Abstract
During leaf senescence, chlorophyll (Chl) is broken down to nonfluorescent chlorophyll catabolites (NCCs). These arise from intermediary fluorescent chlorophyll catabolites (FCCs) by an acid-catalyzed isomerization inside the vacuole. The chemical structures of NCCs from Arabidopsis (Arabidopsis thaliana) indicate the presence of an enzyme activity that demethylates the C132-carboxymethyl group present at the isocyclic ring of Chl. Here, we identified this activity as methylesterase family member 16 (MES16; At4g16690). During senescence, mes16 leaves exhibited a strong ultraviolet-excitable fluorescence, which resulted from large amounts of different FCCs accumulating in the mutants. As confirmed by mass spectrometry, these FCCs had an intact carboxymethyl group, which slowed down their isomerization to respective NCCs. Like a homologous protein cloned from radish (Raphanus sativus) and named pheophorbidase, MES16 catalyzed the demethylation of pheophorbide, an early intermediate of Chl breakdown, in vitro, but MES16 also demethylated an FCC. To determine the in vivo substrate of MES16, we analyzed pheophorbide a oxygenase1 (pao1), which is deficient in pheophorbide catabolism and accumulates pheophorbide in the chloroplast, and a mes16pao1 double mutant. In the pao1 background, we additionally mistargeted MES16 to the chloroplast. Normally, MES16 localizes to the cytosol, as shown by analysis of a MES16-green fluorescent protein fusion. Analysis of the accumulating pigments in these lines revealed that pheophorbide is only accessible for demethylation when MES16 is targeted to the chloroplast. Together, these data demonstrate that MES16 is an integral component of Chl breakdown in Arabidopsis and specifically demethylates Chl catabolites at the level of FCCs in the cytosol.
The degradation of chlorophyll (Chl) is a catabolic process that massively occurs during leaf senescence and fruit ripening (Hörtensteiner and Kräutler, 2011). It aims at the detoxification of this potentially phototoxic pigment; therefore, Chl breakdown is seen as a prerequisite for the degradation of Chl-binding proteins, which are an important nitrogen source for recycling from leaves to storage organs or seeds. This view is supported by the analysis of stay-green (sgr) mutants that are affected in certain steps of Chl breakdown; during senescence, leaves of these mutants retain large quantities of Chl-apoprotein complexes (Hilditch et al., 1989; Kusaba et al., 2007; Park et al., 2007; Morita et al., 2009; Schelbert et al., 2009). The fate of Chl was enigmatic for a long time, but since the identification of a first nonfluorescent chlorophyll catabolite (NCC) as an (end) product of Chl breakdown from barley (Hordeum vulgare; Kräutler et al., 1991), the breakdown pathway has largely been resolved (Kräutler and Hörtensteiner, 2006; Hörtensteiner and Kräutler, 2011).
The first part of the pathway is localized in senescing chloroplasts. It is composed of a series of reactions that are common in higher plants and that lead to the formation of a primary fluorescent chlorophyll catabolite (pFCC; Mühlecker et al., 1997). Thereby, first the central magnesium (Mg) atom is removed from Chl by a heat-stable, low-Mr compound, termed metal-chelating substance (Suzuki et al., 2005), whose molecular nature is so far unknown. The product of Mg dechelation, pheophytin, is then hydrolyzed to pheophorbide (Pheide) and phytol by pheophytinase (PPH; Schelbert et al., 2009). For a long time, phytol removal was thought to be catalyzed by chlorophyllase (Takamiya et al., 2000), but recent analysis of single and double knockout mutants in Arabidopsis (Arabidopsis thaliana) demonstrated chlorophyllase to be dispensable for Chl breakdown during leaf senescence (Schenk et al., 2007). Instead, PPH specifically dephytylates pheophytin, but not Chl, and pph mutants are blocked in Chl breakdown and as a consequence exhibit a stay-green phenotype (Schelbert et al., 2009). After the formation of Pheide, the porphyrin macrocycle is opened by the action of Pheophorbide a Oxygenase (PAO), a Rieske-type monooxygenase (Hörtensteiner et al., 1998), which has an intriguing specificity for Pheide a (Hörtensteiner et al., 1995; Pruzinská et al., 2003; Tanaka et al., 2003). PAO is a key enzyme, because it determines the basic structure of all downstream catabolites. Therefore, this pathway of Chl breakdown is nowadays often called the “PAO pathway.” The substrate specificity of PAO is believed to force Chl b-to-Chl a conversion to occur upstream of PAO to enable Chl b degradation. Recently, nonyellow coloring1 (NYC1), NYC1-like, and 7-hydroxymethyl chlorophyll a reductase were cloned and shown to encode enzymes required to catalyze the two consecutive steps of Chl b-to-Chl a reduction (Kusaba et al., 2007; Horie et al., 2009; Sato et al., 2009; Meguro et al., 2011). Mutants deficient in NYC1 exhibit a stay-green phenotype, indicating that Chl b reduction could initiate the degradation of both Chl and Chl-binding proteins by destabilizing respective pigment-protein complexes (Hörtensteiner, 2006). The final step in pFCC formation is the site-specific and stereoselective reduction of red Chl catabolite (the product of PAO activity) catalyzed by red chlorophyll catabolite reductase (RCCR; Wüthrich et al., 2000; Pruzinská et al., 2007).
pFCC was shown to be exported from senescing chloroplasts in an ATP-dependent manner (Matile et al., 1992). This indicates that the later steps of the PAO pathway (i.e. modifications of different side positions of pFCC) likely occur in the cytosol. While hydroxylation of the C82-ethyl moiety (for atom numbering in FCCs, see Fig. 2D below) seems to be a common reaction in all species from which NCCs have been structurally characterized so far, other modifications occur in a species-specific manner. For example, catabolites with a demethylated C132-carboxymethylester have so far only been found in Arabidopsis, oilseed rape/canola (Brassica napus), and spinach (Spinacia oleracea; Mühlecker and Kräutler, 1996; Berghold et al., 2002; Pruzinská et al., 2005; Hörtensteiner and Kräutler, 2011). Finally, modified FCCs are imported into the vacuole, where they are nonenzymically isomerized to their respective NCCs, because of the acidic pH of the vacuolar sap (Oberhuber et al., 2003). Interestingly, persistent so-called “hypermodified” FCCs have recently been identified in banana (Musa sp.) and peace lily (Spathiphyllum wallisii; Moser et al., 2009; Banala et al., 2010; Kräutler et al., 2010). In these FCCs, the C17 side chain is modified with different ester moieties; thus, FCC-to-NCC isomerization, which requires a free C17-propionyl acid function, is disabled (Oberhuber et al., 2003; Hörtensteiner and Kräutler, 2011).
Figure 2.
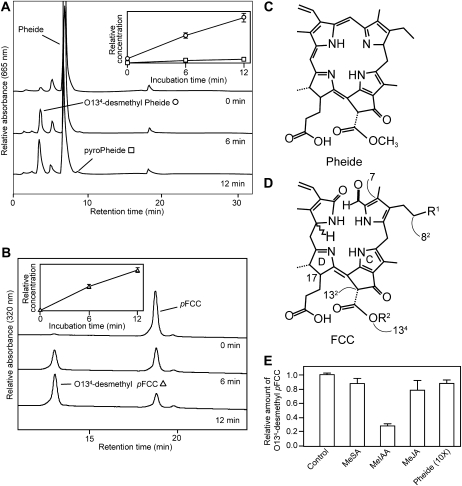
Analysis of recombinant MES16. A and B, HPLC analysis of assays employing E. coli lysate expressing 6xHis-MES16 with Pheide (A) and pFCC (B) as substrate. HPLC traces at A665 (A) or A320 (B) before (0 min) and after 6 and 12 min of incubation at 25°C are shown. For clarity, only a part of the HPLC traces at A320 (B) is shown. The insets show the relative concentrations of formed products (O134-desmethyl Pheide, white circles; pyro-Pheide, white squares; O134-desmethyl pFCC, white triangles). Values are means of three replicates. Error bars indicate sd. C and D, Chemical structures of Pheide a and FCCs, respectively. Relevant carbon atoms and pyrrole rings are indicated in the FCC structure in D. E, Competition assays were performed for 6 min using MeIAA, MeSA, or MeJA at a final concentration of 1.5 mm (100× molar excess to pFCC) and Pheide at a concentration of 150 μm (10×). A relative value of 1 corresponds to 0.05 nmol of O134-desmethyl pFCC being produced during the incubation under the standard conditions as described in “Materials and Methods.” Values are means of three replicates. Error bars indicate sd.
