Abstract
X-ray microcomputed tomography (μCT) is an invaluable tool for visualizing plant root systems within their natural soil environment noninvasively. However, variations in the x-ray attenuation values of root material and the overlap in attenuation values between roots and soil caused by water and organic materials represent major challenges to data recovery. We report the development of automatic root segmentation methods and software that view μCT data as a sequence of images through which root objects appear to move as the x-y cross sections are traversed along the z axis of the image stack. Previous approaches have employed significant levels of user interaction and/or fixed criteria to distinguish root and nonroot material. RooTrak exploits multiple, local models of root appearance, each built while tracking a specific segment, to identify new root material. It requires minimal user interaction and is able to adapt to changing root density estimates. The model-guided search for root material arising from the adoption of a visual-tracking framework makes RooTrak less sensitive to the natural ambiguity of x-ray attenuation data. We demonstrate the utility of RooTrak using μCT scans of maize (Zea mays), wheat (Triticum aestivum), and tomato (Solanum lycopersicum) grown in a range of contrasting soil textures. Our results demonstrate that RooTrak can successfully extract a range of root architectures from the surrounding soil and promises to facilitate future root phenotyping efforts.
Roots represent the hidden half of plant biology (Waisel et al., 2002) since soil makes them difficult to image noninvasively. Understanding the development of roots and their interaction with the soil environment is vital to efforts toward food security. Roots provide anchorage and facilitate acquisition of water and nutrients from the soil. Growing roots explore their local environment to exploit those resources and as such they depend on a wide range of soil properties (Lynch, 1995). However, the complex relationship between roots and soil is not one sided. Plant roots have a large impact on the surrounding soil’s physical and biochemical properties. They stimulate the growth and activity of microorganisms and hence affect the regulation of soil organic matter decomposition (Gregory, 2006a). Furthermore, roots help develop the stability of soil aggregates and thus prevent soil erosion.
Many different methodologies have been used to study the development of roots. Popular techniques include the use of artificial growth media such as semitransparent nutrient agar (Clark et al., 1999; French et al., 2009) or gellan gum (Clark et al., 2011). While this overcomes the major problem of root visibility, it is not representative of a plants’ natural environment. The most common method used to study the root system of plants grown in soil is root washing (Smit et al., 2000; Gregory, 2006b). However, this often leads to the underestimation of fine roots through breakage, while information about the spatial distribution of roots is lost. Rhizotrons and minirhizotrons (Vamerali et al., 1999; Johnson et al., 2001) have also been used extensively but, while roots are grown in soil, rhizotrons artificially restrict the direction of root growth to two dimensions. In addition, observations are limited to the boundary surface, showing only a small fraction of the entire root system.
An alternative approach is the use of x-ray microcomputed tomography (μCT), a nondestructive imaging technique that can visualize the internal structure of opaque objects. A μCT scanner acquires a series of projections from different angles, measuring the attenuation of ionizing radiation passing through the examined object. These projections are used for the reconstruction of the CT data in three-dimensional (3D) space. Data values are expressed in Hounsfield units and are usually mapped to grayscale intensity values for visualization purposes (Mooney, 2002).
X-ray μCT allows observation not only of plant roots, but also of the surrounding soil matrix and associated pore volume. Many researchers have shown that μCT is an efficient tool with which to visualize plant roots growing in soil (Moran et al., 2000; Gregory et al., 2003; Jenneson et al., 2003; Tracy et al., 2010), yet there is still a lack of suitable methods to analyze the resulting data. The limiting obstacle has been the overlap in x-ray attenuation values of plant roots and the organic matter in soil, along with the variations in attenuation of the x-rays caused by water retained in roots and stored in the soil pores. Together these have made the automatic extraction of roots very difficult using current, global-thresholding-based image analysis approaches. This has been stressed by Heeraman et al. (1997), who used a classification approach on a per-voxel basis for the quantification of root material. Selected voxels were assigned to different classes representing the components contained in the scanned specimen, effectively building a single model of the x-ray attenuation data expected from each material. This model is then used to interpret the remainder of the x-ray data. The approach is, however, sensitive to noise and requires a clear differentiation between an object’s grayscale values and the values of its background. The same applies to threshold-based methods, which have been extensively used (Pierret et al., 1999; Lontoc-Roy et al., 2005, 2006; Kaestner et al., 2006; Perret et al., 2007). Thresholding alone rarely provides sufficiently accurate results. Thus, it is often used in combination with additional operations. For instance, Pierret et al. (1999) and Kaestner et al. (2006) both used different morphological operations for postprocessing to determine whether remaining voxels should belong to the final outcome or not. Lontoc-Roy et al. (2006) and Perret et al. (2007), in turn, performed an explicit connectivity check to identify voxels that have fallen within the threshold limits but are not part of root system. The resulting methods operate in a bottom-up fashion, starting with the raw data and moving toward root descriptions through a sequence of operations, each applied to the entire sample.
