treatments the light period in the growth chamber started at 6 am. For short dark treatments the light period in the growth chamber started at midnight.
For Supplemental Figures S2 and S3, plants were grown hydroponically using the experimental set up describes previously (Lejay et al., 1999).
Generation of Transformant Lines
All constructs were made using Gateway cloning technology (Invitrogen) according to the manufacturer’s instructions. The NRT2.1 DNA sequence from the bac clone T6D22 (Arabidopsis Biological Resource Center) was amplified using primers pair: GATE NRT2.1 L, 5′-CACCATGGGTGATTCTACTGGT-3′; GATE NRT2.1 R, 5′-TCAAACATTGTTGGGTGTGTT-3′; and was cloned into the pENTR/d-Topo vector (Invitrogen). One entry clone was fully sequenced before subsequent cloning in the binary Gateway destination vector pGWB2 obtained from Tsuyoshi Nakagawa (Research Institute of Molecular Genetics, Shimane University, Matsue, Japan) by using a Gateway LR clonase enzyme mix (Invitrogen). The pGWB2 vector allows expression of the cloning sequence under the control of the cauliflower mosaic virus 35S promoter. The binary construct was introduced into the Agrobacterium tumefaciens strain GV3101, and the resulting bacterial culture was used to transform the atnrt2.1-2 mutant line by the standard flower-dip method (Clough and Bent, 1998). Transformants (T1) were selected on Murashige and Skoog-2 medium containing hygromycin (30 μg mL−1). Homozygous lines (T3) were obtained from resistance segregation assays. Integrity of the transgene was checked in transgenic lines by PCR analysis using specific primers.
RNA Extraction and Gene Expression Analysis
Root samples were frozen in liquid N2 in 2-mL tubes containing one steel bead (2.5-mm diameter). Tissues were disrupted for 1 min at 30 s−1 in a Retsch mixer mill MM301 homogenizer (Retsch). Total RNA was extracted from tissues using TRIzol reagent (Invitrogen). Subsequently, 40 μg of RNA were treated with DNase (RNase free DNase kit, Qiagen) and purified (RNeasy MinEluteTM cleanup kit, Qiagen) following the manufacturer’s instructions. Reverse transcription was achieved with 4 μg of RNAs in the presence of Moloney murine leukemia virus reverse transcriptase (Promega) after annealing with an anchored oligo(dT)18 primer as described by Wirth et al. (2007). The quality of the cDNA was verified by PCR using specific primers spanning an intron in the gene APTR (At1g27450) forward 5′-CGCTTCTTCTCGACACTGAG-3′; reverse 5′-CAGGTAGCTTCTTGGGCTTC-3′.
Gene expression was determined by quantitative real-time PCR (LightCycler; Roche Diagnostics) with the kit LightCycler FastStart DNA master SYBR green I (Roche Diagnostics) according to the manufacturer’s instructions with 1 μL of cDNA in a total volume of 10 μL. The conditions of amplifications were performed as described previously by Wirth et al. (2007). All the results presented were standardized using the housekeeping gene Clathrin (At4g24550). Gene-specific primer sequences were: NRT2.1 forward, 5′-AACAAGGGCTAACGTGGATG-3′; NRT2.1 reverse, 5′-CTGCTTCTCCTGCTCATTCC-3′; NAR2.1 forward, 5′-GGCCATGAAGTTGCCTATG-3′; NAR2.1 reverse, 5′-TCTTGGCCTTCCTCTTCTCA-3′; NRT2.2 forward, 5′-CAGGTGGAAACAGAGCTGCCATGG-3′; NRT2.2 reverse, 5′-GGACCATAGATACAACGGCAGTGACGAG-3′; NRT1.1 forward, 5′-GCACATTGGCATTAGGCTTT-3′; NRT1.1 reverse, 5′-CTCAATCCCCACCTCAGCTA-3′; Clathrin forward, 5′-AGCATACACTGCGTGCAAAG-3′; Clathrin reverse, 5′-TCGCCTGTGTCACATATCTC-3′.