Besides these main steps of the PAO pathway, additional/alternative reactions of Chl breakdown have been described in the past. These were (mostly) inferred from the identification of different types of Chl degradation products, such as Chl-derived monopyrroles (Suzuki and Shioi, 1999), urobilinogenoidic catabolites (Losey and Engel, 2001), and different pigments with an intact porphyrin ring. Among the latter are pyro (=C132-decarboxymethylated) forms of Pheide and pheophytin, which have been discussed as breakdown products of Chl in algae and higher plants (Schoch and Vielwerth, 1983; Ziegler et al., 1988; Shioi et al., 1991). In support of this finding was the demonstration of Pheide-to-pyro-Pheide conversion in enzyme extracts of different higher plants species, such as satsuma (Citrus unshiu) fruit peel and leaves of goosefoot (Chenopodium album) and different Brassicaceae species (Shimokawa et al., 1990; Suzuki et al., 2002). The enzyme, named pheophorbidase (PPD), was shown to only catalyze demethylation at the C132-carboxymethyl group of Pheide to yield O134-desmethyl Pheide, while the subsequent decarboxylation to form pyro-Pheide occurred spontaneously without the contribution of PPD (Shioi et al., 1996). In contrast, an activity that directly converts Pheide to pyro-Pheide without the occurrence of the O134-desmethyl intermediate was described in Chlamydomonas reinhardtii (Suzuki et al., 2002). PPD was recently purified and cloned from radish (Raphanus sativus [RsPPD]; Suzuki et al., 2006, 2008). RsPPD specifically acts on Pheide and bacterio-Pheide, but not on phytol- and/or Mg-containing- or proto-Chl species. Homologs of RsPPD were identified in other Brassicaceae species (Suzuki et al., 2006). In Arabidopsis, RsPPD is most closely related to MES16 (At4g16690), one of the 20 members of the Arabidopsis methylesterase (MES) protein family (Yang et al., 2008). Structurally, PPD and MES proteins belong to the α/β-hydrolase protein “superfamily” and possess a catalytic Ser-His-Asp triad (Dodson and Wlodawer, 1998). MES16 was shown to hydrolyze two different methylated plant hormones, methyl-indole acetic acid (MeIAA) and methyl-jasmonic acid (MeJA), in vitro (Yang et al., 2008).
Here, we demonstrate that MES16 is involved in Chl breakdown during Arabidopsis leaf senescence. MES16-deficient mutants (mes16) were still able to degrade Chl, but they accumulated FCCs and NCCs with an intact C132-carboxymethyl group. As a consequence, FCC-to-NCC isomerization was compromised and the mutants accumulated large quantities of FCCs, which caused senescent leaves to fluoresce under UV light. Recombinant MES16 protein was able to demethylate both Pheide and pFCC, but in vivo, MES16 specifically acts on FCCs. To prove this, we show the cytosolic localization of MES16 using GFP fusions. In addition, targeting of MES16 to the chloroplast led to an accumulation of pyro-Pheide in a PAO mutant (pao1) background, while pao1 and a mes16pao1 double mutant accumulated Pheide. In summary, we demonstrate that MES16 specifically catalyzes methylester hydrolysis at O134 (O134-demethylation) of FCCs and, thus, that this enzyme is located within the PAO pathway of the breakdown of Chl to NCCs in Arabidopsis.
RESULTS
Arabidopsis MES16 Catalyzes the O134-Demethylation of Pheide and pFCC in Vitro
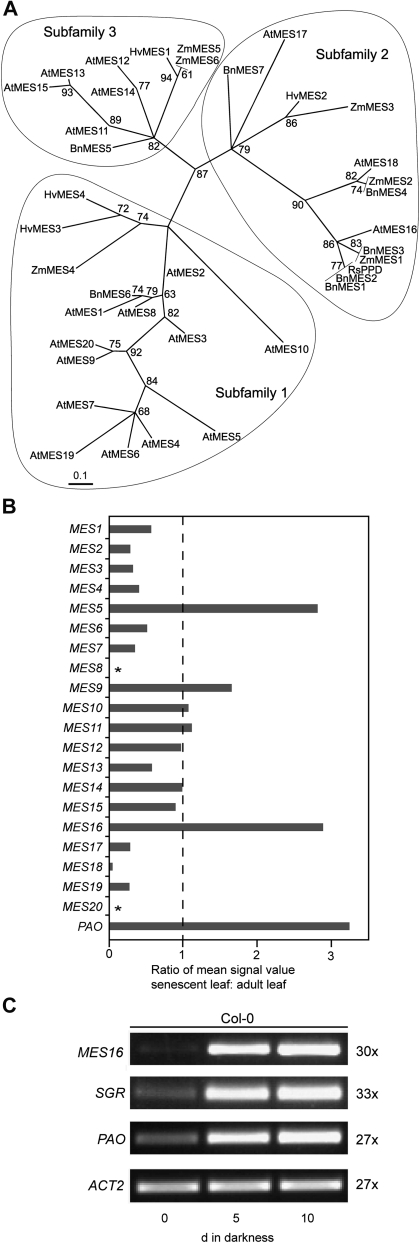
The distribution of PPD activity was shown to correlate with the occurrence of NCCs harboring a free C132-carboxyl group. For example, all investigated Brassicaceae species have PPD activity (Suzuki et al., 2002), and O134-demethylated FCCs and NCCs have been found in canola and Arabidopsis (Mühlecker and Kräutler, 1996; Pruzinská et al., 2005), indicating that PPD could be responsible for this site-specific modification. A protein BLAST search of RsPPD against the Arabidopsis proteome uncovered high homology to members of the MES family of Arabidopsis (Vlot et al., 2008; Yang et al., 2008). Thereby, RsPPD clustered with the members of subfamily 2, as determined by Yang et al. (2008), with highest homology to MES16 (Fig. 1A). The phylogenetic tree of Figure 1A includes further MES-like EST-derived sequences from plant species for which NCC structures have been determined. The tree indicates that subfamily 2 members of the MES protein family are indeed present in other Brassicaceae species, but also in barley and maize (Zea mays), where evidence for hydrolysis of the methylester group at C132 has not been provided (Kräutler et al., 1991; Berghold et al., 2006).
Figure 1.
Identification of MES16. A, Maximum likelihood phylogenetic tree of radish PPD (RsPPD) and MES proteins from Arabidopsis (At), canola (Bn), barley (Hv), and maize (Zm). Branch support values are based on 100 bootstrap replicates and are indicated when higher than 50%. Three subfamilies as determined by Yang et al. (2008) are circled. For sequence accession numbers, see “Materials and Methods.” B, Analysis of senescence-related expression of the Arabidopsis MES family using the Genevestigator Meta-Analyzer tool (Zimmermann et al., 2004). The ratio of mean fluorescence values from senescent leaves (organ no. 44; number of chips, three) and adult leaves (organ no. 42; number of chips, 274) is shown. The value for PAO is shown as a reference. Asterisks indicate that MES8 and MES20 are not represented on the ATH1 chip used for analysis. C, Analysis of gene expression during dark-induced senescence in Col-0. ACT2 was used as a control. Expression was analyzed with nonsaturating numbers of PCR cycles as shown at the right. PCR products were separated on agarose gels and visualized with ethidium bromide.
Analysis of gene expression patterns has proven successful in the past to identify potential Chl catabolic enzymes (Schelbert et al., 2009; Ren et al., 2010). When analyzing microarray expression data of the Arabidopsis MES genes using the Genevestigator tool (Zimmermann et al., 2004), MES5, MES9, and MES16 exhibited increased expression levels in senescent leaves compared with green leaves (Fig. 1B). In addition, using the ATTED tool (Obayashi et al., 2009), MES16 was found in a network of coexpression with other Chl catabolic genes (Supplemental Fig. S1). Using semiquantitative reverse transcription (RT)-PCR, we confirmed the senescence-regulated expression of MES16 in the wild type upon dark-induced senescence (Fig. 1C). Thereby, the expression pattern was very similar to the expression of PAO and SGR. SGR is required for the initiation of Chl breakdown and has been proposed to play a regulatory role in Chl-apoprotein complex destabilization (Hörtensteiner, 2009). From this analysis, MES16 appeared to be the most likely candidate for O134-demethylation in Arabidopsis.
To investigate its enzymatic activity, recombinant MES16 was produced in Escherichia coli in fusion with an N-terminal His tag. MES16 (Fig. 2A) was able to convert Pheide a to pyro-Pheide a in a time-dependent manner. As described for RsPPD (Suzuki et al., 2006), MES16 most likely only catalyzed methylester hydrolysis of Pheide a to O134-desmethyl Pheide a, while the subsequent decarboxylation to pyro-Pheide a occurred spontaneously without the involvement of MES16; however, this was not analyzed in more detail. When pFCC was used as the substrate for recombinant MES16, a new, more polar FCC peak appeared in reverse-phase HPLC (Fig. 2B), which by mass spectrometry (MS) analysis (for MS data, see “Materials and Methods”) was identified as the O134-demethylated form of pFCC (O134-desmethyl pFCC). In contrast, recombinant His-tagged versions of MES17 and MES18, the two other subfamily 2 members (Fig. 1A), and MES5 and MES9, the other senescence-regulated MES proteins (Fig. 1B), were unable to demethylate pFCC (Supplemental Fig. S2, A–C).