Bottom up and top down define different data or information processing strategies. In a top-down approach, a representation of an object is built from previously available data and is used to control detection of further objects of the same class. A top-down, or model-driven, process searches the incoming data for elements that match some model of the target object or class. In a bottom-up or data-driven approach, local image-based criteria are used to define and successively refine groups of pixels that are likely to belong to the same class. This is often implemented as a fixed pipeline comprising multiple processing steps. The RootTrace system of French et al. (2009), for example, takes a top-down approach to the analysis of color images of plant roots, while EZ-RHIZO (Armengaud et al., 2009) takes a bottom-up approach. The bottom-up approach is common in image analysis, but suffers several drawbacks. For example, errors tend to accumulate as each process introduces some inaccuracy. These are typically addressed by introducing interactive error correction tools. Manual correction, however, is often time consuming, as incorrectly classified objects are frequently small and usually distributed across the data set.
We present a new, top-down approach to the segmentation of μCT scans of plant roots growing in soil. We view the volumetric μCT data as a sequence of x-y cross-sectional images aligned along the z axis. As the image stack is traversed, root cross sections appear to move around the image, those movements reflecting the shape of the scanned root. Adopting this view of the data allows methods and strategies developed for use in the visual tracking of moving objects to be applied to the problem of tracing plant roots in three dimensions. The resulting method is less susceptible to classification errors arising from overlaps in attenuation values between roots and soil caused by the presence of water and organic material: The tracking framework focuses analysis on the root. The use of multiple, local models of root grayscale, in the form of histograms of the x-ray attenuation values found within root segments, makes the approach robust to variations in attenuation across the root system architecture. The technique requires only a single mouse click to initialize tracking by indicating, in the first image, the top of the root system.
Testing and validation of the approach are presented and the resulting software, RooTrak, is shown to work well on a range of root architectures from a variety of plant species crucially grown in a range of contrasting soil textures for the first time.
RESULTS AND DISCUSSION
Root Architecture Recovery Using Visual Tracking
The key feature of the proposed method is the level-set segmentation technique (Sethian, 1999). The proposed root tracking algorithm is based upon the level-set method (Sethian, 1999). The level-set method represents the boundary of a given shape as the intersection of some function Φx,y,t with the plane t = 0. Points on the x-y plane for which is negative lie inside the shape, while those with positive values are outside. Changes are made to the boundary curve, not by direct manipulation of the points lying on it, but by changing, or evolving the level-set function Φ. Level-set methods are widely used in image processing and analysis, where the goal is usually to evolve a level-set function until the associated boundary, or front, fits over some target feature(s) in the input image.
In our novel variation of the level-set method, the level-set function evolves on the basis of the Jensen-Shannon (JS) divergence (Lin, 1991). JS divergence provides a measure of the similarity of two data distributions, in this case two histograms of μCT density values. The first of these is a previously constructed model of the distribution of densities expected within a root segment. The second is the distribution found around the front created by a level-set function evolving to fit to an x-y slice of a μCT data volume. Pixels around this boundary are therefore required to be consistent with a model of the distribution of x-ray attenuation values expected from root material.
Once the level-set method has fitted a front to the current slice, identifying the boundary of a section through a root, attention turns to the next image. Since both the position and boundary shape of a root object can vary through the stack, the level-set function needs to evolve from the previous state to fit the new root segment. A new level-set function is derived from the previous one, and the level-set method applied again. This process is applied to each image, moving down the z axis of the stack, simultaneously identifying and tracking moving root sections and so tracing out the 3D root architecture. Root branching is handled by the level set’s ability to adapt to topological changes in the front. Interested readers desiring more details of the level-set method and the modifications made here will find a comprehensive description in the Supplemental Material S1, available online.