NO3− Influx Studies
NO3− influxes were determined by 15N labeling as described by Remans et al. (2006). Liquid media for influx studies contained basic N-free medium supplemented with 0.2 mm K15NO3 (atom % 15N: 99%). Briefly, four plants were transferred to a 5-cm-diameter petri dish containing 0.1 mm CaSO4, with the roots in the solution and the aerial parts outside. This solution was replaced after 1 min with the 0.2 mm 15NO3− solution for 5 min. Plants were then rinsed again for 1 min in 0.1 mm CaSO4 before being harvested and dried at 70°C for 48 h. After determination of their dry weight, the samples were analyzed for total N and atom % 15N using a continuous-flow isotope ratio mass spectrometer (IsoPrime mass spectrometer; GV instruments) coupled to a carbon/N elemental analyzer (EuroVector S.p.A.) as described in Clarkson et al. (1996). Each influx value is the mean of six to 12 replicates.
NAR2.1 and NRT2.1 Immunodetection and Membrane Purification
For plants grown in vitro, microsomes were purified as followed. All procedures were carried out at 4°C. Harvested roots were homogenized with a roller grinder (C. Fauvel, Institut National de la Recherche Agronomique) and 0.25 g/mL of homogenization buffer (50 mm Tris, 500 mm Suc, 10% glycerol, 20 mm EDTA, 20 mm EGTA, 50 mm NaF, 5 mm β-glycerophosphate, 1 mm phenantroline, 0.6% polyvinylpyrrolidone, 10 mm ascorbic acid adjusted to pH 8 with MES 1 m, 1 μm leupeptine, 5 mm dithiothreitol, 1 mm Na2 vanadate, 1 mm phenylmethylsulfonyl fluoride). The homogenate was centrifuged at 2,000 rpm (Eppendorf 5810 R) for 2 min to remove the debris. The supernatant was then centrifuged at 9,000gmax for 12 min and the resulting supernatant was centrifuged again at 50,000gmax to recover the microsomal fraction. To obtain microsomes the pellet was resuspended in a minimal volume of conservation buffer (10 mm Tris, 10 mm borate, 300 mm Suc, 9 mm KCl, 5 mm EDTA, 5 mm EGTA, 50 mm NaF pH 8.3, 4.2 μm leupeptine, 1 mm phenylmethylsulfonyl fluoride, 5 mm dithiothreitol) and frozen at −80°C. For plants grown in hydroponic, microsomes were purified as described by Giannini et al. (1987).
For western blots proteins were separated on denaturing SDS-PAGE followed by an electrotransfer at 4°C onto a polyvinylidene difluoride membrane (0.2 μm, Immobilon, Millipore) according to manufacturer instructions. NAR2.1 was detected using an anti-NAR2.1 antiserum produced by Eurogentec against the synthetic peptide DVTTKPSREGPGVVL. The polyclonal antiserum was affinity purified by Eurogentec. NRT2.1 was detected using the antibody NRT2.1 20 described by Wirth et al. (2007). The immunodetection for both NRT2.1 and NAR2.1 was performed with a chemiluminescent detection system kit (SuperSignal, Pierce).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. NO3− influx, NRT2.1, and NAR2.1 mRNA level in roots of wild type and the L10 transgenic plants in response to 10 mm NH4NO3.
Supplemental Figure S2. Immunoblot for NAR2.1 using microsomes from roots of wild-type and knockout mutants for NRT2.1 and NAR2.1.
Supplemental Figure S3. Immunoblot for NRT2.1 and NAR2.1 using microsomes from roots of wild-type plants grown hydroponically.
Supplemental Figure S4. NRT2.2 and NRT1.1 mRNA level in roots of wild-type (WT) and the transgenic line L5.