As described for RsPPD (Suzuki et al., 2006), MES16 was unable to demethylate CjNCC-1, an NCC isolated from senescent leaves of Cercidiphyllum japonicum (Curty and Engel, 1996; Supplemental Fig. S2D). MES16 has been shown to hydrolyze MeJA and MeIAA (Yang et al., 2008). To obtain insight into the substrate specificity of recombinant MES16, we tested its pFCC esterase activity in the presence of 100-fold molar excesses of MeJA, MeIAA, and methyl salicylic acid (MeSA) or a 10-fold molar excess of Pheide a. As shown in Figure 2E, under these conditions, pFCC demethylation was only impaired to some extent by MeIAA, indicating that in vitro, MES16 has a high preference for an FCC as substrate.
mes16 Mutants Are Compromised in Chl Degradation
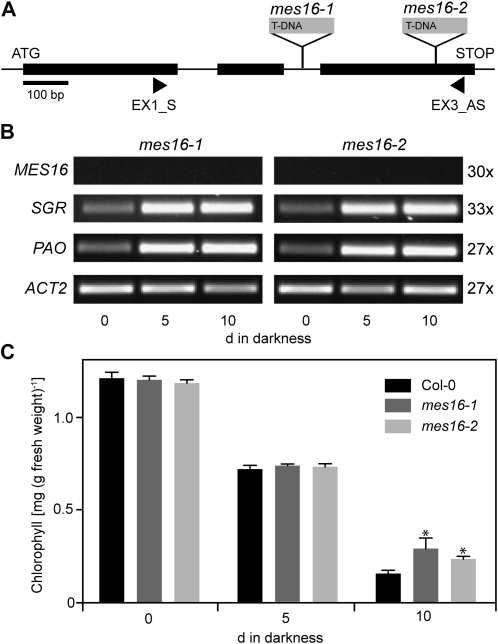
To investigate the in vivo role of MES16 in relation to Chl breakdown, two independent T-DNA insertion lines, named mes16-1 and mes16-2 (Fig. 3A), were obtained from the SALK resource (Alonso et al., 2003). MES16 expression analysis of these mutants indicated both alleles to represent true knockout mutants (Fig. 3B). The absence of full-length MES16 transcripts in these mutants did not affect the senescence-regulated expression of PAO and SGR (Fig. 3B). The mutants did not show any phenotype during normal growth under either short-day or long-day conditions. When senescence was induced in the dark with detached leaves, mes16 mutants did not exhibit a dramatic stay-green phenotype, although, compared with the wild type, Chl was significantly retained at a low level (Fig. 3C).
Figure 3.
Deficiency of MES16 does not affect the first steps of Chl degradation. A, Gene structure of MES16 showing the T-DNA insertion sites of two different mes16 mutants studied here. The sites of insertion were verified by sequencing. The positions of primers used for RT-PCR are shown. B, Analysis of gene expression during dark-induced senescence in mes16-1 and -2. ACT2 was used as a control. Expression was analyzed with nonsaturating numbers of PCR cycles as shown at the right. After 30 PCR cycles, which produced clearly visible bands in Col-0 (Fig. 1C), MES16 expression was not obvious in mes16 mutants. PCR products were separated on agarose gels and visualized with ethidium bromide. C, Chl degradation of mes16 mutants during dark-induced senescence. Data are mean values of a representative experiment with three replicates. Error bars indicate sd. The asterisks indicate the significance of differences between the wild type and mes16 mutants at P < 0.03 as determined using a two-tailed t test.
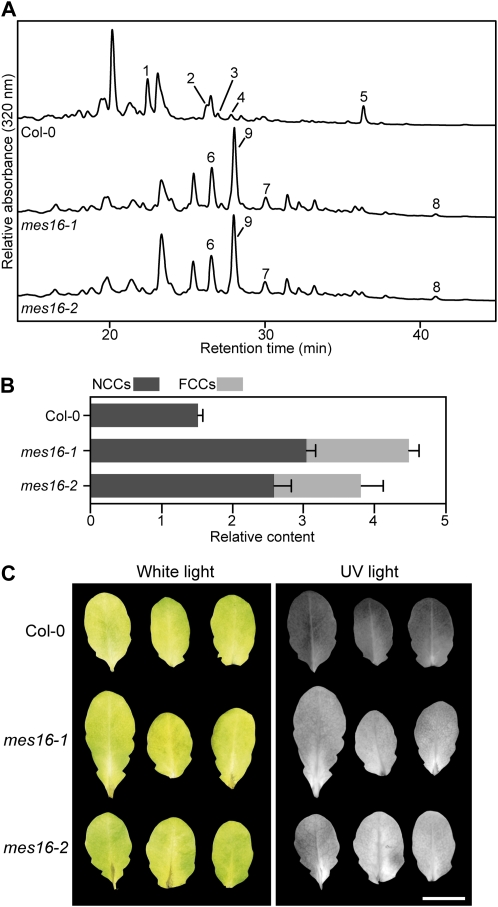
When extracts of senescent leaves of mes16-1 and mes16-2 were analyzed for the presence of FCCs and NCCs using reverse-phase HPLC, major differences were observed in comparison with wild-type leaves (Fig. 4A). Thus, except for two peaks comigrating with pFCC and At-NCC-4 (Pruzinská et al., 2005), all FCCs and NCCs of the mes16 mutants, as detected by their typical fluorescence and/or absorption properties, were likely novel Chl catabolites not found before in Arabidopsis. The three major FCC fractions, tentatively named mes16-FCC-1, mes16-FCC-2, and mes16-FCC-3, and the major NCC fraction (mes16-NCC-1) were isolated from mes16-1 and analyzed by MS (for MS data, see “Materials and Methods”). As seen in Table I, all analyzed catabolites possessed an intact C132-carboxymethyl group. In addition, the respective identities of mes16-FCC-3 and mes16-NCC-1 with pFCC and At-NCC-4, the only O134-methylated catabolites detected in the wild type (Pruzinská et al., 2005), could be confirmed by MS and HPLC. To our surprise, quantification of these catabolites revealed that both mes16 lines accumulated significantly higher overall amounts of FCC and NCC (Fig. 4B). The reason for this higher quantity of Chl catabolites in the mutants is unclear at present. Moreover, while the wild type almost exclusively accumulated NCCs, senescent mes16 leaves contained high proportions of FCCs. As a consequence, senescent mes16 leaves, but not wild-type leaves, exhibited blue fluorescence when excited with UV light (Fig. 4C). Intense fluorescence could also be observed in naturally senesced mes16-1 leaves (Supplemental Fig. S3B). To investigate whether the other MES subfamily 2 members (i.e. MES17 and MES18; Fig. 1A) or other senescence-regulated MES proteins (i.e. MES5 and MES9; Fig. 1B) are involved in Chl breakdown, T-DNA insertion lines of respective genes were isolated and FCCs and NCCs were analyzed after dark-induced senescence (Supplemental Fig. S3C). None of these mutants exhibited a catabolite pattern that was different from the wild type. Together with the above-described absence of Chl catabolite esterase activity of the respective recombinant proteins (Supplemental Fig. S2), this result indicates that MES16 acts nonredundantly toward the formation of Chl catabolites that carry a free carboxylic acid function at C132.
Figure 4.
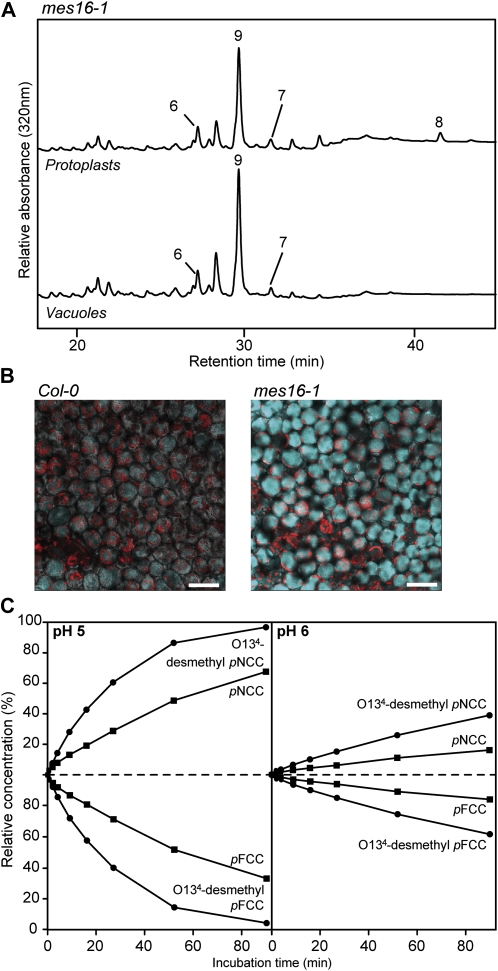
Colorless catabolites of mes16 mutants. A, Colorless catabolites of dark-incubated (10 d) leaves of Col-0 and mes16 mutants were separated by HPLC as described in “Materials and Methods.” A320 is shown. For clarity, only a part of the HPLC traces is shown. For identification and peak numbering of FCCs and NCCs, see Table I. B, Relative contents of NCCs and FCCs. Values are means of three replicates. Error bars indicate sd. C, Photographs of dark-incubated (10 d) Col-0 and mes16 leaves under white light and UV light (366 nm). Bar = 1 cm.