Updating the Root Appearance Model
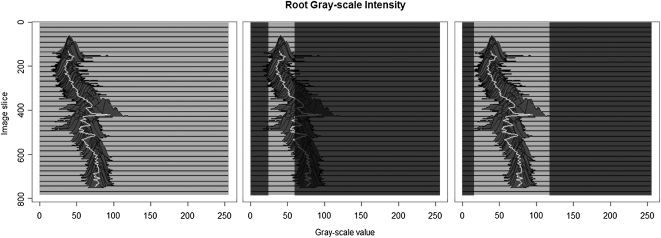
The JS divergence (Lin, 1991), on which the evolution of the level-set function is based, uses an estimate of the probability density function of the root x-ray attenuation values as reference. The easiest way of defining such a model is to select a representative set of voxels corresponding to root material before processing begins and use their grayscale intensity values to create a model that is employed throughout the segmentation. Unfortunately, this approach is not viable as the intensity values can change across the root system architecture (and along individual root branches) as the soil and root moisture content and soil organic mineralogy matrix changes. Figure 1 shows the intensity distribution of a single root obtained at different soil depths. Setting decision criteria based on the appearance of a root object in a single slice is unlikely to result in the entire root system being extracted. On the other hand, if criteria are based upon the minimum and maximum grayscale intensity present at any location of the root system, a wide range of nonroot material will be incorrectly marked as root material. This highlights why methods based on global information, such as thresholds or a single root appearance model (e.g. Heeraman et al., 1997) cannot extract roots accurately: It is necessary to update the root model while tracking root branches through the μCT volume. In addition, each object has also to be differentiated from others found within the same image slice.
Figure 1.
The left image shows histograms of the grayscale intensity of a single root as it appears in each slice of a μCT volume (maize). The middle image shows threshold boundaries based on the grayscale intensities of the first root object. If this criterion were applied the root would only be partially extracted. The right image shows the data that would be considered root if a decision criterion based on the minimum and maximum grayscale intensity of the sample were used. Other materials are likely to be extracted as well.
The root model used to drive the evolution of the interface is built from the grayscale values of those voxels identified as root material in the previous image. It is assumed that those values do not change excessively between two consecutive images, but vary smoothly through the stack (Fig. 1). A special case is the very first image, for which no previous root model exists. Here, the root model is taken from the pixels underlying the user-defined start points. The precise location of start points in the first image does not usually affect the overall segmentation, as long as they are each placed inside a root object.
Each object being tracked must be aware of its grayscale intensity distribution and therefore, to differentiate between multiple root objects, the classical two-pass connected component algorithm (Rosenfeld, 1970) is used to build an intensity distribution model for each root section every time a new image is loaded. By doing so, different root objects are distinguished and so are their interfaces. Thus it is possible to evolve the level-set function using different models for each root object.
Updating the root model is an inevitable step, yet it conceals potential problems. Noise or small areas of nonroot material might be included in the probability density distribution. These errors can accumulate and result in a model that is no longer an appropriate representation of the tracked root. Should this happen, the level-set method will segment the current slice incorrectly. This will manifest itself as a sudden change in the perceived shape of the root cross section. An additional test is therefore performed. The assumption is made that in normal situations the shape of a root object changes only slightly, if at all, between two consecutive images. At the high resolutions achieved when using μCT, we believe this holds; images are typically separated by <50 μm. A Fourier shape descriptor technique (Gonzalez and Woods, 2001) is adopted to compare root sections. Only when the shapes extracted from consecutive slices are considered similar is the root model update accepted, otherwise the tracker continues with the previous model. This is a conservative strategy; RooTrak will not perform a model update that may have caused a significant change in the perceived shape of the root.
Evaluation of RooTrak
Samples from different plant species grown in different soil textural types were scanned using a high-resolution x-ray μCT system (Nanotom, Phoenix x-ray, GE Measurement and Control Systems). The time taken to scan a sample depends on the system settings and operation mode, and in this case varied between approximately 15 min (fast scan mode—used for tomato [Solanum lycopersicum]) and approximately 60 min (normal scan mode—used for wheat [Triticum aestivum]). While fast scans typically require less time as the sample revolves continuously while scanning, the quality of the images produced decreases (through increased noise). The time taken to process the resulting CT data depends on image size, number of images, and amount of root material (i.e. approximately 15 min for the tomato grown in loamy sand and approximately 60 min for the wheat sample). The plants and soil types used in the experiment, as well as the scanner settings for imaging the samples are listed in Table I.
Table I. X-ray μCT scanner settings and image data properties.
