Abstract
Eukaryotic precursor mRNA splicing is a process involving a very complex RNA-protein edifice. Serine/arginine-rich (SR) proteins play essential roles in precursor mRNA constitutive and alternative splicing and have been suggested to be crucial in plant-specific forms of developmental regulation and environmental adaptation. Despite their functional importance, little is known about their origin and evolutionary history. SR splicing factors have a modular organization featuring at least one RNA recognition motif (RRM) domain and a carboxyl-terminal region enriched in serine/arginine dipeptides. To investigate the evolution of SR proteins, we infer phylogenies for more than 12,000 RRM domains representing more than 200 broadly sampled organisms. Our analyses reveal that the RRM domain is not restricted to eukaryotes and that all prototypical SR proteins share a single ancient origin, including the plant-specific SR45 protein. Based on these findings, we propose a scenario for their diversification into four natural families, each corresponding to a main SR architecture, and a dozen subfamilies, of which we profile both sequence conservation and composition. Finally, using operational criteria for computational discovery and classification, we catalog SR proteins in 20 model organisms, with a focus on green algae and land plants. Altogether, our study confirms the homogeneity and antiquity of SR splicing factors while establishing robust phylogenetic relationships between animal and plant proteins, which should enable functional analyses of lesser characterized SR family members, especially in green plants.
In a broad range of eukaryotes, including green plants, most nuclear genes are interrupted by introns that must be accurately excised from precursor mRNA molecules to give rise to functional mature protein-coding mRNAs. Precursor mRNA splicing occurs within a dynamic macromolecular complex known as the spliceosome. The spliceosome is one of the most elaborate edifices in the cell, whose precise assembly at each intron involves five small nuclear ribonucleoprotein particles (snRNPs) associated with snRNP-specific proteins (for review, see Roy and Irimia, 2009; Wahl et al., 2009).
Precursor mRNA alternative splicing (AS) is a regulated mechanism that allows the synthesis of multiple mRNAs from a single gene. AS is widespread in eukaryotes (including unicellular organisms) and has a significant role in expanding transcriptome and proteome diversity (Keren et al., 2010). Recent estimates indicate that approximately 95% of multiexon human genes undergo AS and that most AS events are differentially regulated between tissues (Pan et al., 2008).
Global AS has been investigated in green algae and land plants, and recent deep transcriptome sequencing in the model plant species Arabidopsis (Arabidopsis thaliana) points toward a far greater complexity of AS than previously assumed (Filichkin et al., 2010; Labadorf et al., 2010, and refs. therein). In rice (Oryza sativa), more than 50% of AS-related genes undergo multiple AS events, producing a variety of transcripts from a single gene, highlighting the extremely high complexity of transcriptome regulation. Expression analysis showed that approximately 60% of the AS events were organ specific, suggesting an association of AS events with organ differentiation and plant functional complexity (Chung and Howe, 2009; Zhang et al., 2010).
Constitutive splicing and AS require a large number of non-snRNP-associated proteins acting as positive or negative regulators. The serine/arginine-rich (SR) splicing factors dynamically participate in spliceosome assembly. SR proteins are generally viewed as a phylogenetically highly conserved family of RNA-binding proteins (Long and Caceres, 2009), although this hypothesis has not been formally tested. Metazoan SR proteins were discovered nearly 20 years ago as essential splicing factors that could also regulate AS (for review, see Lin and Fu, 2007). In humans, at least nine SR proteins have been described with sizes ranging from 20 to 75 kD: SRSF1 (ASF/SF2), SRSF2 (SC35), SRSF3 (SRp20), SRSF4 (SRp75), SRSF5 (SRp40), SRSF6 (SRp55), SRSF7 (9G8), SRSF9 (SRp30c), and SRSF11 (SRp54; Long and Caceres, 2009; Manley and Krainer, 2010). Prototypical SR proteins have a modular architecture consisting of one or two N-terminal RNA recognition motifs (RRMs) and a C-terminal RS domain of low complexity enriched in Arg-Ser (or Ser-Arg) repeats (Haynes and Iakoucheva, 2006). Some SR proteins (such as SRSF7) contain an RNA-binding CCHC zinc-knuckle (ZnK) motif located between the RRM and RS domains. A few criteria have been proposed to define bona fide SR proteins. Beyond their minimal structural organization (at least one RRM and one RS domain), they share several immunological and biochemical properties. SR proteins (1) contain a common phosphoepitope; (2) complement splicing in HeLa cell S100 extracts deficient in SR factors; and (3) can be precipitated in specific salt buffers (Bourgeois et al., 2004). However, the atypical SR splicing factor SRSF10/SRp38 is unable to activate splicing in S100 extracts, whereas it has been characterized as a general splicing repressor when dephosphorylated and as a sequence-dependent splicing activator when phosphorylated (Shin et al., 2004, 2005; Feng et al., 2009). Several SR proteins exhibit multifunctionality, playing additional roles in mRNA metabolism (Li and Manley, 2005; Xiao et al., 2007; Loomis et al., 2009).
In addition to the prototypical SR family, many other RS domain-containing proteins, which may or may not contain a RRM domain, have been identified and are collectively referred to as SR-related proteins (Boucher et al., 2001; Lin and Fu, 2007; Long and Caceres, 2009; Shepard and Hertel, 2009). Because of the functional and structural diversity among SR, SR-related, and RS domain-containing proteins, a simple definition of SR proteins and a unified nomenclature have recently been proposed for mammals (and vertebrates; Manley and Krainer, 2010). SR proteins are defined only according to their structural and sequence features (i.e. one or two N-terminal RRMs followed by a downstream RS domain of at least 50 amino acids with more than 40% RS content characterized by consecutive RS or SR repeats). This precise definition allowed the identification of 12 human SR proteins (Manley and Krainer, 2010).
Analyses of Arabidopsis and rice genomes yielded at least 19 and 24 SR protein-encoding genes, respectively. Some SR proteins are homologous to human prototypes SRSF2 (one RRM), SRSF1 (two RRMs), and SRSF7 (one RRM and one ZnK), while others are reported to be specific to green plants (Reddy, 2007, and refs. therein; Barta et al., 2008). For example, members of the RS2Z subfamily are characterized by the presence of two adjacent ZnKs, and the plant-specific SR45 displays atypical structural features with a single RRM located between two distinct N- and C-terminal RS domains (Tanabe et al., 2009; Zhang and Mount, 2009). Following the newly revised nomenclature of the mammalian SR proteins, Barta et al. (2010) have proposed a unified nomenclature for plant SR proteins that takes into account a number of plant-specific properties.
Even if SR splicing factors have been detected in a few model organisms besides animals and green plants (Portal et al., 2003; Collins and Penny, 2005; Barbosa-Morais et al., 2006; Plass et al., 2008), these proteins have been little studied in a broader evolutionary perspective. Significantly, the two recent nomenclature efforts (Barta et al., 2010; Manley and Krainer, 2010) were carried out independently and ended up being relatively discordant. The proposed nomenclatures notably do not account for orthology relationships between animal and plant proteins that have been suspected for years (Birney et al., 1993; Maruyama et al., 1999; Bourgeois et al., 2004; Barta et al., 2008, 2010). Although recent studies have focused on the history of splicing factors (Barbosa-Morais et al., 2006; Plass et al., 2008; Richardson et al., 2011), a reliable phylogenetic framework has not yet been established for SR proteins, partly due to global approaches poorly suited to their multiple architectures (Shepard and Hertel, 2009). In this respect, it remains to be determined whether they all genuinely belong to a single protein family or have acquired their similar structural and functional features by convergence.
Here, we mine about 700 complete proteomes from archaea, bacteria, eukaryotes, and viruses for RRM-containing proteins to investigate the origin and subsequent diversification of SR splicing factors using the widespread RRM domain as their only shared attribute of phylogenetic utility. In spite of the small size of the RRM domain, our unbiased genome-wide strategy provides evidence for a single ancient origin of all prototypical SR proteins among RRM-containing proteins, probably tracing back to the last common ancestor of extant eukaryotes. Then, based on a series of refined analyses focusing on SR proteins only, we propose a hypothetical scenario for their diversification into four natural families and a dozen subfamilies, of which we profile sequence conservation and composition. Finally, we assemble curated inventories of SR splicing factors for 20 proteomes, with emphasis on green algae and land plants. Altogether, this study establishes SR proteins as members of a genuine protein family and defines operational criteria for both the computational discovery and the classification of uncharacterized SR proteins. Furthermore, through the establishment of robust orthology relationships with domains and proteins studied in animals, it will help to generate functional hypotheses for their green plant counterparts.
RESULTS
The RRM Domain as a Phylogenetic Marker
Due to their modular organization that prevents the meaningful alignment of full-length sequences and to the low complexity of the RS domain that limits their information content, SR proteins do not readily lend themselves to phylogenetic analysis. In contrast, the RRM domain is a feature shared by all these splicing factors and is the most common and widespread eukaryotic RNA-binding domain (Lorković and Barta, 2002; Lunde et al., 2007). It is composed of about 80 amino acids that form a four-stranded antiparallel β-sheet packed against two α-helices. The two more central β-strands contain the highly conserved motifs RNP1 and RNP2, consisting of predominantly aromatic and hydrophobic residues (Maris et al., 2005). Using the RRM domain as a proxy to gain insight into the evolutionary history of SR proteins may solve the issues raised by their multiple architectures, provided it appeared only once, which is a reasonable assumption. Moreover, this domain occurs in more than 40 distinct orthologous groups of proteins predating the eukaryotic radiation (Anantharaman et al., 2002), thus providing a large number of outgroup sequences to test the hypothesis of a single origin for all known SR splicing factors. Our prediction is as follows: in a broadly sampled phylogeny of the RRM domain, RRMs extracted from SR proteins should group together if the latter proteins indeed share a common ancestor, whereas polyphyletic SR proteins resulting from convergent evolution would display RRM domains that are more scattered across the tree. Considering the short size of the RRM domain, the phylogenetic resolution as measured by statistical support values (e.g. bootstraps) is expected to be low, in line with other works (Birney et al., 1993; Fukami-Kobayashi et al., 1993; Maruyama et al., 1999; Barbosa-Morais et al., 2006; Plass et al., 2008; Richardson et al., 2011). Therefore, the actual robustness of our analyses will require assessment through alternative phylogenetic approaches, such as the careful comparison of multiple trees obtained with different sequence samplings and inference methods (for review, see Delsuc et al., 2005; Philippe et al., 2005).