Table I. FCCs and NCCs occurring in Col-0 and mes16 mutants during chlorophyll breakdown as identified in this work.
| Name | IDa | R1b | R2b | Identificationc | Identity with |
| At-NCC-1 | 1 | O-Glucosyl | H | s | – |
| At-NCC-2 | 2 | OH | H | s | – |
| At-NCC-3d | 3 | H | H | s | – |
| At-NCC-4 | 4 | O-Glucosyl | CH3 | s | mes16-NCC-1 |
| At-NCC-5 | 5 | H | H | s | – |
| mes16-FCC-1 | 6 | O-Glucosyl | CH3 | s, m | – |
| mes16-FCC-2 | 7 | OH | CH3 | s, m | – |
| mes16-FCC-3 | 8 | H | CH3 | s, m | pFCC |
| mes16-NCC-1 | 9 | O-Glucosyl | CH3 | s, m | At-NCC-4 |
R1 and R2 indicate residues at C82 and O134 side positions, respectively, of FCCs or NCCs (for atom labeling, see Fig. 2D).
Peak identification by UV/visible spectra (s) or MS (m).
At-NCC-3 carries a hydroxymethyl group instead of a methyl group at C7.
FCC-to-NCC Isomerization Is Affected in mes16 Mutants
To analyze why mes16 mutants accumulated rather high amounts of FCCs, we first investigated the subcellular localization of these Chl catabolites. To this end, Chl catabolites extracted from isolated vacuoles of senescent mes16-1 leaf material were analyzed by reverse-phase HPLC and compared with the catabolites of protoplasts (Fig. 5A). The pattern of FCCs and NCCs was identical in the two fractions and, given a rather low contamination of the vacuolar fraction with extravacuolar compartments, also the relative amounts of different catabolites were comparable between vacuoles and protoplasts. The major difference between these two fractions was pFCC (=mes16-FCC-3), which was absent from vacuoles. This indicated that most of the FCCs and NCCs found in mes16-1 accumulated in the vacuolar sap, as is known for wild-type Arabidopsis and other plant species (Matile et al., 1988; Hinder et al., 1996; Pruzinská et al., 2007). Using confocal microscopy, vacuolar localization of mes16-FCCs was confirmed in senescent mes16-1 leaves; when excited with a 355-nm laser, mutant cells of the palisade mesophyll exhibited a strong blue fluorescence that covered the entire cellular space, indicating vacuolar origin (Fig. 5B). In contrast, wild-type mesophyll tissue only weakly fluoresced under these conditions.
Figure 5.
mes16 mutants retain FCCs in the vacuoles. A, Protoplasts and vacuoles were isolated from mes16-1, and colorless catabolites were separated by HPLC as described in “Materials and Methods.” For clarity, only a part of the HPLC traces is shown. For identification and peak numbering of FCCs and NCCs, see Table I. B, Palisade mesophyll of dark-induced leaves (4 d) of Col-0 and mes16-1 observed with a laser scanning confocal microscope. FCC fluorescence was induced with an excitation wavelength of 355 nm, and the emission signal (blue) was recovered between 430 and 470 nm. Red is Chl autofluorescence. Bars = 50 μm. C, FCC-to-NCC isomerization assays performed with pFCC (black squares) or O134-desmethyl pFCC (black circles). Relative concentrations of FCCs and corresponding NCCs are plotted for pH 5 and 6. For more details, see “Materials and Methods.” Calculated half-lives of FCCs are listed in Table II.
These data let us assume that the isomerization of FCCs to NCCs was compromised in mes16 mutants. We speculated that this phenomenon could result from the possibility that the presence of an intact C132-carboxymethyl group in the mes16-FCCs could affect the efficiency of their isomerization to the respective NCCs. To test this, we compared the rates of FCC-to-NCC isomerization at different pH values using either pFCC (with an intact C132-carboxymethyl group) or the product of its hydrolysis with recombinant MES16, O134-desmethyl pFCC (Fig. 5C). Both FCCs were converted to the respective NCCs, but the rates of isomerization differed considerably, as judged from the half-lives of the FCC substrates under the given pH conditions. Thus, both at pH 5 and 6, the conversion of O134-desmethyl pFCC was about three times faster than that of pFCC (Table II). This delay of isomerization is likely, at least in part, responsible for the observed retention of FCCs in the mes16 mutants. A second (additional) possibility (i.e. that the vacuolar pH could be higher in mes16 and, therefore, the acid-catalyzed FCC-to-NCC isomerization could be slower as compared with the wild type) was not experimentally tested here.
Table II.
4Half-lives of O134-desmethyl-pFCC and pFCC
| Sample | pH | t1/2a | ka | R2b |
| min | min−1 | |||
| O134-desmethyl-pFCC | 5 | 18 | −0.0382 | 0.9989 |
| pFCC | 5 | 57 | −0.0122 | 0.9995 |
| O134-desmethyl-pFCC | 6 | 128 | −0.0054 | 0.9981 |
| pFCC | 6 | 365 | −0.0019 | 0.993 |
Half-lives (t1/2) and reaction rate constants (k) were determined using the following formula: ln[FCC]t = ln[FCC]0 – kt; t1/2 = ln(2)/k.
Coefficient of determination of the linear regression of x = t (min) and y = ln[FCC]t.
Pheide a Is Not an in Vivo Substrate for MES16
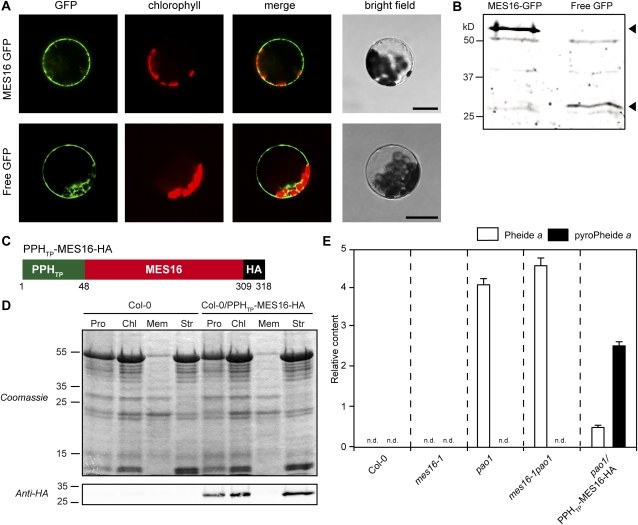
As shown above, the absence of MES16 in mes16 mutants abolished the O134-demethylation of Chl catabolites, as seen in the FCCs and NCCs, the (final) products of degradation. At the same time, recombinant MES16 was able to demethylate both Pheide and pFCC. Therefore, the question arose, what is the in vivo substrate of MES16? To answer this, we first investigated the subcellular localization of MES16 by transiently expressing a MES16-GFP fusion protein in wild-type protoplasts and analyzing GFP fluorescence with confocal microscopy (Fig. 6A). Similar to a free GFP control, MES16-GFP localized to the cytosol. The integrity of the fusion protein was analyzed on immunoblots of transformed protoplasts using an anti-GFP antiserum (Fig. 6B). This localization excluded Pheide a as a likely substrate for MES16, because PAO, the exclusive Pheide a-catabolizing enzyme of higher plants, has been localized to the chloroplast (Matile and Schellenberg, 1996; Joyard et al., 2009) and export of Pheide from senescent chloroplasts followed by reimport of O134-desmethyl Pheide is unlikely to occur (Hörtensteiner, 2006). To confirm that the in vivo activity of MES16 is restricted to the cytosol, we mistargeted C-terminally hemagglutinin (HA)-tagged MES16 to the chloroplast in transgenic ecotype Columbia (Col-0) and pao1. To direct MES16-HA to the chloroplast, we N-terminally fused it to the 48 N-terminal amino acids of Arabidopsis PPH, which represent the predicted chloroplast transit peptide of PPH (Schelbert et al., 2009; Fig. 6C). Stroma localization of the chimeric construct (PPHTP-MES16-HA) in Col-0 (Col-0/PPHTP-MES16-HA) lines could be confirmed on immunoblots of protein extracts of protoplasts, chloroplasts, and subchloroplast fractions using anti-HA antibodies (Fig. 6D). We then compared the patterns of accumulating Pheide pigments in senescent leaves of the wild type, pao1/PPHTP-MES16-HA, mes16-1, and a mes16-1pao1 double mutant (Fig. 6E). While the wild type, as expected, was devoid of Pheide a, this Chl breakdown intermediate accumulated to a similar extent in pao1 and mes16-1pao1. In contrast, when targeting MES16 to the chloroplast in the pao1 background, a large proportion of the accumulating Pheide a had been converted to pyro-Pheide a. This result indicated that under natural conditions, MES16 does not have access to Pheide a, implying that the likely in vivo substrate of MES16 (in the cytosol) is an FCC.