Experimental set up: plant species, soil textural type, and x-ray μCT scanner settings. Plants were grown in an environmentally controlled growth room with 16-/8-h light cycle (*, greenhouse). All plants were imaged in soil with water content approximately at field capacity.
| Plant Species | Wheat | Wheat | Wheat | Maize* | Tomato | Tomato |
| Soil textural type | Sand | Sand | Sand | Loamy sand | Clay loam | Loamy sand |
| Voltage | 130 kV | 130 kV | 130 kV | 80 kV | 110 kV | 110 kV |
| Current | 90 μA | 90 μA | 110 μA | 160 μA | 180 μA | 180 μA |
| Filter | 1 mm aluminum | 1 mm aluminum | 1 mm aluminum | None | 0.1 mm copper | 0.1 mm copper |
| No. of projections | 1,440 | 1,440 | 1,440 | 1,200 | 1,200 | 1,200 |
| Resolution (per voxel) | 28.75 μm | 28.75 μm | 28.75 μm | 44.23 μm | 23.91 μm | 23.91 μm |
| Detector exposure time | 1,000 ms | 1,000 ms | 1,000 ms | 250 ms | 1,000 ms | 1,000 ms |
| Binning mode | 1 × 1 | 1 × 1 | 1 × 1 | 2 × 2 | 1 × 1 | 1 × 1 |
| Signal averaging | 4 (1 skip) | 4 (1 skip) | 4 (1 skip) | 6 (3 skip) | 1 (0 skip) | 1 (0 skip) |
| Images in stack after reconstruction | 1,800 | 1,800 | 2,000 | 771 | 1,313 | 1,698 |
| Image dimension after reconstruction (pixels) | 864 × 864 | 864 × 864 | 876 × 872 | 678 × 668 | 1,388 × 1,404 | 1,404 × 1,380 |
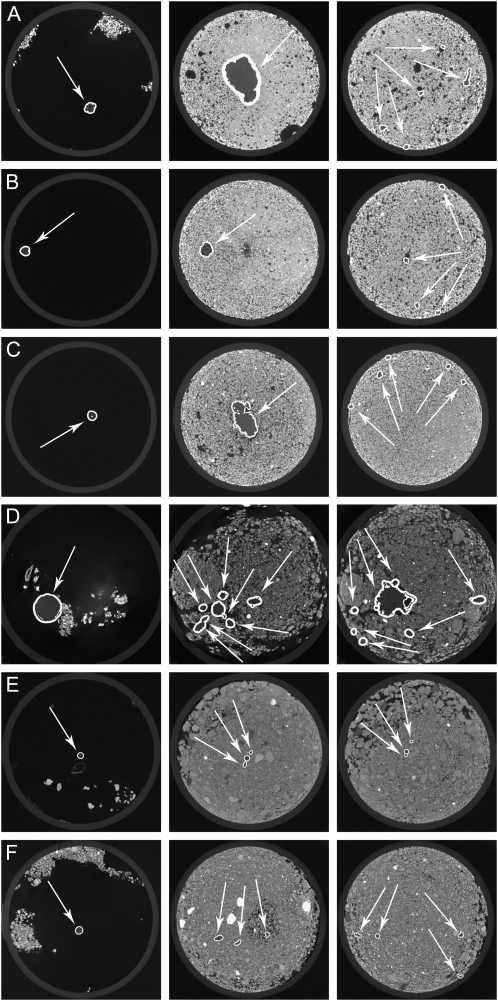
Figure 2 shows sample frames extracted from the μCT data summarized in Table I. Root material identified by RooTrak is highlighted, and the differences between soil types are clear from the surrounding intensity values. The root descriptions obtained by RooTrak are given as stacks of image segments. However, it is difficult to get a clear idea of the extracted root system by traversing the sequence of images. A volume-rendering technique, known as volume ray casting and based on the method described in Krüger and Westermann (2003) was therefore implemented.
Figure 2.
Root sections recovered from using RooTrak. Highlighted regions represent identified plant roots. The very first image frame in each sequence shows the stem of the plant, shortly before entering the soil where the plant’s root system starts. A, Wheat in sand—frame: 10 (0.28 mm), 310 (8.91 mm), 610 (17.53 mm). B, Wheat in sand—frame: 10 (0.28 mm), 310 (8.91 mm), 610 (17.53 mm). C, Wheat in sand—frame: 10 (0.28 mm), 610 (17.53 mm), 1,210 (34.78 mm). D, Maize in loamy sand—frame: 10 (0.44 mm), 110 (4.84 mm), 210 (9.24 mm). E, Tomato in clay loam—frame: 10 (0.24 mm), 260 (6.24 mm), 510 (12.24 mm). F, Tomato in loamy sand—frame: 10 (0.24 mm), 260 (6.24 mm), 510 (12.24 mm).