RRM Distribution across the Tree of Life
To address the origin of SR splicing factors, we searched for RRM-containing proteins in 704 complete proteomes (including 77 eukaryotes) with a hidden Markov model (HMM) of the RRM domain computed from the corresponding Pfam alignment (Supplemental Text S1; Supplemental Fig. S1). We elected to use a very generic HMM rather than specific BLAST searches both to maximize detection sensitivity and to minimize sampling biases that might be caused by nonrandom selection of a limited number of query sequences. At the E-value threshold of 1e-10, we retrieved a total of 12,023 RRM domains extracted from 8,042 proteins (Supplemental Table S1).
Previous studies conducted on a few prokaryotes either suggested that the rare RRM domains found in archaea and bacteria probably originated from horizontal gene transfer (HGT) from eukaryotes (Anantharaman et al., 2002) or concluded that prokaryotic and eukaryotic RRMs shared a common origin (Maruyama et al., 1999). Our study identified 259 RRM-containing proteins in two closely related archaea and in 124 proteomes belonging to a wide array of bacterial lineages (Fig. 1A). As will be shown in our phylogenetic analysis, most prokaryotic RRMs are resolved as three successive clades, with a few exceptions that might be interpreted as HGT (Supplemental Figs. S5–S9). Prokaryotic RRM domains display the classical RNP1 and RNP2 motifs (Supplemental Fig. S22). These results thus suggest that the RRM is a bona fide prokaryotic structure tracing back to the last common ancestor of the three domains of life. Although the lack of RRM-containing proteins in many of the surveyed proteomes might point to an incomplete detection in prokaryotes, we consider this unlikely, as more sensitive searches did not yield additional RRMs beyond the canonical domains already predicted (Supplemental Table S1). This absence could be genuine and stem from multiple losses among prokaryotic lineages following the secondary simplification of modern prokaryotes from a more complex last common ancestor of the three domains of life (Forterre and Philippe, 1999; Kurland et al., 2006). Furthermore, the abundance of the RRM domain could be underestimated due to the biased sampling of sequenced prokaryotic genomes toward reduced and pathogenic organisms (Wu et al., 2009). On the other hand, members of the mainly free-living and quite complex cyanobacterial lineage possess the highest number of RRM-containing proteins among prokaryotes (Fig. 1A).
Figure 1.
Effect of taxonomy and lifestyle on RRM domain occurrence and the discriminative power of RS/SR dipeptides. A, Distribution of the number of RRM-containing proteins per proteome within archaeal and bacterial lineages. All surveyed lineages are included, even those that did not yield any RRM (96% of archaeal and 77% of bacterial proteomes). B, Number of RRM-containing proteins as a function of proteome size (log-log scale). In eukaryotes, these numbers are correlated (r = 0.924) and the correlation depends on both cellularity (F test P = 1.24e-05) and energetic metabolism (P = 1.44e-04). In particular, multicellular eukaryotes (y = 1.112x − 2.719) possess relatively more RRM-containing proteins than unicellular eukaryotes (y = 0.666x − 1.062). Furthermore, for any proteome size, photosynthetic eukaryotes (y = 1.031x − 2.551) always have about 1.5 times less RRM-containing proteins than heterotrophs (y = 1.038x − 2.392). Finally, a similar correlation is observed for (photosynthetic) cyanobacteria (r = 0.789; y = 0.752x − 2.051), which is not the case for bacteria considered as a single class (r = 0.115; y = 0.166x − 0.343). C, Comparative distribution of RS/SR dipeptide counts in sequences of 8,042 RRM-containing proteins (gray bars) and in a subset of 196 proteins annotated as SR splicing factors (red bars). Classes have a width of four except the last one, which includes all counts of 48 or more dipeptides. The inset shows the corresponding cumulative curves.
All eukaryotes feature at least a dozen RRM-containing proteins (Supplemental Table S1). The actual number directly depends on the total number of proteins, further modulated by the organism being (1) unicellular or multicellular and (2) photosynthetic or not (Fig. 1B). In humans, we discovered 556 RRM domains belonging to 353 RRM-containing proteins (out of 47,547), of which 64 display at least one occurrence of either RSRS or SRSR tetrapeptides (Boucher et al., 2001), whereas for the Arabidopsis proteome, the corresponding numbers are 363, 251 (out of 31,711), and 47, respectively (Supplemental Table S1).
Although SR splicing factors are expected to contain a downstream domain of at least 50 amino acids and a minimum of 20% to 40% RS content (Barta et al., 2010; Manley and Krainer, 2010), some genuine SR/SRrp proteins exhibit a lower RS content (Fig. 1C). Therefore, we selected the minimal threshold of at least one tetrapeptide to tag putative SR proteins among RRM-containing proteins. It is noteworthy that the SR tag was only used for manual curation and that no protein was excluded due to a lack of RS/SR dipeptides. Our goal was again to minimize sampling biases while accounting for inefficient handling of this kind of repetitive protein motif by gene prediction algorithms (Barbosa-Morais et al., 2006). Nevertheless, eukaryotic proteins annotated as SR splicing factors generally contained many more RS/SR dipeptides than most RRM-containing proteins (Fig. 1C, inset). In comparison, none of the cyanobacterial RRM-containing proteins fulfilled our minimal threshold (Supplemental Table S1).
A Single Origin for Prototypical SR Splicing Factors
To allow phylogenetic analysis, the 12,023 RRM domains retrieved above were reduced to 1,266 slowly evolving representative domains through clustering based on sequence similarity (Supplemental Figs. S2 and S3). This more tractable data set was aligned against the HMM profile to limit the number of gaps introduced by sequence-specific insertions, yielding an alignment of 72 amino acid positions. Phylogenetic inference using different approaches (maximum parsimony versus maximum likelihood [ML]), evolutionary models (WAG+Γ4 versus LG+F+Γ4; Yang, 1993; Whelan and Goldman, 2001; Le and Gascuel, 2008), and sequence samples (1,266 versus an enlarged data set of 1,831 clusters assembled from two additional data sources) was then applied (Supplemental Figs. S5–S9). Trees were annotated using the Eukaryotic “Clusters of Orthologous Groups” (KOG) database (Tatusov et al., 2003) and a corpus of reference RRM-containing proteins (Supplemental Fig. S4; Supplemental Table S2; for details, see Supplemental Text S1).
Non-SR RRM-containing proteins that consistently associated in the five trees generally shared similar functional annotations (Supplemental Fig. S10; Supplemental Table S3), which confirms that the RRM domain carries relevant phylogenetic information in spite of its short size. Similarly, the unique RRM of single-RRM SR proteins and the first (or N-terminal) RRM of dual-RRM SR proteins (both hereafter referred to as “RRM1”) displayed a limited scattering. However, their recovery as a single subtree was never obtained; instead, RRM1 domains fell into four basic clades of mutual affinities, which were sensitive to both domain sampling and the reconstruction method. Four trees (Supplemental Figs. S5, S6, S8, and S9) yielded three different combinations of three versus one SR architectures, whereas the last tree (Supplemental Fig. S7) only grouped two architectures (Supplemental Fig. S11; Supplemental Table S4). To ascertain that these relatively stable associations were indicative of a common ancestry for SR splicing factors, an additional ML tree was computed on the 152 largest RRM clusters (representing 10,101 [84%] out of 12,023 retrieved domains) using the LG model (Fig. 2; Supplemental Fig. S12). Remarkably, the removal of hundreds of minor clusters much improved the resolution of the RRM tree, where the RRM1 domains of most SR proteins were resolved as a single clade (i.e. single-RRM SR proteins, single-RRM ZnK-like SR proteins, and dual-RRM SR proteins), sister to a smaller group corresponding to the atypical RNPS1/SR45 proteins. Altogether, our phylogenetic analyses of the extant diversity of the RRM domain thus support a single origin for prototypical SR protein architectures.
Figure 2.
Single origin of SR splicing factors among RRM-containing proteins. The tree was obtained with RAxML (LG+F+Γ4 model) from the analysis of an alignment of 72 amino acids × 152 slowly evolving RRM domains representative of all multispecies RRM clusters with at least eight members and was rooted using prokaryotic RRM clusters as outgroups. Leaves are color coded as follows: RRM1 of single-RRM SR proteins (green); RRM1 of single-RRM ZnK-like SR proteins (blue); RRM1 of dual-RRM SR proteins (red); RRM1 of RNPS1/SR45 proteins (orange); RRM2 of nonplant dual-RRM SR proteins (violet); RRM2 of the plant-specific RS group of SR proteins (brown); RRMx of non-SR proteins (light gray); prokaryotic RRM (dark gray). Nonzero bootstrap proportions are shown. The scale bar at the bottom gives the number of substitutions per site. The white arrowhead points to a conservative position for the origin of all SR splicing factors, whereas the black arrowhead hypothesizes that the SR clade is affected by an imperfect reconstruction. In both cases, we assume that node 22b is incorrectly placed in this particular tree. The fully annotated tree is shown in Supplemental Figure S12.