Supplementary Material
References
- Castaings L, Camargo A, Pocholle D, Gaudon V, Texier Y, Boutet-Mercey S, Taconnat L, Renou JP, Daniel-Vedele F, Fernandez E, et al. (2009) The nodule inception-like protein 7 modulates nitrate sensing and metabolism in Arabidopsis. Plant J 57: 426–435 [DOI] [PubMed] [Google Scholar]
- Cerezo M, Tillard P, Filleur S, Muños S, Daniel-Vedele F, Gojon A. (2001) Major alterations of the regulation of root NO(3)(-) uptake are associated with the mutation of Nrt2.1 and Nrt2.2 genes in Arabidopsis. Plant Physiol 127: 262–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopin F, Wirth J, Dorbe MF, Lejay L, Krapp A, Gojon A, Daniel-Vedele F. (2007) The Arabidopsis nitrate transporter AtNRT2.1 is targeted to the root plasma membrane. Plant Physiol Biochem 45: 630–635 [DOI] [PubMed] [Google Scholar]
- Clarkson DT, Gojon A, Saker LR, Wiersema PK, Purves JV, Tillard P, Arnold GM, Paans AJM, Vaalburg W, Stulen I. (1996) Nitrate and ammonium influxes in soybean (Glycine max) roots: direct comparison of 13N and 15N tracing. Plant Cell Environ 19: 859–868 [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Connolly EL, Fett JP, Guerinot ML. (2002) Expression of the IRT1 metal transporter is controlled by metals at the levels of transcript and protein accumulation. Plant Cell 14: 1347–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox WJ, Reisenauer HM. (1973) Growth and ion uptake by wheat supplied nitrogen as nitrate, or ammonium, or both. Plant Soil 38: 363–380 [Google Scholar]
- Crawford NM, Glass ADM. (1998) Molecular and physiological aspects of nitrate uptake in plants. Trends Plant Sci 3: 389–395 [Google Scholar]
- Filleur S, Daniel-Vedele F. (1999) Expression analysis of a high-affinity nitrate transporter isolated from Arabidopsis thaliana by differential display. Planta 207: 461–469 [DOI] [PubMed] [Google Scholar]
- Filleur S, Dorbe MF, Cerezo M, Orsel M, Granier F, Gojon A, Daniel-Vedele F. (2001) An Arabidopsis T-DNA mutant affected in Nrt2 genes is impaired in nitrate uptake. FEBS Lett 489: 220–224 [DOI] [PubMed] [Google Scholar]
- Forde BG. (2000) Nitrate transporters in plants: structure, function and regulation. Biochim Biophys Acta 1465: 219–235 [DOI] [PubMed] [Google Scholar]
- Forde BG. (2002) Local and long-range signaling pathways regulating plant responses to nitrate. Annu Rev Plant Biol 53: 203–224 [DOI] [PubMed] [Google Scholar]
- Fraisier V, Gojon A, Tillard P, Daniel-Vedele F. (2000) Constitutive expression of a putative high-affinity nitrate transporter in Nicotiana plumbaginifolia: evidence for post-transcriptional regulation by a reduced nitrogen source. Plant J 23: 489–496 [DOI] [PubMed] [Google Scholar]
- Gansel X, Muños S, Tillard P, Gojon A. (2001) Differential regulation of the NO3- and NH4+ transporter genes AtNrt2.1 and AtAmt1.1 in Arabidopsis: relation with long-distance and local controls by N status of the plant. Plant J 26: 143–155 [DOI] [PubMed] [Google Scholar]
- Giannini JL, Gildensoph LH, Reynolds-Niesman I, Briskin DP. (1987) Calcium Transport in Sealed Vesicles from Red Beet (Beta vulgaris L.) Storage Tissue : I. Characterization of a Ca-Pumping ATPase Associated with the Endoplasmic Reticulum. Plant Physiol 85: 1129–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girin T, El-Kafafi S, Widiez T, Erban A, Hubberten HM, Kopka J, Hoefgen R, Gojon A, Lepetit M. (2010) Identification of Arabidopsis mutants impaired in the systemic regulation of root nitrate uptake by the nitrogen status of the plant. Plant Physiol 153: 1250–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girin T, Lejay L, Wirth J, Widiez T, Palenchar PM, Nazoa P, Touraine B, Gojon A, Lepetit M. (2007) Identification of a 150 bp cis-acting element of the AtNRT2.1 promoter involved in the regulation of gene expression by the N and C status of the plant. Plant Cell Environ 30: 1366–1380 [DOI] [PubMed] [Google Scholar]