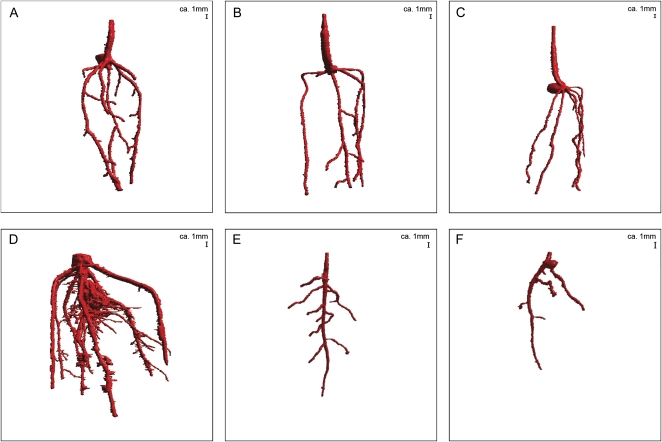
Figure 3 shows a visualization of the root system architecture of a variety of plant species in 3D space, using the above-mentioned volume-rendering technique. RooTrak is able to visualize the contrasting fibrous and herring-bone root systems of monocot and dicot species, respectively. It is worth noting that the soils used in this study are typical United Kingdom field soils and not artificial media such as washed sands that have been used in many previous studies. Importantly we observed no difference in RooTrak root segmentation efficiency depending on soil type, which might have been expected since soil water content, a previously reported limitation of this technique, is a function of soil texture. It should be noted that roots do not appear perfectly tubular all the time. This is due to the direction in which the cross section is taken and the complexity of branching structures, but also because of the nature of data representation. A voxel can sample two or more different components and as such the resulting intensity value is an average of all included values. The resolution at which data are captured or displayed also has a great influence, be it the two-dimensional image stack of μCT data or the 3D visualization of an object. Down-sampled data usually looks smoother than high-resolution data, as details disappear. Note that the quality of the rendered data can vary significantly between different visualization techniques, introducing visual artifacts or leaving out important details, yet the 3D structure and complexity of the extracted root system architecture is still captured. It should be stressed that extraction of the descriptions shown in Figure 3 each required only a single mouse click from the user.
Figure 3.
3D visualization using volume ray casting of data extracted by RooTrak. A, Wheat in sand (α = 0.06; β = 0.32; Supplementary Video S1). B, Wheat in sand (α = 0.64; β = 0.36; Supplementary Video S2). C, Wheat in sand (α = 0.64; β = 0.36; Supplementary Video S3). D, Maize in loamy sand (α = 0.92; β = 0.42; Supplementary Video S4). E, Tomato in clay loam (α = 0.61; β = 0.47; Supplementary Video S5). F, Tomato in loamy sand (α = 0.68; β = 0.45; Supplementary Video S6).
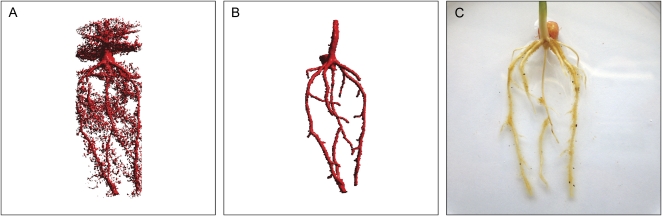
To determine the success of RooTrak, its output should be compared to that obtained from other methods. Unfortunately, none of the previously reported software tools discussed here have been made publicly available. However, to form a point of comparison for the proposed tracking method, global thresholding was applied to each of the samples shown in Figure 3. Threshold boundaries were selected manually (compare with Lontoc-Roy et al., 2005, 2006), the operator trying to include as much root material as possible while at the same time reducing the amount of nonroot material extracted. Global thresholding is a very basic operation and therefore, in addition, a connectivity constraint based on a 26 neighborhood (compare with Lontoc-Roy et al., 2006; Perret et al., 2007) was applied. We believe that together these operations form a common denominator of previous root extraction methods from μCT data, though they do not correspond exactly to any given published technique. Root voxels were counted to compare the volumes extracted by both methods. The results are listed in Table II. Figure 4 provides a visual representation of a wheat plant root system extracted using global thresholding with connectivity checking and RooTrak, alongside an image of the root system obtained by washing the plant free from the surrounding soil after the μCT scan. The result of global thresholding is typical: While the root architecture is present in the segmentation result, it is masked by a great many incorrectly labeled voxels. In Figure 4 these voxels may seem to be disconnected from the plant root system. This, however, comes from the rendering process; in reality all voxels are connected to each other due to the applied connectivity constraint.
Table II. Measured volume using global thresholding with 26-neighbor connectivity constraint and RooTrak.