Figure 6.
FCCs are the in vivo substrates of MES16. A, Transient expression of MES16-GFP and free GFP in Arabidopsis mesophyll protoplasts. GFP fluorescence (GFP) and Chl autofluorescence (chlorophyll) were examined by confocal laser scanning microscopy. The merge panels show overlays of GFP and autofluorescence. Bars = 20 μm. B, Anti-GFP immunoblotting of proteins from protoplasts expressing MES16-GFP and free GFP. The arrowheads indicate the predicted sizes of transiently expressed proteins. C, The chimeric construct used to target MES16 to the chloroplast. PPHTP, Amino acids 1 to 48 from PPH, representing the chloroplasts transit peptide; HA, HA tag. D, Verification of MES16 targeting to the chloroplast. Leaves of Col-0 and Col-0/PPHTP-MES16-HA were fractionated into protoplasts (Pro) and chloroplasts (Chl) and chloroplast subfractions (Mem, chloroplast membranes; Str, stroma). Gel loadings of protoplast and chloroplast fractions are based on equal amounts of chlorophyll. Anti-HA antibodies were used for detection of the chimeric protein. E, Quantification of Pheide a (white bars) and pyro-Pheide a (black bars) in dark-induced (5 d) Col-0, mes16-1, pao1, mes16-1pao1, and pao1/PPHTP-MES16-HA. Values are means of three replicates, and error bars represent sd. n.d., Not detected.
DISCUSSION
Leaf senescence in higher plants is associated with the disappearance of green color and the unmasking of yellow-colored carotenoids. Thereby, Chl is degraded in the so-called PAO pathway to colorless linear tetrapyrroles (i.e. breakdown proceeds beyond the level of colored species with an intact porphyrin macrocycle; Hörtensteiner and Kräutler, 2011). Thus, green porphyrinic pigments identified as Chl breakdown products in the past (Brown et al., 1991) most likely merely represent experimental artifacts or intermediates of the PAO pathway. This pathway is considered to be active in all higher plant species (Hörtensteiner and Kräutler, 2011), and PAO genes are widely distributed in angiosperms, are present in mosses, and probably even appear in algae and cyanobacteria (Gray et al., 2004).
Among the NCCs, which are modified at side groups of the tetrapyrrolic backbone, are O134-demethylated catabolites. PPD from radish has been demonstrated to catalyze O134-demethylation of Pheide in vitro; however, this demethylation is followed by a spontaneous decarboxylation resulting in pyro-Pheide (Suzuki et al., 2006). Likewise, we could show here that MES16, the Arabidopsis protein most closely related to RsPPD (Fig. 1A), catalyzes the same reaction (Fig. 2A). It had been assumed that PPD is involved in Chl breakdown during leaf senescence, and pyro-Pheide was suggested to feed into the main (PAO) pathway leading to the formation of FCCs and NCCs (Suzuki et al., 2006). However, none of the known linear tetrapyrrolic breakdown products of Chl exhibit a pyro-type structure at C132 (Kräutler, 2008; Hörtensteiner and Kräutler, 2011). When targeting MES16 to the chloroplast in the pao1 background, pyro-Pheide accumulated (Fig. 6E), indicating that also in vivo demethylation of Pheide is followed by decarboxylation. In contrast, in vitro MES16 assays with pFCC as substrate exclusively produced O134-desmethyl pFCC and not pyro-pFCC (Fig. 2B). Furthermore, O134-demethylated FCCs and NCCs, but no pyro-pigments, accumulate in senescent wild-type Arabidopsis leaves (Pruzinská et al., 2005), while in mes16 mutants, all catabolites have an intact C132-carboxymethylester (Table I). Finally, like RsPPD (Suzuki et al., 2006), MES16 did not accept an NCC as substrate for demethylation. This substrate selectivity can be rationalized by comparison of the chemical constitution around the isocyclic ring and pyrrole rings C and D, which is identical in Pheide and pFCC but different in NCCs. This causes a significant difference in the three-dimensional shape around the C132 position, which results in the selectivity. Furthermore, the rapid spontaneous decarboxylation that specifically occurs in the O134-demethylation product of Pheide but not in pFCC results from an additional stabilization by the delocalized π system spanning the entire porphyrin macrocycle of Pheide, which is absent in pFCC.
In summary, the observations regarding the in vitro and in vivo methylesterase activity of MES16 strongly indicate that FCCs, but not Pheide or NCCs, are the in vivo substrate for demethylation. Further support for this was obtained from the subcellular localization of MES16 in the cytosol (Fig. 6A). Pheide a formation and further degradation to pFCC exclusively occur inside the chloroplast (Hörtensteiner, 2006); thus, only after export from the chloroplast are Chl degradation products accessible to MES16. Thus, the fact that Pheide a but not pyro-Pheide a was found in pao1 (Fig. 6E) indicates that in this mutant, Pheide a accumulates inside the plastids and is not exported to the cytosol. In line with this is the chloroplast localization of MES16 in the pao1 background, which enabled Pheide a demethylation (and decarboxylation; Fig. 6E). Interestingly, however, expressing MES16 in the chloroplast in the wild type did not alter the FCC/NCC catabolite pattern (i.e. no pyro-catabolites were formed; Supplemental Fig. S4), indicating that in a PAO-containing background, chloroplast-localized MES16 is unable to access Pheide and to convert it to pyro-Pheide. This may hint at the existence in chloroplasts of a metabolic channeling mechanism for Chl breakdown intermediates. Metabolic channeling and physical interaction have been shown for PAO and RCCR (Rodoni et al., 1997; Pruzinská et al., 2007), but these might also involve further (upstream) reactions. In such a scenario, lines that contain the entire chloroplast-located degradation machinery (e.g. mes16-1 or the wild type) could shield Pheide a from chloroplast-targeted MES16. In contrast, in the pao1 background, this intermediate is likely released from the degradation machinery and is accessible for chloroplast-localized MES16.
Recently, it was shown that the salicylic acid-binding protein 2 (SABP2) from tobacco (Nicotiana tabacum) is able to hydrolyze MeSA to salicylic acid (Forouhar et al., 2005). Since the MES proteins of Arabidopsis are homologous to SABP2, members of this family have been tested in vitro as possible hydrolases for methylesters of different plant hormones. All three members of subfamily 2, MES16, MES17, and MES18 (Fig. 1A), were shown to hydrolyze MeIAA, and MES16 also hydrolyzed MeJA, but not MeSA and the methylesters of two different gibberellic acids (Yang et al., 2008). In our inhibition studies, pFCC-hydrolyzing activity was inhibited to some extent by MeIAA but not by MeJA (Fig. 2E), confirming MeIAA to be a potential substrate of MES16. However, when analyzing mes16 mutants (the identical alleles also used in our work) for auxin-related phenotypes, Yang et al. (2008) could not confirm a relation between MES16 activity and auxin function. In contrast, analysis of mutants deficient in MES17 demonstrated the capability of MES17 to hydrolyze MeIAA in vivo (Yang et al., 2008). On the other hand, neither MES17 nor MES18 exhibited FCC-demethylating activity both in vitro and in vivo (Supplemental Figs. S2 and S3). A predicted MES protein from maize (tentatively named ZmMES1) clustered together with Arabidopsis MES16 and RsPPD (Fig. 1A); however, all maize NCCs identified to date have an intact C132-carboxymethyl group, indicating that ZmMES1 is not involved in Chl breakdown in maize. In summary, these data imply that MES proteins of subfamily 2 have different functions and that function does not necessarily relate to the primary amino acid composition of these proteins. Furthermore, MES16 has a predominant (and possibly exclusive) role as an FCC methylesterase during Chl breakdown.