Subsequent Diversification of SR Splicing Factors
Based on shared features, parsimony leads us to infer that the common ancestor of SR proteins consisted of a single RRM domain followed by a RS domain. To investigate the further evolution of prototypical SR proteins, a second round of phylogenetic analysis taking advantage of the full sequence diversity available before clustering was carried out. Two subtrees of SR-associated RRM domains (shown in pink in Supplemental Fig. S6) were thus selected: (1) RRM1 domains of single-RRM (nodes 22 and 23), single-RRM ZnK-like (node 17), and dual-RRM (node 16) SR proteins, and (2) RRM1 domains of the atypical RNPS1/SR45 proteins (nodes 26 and 27). The tree in Supplemental Figure S6 was chosen as a source because its topology of SR-associated domains was the closest to the topology shown in Figure 2. In addition, the evolution of the second (or internal) RRM (“RRM2”) of dual-RRM SR proteins of animals and fungi (node 29; third pink subtree in Supplemental Fig. S6) was also examined, whereas plant RRM2 domains were not investigated, as the corresponding subtree for RS proteins (node 56) was too small (Supplemental Fig. S6) and since RRM2s of SR (ASF-like) proteins were missing from our large trees.
For these refined analyses, where outgroup domain sequences were also included for rooting (e.g. nodes 24, 30, and 65), data sets were aligned as above, except that insertions were allowed to preserve more phylogenetic signal. Small trees were inferred by ML (Supplemental Figs. S13–S18). Due to the poor resolution of the exhaustive data set of SR-associated RRM1 domains (Supplemental Figs. S11 and S12), variants restricted to slowly evolving domains and optionally omitting unstable subfamilies (i.e. RS and/or RS2Z) were also studied (Table I; Supplemental Figs. S19 and S20).
Table I.
Comparative support values for relationships between SR families and subfamilies
| Bootstrap Proportions for Nodes in Figure 3 |
||||||||||
| Data Seta | Sequence No. × Amino Acid No. | Heuristic | Model | Tree | N1 | N2 | N3 | N4 | N5 | N6 |
| Exhaustive | 434 × 93 | RAxML | WAG+Γ4 | Supplemental Figure S13 | –b | <50 | <50 | <50c | <50d | <50 |
| TreeFinder | WAG+Γ4 | Supplemental Figure S14 | –b | <50 | <50 | <50c | <50d | <50 | ||
| Slow evolving | 304 × 87 | RAxML | WAG+Γ4 | Supplemental Figure S19 | <50 | 68 | 74 | 64 | <50d | 87 |
| TreeFinder | WAG+Γ4 | Supplemental Figure S20 | <50 | 62 | 85 | 84 | 55d | 87 | ||
| Slow-evolving−RS | 292 × 85 | RAxML | WAG+Γ4 | /e | <50 | 58 | 69 | 62 | 51d | 84 |
| TreeFinder | WAG+Γ4 | /e | –b | 64 | 69 | 88 | <50 | 94 | ||
| Slow-evolving−RS2Z | 297 × 87 | RAxML | WAG+Γ4 | /e | 53 | 60 | 62 | 59 | 57 | 71 |
| TreeFinder | WAG+Γ4 | /e | <50 | 58 | 61 | 89 | 68 | 89 | ||
| Slow-evolving−RS-RS2Z | 285 × 85 | RAxML | WAG+Γ4 | /e | <50 | 53 | 56 | 63 | 59 | 85 |
| TreeFinder | WAG+Γ4 | /e | –b | 62 | 67 | 83 | 58 | 97 | ||
| Inventory, all | 270 × 82 | RAxML | WAG+Γ4 | /e | –b | <50 | –f | <50c | <50d | <50 |
| TreeFinder | WAG+Γ4 | /e | 58 | <50 | –f | –f | –f | <50 | ||
| Inventory, no gap | 270 × 64 | RAxML | WAG+Γ4 | Supplemental Figure S26 | <50 | <50 | <50 | <50c | –f | <50c |
| TreeFinder | WAG+Γ4 | Supplemental Figure S27 | 64 | <50 | <50 | <50c | –f | 62c | ||
| RAxML | LG+F+Γ4 | Supplemental Figure S28 | <50 | <50 | –f | <50c | <50d | <50c | ||
| PhyloBayes | CAT+Γ4 | Supplemental Figure S29 | 92 | –f | –f | –f | <50d | <50c | ||
Data sets are described in the text. The relationships are based on the analysis of RRM1 domains.
Paraphyletic group due to outgroup domains.
Except for a few fast-evolving domains.
Actually paraphyletic due to RS2Z domains.
Tree not shown.
Node not recovered in the tree (i.e. the group is polyphyletic).
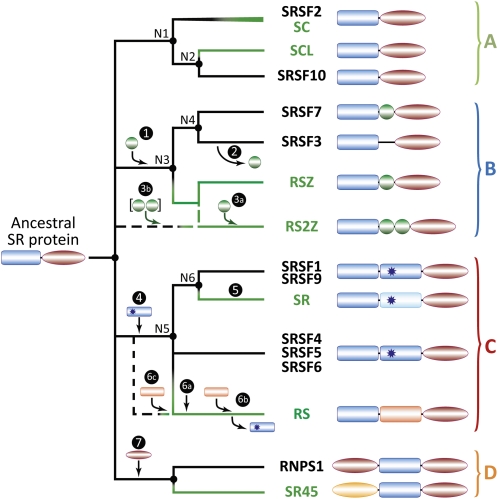
Taken as a whole, our phylogenetic analysis allowed us to propose an evolutionary scenario for SR proteins leading to four natural families (Fig. 3). Unsurprisingly, these families are congruent with the main SR protein architectures. The first family corresponds to single-RRM SR proteins that probably retained the basic architecture of the common ancestor. It is composed of two groups: (1) SRSF2 (SC35) proteins found in both plants (SC subfamily, including red algae) and animals, and (2) plant-specific SC35-like proteins (SCL subfamily) associated with animal SR-repressor proteins (SRSF10/SRrp; Cowper et al., 2001). The second family consists of single-RRM ZnK-like SR proteins. The prototype is the human SRSF7/9G8 protein (Cavaloc et al., 1994), and its plant counterparts are the RSZ proteins (Golovkin and Reddy, 1998). In addition to these members containing precisely one ZnK, the family also includes proteins either having secondarily lost the ZnK (SRSF3/SRp20; Zahler et al., 1992; Cavaloc et al., 1999) or possessing an additional ZnK (plant-specific RS2Z proteins; Lopato et al., 2002). The mode of acquisition of the second ZnK could not be determined from the analysis of the RRM domain (Table I; Supplemental Figs. S13, S14, S19, and S20) or of the ZnK domain (Supplemental Fig. S23). In the absence of compelling evidence for the creation of a fifth natural family, RS2Z proteins were parsimoniously classified within ZnK-like SR proteins (Fig. 3). The third family groups all dual-RRM SR proteins, which include (1) well-known ASF-like proteins found in both plants (SR subfamily) and animals (SRSF1-ASF/SF2 and SRSF9/SRp30c), (2) animal SRSF5-6-4/SRp40-55-75 proteins, and (3) plant-specific RS proteins (Kalyna et al., 2006). Except for plants, the RRM2 of these proteins appears to be related to the three RRMs of hnRNP-M proteins (Supplemental Table S3; Supplemental Figs. S10, S17, and S18) and shares with them the SWQDLKD motif. The RRM2 of green plant SR/ASF-like proteins also retain this motif (Fig. 4), even if this domain is not in our trees due to evolutionary divergence. In contrast, this motif is lacking in the RRM2 of RS proteins, which are present but branch independently in our trees. Nevertheless, detailed sequence comparison of the two RRM domains of RS proteins indicates that their RRM2 might have originated from an internal duplication of the RRM1, which is also supported by some of our refined phylogenetic analyses (Figs. 2 and 4; Supplemental Figs. S27 and S29). Strikingly, both RRM1 and RRM2 of red algal RS proteins branch with the corresponding domains from green plants (Supplemental Figs. S26 to S29), which implies that this subfamily evolved before the divergence of green plants and red algae. The fourth family is composed of animal and fungal RNPS1 and plant-specific SR45 proteins, both featuring an additional (N-terminal) RS domain before the RRM. Although considering SR45 as a genuine SR splicing factor has been questioned (Zhang and Mount, 2009; Barta et al., 2010), it appears nonetheless related to prototypical SR proteins.
Figure 3.