Measured volume using global thresholding with a 26-neighbor connectivity constraint and RooTrak. The volume was calculated by counting the number of voxels and multiplying by the voxels’ size cubed.
| Plant Species | Thresholding + Connectivity (Volume) | RooTrak (Volume) |
| mm3 | ||
| Wheat | 573.69 | 120.88 |
| Wheat | 558.75 | 76.94 |
| Wheat | 693.07 | 147.53 |
| Maize | 3,600.64 | 378.37 |
| Tomato | 270.24 | 22.58 |
| Tomato | 836.29 | 33.92 |
Figure 4.
Results produced from the wheat scan by global thresholding with a 26-neighborhood connectivity constraint (A) and RooTrak (B). C, Image of washed root for comparison.
Extracting Quantitative Information about Root System Architecture Using RooTrak
Once the root system is successfully extracted from the surrounding soil, the resulting volumetric representation of root system architecture can be analyzed to quantify root characteristics. Extensive lists of measurable traits are presented in Iyer-Pascuzzi et al. (2010) and Clark et al. (2011). Some of these measures can be extracted directly from RooTrak segmentations and have been computed to obtain biologically meaningful parameters. Root volume can be estimated from a simple count of the number of root voxels, and gives an idea of the total mass of the root system. Root surface area can be calculated by representing the isosurface of the segmented root system as a mesh of triangles, using an algorithm known as Marching Cubes (Lorensen and Cline, 1987). The surface area is then determined by summing the areas of all triangles in the mesh. This approximates the area that is in direct contact with the soil environment. The convex hull of the root system is obtained using the QuickHull (Barber et al., 1996) algorithm and its volume estimated using Monte Carlo Integration (Rubinstein, 1981). The convex hull of a root system can be used to compare and distinguish root systems from different plants (Iyer-Pascuzzi et al., 2010). The depth of the root system is calculated by counting the number of vertical voxels between the first and the last voxel of the extracted root system. Using Welzl’s algorithm for the minimum enclosing circle (Welzl, 1991), it is possible to determine the root system’s maximum width, reflecting the maximum horizontal distance a root system traveled when exploring its environment for resources. In addition, the centroid of the root voxels is determined, which is the geometric center of an object and corresponds to its center of mass, if the mass per unit volume is constant throughout. Figure 5 shows some of these measurements applied to RooTrak’s segmentation of the root system of the wheat plant shown in Figure 3A.
Figure 5.
Visualization of quantified root characteristics on an extracted wheat root system. A, Convex hull. B, Maximum root system depth. C, Maximum root system width. D, Centroid and its vertical center line.
CONCLUSION
We have presented a new strategy for the extraction of root systems from x-ray μCT images. The RooTrak approach exploits visual-tracking techniques, employing multiple models of the appearance of root material in μCT data. Models constructed from root sections identified in one frame are used to seek root material in the next. The technique relies upon a novel combination of level-set tracking with the JS divergence. A key advantage of the level-set approach is its ability to deal appropriately with targets (in this case root sections) splitting and merging, while the JS test compares model and image data in a principled fashion. The resulting root extraction method requires minimal user interaction and is able to adapt to local variations in x-ray attenuation.
RooTrak was tested on a variety of plant species with monocot and dicot root architectures growing in different soil textural types that represent a large portion of all United Kingdom soil types. Nevertheless, the proposed local, adaptive tracking method (instead of global voxel classification) was shown to be suitable for this task. Our algorithm was able to extract both coarse and fine roots, limited only by the resolution of the scanner.
The size of the smallest possible root that can be extracted using RooTrak is primarily data dependent. If very fine roots are visible in the image stack, then they can reasonably be expected to be recovered by the presented tracking method. Thus, the smallest size depends on the resolution at which the sample is scanned. Modern x-ray μCT scanners have the capability of imaging data with a resolution <0.5 μm, thus making it theoretically possible to detect even root hairs. Such a high resolution, however, comes with the cost of a limited sample size, which otherwise would exceed the data volumes that can be handled by today’s technology. A cubic sample of 1-cm side length, scanned using a resolution of 0.5 μm, would consist of 20,0003 voxels, requiring several terabytes of storage, not to mention the great amount of time needed for processing.
Global thresholding with 26-neighbor connectivity was applied to the collected dataset and the resulting segmentations compared to those provided by RooTrak. Thresholding and connectivity constraints resulted in a much larger volume of the sample being considered root material, compared to the volume obtained by the proposed technique. The reason for this is that the variation in μCT data values associated with root material (Fig. 1) forces the user to specify that quite a wide range of gray values be considered root to avoid producing only a partial segmentation. When thresholds are set to encompass all values that arise from root material at some point in the data volume, they invariably capture nonroot voxels at other locations. This emphasizes the advantages of the top-down, adaptive segmentation strategy that arises from adopting a visual-tracking approach.