- Glass AD, Britto DT, Kaiser BN, Kinghorn JR, Kronzucker HJ, Kumar A, Okamoto M, Rawat S, Siddiqi MY, Unkles SE, et al. (2002) The regulation of nitrate and ammonium transport systems in plants. J Exp Bot 53: 855–864 [DOI] [PubMed] [Google Scholar]
- Glass ADM, Siddiqi MY. (1995) Nitrogen absorption in higher plants. Srivastava HS, Singh RP, , Nitrogen Nutrition in Higher Plants. Associated Publishing, New Delhi, India, pp 21–55 [Google Scholar]
- Gojon A, Nacry P, Davidian JC. (2009) Root uptake regulation: a central process for NPS homeostasis in plants. Curr Opin Plant Biol 12: 328–338 [DOI] [PubMed] [Google Scholar]
- Good AG, Johnson SJ, Pauw MD, Carroll RT, Savidov N, Vidmar J, Lu Z, Taylor G, Stroeher V. (2007) Engineering nitrogen use efficiency with alanine aminotransferase. Can J Bot 85: 252–262 [Google Scholar]
- Ho CH, Lin SH, Hu HC, Tsay YF. (2009) CHL1 functions as a nitrate sensor in plants. Cell 138: 1184–1194 [DOI] [PubMed] [Google Scholar]
- Hong Y, Devaiah SP, Bahn SC, Thamasandra BN, Li M, Welti R, Wang X. (2009) Phospholipase D epsilon and phosphatidic acid enhance Arabidopsis nitrogen signaling and growth. Plant J 58: 376–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa S, Ito Y, Sato Y, Fukaya Y, Takahashi M, Morikawa H, Ohtake N, Ohyama T, Sueyoshi K. (2009) Two-component high-affinity nitrate transport system in barley: membrane localization, protein expression in roots and a direct protein-protein interaction. Plant Biotechnol 26: 197–205 [Google Scholar]
- Krouk G, Tillard P, Gojon A. (2006) Regulation of the high-affinity NO3− uptake system by NRT1.1-mediated NO3− demand signaling in Arabidopsis. Plant Physiol 142: 1075–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanquar V, Loqué D, Hörmann F, Yuan L, Bohner A, Engelsberger WR, Lalonde S, Schulze WX, von Wirén N, Frommer WB. (2009) Feedback inhibition of ammonium uptake by a phospho-dependent allosteric mechanism in Arabidopsis. Plant Cell 21: 3610–3622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RB, Drew MC. (1989) Rapid, reversible inhibition of NO3− influx in barley by ammonium. J Exp Bot 40: 741–752 [Google Scholar]
- Lejay L, Gansel X, Cerezo M, Tillard P, Müller C, Krapp A, von Wirén N, Daniel-Vedele F, Gojon A. (2003) Regulation of root ion transporters by photosynthesis: functional importance and relation with hexokinase. Plant Cell 15: 2218–2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejay L, Tillard P, Lepetit M, Olive F, Filleur S, Daniel-Vedele F, Gojon A. (1999) Molecular and functional regulation of two NO3- uptake systems by N- and C-status of Arabidopsis plants. Plant J 18: 509–519 [DOI] [PubMed] [Google Scholar]