Demethylated Chl catabolites have so far only been found in a few species, raising the question about the significance of O134-demethylation. Interestingly, senescent mes16 mutants exhibited a strong UV-excitable fluorescence (Figs. 4 and 5), which was due to large increases of FCCs accumulating in the mutants. This resulted, at least in part, from slower FCC isomerization to the respective NCC in the presence of an intact C132-carboxymethylester compared with a free carboxylic acid group (Fig. 5C; Oberhuber et al., 2008). However, in contrast to Arabidopsis mes16, several plant species in which O134-demethylation has not been observed, such as C. japonicum or Nicotiana rustica, exclusively accumulate NCCs during leaf senescence (Curty and Engel, 1996; Oberhuber et al., 2003; Berghold et al., 2004). This is likely explained by differences in the vacuolar pH, which determines the velocity of FCC-to-NCC isomerization (Table II). Thus, whether a plant accumulates FCCs or NCCs might depend on the presence/absence of O134-demethylation and/or the vacuolar pH. Nevertheless, hypermodified FCCs, which are esterified at the C17-propionic acid side chain and therefore are not isomerized to NCCs (i.e. they persist as FCCs), have recently been identified in some senescing leaves and ripening banana fruits (Moser et al., 2009; Banala et al., 2010; Kräutler et al., 2010). The resulting strong fluorescence in these tissues might, as suggested for the bright colors of many autumnal leaves (Archetti and Brown, 2004), play an important ecological function (Moser et al., 2009). Noteworthy is the finding that the Arabidopsis Landsberg erecta (Ler) ecotype exhibits a MES16 mutant phenotype (B. Christ and S. Hörtensteiner, unpublished data). Most probably, this is due to a single nucleotide polymorphism present in the Ler MES16 gene (http://polymorph-clark20.weigelworld.org/cgi-bin/retrieve_cds_snp.cgi), which alters a Val residue within the MES16 lipase motif (PROSITE motif 00120) that is fully conserved in all MES proteins (Yang et al., 2008) to Ala. Thus, Ler likely is a natural mutant in MES16, and it will be interesting to examine in the future whether MES16 absence in this accession has any significance in an ecological context.
MATERIALS AND METHODS
Plant Material and Senescence Induction
Arabidopsis (Arabidopsis thaliana) Col-0 was used as the wild type. Ecotype Wassilewskija was also analyzed in experiments including mes18-1 (Wassilewskija background). T-DNA insertion lines were from the following collections: SALK lines (Alonso et al., 2003): AT4G16690-1 (mes16-1), SALK_139756; AT4G16690-2 (mes16-2), SALK_151578; AT4G37150-1 (mes9-1), SALK_030442; SAIL lines (Sessions et al., 2002): AT3G10870-1 (mes17-1), SAIL_503_C03; GABI lines (Rosso et al., 2003): AT5G10300-1 (mes5-1), GABI_453E01; FLAG lines (Balzergue et al., 2001): AT5G58310-1 (mes18-1), FLAG_271B02. SALK and SAIL lines were obtained from the European Arabidopsis Stock Center. The GABI line was obtained from GABI Kat, Max Planck Institute for Plant Breeding Research. The FLAG line was obtained from the Arabidopsis Resource Centre for Genomics. Homozygous plants were identified by PCR using T-DNA-, transposon-, and gene-specific primers as listed in Supplemental Table S1. Likewise, a homozygous mes16-1pao-1 double mutant was identified by PCR.
Plants were grown on soil either in short days (8 h of light/16 h of dark) under fluorescent light of 60 to 120 μmol photons m−2 s−1 at 22°C or in long days (16 h of light/8 h of dark) in a greenhouse with fluence rates of 100 to 200 μmol photons m−2 s−1 at 22°C. For senescence induction, leaves from 8-week-old (short-day) plants were excised and incubated in permanent darkness on wet filter paper for up to 10 d at ambient temperature.
Biocomputational Methods and Phylogenetic Analysis
Methylesterase homologs from canola (Brassica napus), barley (Hordeum vulgare), and maize (Zea mays) were identified by TBLASTN searches (Altschul et al., 1997) of MES16 with the est_others database of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov). EST sequences were conceptually translated. MES protein sequences were aligned and subjected to a maximum likelihood phylogenetic analysis using phylogeny.fr (http://www.phylogeny.fr/version2_cgi/index.cgi). Phylogeny.fr (Dereeper et al., 2008) employs MUSCLE for multiple sequence alignments, Gblocks for alignment curation, PhyML for phylogeny analysis, and TreeDyn for tree drawing and was used with the default settings. Bootstrap analysis was performed with 100 replicates.
GFP Fusion Protein Production and Confocal Microscopy
MES16 was amplified by PCR with the Expand High Fidelity PCR system (Roche Applied Science) using the primers listed in Supplemental Table S1, introducing SmaI/SpeI restriction sites at the ends. The PCR fragment was then cloned into the pGEM-T-easy vector (Promega) and, after SmaI/Spel digestion, subcloned into pUC18-spGFP6 (Meyer et al., 2006), thereby producing a fusion of MES16 with the N terminus of GFP. Arabidopsis mesophyll protoplasts were isolated from 6-week-old short-day-grown Col-0 plants according to published procedures (Endler et al., 2006; Schelbert et al., 2009). Cell numbers were quantified with a Neubauer chamber and adjusted to a density of 2 to 3 × 106 protoplasts mL−1. Protoplasts were transformed by 20% polyethylene glycol transformation (Meyer et al., 2006). Transformed cells were incubated for 48 h in the dark at room temperature before laser scanning confocal microscopic analysis (DM IRE2; Leica Microsystems). GFP fluorescence was imaged at an excitation wavelength of 488 nm, and the emission signal was recovered between 495 and 530 nm. Free GFP expressed from empty pUC18-spGFP6 was used as a control for cytosolic localization. Proteins of fractions corresponding to 1 to 2 × 106 protoplasts were precipitated with chloroform-methanol (Wessel and Flügge, 1984) and analyzed by SDS-PAGE and anti-GFP immunoblotting.
For confocal microscopy of leaves, the lower epidermis and spongy mesophyll were removed from senescent leaves of Col-0 and mes16-1 using fine sandpaper. The palisade mesophyll was then observed with a laser scanning confocal microscope (TCS SP5; Leica Microsystems). FCC fluorescence was induced with an excitation wavelength of 355 nm, and the emission signal was recovered between 430 and 470 nm.
Targeting MES16 to the Chloroplast
A fusion protein consisting of the PPH transit peptide (amino acids 1–48; Schelbert et al., 2009), MES16, and an HA tag (PPHTP-MES16-HA) was produced using a two-step PCR (Expand High Fidelity PCR system) with the primers listed in Supplemental Table S1. The PCR fragment was cloned into pGEM-T-easy (Promega) and, after digestion with EcoRI/HindIII, subcloned into pHannibal (Wesley et al., 2001). The pHannibal cassette containing the cauliflower mosaic virus 35S promoter, the fusion open reading frame, and an octopine synthase terminator was excised with NotI and introduced into NotI-restricted pGreen0179 (Hellens et al., 2000). Arabidopsis Col-0 and pao-1 plants were transformed by the floral-dip method (Clough and Bent, 1998). Transformants were selected on hygromycin, and resistant T2 plants were used for further analysis. To verify that PPHTP-MES16-HA was targeted to the chloroplasts, protoplasts were isolated from Col-0 and Col-0/PPHTP-MES16-HA as described above. Chloroplasts were then isolated from protoplasts following published procedures (Kubis et al., 2008; Schelbert et al., 2009). Chl concentration was measured for each fraction as described below. Proteins from aliquots corresponding to 15 μg of Chl were precipitated with chloroform-methanol (Wessel and Flügge, 1984). In parallel, an aliquot of chloroplasts was fractionated into soluble and membrane fractions (Smith et al., 2002), and proteins were precipitated as well. Proteins from the different fractions were resolved by SDS-PAGE and analyzed by anti-HA immunoblotting.
RNA Isolation and RT-PCR
RNA was isolated using the RNeasy Plant kit (Qiagen). After DNA digestion with RQ1 DNase (Promega), first-strand cDNA was synthesized using the Moloney murine leukemia virus reverse transcriptase (Promega). PCR was performed with a nonsaturating number of amplification cycles as shown in the figures using gene-specific primers as listed in Supplemental Table S1.
Heterologous Expression of MES Proteins and Activity Determination
MES5, MES9, MES16, and MES17 cloned in pET28a (Vlot et al., 2008) were subcloned in pProEX Hta (Invitrogen) using EcoRI (MES5, MES16, and MES17) or EcoRI/XhoI (MES9) restriction sites. The MES18 coding sequence was amplified by PCR (Expand High Fidelity PCR system) from clone U50042 (Arabidopsis Biological Resource Center) using the primers listed in Supplemental Table S1 and cloned into pProEX Hta via EcoRI. After sequencing, the plasmids were transformed into BL21-CodonPlus (DE3)-RIL (Stratagene). At an optical density at 600 nm of 0.6, protein expression was induced with 1.0 mm isopropylthio-β-galactoside and cells were grown at 30°C overnight. Cells were centrifuged and resuspended in 20 mm Tris-HCl, pH 8, complemented with a protease inhibitor cocktail (Complete; Roche Applied Science) before lysis with a high-pressure cell breaker (Constant Cell Disruption System; Constant Systems) at a pressure of 150 MPa. The cell lysates were examined by SDS-PAGE and anti-His immunoblotting to quantify relative levels of MES protein expression.