Hypothetical scenario for the evolution of SR protein families. Starting from an ancestral protein with a single RRM domain and an RS domain that predates the separation of animal and plant lineages, a series of key events account for most of the diversity of SR architectures and proteins: 1, acquisition of a ZnK; 2, loss of the ZnK; 3, acquisition of a second ZnK (3a) or independent acquisition of two ZnKs (3b); 4, acquisition of a second RRM (RRM2); 5, evolutionary divergence of the RRM2 with conservation of the SWQDLKD motif; 6, evolutionary divergence (6a), secondary replacement (6b), or independent acquisition (6c) of the RRM2; 7, acquisition of an additional (N-terminal) RS domain. Together, these events eventually led to the emergence of four natural families of SR splicing factors: A, single-RRM proteins (SRSF2/SC35, SRSF10/SRrp, and SCL subfamily); B, single-RRM ZnK-like proteins (SRSF7/9G8, SRSF3/SRp20, RSZ subfamily, and RS2Z subfamily); C, dual-RRM SR proteins (SRSF1-9/ASF-like, SRSF5-6-4/SRp40-55-75, SR subfamily, and RS subfamily); and D, RNPS1-like proteins (RNPS1 and SR45). RRM domains are represented by rectangles, ZnKs by circles, and RS domains by ovals. The star indicates the SWQDLKD motif in RRM2. Plant-specific branches are shown in green. Black dots denote nodes considered robust after the integration of all analyses. Bootstrap proportions for nodes labeled N1 to N6 are provided in Table I.
Figure 4.
Sequence conservation in the second RRM domain of SR proteins. RRM sequence logos are aligned based on secondary structure (bottom). At a given position, the height of any residue is proportional to its frequency, while overall stack height corresponds to sequence conservation. Error bars reflect the uncertainty of conservation estimates. RNP1, RNP2, and SWQDLKD motifs are shaded, as are conserved aromatic residues in these motifs. The RRM1 of RS proteins has been realigned on the corresponding RRM2 domain to highlight their similarities. β1-4, α1-2, and L1-5 stand for β-sheets, α-helices, and linkers of the RRM secondary structure, respectively.
Profiling of Sequence Features in SR Families and Subfamilies
Based on the RRM1 phylogeny, we have defined four natural SR families and a maximum of 13 subfamilies, including RNPS1 and SR45-related proteins (Fig. 3). As these natural families match the main SR architectures, any uncharacterized SR protein should be fairly easy to classify according to its domain organization. However, a major shortcoming of this approach is tied to secondary domain loss. For example, SRSF3/SRp20 belongs to the single-RRM ZnK-like family, even though it has no ZnK domain. Therefore, we explored whether specific sequence features could be identified to discriminate subfamilies as an alternative to the accurate but time-consuming phylogenetic approach.
To this end, we computed sequence logos for the RRM1 domain of each subfamily (Supplemental Fig. S21). Logos were generated from subfamily-specific structural alignments and confirmed the conservation of both RNP1 and RNP2 motifs. However, logos did not provide diagnostic features for subfamily affiliation beyond the expected congruence with the RRM1 phylogeny. This prompted us to investigate the potential of the corresponding full-length proteins, even though SR proteins have been described as intrinsically disordered, owing to their low-complexity RS domains (Haynes and Iakoucheva, 2006).
First, sequence conservation was measured within each subfamily to localize conserved regions other than RRM and ZnK domains. Second, sequence composition was profiled by sliding a window on individual SR proteins belonging to each subfamily in order to count the occurrences of all possible words with a size of one to three amino acids. This blind analysis led to the identification of an array of compositional features that appear specific to one or more subfamilies (Supplemental Fig. S24; Supplemental Table S5). In addition to the characteristic Ser/Arg enrichment in the C-terminal part of SR proteins, a number of additional features have been detected (Fig. 5; Supplemental Fig. S25). Among these, a Gly-rich region is observed between the first RRM and the following domain (ZnK or RRM2) in most multidomain subfamilies except plant RS proteins. Another example is the enrichment in Lys/Ser dipeptides after the last RRM in SRSF2/SC (SC35), SRSF10/SRrp, plant SR/ASF-like, and SFRS5-6-4/SRp40-55-75 subfamilies. Altogether, these profiling analyses allowed us to design a tentative determination key for SR families and subfamilies (Supplemental Table S6).
Figure 5.
Architecture, conservation, and compositional profile of SR natural families. Each natural family is represented by one of its subfamilies (for all subfamilies, see Supplemental Fig. S25). Within each panel, a representative protein was selected for compositional profiling, while conservation was computed on the alignment of all subfamily sequences. Only informative features (Supplemental Table S5) are shown, with discriminating features used in the key (Supplemental Table S6) in solid lines and secondary features in dashed lines. A, Single RRM: human SRSF2/SC35. B, Single RRM ZnK-like: Arabidopsis At-RSZ21. C, Dual RRM: rice Os-SR32 (ASF/SF2). D, RNPS1-like: Arabidopsis SR45.
Inventory of SR Proteins in Selected Organisms
To test whether the genomic approach developed so far would improve our ability to specifically discover and classify SR splicing factors in uncharacterized proteomes, we reanalyzed the proteomes of common model organisms along with those of a set of green algae and land plants not included in our initial data sets. In addition to compiling an exhaustive inventory, we were also interested in assessing the accuracy of our determination key compared with phylogenetic inference.
Due to the universality of the HMM profile used for the initial data mining, the largest part of the retrieved RRM-containing proteins was non-SR proteins, which needed to be filtered out before applying any classification. To address this issue, we generated six SR-enriched HMM profiles based on RRM1 subtrees (Supplemental Figs. S13 and S15) and structurally aligned RRM1 logos (Supplemental Fig. S21). The 20 selected proteomes were mined using these new profiles and yielded 319 RRM domains belonging to 247 candidate SR proteins. These predicted domains were extracted and automatically aligned on the most closely related subfamily consensus sequence (Supplemental Text S1), thus resulting in a high-quality RRM alignment that was used in phylogenetic analyses as above, additionally including Bayesian inference with the CAT model (Lartillot and Philippe, 2004; Lartillot et al., 2009; Supplemental Figs. S26–S29). In parallel, the corresponding full-length proteins were submitted to (1) architecture prediction using the National Center for Biotechnology Information (NCBI) Conserved Domain Database, (2) compositional profiling, and (3) classification using our determination key (Supplemental Data Set S3).
Computational results from all four approaches were manually evaluated to produce the curated inventory presented in Table II (for accession numbers and sequences, see Supplemental Data Set S2). We found candidate SR splicing factors in all proteomes investigated except the fungus Aspergillus fumigatus. New HMM profiles for SR-associated RRM domains proved to be very effective, as 244 out of 247 candidates were eventually confirmed as genuine SR splicing factors or isoforms, which corresponds to a specificity of 98.8%. Regarding sensitivity, all previously described SR proteins from model organisms (Homo sapiens, Caenorhabditis elegans, Arabidopsis, and rice; Supplemental Table S2) were retrieved (100% sensitivity), except human SRSF11/SRp54 and its Caenorhabditis ortholog rsp-7 (Cep54). For the interested reader, these HMM profiles are provided in Supplemental Data Set S1. The determination key efficiency was lower, since 128 SR proteins were successfully classified (66.7%), although this ratio increased to 79% when 30 truncated proteins (mismodeled by gene prediction algorithms) were not considered (Supplemental Table S7). Moreover, unambiguous subfamily affiliation was not always possible, especially for organisms with fast-evolving proteins leading to long branches difficult to position with accuracy.
Table II.
Curated inventory of SR splicing factors in 20 (excluding S. pombe) selected proteomes
| Single RRM |
Single RRM ZnK-Like |
Dual RRM |
RNPS1-Like |
|||||||||||
| Species | Taxon | SRSF2/SC | SCL | SRSF10 | SRSF3 | SRSF7 | RSZ | RS2Z | SRSF1/9 | SR | SRSF4/5/6 | RS | RNPS1 | SR45 |
| H. sapiens | Eutheria | 2 | – | 2 | 1 | 1 | – | – | 2 | – | 3 | – | 1 | – |
| Mus musculus | Eutheria | 1 | – | 2 | 2 | 1 | – | – | 2 | – | 3 | – | 1 | – |
| Drosophila melanogaster | Insecta | 1 | – | – | 3 | 1 | – | – | 1 | – | 1 | – | 2 | – |
| C. elegans | Nematoda | 2 | – | – | 1 | – | – | – | 1 | – | 2 | – | 1 | – |
| A. fumigatus | Ascomycota | – | – | – | – | – | – | – | – | – | – | – | – | – |
| S. pombea | Ascomycota | 1 | – | – | – | – | – | – | – | – | 1 | – | 1 | – |
| Arabidopsis | Eudicotyledons | 1 | 4 | – | – | – | 3 | 2 | – | 4 | – | 4 | – | 1 |
| Populus trichocarpa | Eudicotyledons | 4 | 6 | – | – | – | 4 | 2 | – | 6 | – | 7 | – | 1 |
| Rice | Liliopsida | 3 | 5 | – | – | – | 3 | 4 | – | 3 | – | 2 | – | 2 |
| Sorghum bicolor | Liliopsida | 2 | 5 | – | – | – | 2 | 6 | – | 2 | – | 3 | – | 1 |
| Selaginella moellendorfii | Lycopodiophyta | 2 | 3 | – | – | – | 1 | – | – | 2 | – | 1 | – | 1 |
| Physcomitrella patens | Bryophyta | 2 | 2 | – | – | – | 2 | – | – | 3 | – | 1 | – | 2 |
| Chlamydomonas reinhardtii | Chlorophyta | – | 3 | 1 | – | – | – | – | – | – | – | – | – | |
| Volvox carteri | Chlorophyta | – | 3 | – | – | 1 | – | – | – | – | – | – | – | |
| Chlorella sp. NC64A | Chlorophyta | – | 2 | – | 1 | – | 1 | – | – | 1 | – | 1 | – | 1 |
| Micromonas pusilla CCMP1545 | Chlorophyta | 1 | 1 | – | – | – | – | – | – | – | – | – | – | 1 |
| M. pusilla RCC299 | Chlorophyta | 1 | 1 | – | – | – | – | – | – | – | – | – | – | 1 |
| Ostreococcus sp. RCC809 | Chlorophyta | 2 | – | – | – | – | – | – | – | 1 | – | – | – | – |
| Ostreococcus lucimarinus | Chlorophyta | 2 | – | – | – | – | – | – | – | 1 | – | – | – | – |
| Ostreococcus tauri | Chlorophyta | 1 | – | – | – | – | – | – | – | – | – | – | – | – |
| C. merolae | Rhodophyta | 1 | – | – | – | – | – | – | – | – | – | 1 | – | – |
Data taken from initial analyses, as S. pombe was not part of the curated inventory.