- Lejay L, Wirth J, Pervent M, Cross JM, Tillard P, Gojon A. (2008) Oxidative pentose phosphate pathway-dependent sugar sensing as a mechanism for regulation of root ion transporters by photosynthesis. Plant Physiol 146: 2036–2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Wang Y, Okamoto M, Crawford NM, Siddiqi MY, Glass AD. (2007) Dissection of the AtNRT2.1:AtNRT2.2 inducible high-affinity nitrate transporter gene cluster. Plant Physiol 143: 425–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little DY, Rao H, Oliva S, Daniel-Vedele F, Krapp A, Malamy JE. (2005) The putative high-affinity nitrate transporter NRT2.1 represses lateral root initiation in response to nutritional cues. Proc Natl Acad Sci USA 102: 13693–13698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loqué D, Lalonde S, Looger LL, von Wirén N, Frommer WB. (2007) A cytosolic trans-activation domain essential for ammonium uptake. Nature 446: 195–198 [DOI] [PubMed] [Google Scholar]
- Malagoli P, Lainé P, Le Deunff E, Rossato L, Ney B, Ourry A. (2004) Modeling nitrogen uptake in oilseed rape cv Capitol during a growth cycle using influx kinetics of root nitrate transport systems and field experimental data. Plant Physiol 134: 388–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschner H. (1995) Mineral Nutrition of Higher Plants, Ed 2. Academic Press, London [Google Scholar]
- Miller AJ, Fan X, Orsel M, Smith SJ, Wells DM. (2007) Nitrate transport and signalling. J Exp Bot 58: 2297–2306 [DOI] [PubMed] [Google Scholar]
- Muños S, Cazettes C, Fizames C, Gaymard F, Tillard P, Lepetit M, Lejay L, Gojon A. (2004) Transcript profiling in the chl1-5 mutant of Arabidopsis reveals a role of the nitrate transporter NRT1.1 in the regulation of another nitrate transporter, NRT2.1. Plant Cell 16: 2433–2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro FJ, Martín Y, Siverio JM. (2008) Phosphorylation of the yeast nitrate transporter Ynt1 is essential for delivery to the plasma membrane during nitrogen limitation. J Biol Chem 283: 31208–31217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazoa P, Vidmar JJ, Tranbarger TJ, Mouline K, Damiani I, Tillard P, Zhuo D, Glass AD, Touraine B. (2003) Regulation of the nitrate transporter gene AtNRT2.1 in Arabidopsis thaliana: responses to nitrate, amino acids and developmental stage. Plant Mol Biol 52: 689–703 [DOI] [PubMed] [Google Scholar]
- Neuhäuser B, Dynowski M, Mayer M, Ludewig U. (2007) Regulation of NH4+ transport by essential cross talk between AMT monomers through the carboxyl tails. Plant Physiol 143: 1651–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieves-Cordones M, Miller AJ, Alemán F, Martínez V, Rubio F. (2008) A putative role for the plasma membrane potential in the control of the expression of the gene encoding the tomato high-affinity potassium transporter HAK5. Plant Mol Biol 68: 521–532 [DOI] [PubMed] [Google Scholar]
- Nunes-Nesi A, Fernie AR, Stitt M. (2010) Metabolic and signaling aspects underpinning the regulation of plant carbon nitrogen interactions. Mol Plant 3: 973–996 [DOI] [PubMed] [Google Scholar]
- Okamoto M, Kumar A, Li W, Wang Y, Siddiqi MY, Crawford NM, Glass AD. (2006) High-affinity nitrate transport in roots of Arabidopsis depends on expression of the NAR2-like gene AtNRT3.1. Plant Physiol 140: 1036–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsel M, Chopin F, Leleu O, Smith SJ, Krapp A, Daniel-Vedele F, Miller AJ. (2006) Characterization of a two-component high-affinity nitrate uptake system in Arabidopsis: physiology and protein-protein interaction. Plant Physiol 142: 1304–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsel M, Eulenburg K, Krapp A, Daniel-Vedele F. (2004) Disruption of the nitrate transporter genes AtNRT2.1 and AtNRT2.2 restricts growth at low external nitrate concentration. Planta 219: 714–721 [DOI] [PubMed] [Google Scholar]