Standard pFCC assays with MES16 (total volume of 20 μL) consisted of 0.25 μg of crude protein extracts and 15 μm pFCC. Pheide assays contained 0.85 mm Pheide and 100 μg of crude protein extracts in a total volume of 30 μL. For assays with an NCC, CjNCC-1 (Moser et al., 2008) was used at 15 μm. After incubation at 25°C in darkness for different periods of time as indicated in the figures, the reactions were stopped by the addition of methanol (pFCC assay) or acetone (Pheide assay) to final concentrations of 60% or 50%, respectively. Crude protein extracts produced with empty pProEX Hta were used as controls. After centrifugation (2 min at 16,000g), samples were analyzed by HPLC as described below. For competition experiments, inhibitors were added to the pFCC assay at a final concentration of 1.5 mm (100×) for MeIAA, MeSA, and MeJA or 150 μm (10×) for Pheide. pFCC assays with MES5, MES9, MES17, and MES18 (total volume of 40 μL) consisted of 7.5 μm pFCC and a volume of Escherichia coli lysates containing an amount of recombinant protein equivalent to the amount of MES16 present in 0.25 μg of crude MES16 extract.
PAO and RCCR Extraction from Red Pepper and Synthesis of pFCC
PAO and RCCR were extracted from red pepper (Capsicum annuum) fruits as follows. Exocarp tissue was blended in a Sorvall mixer three times for 5 s each in a solution (2–3 mL g−1 fresh weight) containing 400 mm Suc, 50 mm Tris-MES, pH 8, 2 mm EDTA, 10 mm polyethylene glycol 4000, 5 mm dithiothreitol, and 5 mm l(+)-ascorbic acid. The homogenate was filtered through two layers of Miracloth and centrifuged (10 min at 10,000g). The pellet was resuspended (25 μL g−1 fresh exocarp tissue) in 25 mm Tris-MES, pH 8, 5 mm l(+)-ascorbic acid, and 1% (v/v) Triton X-100 and incubated during 30 min in the dark at 4°C on a shaking plate (15 rpm) to solubilize membrane proteins. After ultracentrifugation (1 h at 100,000g), aliquots of the supernatant containing RCCR and solubilized PAO were frozen in liquid nitrogen and stored at −80°C before use. pFCC was synthesized as described previously (Pruzinská et al., 2007). Briefly, assays contained 80% (v/v) PAO/RCCR extract from red pepper (see above), 0.5 mm Pheide (Hörtensteiner et al., 1995), 0.2 mg mL−1 ferrodoxin (Fd), and a Fd-reducing system consisting of 2 mm Glc-6-P, 1 mm NADPH, 1 milliunit μL−1 Glc-6-P dehydrogenase, 0.1 milliunit μL−1 Fd-NADPH-oxidoreductase, and 0.1 milliunit μL−1 catalase. After 1 h of incubation at 25°C, the reaction was terminated by the addition of methanol to a final concentration of 60% (v/v). The assays were then cleaned and concentrated using a C18-SepPak cartridge (Waters) prior to purification of pFCC by HPLC (see below). Pure pFCC fractions were stored in liquid nitrogen before use.
Vacuole Isolation
Arabidopsis mesophyll protoplasts were isolated from 6-week-old short-day-grown Col-0 and mes16-1 plants after dark incubation of detached leaves for 4 d. Recovered protoplasts were digested in 10 volumes of lysis solution as described (Frelet-Barrand et al., 2008). The progression of vacuole release was continuously controlled using the microscope, and vacuoles were purified and concentrated by centrifugation (8 min at 1,400g) using a step gradient as follows: lower phase, 1 volume of lysate; middle phase, 1 volume of a 1:1 mixture of lysis solution and betaine buffer (0.4 m betaine, 30 mm potassium-gluconate, 20 mm HEPES-imidazole, pH 7.2, 1 mg mL−1 bovine serum albumin, and 1 mm dithiothreitol); upper phase, one-third volume of betaine buffer. Vacuoles were collected from the interface between the middle and upper phases. Contamination of Arabidopsis vacuoles with Chl was determined as described below. The activity of soluble vacuolar 3-N-acetyglucosaminidase was determined using p-nitrophenyl-N-acetyl-β-d-glucosaminide as described previously (Gao and Schaffer, 1999). Briefly, 10 μL of protoplast and vacuole fractions was incubated with 90 μL of 100 mm HEPES-KOH, pH 7.5, containing 3 mm p-nitrophenyl-N-acetyl-β-d-glucosaminide. The assay mixtures were incubated on a 96-well microtiter plate at room temperature, and the reaction was stopped by the addition of 160 μL of 1 m Na2CO3. The absorbance of formed p-nitrophenol was read with a plate reader spectrophotometer at 405 nm (Fusion Universal Microplate Analyzer; Packard). NCCs and FCCs of protoplast and vacuole fractions corresponding to equal activities of 3-N-acetyglucosaminidase were extracted with methanol and concentrated on a C18-SepPak cartridge prior to analysis by HPLC as described below for plant material.
Immunoblot Analysis and UV Analysis of Leaves
After separation by SDS-PAGE, proteins were transferred to nitrocellulose membranes according to standard procedures. Proteins were labeled with monoclonal antibodies against the poly-His tag (1:5,000) or GFP (1:2,000) or with polyclonal antibodies against the HA tag (1:5,000; all from Sigma). Thereafter, primary antibodies were labeled with horseradish peroxidase-conjugated secondary antibodies and proteins were visualized on a ChemiDoc XRS station (Bio-Rad) using the Immun-Star HRP Chemifluorescence kit (Bio-Rad). FCC fluorescence in senescent leaves of Col-0 and mes16-1 was visualized under UV light (366 nm) with the ChemiDoc XRS setup.
Analysis of Chl and Chl Catabolites
HPLC Analysis of Pheide, O134-Desmethyl Pheide, and Pyro-Pheide, and Quantification of Chl
Pigments were extracted from liquid nitrogen-homogenized tissue during 2 h at −20°C in 10% (v/v) 0.2 m Tris-HCl, pH 8, in acetone precooled to –20°C (5 mL g−1 fresh weight). After two centrifugation steps (4 min, 16,000g, 4°C), supernatants were analyzed by reverse-phase HPLC as described (Pruzinská et al., 2005). The same conditions were used to analyze the products of in vitro MES assays with Pheide as substrate. Pigments were identified by their absorption spectra and quantified using peak areas at 665 nm. For Chl quantification, supernatants were analyzed spectrophotometrically (Strain et al., 1971).
Chl quantification of protoplast, chloroplast, and vacuole fractions was performed as follows. An aliquot of the fractions was 333-fold diluted with 20% (v/v) 20 mm Tris-HCl, pH 8, in acetone and mixed vigorously. After centrifugation for 1 min at 16,000g, Chl concentrations of the supernatants were determined spectrophotometrically according to Arnon et al. (1959).
Colorless Chl Catabolites
Plant material was ground in liquid nitrogen, and colorless catabolites were extracted with 3 volumes (w/v) of 50 mm phosphate buffer, pH 7:methanol (1:3, v/v). The reverse-phase HPLC system consisted of a C18 Hypersil ODS column (250 × 4.6 mm; Thermo Electron), which was developed with a gradient (flow rate of 1.0 mL min−1) of solvent B (100% methanol) in solvent A (50 mm potassium phosphate, pH 7.0) as follows (all v/v): 20% to 60% in 30 min, 60% during 10 min, to 100% in 2 min, and 100% during 5 min.
For HPLC analysis of in vitro pFCC assays (esterase and isomerization assays), the column was developed with a gradient (flow rate of 1.0 mL min−1) of solvent B in solvent A as follows (all v/v): 35% to 75% in 19 min, to 100% in 1 min, and 100% during 4 min.
Peak detection was performed with sequential monitoring using a PA-100 photodiode array detector (200–700 nm; Dionex) and a RF2000 fluorescence detector (excitation at 320 nm, emission at 450 nm; Dionex). Chl catabolites were identified by their absorption (FCCs and NCCs) and fluorescence (FCCs) properties. Relative amounts of FCCs and NCCs were determined using peak areas at 320 nm.
MS
For MS analysis, catabolite-containing HPLC fractions derived from MES16 enzyme assays or mes16-1 leaf extracts were mixed with 1 volume of a saturated solution of 2,5-dihydroxybenzoic acid in acetonitrile and were spotted onto stainless-steel targets. MS was performed with an Ultraflex matrix-assisted laser-desorption ionization time of flight mass spectrometer (Bruker).