Image quality, such as variations in contrast and the amount of noise perturbing the image, plays an important role in the extraction of roots. There are several factors that can influence the quality of images. These include the composition of the sample (plant species, soil type), its condition (compaction level, soil moisture content), and the wide range of possible scanning settings (x-ray energy, number of projection images). The success of the method is not guaranteed, but our testing has demonstrated, as with most image analysis procedures, that the better the image quality, the more reliable the (root) segmentation process.
In summary, our results demonstrate that RooTrak can successfully, and with minimal user intervention, extract a range of root architectures from the surrounding soil. RooTrak supports the computation of a range of quantitative measures and promises to facilitate future root phenotyping for trait-based crop breeding efforts.
MATERIALS AND METHODS
Plant Growth
Wheat (Triticum aestivum), maize (Zea mays), and tomato (Solanum lycopersicum) were germinated in petri dishes and after 2 d planted in plastic columns filled with sand, loamy sand, or clay loam sieved to <2 mm. All plants grew in environmental controlled growth rooms with 16-/8 light cycle at a temperature of 26°C and were scanned 10 d after germination. The only exception is the maize plant used in the experiment. The maize grew in a greenhouse under natural photoperiods and was scanned 21 d after germination. The water status of the samples at the point of imaging was approximately at field capacity.
Imaging
All μCT data were acquired at the University of Nottingham using a Nanotom, Phoenix x-ray scanner. Scanning resolution varied between 23.91 and 44.23 μm, voltage from 80 to 130 kV, and current from 90 to 180 μA. At least 1,200 projections were used in each case. Details of each scan are provided in Table I.
Access to RooTrak Software
The RooTrak software and sample data sets may be obtained from www.rootrak.net.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Material S1. Detailed description of the level-set method and its modification for visual tracking as implemented and used by the RooTrak software tool.
Supplemental Video S1. Video showing the segmentation of a wheat root from sand (Fig. 3A) using RooTrak.
Supplemental Video S2. Video showing the segmentation of a wheat root from sand (Fig. 3B) using RooTrak.
Supplemental Video S3. Video showing the segmentation of a wheat root from sand (Fig. 3C) using RooTrak.
Supplemental Video S4. Video showing the segmentation of a maize root from loamy sand (Fig. 3D) using RooTrak.
Supplemental Video S5. Video showing the segmentation of a tomato root from clay loam (Fig. 3E) using RooTrak.
Supplemental Video S6. Video showing the segmentation of a tomato root from loamy sand (Fig. 3F) using RooTrak.
Supplementary Material
References
- Armengaud P, Zambaux K, Hills A, Sulpice R, Pattison RJ, Blatt MR, Amtmann A. (2009) EZ-Rhizo: integrated software for the fast and accurate measurement of root system architecture. Plant J 57: 945–956 [DOI] [PubMed] [Google Scholar]
- Barber CB, Dobkin DP, Huhdanpaa H. (1996) The quickhull algorithm for convex hulls. ACM Trans Math Softw 22: 469–483 [Google Scholar]
- Clark L, Whalley W, Leigh R, Dexter A, Barraclough P. (1999) Evaluation of agar and agarose gels for studying mechanical impedance in rice roots. Plant Soil 207: 37–43 [Google Scholar]
- Clark RT, MacCurdy RB, Jung JK, Shaff JE, McCouch SR, Aneshansley DJ, Kochian LV. (2011) Three-dimensional root phenotyping with a novel imaging and software platform. Plant Physiol 156: 455–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French A, Ubeda-Tomás S, Holman TJ, Bennett MJ, Pridmore T. (2009) High-throughput quantification of root growth using a novel image-analysis tool. Plant Physiol 150: 1784–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez RC, Woods RE. (2001) Digital Image Processing, Ed 2. Addison-Wesley Longman Publishing Co., Inc., Boston [Google Scholar]
- Gregory PJ. (2006a) Roots, rhizosphere and soil: the route to a better understanding of soil science? Eur J Soil Sci 57: 2–12 [Google Scholar]
- Gregory PJ. (2006b) Plant Roots: Their Growth, Activity, and Interaction with Soils. Blackwell Publishing, Oxford [Google Scholar]