In several species (Caenorhabditis, Drosophila, Chlamydomonas, and Chlorella), SRSF3/SRp20 proteins were annotated (based on their RRM sequence) as 9G8 or RSZ, while SRSF7/9G8 proteins were conversely annotated as SRp20. This incongruence between structural and sequence-based classifications highlights the close relationship between single-RRM ZnK-like SRp20 proteins (secondarily lacking a ZnK domain) and “true” single-RRM ZnK proteins (Supplemental Figs. S13, S14, and S26–S29) and raises the issue of a possible paraphyly of SRp20 proteins. Similarly, some proteins belonging to either one of the orthologous SRSF10/SRrp and SCL subfamilies could not be reliably associated with one or the other subfamily (e.g. in Chlamydomonas and Volvox; Supplemental Figs. S13, S14, and S26–S29), hence their folding in a single subfamily for these species (Table II). In Caenorhabditis, the single-RRM protein CeSC35-2 (Longman et al., 2000) was resolved as a dual-RRM SRSF5-6-4/SRp40-55-75 protein (Supplemental Figs. S13, S14, and S26–S29). If this is not a phylogenetic artifact, it might point to a secondary loss of the RRM2. Finally, although several species and/or strains were examined for the two picoeukaryotic genera Micromonas and Ostreococcus, the analysis of candidate SR proteins was not very fruitful, mainly due to the large number of (probably artifactually) truncated protein models combined to an extensive evolutionary divergence. Consequently, our inventory of SR splicing factors in these ultrasmall green microalgae should be considered as preliminary.
DISCUSSION
SR proteins are involved in constitutive splicing/AS and nonsplicing events and serve as essential regulators of gene expression. Prototypical SR splicing factors contain at least one RRM domain and one RS domain (Manley and Krainer, 2010). Herein, using a genome-wide phylogenetic analysis of more than 12,000 RRM domains detected in over 200 species, we gained insight into the origins of SR splicing factors and tentatively unraveled the evolutionary relationships between SR protein families. Our focus on the only informative feature shared by all prototypical SR proteins (i.e. the RRM domain) enabled a large-scale and unbiased approach that considerably extended previous phenetic (BLAST-based) studies (Collins and Penny, 2005) or (mostly) small-scale phylogenetic studies (Birney et al., 1993; Fukami-Kobayashi et al., 1993; Maruyama et al., 1999; Barbosa-Morais et al., 2006; Plass et al., 2008; Richardson et al., 2011). Although the RS domain is a distinctive feature of SR family proteins, it is also present in SR-related proteins. RS domains have been involved in both protein-RNA and protein-protein interactions. Unlike prototypical SR splicing factors, SR-related proteins may lack a RRM domain and/or contain other distinct domains, such as a PWI motif, a DEAD/H box, or a kinase domain. SR-related family members include both U2AF65 and U2AF35 subunits, U1-70K, SRm 160/300, the RNA helicase hPRP16, and many various proteins (for review, see Lin and Fu, 2007; Long and Caceres, 2009). While the mechanistic function of SR-related proteins in splicing is of importance, their in-depth study was beyond the scope of this work.
Our survey strongly suggests that the RRM domain is likely to be a very ancient structure, tracing back to the common ancestor of modern bacteria and eukaryotes. Depending on the tree-of-life model considered (e.g. eukaryotes are one of the three domains of life versus they result from a hypothetical fusion between two or more prokaryotic partners; for review, see Forterre and Philippe, 1999; Embley and Martin, 2006; Poole, 2006; Forterre, 2011), the scarcity of the RRM domain in investigated archaeal proteomes could be either interpreted as a secondary loss or as a genuinely primitive feature. In the latter case, the few occurrences detected in archaea would result from HGT events from promiscuous bacterial donors.
Our large trees showed a recurrent association of the RRM1 of all analyzed SR proteins, despite a low phylogenetic resolution, as expected from the short length of the RRM domain that we analyzed (Birney et al., 1993; Fukami-Kobayashi et al., 1993; Maruyama et al., 1999; Barbosa-Morais et al., 2006; Plass et al., 2008; Richardson et al., 2011), while a tree focusing on the largest RRM clusters resolved all major SR protein architectures as a single clade. Considering the potentially overwhelming competition with non-SR sequences, these results are consistent with a single origin for most SR splicing factors. Interestingly, the latter also includes the atypical plant-specific SR45 protein but maybe not the SRSF11/SRp54 protein described in animals (see below). This conclusion does not imply a strict monophyly of all SR splicing factors, since some of the non-SR protein families intermingled with SR proteins in our trees (e.g., RBM8 [node 20] and CBP20 [node 24] families in Supplemental Figs. S10 and S11), which might point to a paraphyletic origin. If so, the distribution of SR-specific features and functions (e.g. RS/SR dipeptides) would then result from parallel evolution, secondary loss, or both. Our opinion is that this apparent paraphyly is more likely to originate in stochastic errors affecting phylogenetic reconstruction due to the short size of the RRM domain. Such an interpretation is supported by the observation that several “contaminating” non-SR protein families differ between analyses (Supplemental Figs. S5–S9 and S11) and by the lack of statistical support obtained in Bayesian analyses of the same data set using the CAT model.
For the subsequent diversification of this ancestral SR splicing factor, our analyses support a scenario in which each of the main SR architectures (RRM1-RS, RRM1-RRM2-RS, RRM1-ZnK-RS, and RS-RRM1-RS) corresponds to a natural family tracing back to a single ancestor. As both animal and plant (including green algae) lineages are represented within each natural family, the origin and architectural diversification of SR splicing factors likely predate the radiation of most, or even all, extant eukaryotes, depending on the position of the eukaryotic root. Indeed, the consensus is that plants and animals belong to distinct supergroups of eukaryotes that diverged very early on (Roger and Simpson, 2009; Walker et al., 2011; but see Stiller, 2007). An ancient origin of SR proteins would be in line with the last common ancestor of extant eukaryotes already featuring a sophisticated spliceosomal machinery (Collins and Penny, 2005), along with moderate to high spliceosomal intron density and maybe even AS (for review, see Roy and Irimia, 2009). Although likely, this possibility still remains to be formally demonstrated (Collins and Penny, 2005), as the lack of experimentally validated reference sequences complicates the in silico assessment of candidate SR splicing factors identified in “protist” proteomes (e.g. SR-like RRM domains from stramenopiles and alveolates in Supplemental Figs. S13 and S14 and also from amoebozoa, rhizaria, and metamonads in Supplemental Fig. S9). Ironically, the only “exotic” SR splicing factor described in the literature (the dual-RRM TcSR protein from Trypanosoma cruzi; Portal et al., 2003) was not included in our trees due to its RRM domains being too divergent from the HMM profile (E > 1e-5 for the T. brucei ortholog XP_846927). Nonetheless, its mere existence argues for an early emergence of SR proteins, since euglenozoa (the eukaryotic phylum including trypanosomes) are very distant from animals and fungi, yet less from plants (Roger and Simpson, 2009; Cavalier-Smith, 2010; Walker et al., 2011).
The RS, SCL, and RS2Z subfamilies of SR splicing factors are plant specific, but their RRM1 indicates that they belong to the same radiation as other SR proteins. As suggested by their architecture, RS proteins are specifically related to other dual-RRM SR proteins, while our analyses further revealed that SCL proteins are orthologous to animal SRrp proteins such as SRSF10/SRp38, which acts as a general splicing repressor when dephosphorylated (Shin et al., 2005). Obviously, this relationship opens interesting avenues for the investigation of their function in plant cells. In contrast, the exact affiliation of RS2Z proteins could not be established with confidence. Compared with other ZnK-like SR splicing factors, their RRM is quite different, which contributes to their unclear phylogenetic position.
Our study further indicates that the plant-specific SR45 did not originate independently from the three other SR protein families and confirms that it is orthologous to animal RNPS1, as previously suggested by sequence similarity (Wang and Brendel, 2004; Koroleva et al., 2009; Zhang and Mount, 2009). Whereas some authors question whether SR45 can be considered as a prototypical SR protein (Zhang and Mount, 2009), which led it to be excluded from the recent nomenclatural revision (Barta et al., 2010), our findings show that SR45 and RNPS1 define a fourth natural family within the SR radiation.
Although the animal SRSF11/SRp54 is currently considered as a prototypical SR protein (Cowper et al., 2001), it could not be included in our phylogenetic analyses due to its divergent RRM (E > 1e-3 for the human ortholog ENSG00000116754), which prevented any conclusion about its evolutionary history (Cowper et al., 2001; Bourgeois et al., 2004). Consistent with our findings, SRSF11/SRp54 has been described earlier as the most divergent family member, stimulating exon skipping and having significant roles in splicing repression (Zhang and Wu, 1996; Wu et al., 2006; Lin and Fu, 2007). Therefore, the cases of SR45 and SRSF11/SRp54 both illustrate the fact that even if protein nomenclature should ideally reflect evolutionary relationships, incongruences between functional and phylogenetic criteria are not uncommon.