- Quesada A, Galván A, Fernández E. (1994) Identification of nitrate transporter genes in Chlamydomonas reinhardtii. Plant J 5: 407–419 [DOI] [PubMed] [Google Scholar]
- Remans T, Nacry P, Pervent M, Girin T, Tillard P, Lepetit M, Gojon A. (2006) A central role for the nitrate transporter NRT2.1 in the integrated morphological and physiological responses of the root system to nitrogen limitation in Arabidopsis. Plant Physiol 140: 909–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano J, Miwa K, Yuan L, von Wirén N, Fujiwara T. (2005) Endocytosis and degradation of BOR1, a boron transporter of Arabidopsis thaliana, regulated by boron availability. Proc Natl Acad Sci USA 102: 12276–12281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Y, Zhou JJ, Li Z, Miller AJ. (2005) A two-component high-affinity nitrate uptake system in barley. Plant J 41: 442–450 [DOI] [PubMed] [Google Scholar]
- Tsay YF, Chiu CC, Tsai CB, Ho CH, Hsu PK. (2007) Nitrate transporters and peptide transporters. FEBS Lett 581: 2290–2300 [DOI] [PubMed] [Google Scholar]
- Ullrich WR, Larsson M, Larsson CM, Lesch S, Novacky A. (1984) Ammonium uptake in Lemna gibba G1, related membrane potential changes and inhibition of anion uptake. Physiol Plant 61: 369–376 [Google Scholar]
- Wang MY, Glass ADM, Shaff JE, Kochian LV. (1994) Ammonium uptake by rice roots (III. Electrophysiology). Plant Physiol 104: 899–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang MY, Siddiqi MY, Glass ADM. (1996) Interactions between K+ and NH4+ effects on ion uptake by rice roots. Plant Cell Environ 19: 1037–1046 [Google Scholar]
- Wang R, Xing X, Wang Y, Tran A, Crawford NM. (2009) A genetic screen for nitrate regulatory mutants captures the nitrate transporter gene NRT1.1. Plant Physiol 151: 472–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li W, Siddiqi Y, Kinghorn JR, Unkles SE, Glass AD. (2007) Evidence for post-translational regulation of NrtA, the Aspergillus nidulans high-affinity nitrate transporter. New Phytol 175: 699–706 [DOI] [PubMed] [Google Scholar]
- Widiez T, El Kafafi S, Girin T, Berr A, Ruffel S, Krouk G, Vayssières A, Shen WH, Coruzzi GM, Gojon A, et al. (2011) High nitrogen insensitive 9 (HNI9)-mediated systemic repression of root NO3- uptake is associated with changes in histone methylation. Proc Natl Acad Sci USA 108: 13329–13334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth J, Chopin F, Santoni V, Viennois G, Tillard P, Krapp A, Lejay L, Daniel-Vedele F, Gojon A. (2007) Regulation of root nitrate uptake at the NRT2.1 protein level in Arabidopsis thaliana. J Biol Chem 282: 23541–23552 [DOI] [PubMed] [Google Scholar]
- Yong Z, Kotur Z, Glass AD. (2010) Characterization of an intact two-component high-affinity nitrate transporter from Arabidopsis roots. Plant J 63: 739–748 [DOI] [PubMed] [Google Scholar]
- Yoshimoto N, Inoue E, Watanabe-Takahashi A, Saito K, Takahashi H. (2007) Posttranscriptional regulation of high-affinity sulfate transporters in Arabidopsis by sulfur nutrition. Plant Physiol 145: 378–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L, Loqué D, Ye F, Frommer WB, von Wirén N. (2007) Nitrogen-dependent posttranscriptional regulation of the ammonium transporter AtAMT1;1. Plant Physiol 143: 732–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuo D, Okamoto M, Vidmar JJ, Glass AD. (1999) Regulation of a putative high-affinity nitrate transporter (Nrt2;1At) in roots of Arabidopsis thaliana. Plant J 17: 563–568 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.