- Gregory PJ, Hutchison DJ, Read DB, Jenneson PM, Gilboy WB, Morton EJ. (2003) Non-invasive imaging of roots with high resolution x-ray micro-tomography. Plant Soil 255: 351–359 [Google Scholar]
- Heeraman D, Hopmans J, Clausnitzer V. (1997) Three dimensional imaging of plant roots in situ with x-ray computed tomography. Plant Soil 189: 167–179 [Google Scholar]
- Iyer-Pascuzzi AS, Symonova O, Mileyko Y, Hao Y, Belcher H, Harer J, Weitz JS, Benfey PN. (2010) Imaging and analysis platform for automatic phenotyping and trait ranking of plant root systems. Plant Physiol 152: 1148–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenneson PM, Gilboy WB, Morton EJ, Gregory PJ. (2003) An x-ray micro-tomography system optimised for the low-dose study of living organisms. Appl Radiat Isot 58: 177–181 [DOI] [PubMed] [Google Scholar]
- Johnson MG, Tingey DT, Phillips DL, Storm MJ. (2001) Advancing fine root research with minirhizotrons. Environ Exp Bot 45: 263–289 [DOI] [PubMed] [Google Scholar]
- Kaestner A, Schneebeli M, Graf F. (2006) Visualizing three-dimensional root networks using computed tomography. Geoderma 136: 459–469 [Google Scholar]
- Krüger J, Westermann R. (2003) Acceleration techniques for gpu-based volume rendering. 14th IEEE Visualization 2003, pp 38 [Google Scholar]
- Lin J. (1991) Divergence measures based on the Shannon entropy. IEEE Trans Inf Theory 37: 145–151 [Google Scholar]
- Lontoc-Roy M, Dutilleul P, Prasher SO, Liwen H, Brouillet T, Smith DL. (2006) Advances in the acquisition and analysis of ct scan data to isolate a crop root system from the soil medium and quantify root system complexity in 3-d space. Geoderma 137: 231–241 [Google Scholar]
- Lontoc-Roy M, Dutilleul P, Prasher SO, Liwen H, Smith DL. (2005) Computed tomography scanning for three-dimensional imaging and complexity analysis of developing root systems. Can J Bot 83: 1434–1442 [Google Scholar]
- Lorensen WE, Cline HE. (1987) Marching cubes: a high resolution 3D surface construction algorithm. SIGGRAPH Computer Graphics 21: 163–169 [Google Scholar]
- Lynch J. (1995) Root architecture and plant productivity. Plant Physiol 109: 7–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney SJ. (2002) Three-dimensional visualization and quantification of soil macroporosity and water flow patterns using computed tomography. Soil Use Manage 18: 142–151 [Google Scholar]
- Moran CJ, Pierret A, Stevenson AW. (2000) X-ray absorption and phase contrast imaging to study the interplay between plant roots and soil structure. Plant Soil 223: 101–117 [Google Scholar]
- Perret J, Al-Belushi M, Deadman M. (2007) Non-destructive visualization and quantification of roots using computed tomography. Soil Biol Biochem 39: 391–399 [Google Scholar]
- Pierret A, Capowiez Y, Moran CJ, Kretzschmar A. (1999) X-ray computed tomography to quantify tree rooting spatial distributions. Geoderma 90: 307–326 [Google Scholar]
- Rosenfeld A. (1970) Connectivity in digital pictures. J ACM 17: 146–160 [Google Scholar]
- Rubinstein RY. (1981) Simulation and the Monte Carlo Methods. John Wiley and Sons, Inc., San Francisco [Google Scholar]
- Sethian JA. (1999) Level Set Methods and Fast Marching Methods: Evolving Interfaces in Computational Geometry, Fluid Mechanics, Computer Vision, and Materials Science (Cambridge ... on Applied and Computational Mathematics), Ed 2. Cambridge University Press, New York [Google Scholar]
- Smit AL, Bengough AG, Engels C, van Noordwijk M, Pellerin S, van de Geijn SC. (2000) Root Methods: A Handbook. Springer-Verlag, Berlin [Google Scholar]
- Tracy SR, Roberts JA, Black CR, McNeill A, Davidson R, Mooney SJ. (2010) The x-factor: visualizing undisturbed root architecture in soils using x-ray computed tomography. J Exp Bot 61: 311–313 [DOI] [PubMed] [Google Scholar]
- Vamerali T, Ganis A, Bona S, Mosca G. (1999) An approach to minirhizotron root image analysis. Plant Soil 217: 183–193 [Google Scholar]
- Waisel Y, Eshel A, Beekman T, Kafkafi U. (2002) Plant Roots: The Hidden Half, Ed 3. CRC Press, New York [Google Scholar]
- Welzl E. (1991) Smallest enclosing disks (balls and ellipsoids). In H. Maurer, ed, New Results and New Trends in Computer Science, Lecture Notes in Computer Science, Vol 555. Springer-Verlag, Berlin, pp 359–370 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.