Our study also sheds light on the origin of the internal RRM (RRM2) of dual-RRM SR proteins. ASF-like RRM2 and hnRNP-M RRM domains are closely related and share the presence of a SWQDLKD motif. Initially identified in human spliceosomes (Rappsilber et al., 2002), hnRNP-M acts as a splicing regulator in animals (Hovhannisyan and Carstens, 2007). Significantly, the RRM2 domain of green plant ASF-like proteins (SR subfamily) retains the SWQDLKD motif in spite of an overall sequence divergence that hindered its phylogenetic analysis (E > 1e-5 versus E < 1e-20 for the RRM1). This conservation indicates that it might ensure similar functions and molecular interactions. In contrast, the origin of the RRM2 of plant-specific RS proteins remains uncertain, even though its sequence is more canonical (E ∼ 1e-15, similar to the RRM1). We hypothesize that it arose from a duplication of the corresponding RRM1 in a common ancestor of red algae and green plants before undergoing extensive sequence divergence.
Although our scenario accounts for the main SR architectures without postulating evolutionary convergence, exceptions are very likely to exist. Hence, the genome of the slime mold Dictyostelium discoideum (amoebozoa) contains an RNA-binding protein (accession no. EAL67423) that is structurally very similar to plant-specific RS2Z proteins. On the other hand, some lineages have secondarily reduced their complement of SR splicing factors, such as the thermoacidophilic red alga Cyanidioschizon merolae (two SR proteins; Table II) and the budding yeast Saccharomyces cerevisiae (no SR protein). In the latter case, this is associated with a low complexity of the splicing machinery owing to genes with reduced intron density (Aravind et al., 2000). Whereas A. fumigatus appears to be similarly devoid of SR splicing factors, other fungi contain several proteins belonging to the SR radiation. This is the case for the fission yeast Schizosaccharomyces pombe (Table II; Käufer and Potashkin, 2000), which features one single-RRM SR protein (Srp1; accession no. NP_596398) unexpectedly showing some affinity to dual-RRM SR proteins (Supplemental Figs. S5, S8, and S9) and one true dual-RRM SR protein (Srp2; accession no. NP_594570), of which both RRM1 and RRM2 are unambiguously orthologous to the corresponding domains of SRSF5-6-4/Srp40-50-75 proteins (Supplemental Figs. S13, S14, S17, and S18). Furthermore, S. pombe and a number of other fungi include a clear ortholog of RNPS1/SR45 (Supplemental Figs. S15 and S16). Concerning the candidate SR protein Npl3 found in S. cerevisiae (accession no. NP_984279) and related yeast species (Kress et al., 2008), it is present in our trees but does not appear to be part of genuine SR splicing factors (Supplemental Figs. S5–S9).
To summarize, our comprehensive study provides a sound evolutionary framework for the classification of SR splicing factors. It suggests that the four main architectures derive from a single ancestral protein and either robustly confirms or reveals that some SR proteins thought to be plant specific are actually orthologous to splicing factors or repressors already described in animals. Along with the proximity between hnRNP-M and the RRM2 of dual-RRM SR proteins, these relationships will help to generate functional hypotheses. Finally, the lack of functional data on species other than animals and land plants somewhat precluded harnessing the full potential of our otherwise broadly sampled analyses. In our opinion, this calls for functional studies on exotic model species, such as so-called “algae” that are scattered across the eukaryotic tree of life.
MATERIALS AND METHODS
For the sake of space, these are abridged methods only. For a detailed description of the computational procedures, see Supplemental Text S1.
Data Set Assembly
To assemble the original data set, complete proteomes were downloaded from NCBI, Department of Energy Joint Genome Institute, The Institute for Genomic Research (now J. Craig Venter Institute), and specific project FTP servers. NCBI RefSeq release 26 (Pruitt et al., 2007) and SMART 6 (Letunic et al., 2009) databases were mined as additional resources for an enlarged data set. RRM and ZnK domains were predicted using HMMER (http://hmmer.janelia.org/; Durbin et al., 1998) and broad HMM profiles computed from their respective PFAM multiple alignment (pfam00076 and pfam00098; Finn et al., 2010). RRM domains with an E < 1e-10 were extracted and clustered on similarity using BLAST scores (Altschul et al., 1997) to yield two data sets suitable for phylogenetic analyses. Briefly, for each cluster, the RRM domain showing the highest average similarity with noncluster RRMs was selected as the most slowly evolving representative of the cluster (Roure et al., 2007). In parallel, the corresponding nonredundant set of RRM-containing proteins was assembled to allow for full-length analyses (e.g. compositional profiling). Smaller data sets (both RRM and full length) were derived from the original data set by focusing on three SR-associated subtrees of RRM1 and RRM2 domains. These SR-associated data sets were used to study the subsequent diversification of SR splicing factors and to design a determination key for SR families and subfamilies. Finally, a curated inventory of SR proteins was assembled, starting anew from 20 raw proteomes using HMM profiles optimized for SR-associated RRM domains (for HMM profiles and for accession numbers and sequences, see Supplemental Data Sets S1 and S2).
Alignments
For the two large data sets, RRM domains were aligned using a constrained HMM-based approach with HMMER, followed by visual inspection to ensure that alignments (1,266 or 1,831 sequences × 72 amino acids) were acceptable for large-scale analysis. For refined analyses of SR-associated domains (RRM1, 285–434 RRMs; SR45/RNPS1, 42 RRMs; RRM2, 349 RRMs), selected RRMs were realigned on the same HMM profile but this time allowing for sequence-specific insertions, which resulted in wider alignments (82–93 amino acids). These alignments were cursorily optimized by hand using ed (MUST software package; Philippe, 1993). For RRM logos, slowly evolving domains belonging to each of the newly defined SR subfamilies were separately realigned by hand using the secondary structure as a guide. SR subfamily alignments were then merged by manually aligning their consensus sequences (again with ed, which supports consensus-based alignment). This step yielded a single structural alignment of the slowly evolving RRM1 and RRM2 domains of all SR subfamilies. Finally, for the curated inventory, baba (also from the MUST package) was used to align each RRM domain to the most similar subfamily consensus sequence present in the high-quality structural alignment. Before proceeding to phylogenetic inference, these instrumental consensus sequences were of course removed. All alignments are available in FASTA format as shown in Supplemental Data Set S4.
Phylogenetic Analyses
The original data set (1,266 × 72 amino acids) was analyzed by maximum parsimony using PAUP* (Swofford, 2002), by ML using either RAxML (Stamatakis et al., 2005) or TreeFinder (Jobb et al., 2004) with both WAG (Whelan and Goldman, 2001) and LG+F (Le and Gascuel, 2008) models, and by Bayesian inference using PhyloBayes with the CAT model (Lartillot and Philippe, 2004; Lartillot et al., 2009). In probabilistic settings, rate heterogeneity was modeled using a Γ distribution with four categories (Γ4; Yang, 1993). In all cases but Bayesian inference, robustness was assessed by generating and analyzing 100 bootstrap replicates (Felsenstein, 1985) using seqboot and consense (PHYLIP software package; Felsenstein, 2005). Because of its large size (1,831 × 72 amino acids), the enlarged data set was only analyzed using RAxML and the WAG+Γ4 model. In contrast, all SR-associated data sets were analyzed using both RAxML and TreeFinder with the same model, including variants from which fast-evolving sequences had been excluded. Candidate RRM domains for the curated inventory were further analyzed using both RAxML with the LG+F+Γ4 model and PhyloBayes with the CAT+Γ4 model. All trees were drawn using treeplot (also from the MUST package) and automatically annotated using the KOG database (Tatusov et al., 2003) and reference RRM-containing proteins, among other sources of information.
Sequence Logos
Logos (Schneider and Stephens, 1990) for the RRM1 and RRM2 domains of each SR subfamily and for prokaryotic RRMs were computed with WebLogo (Crooks et al., 2004) from the separate structural alignments optimized by hand using the MUST ed. ZnK logos were computed similarly except that no alignment was required.
Full-Length Analyses
RRM-containing proteins were tagged as “putative SR” when featuring at least one RSRS or SRSR quadripeptide (Boucher et al., 2001). In Figure 1C, RS/SR dipeptide counts are nonoverlapping, as generally expected. This contrasts with the unbiased compositional profiling of unaligned full-length SR proteins, where we tracked the density of all possible overlapping words of one to three amino acids in a sliding window of 24 amino acids. Compositional analyses were carried out using custom software, while conservation analyses of aligned SR subfamilies further required ClustalW (Thompson et al., 1994) and plotcon (EMBOSS software package; Rice et al., 2000). In the curated inventory, candidate SR protein architectures were determined by automatically querying the NCBI Conserved Domain Database Web server (Marchler-Bauer et al., 2009).
Other Analyses
Statistical analyses (e.g. F tests in Fig. 1B) were performed using the R statistical software (R Development Core Team, 2010). All automation relied on Perl and shell scripting. The corresponding programs are freely available upon request to D.B.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Scheme of the analysis pipeline.
Supplemental Figure S2. Optimization of clustering parameters for RRM domains.
Supplemental Figure S3. Evaluation of the selection algorithm for cluster representatives.
Supplemental Figure S4. Color key for KOG annotation.
Supplemental Figure S5. Maximum parsimony tree of representative RRM domains.
Supplemental Figure S6. RAxML tree of representative RRM domains obtained under the WAG+Γ4 model.
Supplemental Figure S7. TreeFinder tree of representative RRM domains obtained under the WAG+Γ4 model.
Supplemental Figure S8. RAxML tree of representative RRM domains obtained under the LG+F+Γ4 model.
Supplemental Figure S9. RAxML tree of the enlarged RRM data set obtained under the WAG+Γ4 model.
Supplemental Figure S10. Qualitative consensus of phylogenetic analyses of representative RRM domains.
Supplemental Figure S11. Mutual affinities of SR protein RRM domains in the five large trees.
Supplemental Figure S12. RAxML tree of the largest RRM clusters obtained under the LG+F+Γ4 model.
Supplemental Figure S13. Exhaustive RAxML tree of SR-associated RRM1 domains (WAG+Γ4 model).
Supplemental Figure S14. Exhaustive TreeFinder tree of SR-associated RRM1 domains (WAG+Γ4 model).
Supplemental Figure S15. RAxML tree of RRM1 domains from RNPS1-like proteins (WAG+Γ4 model).
Supplemental Figure S16. TreeFinder tree of RRM1 domains from RNPS1-like proteins (WAG+Γ4 model).
Supplemental Figure S17. RAxML tree of RRM2 domains from dual-RRM SR proteins (WAG+Γ4 model).
Supplemental Figure S18. TreeFinder tree of RRM2 domains from dual-RRM SR proteins (WAG+Γ4 model).
Supplemental Figure S19. RAxML tree of slowly evolving SR-associated RRM1 domains (WAG+Γ4 model).
Supplemental Figure S20. TreeFinder tree of slowly evolving SR-associated RRM1 domains (WAG+Γ4 model).
Supplemental Figure S21. Sequence conservation in the first RRM domain of SR proteins.
Supplemental Figure S22. Sequence conservation across prokaryotic RRM domains.
Supplemental Figure S23. Sequence conservation across ZnK domains of SR proteins.
Supplemental Figure S24. Distributions of relevant compositional features within SR subfamilies.
Supplemental Figure S25. Architecture, conservation, and compositional profile of SR subfamilies.
Supplemental Figure S26. RAxML RRM tree of candidate SR proteins from selected proteomes (WAG+Γ4 model).
Supplemental Figure S27. TreeFinder RRM tree of candidate SR proteins from selected proteomes (WAG+Γ4 model).
Supplemental Figure S28. RAxML tree of candidate SR proteins from selected proteomes (LG+F+Γ4 model).
Supplemental Figure S29. PhyloBayes tree of candidate SR proteins from selected proteomes (CAT+Γ4 model).
Supplemental Table S1. Taxonomic distribution of RRM-containing proteins.
Supplemental Table S2. Hand-curated corpus of RRM-containing proteins collected in the primary literature.
Supplemental Table S3. Qualitative comparison of the five large RRM domain trees.
Supplemental Table S4. Summary of the groupings observed for SR protein RRM1 domains.
Supplemental Table S5. Compositional features specific to one or more SR subfamilies.
Supplemental Table S6. Determination key for SR families and subfamilies.
Supplemental Table S7. Statistics for the curated inventory of SR proteins in 20 selected proteomes.
Supplemental Text S1. Detailed description of computational procedures.
Supplemental Data Set S1. HMM profiles for SR-associated RRM domains.
Supplemental Data Set S2. Accession numbers and sequences of SR proteins identified in the inventory.
Supplemental Data Set S3. Architectures and amino acid word densities of candidate SR proteins from selected proteomes.
Supplemental Data Set S4. Alignments in FASTA format.
Supplementary Material
Acknowledgments
T. Druet and P. Geurts are thanked for advice on statistical tests and optimization of clustering parameters, respectively.
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman V, Koonin EV, Aravind L. (2002) Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic Acids Res 30: 1427–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Watanabe H, Lipman DJ, Koonin EV. (2000) Lineage-specific loss and divergence of functionally linked genes in eukaryotes. Proc Natl Acad Sci USA 97: 11319–11324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa-Morais NL, Carmo-Fonseca M, Aparício S. (2006) Systematic genome-wide annotation of spliceosomal proteins reveals differential gene family expansion. Genome Res 16: 66–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barta A, Kalyna M, Lorković ZJ. (2008) Plant SR proteins and their functions. Curr Top Microbiol Immunol 326: 83–102 [DOI] [PubMed] [Google Scholar]
- Barta A, Kalyna M, Reddy AS. (2010) Implementing a rational and consistent nomenclature for serine/arginine-rich protein splicing factors (SR proteins) in plants. Plant Cell 22: 2926–2929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Kumar S, Krainer AR. (1993) Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res 21: 5803–5816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher L, Ouzounis CA, Enright AJ, Blencowe BJ. (2001) A genome-wide survey of RS domain proteins. RNA 7: 1693–1701 [PMC free article] [PubMed] [Google Scholar]
- Bourgeois CF, Lejeune F, Stévenin J. (2004) Broad specificity of SR (serine/arginine) proteins in the regulation of alternative splicing of pre-messenger RNA. Prog Nucleic Acid Res Mol Biol 78: 37–88 [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. (2010) Kingdoms Protozoa and Chromista and the eozoan root of the eukaryotic tree. Biol Lett 6: 342–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaloc Y, Bourgeois CF, Kister L, Stévenin J. (1999) The splicing factors 9G8 and SRp20 transactivate splicing through different and specific enhancers. RNA 5: 468–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaloc Y, Popielarz M, Fuchs JP, Gattoni R, Stévenin J. (1994) Characterization and cloning of the human splicing factor 9G8: a novel 35 kDa factor of the serine/arginine protein family. EMBO J 13: 2639–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HS, Howe GA. (2009) A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell 21: 131–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins L, Penny D. (2005) Complex spliceosomal organization ancestral to extant eukaryotes. Mol Biol Evol 22: 1053–1066 [DOI] [PubMed] [Google Scholar]
- Cowper AE, Cáceres JF, Mayeda A, Screaton GR. (2001) Serine-arginine (SR) protein-like factors that antagonize authentic SR proteins and regulate alternative splicing. J Biol Chem 276: 48908–48914 [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. (2004) WebLogo: a sequence logo generator. Genome Res 14: 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delsuc F, Brinkmann H, Philippe H. (2005) Phylogenomics and the reconstruction of the tree of life. Nat Rev Genet 6: 361–375 [DOI] [PubMed] [Google Scholar]
- Durbin R, Eddy S, Krogh A, Mitchinson G. (1998) Biological Sequence Analysis: Probabilistic Models of Proteins and Nucleic Acids. Cambridge University Press, Cambridge, UK [Google Scholar]
- Embley TM, Martin W. (2006) Eukaryotic evolution, changes and challenges. Nature 440: 623–630 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (2005) PHYLIP (Phylogeny Inference Package) Version 3.6. Department of Genome Sciences, University of Washington, Seattle [Google Scholar]
- Feng Y, Valley MT, Lazar J, Yang AL, Bronson RT, Firestein S, Coetzee WA, Manley JL. (2009) SRp38 regulates alternative splicing and is required for Ca(2+) handling in the embryonic heart. Dev Cell 16: 528–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filichkin SA, Priest HD, Givan SA, Shen R, Bryant DW, Fox SE, Wong WK, Mockler TC. (2010) Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res 20: 45–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. (2010) The Pfam protein families database. Nucleic Acids Res 38: D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forterre P. (2011) A new fusion hypothesis for the origin of Eukarya: better than previous ones, but probably also wrong. Res Microbiol 162: 77–91 [DOI] [PubMed] [Google Scholar]
- Forterre P, Philippe H. (1999) Where is the root of the universal tree of life? Bioessays 21: 871–879 [DOI] [PubMed] [Google Scholar]
- Fukami-Kobayashi K, Tomoda S, Go M. (1993) Evolutionary clustering and functional similarity of RNA-binding proteins. FEBS Lett 335: 289–293 [DOI] [PubMed] [Google Scholar]
- Golovkin M, Reddy AS. (1998) The plant U1 small nuclear ribonucleoprotein particle 70K protein interacts with two novel serine/arginine-rich proteins. Plant Cell 10: 1637–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes C, Iakoucheva LM. (2006) Serine/arginine-rich splicing factors belong to a class of intrinsically disordered proteins. Nucleic Acids Res 34: 305–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovhannisyan RH, Carstens RP. (2007) Heterogeneous ribonucleoprotein m is a splicing regulatory protein that can enhance or silence splicing of alternatively spliced exons. J Biol Chem 282: 36265–36274 [DOI] [PubMed] [Google Scholar]
- Jobb G, von Haeseler A, Strimmer K. (2004) TREEFINDER: a powerful graphical analysis environment for molecular phylogenetics. BMC Evol Biol 4: 18. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kalyna M, Lopato S, Voronin V, Barta A. (2006) Evolutionary conservation and regulation of particular alternative splicing events in plant SR proteins. Nucleic Acids Res 34: 4395–4405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käufer NF, Potashkin J. (2000) Analysis of the splicing machinery in fission yeast: a comparison with budding yeast and mammals. Nucleic Acids Res 28: 3003–3010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren H, Lev-Maor G, Ast G. (2010) Alternative splicing and evolution: diversification, exon definition and function. Nat Rev Genet 11: 345–355 [DOI] [PubMed] [Google Scholar]
- Koroleva OA, Calder G, Pendle AF, Kim SH, Lewandowska D, Simpson CG, Jones IM, Brown JW, Shaw PJ. (2009) Dynamic behavior of Arabidopsis eIF4A-III, putative core protein of exon junction complex: fast relocation to nucleolus and splicing speckles under hypoxia. Plant Cell 21: 1592–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress TL, Krogan NJ, Guthrie C. (2008) A single SR-like protein, Npl3, promotes pre-mRNA splicing in budding yeast. Mol Cell 32: 727–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurland CG, Collins LJ, Penny D. (2006) Genomics and the irreducible nature of eukaryote cells. Science 312: 1011–1014 [DOI] [PubMed] [Google Scholar]
- Labadorf A, Link A, Rogers MF, Thomas J, Reddy AS, Ben-Hur A. (2010) Genome-wide analysis of alternative splicing in Chlamydomonas reinhardtii. BMC Genomics 11: 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot N, Lepage T, Blanquart S. (2009) PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics 25: 2286–2288 [DOI] [PubMed] [Google Scholar]
- Lartillot N, Philippe H. (2004) A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol Biol Evol 21: 1095–1109 [DOI] [PubMed] [Google Scholar]
- Le SQ, Gascuel O. (2008) An improved general amino acid replacement matrix. Mol Biol Evol 25: 1307–1320 [DOI] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. (2009) SMART 6: recent updates and new developments. Nucleic Acids Res 37: D229–D232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Manley JL. (2005) Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell 122: 365–378 [DOI] [PubMed] [Google Scholar]
- Lin S, Fu XD. (2007) SR proteins and related factors in alternative splicing. Adv Exp Med Biol 623: 107–122 [DOI] [PubMed] [Google Scholar]
- Long JC, Caceres JF. (2009) The SR protein family of splicing factors: master regulators of gene expression. Biochem J 417: 15–27 [DOI] [PubMed] [Google Scholar]
- Longman D, Johnstone IL, Cáceres JF. (2000) Functional characterization of SR and SR-related genes in Caenorhabditis elegans. EMBO J 19: 1625–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loomis RJ, Naoe Y, Parker JB, Savic V, Bozovsky MR, Macfarlan T, Manley JL, Chakravarti D. (2009) Chromatin binding of SRp20 and ASF/SF2 and dissociation from mitotic chromosomes is modulated by histone H3 serine 10 phosphorylation. Mol Cell 33: 450–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopato S, Forstner C, Kalyna M, Hilscher J, Langhammer U, Indrapichate K, Lorković ZJ, Barta A. (2002) Network of interactions of a novel plant-specific Arg/Ser-rich protein, atRSZ33, with atSC35-like splicing factors. J Biol Chem 277: 39989–39998 [DOI] [PubMed] [Google Scholar]
- Lorković ZJ, Barta A. (2002) Genome analysis: RNA recognition motif (RRM) and K homology (KH) domain RNA-binding proteins from the flowering plant Arabidopsis thaliana. Nucleic Acids Res 30: 623–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunde BM, Moore C, Varani G. (2007) RNA-binding proteins: modular design for efficient function. Nat Rev Mol Cell Biol 8: 479–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley JL, Krainer AR. (2010) A rational nomenclature for serine/arginine-rich protein splicing factors (SR proteins). Genes Dev 24: 1073–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, et al. (2009) CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res 37: D205–D210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maris C, Dominguez C, Allain FH. (2005) The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J 272: 2118–2131 [DOI] [PubMed] [Google Scholar]
- Maruyama K, Sato N, Ohta N. (1999) Conservation of structure and cold-regulation of RNA-binding proteins in cyanobacteria: probable convergent evolution with eukaryotic glycine-rich RNA-binding proteins. Nucleic Acids Res 27: 2029–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. (2008) Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40: 1413–1415 [DOI] [PubMed] [Google Scholar]
- Philippe H. (1993) MUST, a computer package of Management Utilities for Sequences and Trees. Nucleic Acids Res 21: 5264–5272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H, Delsuc F, Brinkmann H, Lartillot N. (2005) Phylogenomics. Annu Rev Ecol Evol Syst 36: 541–562 [Google Scholar]
- Plass M, Agirre E, Reyes D, Camara F, Eyras E. (2008) Co-evolution of the branch site and SR proteins in eukaryotes. Trends Genet 24: 590–594 [DOI] [PubMed] [Google Scholar]
- Poole AM. (2006) Getting from an RNA world to modern cells just got a little easier. Bioessays 28: 105–108 [DOI] [PubMed] [Google Scholar]
- Portal D, Espinosa JM, Lobo GS, Kadener S, Pereira CA, De La Mata M, Tang Z, Lin RJ, Kornblihtt AR, Baralle FE.et al (2003) An early ancestor in the evolution of splicing: a Trypanosoma cruzi serine-arginine-rich protein (TcSR) is functional in cis-splicing. Mol Biochem Parasitol 127: 37–46 [DOI] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR. (2007) NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res 35: D61–D65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2010) A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna [Google Scholar]
- Rappsilber J, Ryder U, Lamond AI, Mann M. (2002) Large-scale proteomic analysis of the human spliceosome. Genome Res 12: 1231–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy AS. (2007) Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annu Rev Plant Biol 58: 267–294 [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. (2000) EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16: 276–277 [DOI] [PubMed] [Google Scholar]
- Richardson DN, Rogers MF, Labadorf A, Ben-Hur A, Guo H, Paterson AH, Reddy AS. (2011) Comparative analysis of serine/arginine-rich proteins across 27 eukaryotes: insights into sub-family classification and extent of alternative splicing. PLoS ONE 6: e24542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roger AJ, Simpson AG. (2009) Evolution: revisiting the root of the eukaryote tree. Curr Biol 19: R165–R167 [DOI] [PubMed] [Google Scholar]
- Roure B, Rodriguez-Ezpeleta N, Philippe H. (2007) SCaFoS: a tool for selection, concatenation and fusion of sequences for phylogenomics. BMC Evol Biol (Suppl 1) 7: S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy SW, Irimia M. (2009) Splicing in the eukaryotic ancestor: form, function and dysfunction. Trends Ecol Evol 24: 447–455 [DOI] [PubMed] [Google Scholar]
- Schneider TD, Stephens RM. (1990) Sequence logos: a new way to display consensus sequences. Nucleic Acids Res 18: 6097–6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard PJ, Hertel KJ. (2009) The SR protein family. Genome Biol 10: 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin C, Feng Y, Manley JL. (2004) Dephosphorylated SRp38 acts as a splicing repressor in response to heat shock. Nature 427: 553–558 [DOI] [PubMed] [Google Scholar]
- Shin C, Kleiman FE, Manley JL. (2005) Multiple properties of the splicing repressor SRp38 distinguish it from typical SR proteins. Mol Cell Biol 25: 8334–8343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A, Ludwig T, Meier H. (2005) RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics 21: 456–463 [DOI] [PubMed] [Google Scholar]
- Stiller JW. (2007) Plastid endosymbiosis, genome evolution and the origin of green plants. Trends Plant Sci 12: 391–396 [DOI] [PubMed] [Google Scholar]
- Swofford DL. (2002) PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4. Sinauer Associates, Sunderland, MA [Google Scholar]
- Tanabe N, Kimura A, Yoshimura K, Shigeoka S. (2009) Plant-specific SR-related protein atSR45a interacts with spliceosomal proteins in plant nucleus. Plant Mol Biol 70: 241–252 [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al. (2003) The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Lührmann R. (2009) The spliceosome: design principles of a dynamic RNP machine. Cell 136: 701–718 [DOI] [PubMed] [Google Scholar]
- Walker G, Dorrell RG, Schlacht A, Dacks JB. (2011) Eukaryotic systematics: a user’s guide for cell biologists and parasitologists. Parasitology 138: 1638–1663 [DOI] [PubMed] [Google Scholar]
- Wang BB, Brendel V. (2004) The ASRG database: identification and survey of Arabidopsis thaliana genes involved in pre-mRNA splicing. Genome Biol 5: R102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan S, Goldman N. (2001) A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol 18: 691–699 [DOI] [PubMed] [Google Scholar]
- Wu D, Hugenholtz P, Mavromatis K, Pukall R, Dalin E, Ivanova NN, Kunin V, Goodwin L, Wu M, Tindall BJ, et al. (2009) A phylogeny-driven genomic encyclopaedia of bacteria and archaea. Nature 462: 1056–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JY, Kar A, Kuo D, Yu B, Havlioglu N. (2006) SRp54 (SFRS11), a regulator for tau exon 10 alternative splicing identified by an expression cloning strategy. Mol Cell Biol 26: 6739–6747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao R, Sun Y, Ding JH, Lin S, Rose DW, Rosenfeld MG, Fu XD, Li X. (2007) Splicing regulator SC35 is essential for genomic stability and cell proliferation during mammalian organogenesis. Mol Cell Biol 27: 5393–5402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. (1993) Maximum-likelihood estimation of phylogeny from DNA sequences when substitution rates differ over sites. Mol Biol Evol 10: 1396–1401 [DOI] [PubMed] [Google Scholar]
- Zahler AM, Lane WS, Stolk JA, Roth MB. (1992) SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev 6: 837–847 [DOI] [PubMed] [Google Scholar]
- Zhang G, Guo G, Hu X, Zhang Y, Li Q, Li R, Zhuang R, Lu Z, He Z, Fang X, et al. (2010) Deep RNA sequencing at single base-pair resolution reveals high complexity of the rice transcriptome. Genome Res 20: 646–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WJ, Wu JY. (1996) Functional properties of p54, a novel SR protein active in constitutive and alternative splicing. Mol Cell Biol 16: 5400–5408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XN, Mount SM. (2009) Two alternatively spliced isoforms of the Arabidopsis SR45 protein have distinct roles during normal plant development. Plant Physiol 150: 1450–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.