Matrix-assisted laser-desorption ionization time of flight MS data (percentage relative intensity, molecular formula, and type of ion) are as follows: mes16-FCC-1: 883.09 (33, C41H49K2N4O13, [M+2K]+), 845.13 (100, C41H50K1N4O13, [M+K]+), 807.16 (56, C41H51N4O13, [M+H]+); mes16-FCC-2: 683.26 (45, C35H40K1N4O8, [M+K]+), 645.30 (100, C35H41N4O8, [M+H]+); mes16-FCC-3: 667.23 (19, C35H40K1N4O7, [M+K]+), 629.25 (100, C35H41N4O7, [M+H]+); mes16-NCC-1: 883.33 (16, C41H49K2N4O13, [M+2K]+), 845.36 (100, C41H50K1N4O13, [M+K]+), 807.38 (50, C41H51N4O13, [M+H]+); O134-desmethyl pFCC: 647.14 (25, C33H37K2N4O5, [M+2K-CO2]+), 615.22 (33, C34H39N4O7, [M+H]+), 609.17 (80, C33H38K1N4O5, [M+1K-CO2]+), 571.19 (100, C33H39N4O5, [M+H-CO2]+).
In Vitro FCC-to-NCC Isomerization
O134-desmethyl pFCC was produced from pure pFCC fractions using heterologously expressed MES16 and purified by HPLC. The isomerization assays consisted of 11.5 μm FCC (pFCC or O134-desmethyl pFCC) and 70 mm phosphate buffer, pH 5 or 6. Aliquots were taken after the incubation times indicated in Figure 5C, and isomerization was stopped by adding Tris-HCl, pH 8, to a concentration of 300 mm. Methanol was added to a final concentration of 35% (v/v) prior to analysis by HPLC as described above.
GenBank identification numbers for the DNA/protein sequences used in this work are as follows: Arabidopsis: AtMES1, 48310671 (AT2G23620); AtMES2, 15227863 (AT2G23600); AtMES3, 50198958 (AT2G23610); AtMES4, 34146844 (AT2G23580); AtMES5, 332004135 (AT5G10300); AtMES6, 330252371 (AT2G23550); AtMES7, 46402456 (AT2G23560); AtMES8, 330252376 (AT2G23590); AtMES9, 30017285 (AT4G37150); AtMES10, 332645146 (AT3G50440); AtMES11, 27808602 (AT3G29770); AtMES12, 332657411 (AT4G09900); AtMES13, 332192561 (AT1G26360); AtMES14, 94442411 (AT1G33990); AtMES15, 332196779 (AT1G69240); AtMES16, 332658384 (AT4G16690); AtMES17, 332641444 (AT3G10870); AtMES18, 332009649 (AT5G58310); AtMES19, 330252373 (AT2G23570); AtMES20, 332661358 (AT4G37140); PAO, 15230543 (AT3G44880); PPH, 15240707 (AT5G13800); canola: BnMES1, 151016294 (fragment); BnMES2, 151011122 (fragment); BnMES3, 125936604 (fragment); BnMES4, 151324766 (fragment); BnMES5, 242292656 (fragment); BnMES6, 150871604; BnMES7, 29690059 (fragment); barley: HvMES1, 60272487 (fragment); HvMES2, 94339163 (fragment); HvMES3, 24242376 (fragment); HvMES4, 24285085 (fragment); radish (Raphanus sativus): PPD, 122209128; maize: ZmMES1, 211364330 (fragment); ZmMES2, 211406694 (fragment); ZmMES3, 21481135 (fragment); ZmMES4, 149089611; ZmMES5, 78077695 (fragment); ZmMES6, 87153297 (fragment).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Coexpression network around PAO, PPH, SGR, NYC1, and MES16.
Supplemental Figure S2. Analysis of recombinant MES5, MES9, MES16, MES17, and MES18.
Supplemental Figure S3. Colorless catabolites occurring in mes16-1 mutants during natural senescence and in mutants of other closely related MES family members after dark incubation.
Supplemental Figure S4. Colorless catabolites of Col-0/PPHTP-MES16-HA.
Supplemental Table S1. List of primers used in this study.
Supplementary Material
Acknowledgments
We thank Dr. Daniel F. Klessig at the Boyce Thompson Institute for Plant Research at Cornell University for providing E. coli expression clones (pET28a) for MES5, MES9, MES16, and MES17.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archetti M, Brown SP. (2004) The coevolution theory of autumn colours. Proc Biol Sci 271: 1219–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon DI, Whatley FR, Allen MB. (1959) Photosynthesis by isolated chloroplasts. VIII. Photosynthetic phosphorylation and the generation of assimilatory power. Biochim Biophys Acta 32: 47–57 [DOI] [PubMed] [Google Scholar]
- Balzergue S, Dubreucq B, Chauvin S, Le-Clainche I, Le Boulaire F, de Rose R, Samson F, Biaudet V, Lecharny A, Cruaud C, et al. (2001) Improved PCR-walking for large-scale isolation of plant T-DNA borders. Biotechniques 30: 496–498, 502, 504 [DOI] [PubMed] [Google Scholar]
- Banala S, Moser S, Müller T, Kreutz CR, Holzinger A, Lütz C, Kräutler B. (2010) Hypermodified fluorescent chlorophyll catabolites: source of blue luminescence in senescent leaves. Angew Chem Int Ed Engl 49: 5174–5177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berghold J, Breuker K, Oberhuber M, Hörtensteiner S, Kräutler B. (2002) Chlorophyll breakdown in spinach: on the structure of five nonfluorescent chlorophyll catabolites. Photosynth Res 74: 109–119 [DOI] [PubMed] [Google Scholar]
- Berghold J, Eichmüller C, Hörtensteiner S, Kräutler B. (2004) Chlorophyll breakdown in tobacco: on the structure of two nonfluorescent chlorophyll catabolites. Chem Biodivers 1: 657–668 [DOI] [PubMed] [Google Scholar]
- Berghold J, Müller T, Ulrich M, Hörtensteiner S, Kräutler B. (2006) Chlorophyll breakdown in maize: on the structure of two nonfluorescent chlorophyll catabolites. Monatsh Chem 137: 751–763 [Google Scholar]
- Brown SB, Houghton JD, Hendry GAF. (1991) Chlorophyll breakdown. Scheer H, ed, Chlorophylls. CRC Press, Boca Raton, FL, pp 465–489 [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Curty C, Engel N. (1996) Detection, isolation and structure elucidation of a chlorophyll a catabolite from autumnal senescent leaves of Cercidiphyllum japonicum. Phytochemistry 42: 1531–1536 [Google Scholar]
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard J-F, Guindon S, Lefort V, Lescot M, et al. (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36: W465–W469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson G, Wlodawer A. (1998) Catalytic triads and their relatives. Trends Biochem Sci 23: 347–352 [DOI] [PubMed] [Google Scholar]
- Endler A, Meyer S, Schelbert S, Schneider T, Weschke W, Peters SW, Keller F, Baginsky S, Martinoia E, Schmidt UG. (2006) Identification of a vacuolar sucrose transporter in barley and Arabidopsis mesophyll cells by a tonoplast proteomic approach. Plant Physiol 141: 196–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forouhar F, Yang Y, Kumar D, Chen Y, Fridman E, Park SW, Chiang Y, Acton TB, Montelione GT, Pichersky E, et al. (2005) Structural and biochemical studies identify tobacco SABP2 as a methyl salicylate esterase and implicate it in plant innate immunity. Proc Natl Acad Sci USA 102: 1773–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frelet-Barrand A, Kolukisaoglu HU, Plaza S, Rüffer M, Azevedo L, Hörtensteiner S, Marinova K, Weder B, Schulz B, Klein M. (2008) Comparative mutant analysis of Arabidopsis ABCC-type ABC transporters: AtMRP2 contributes to detoxification, vacuolar organic anion transport and chlorophyll degradation. Plant Cell Physiol 49: 557–569 [DOI] [PubMed] [Google Scholar]
- Gao Z, Schaffer AA. (1999) A novel alkaline α-galactosidase from melon fruit with a substrate preference for raffinose. Plant Physiol 119: 979–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray J, Wardzala E, Yang M, Reinbothe S, Haller S, Pauli F. (2004) A small family of LLS1-related non-heme oxygenases in plants with an origin amongst oxygenic photosynthesizers. Plant Mol Biol 54: 39–54 [DOI] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM. (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Hilditch PI, Thomas H, Thomas BJ, Rogers LJ. (1989) Leaf senescence in a non-yellowing mutant of Festuca pratensis: proteins of photosystem II. Planta 177: 265–272 [DOI] [PubMed] [Google Scholar]
- Hinder B, Schellenberg M, Rodoni S, Ginsburg S, Vogt E, Martinoia E, Matile P, Hörtensteiner S. (1996) How plants dispose of chlorophyll catabolites: directly energized uptake of tetrapyrrolic breakdown products into isolated vacuoles. J Biol Chem 271: 27233–27236 [DOI] [PubMed] [Google Scholar]
- Horie Y, Ito H, Kusaba M, Tanaka R, Tanaka A. (2009) Participation of chlorophyll b reductase in the initial step of the degradation of light-harvesting chlorophyll a/b-protein complexes in Arabidopsis. J Biol Chem 284: 17449–17456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörtensteiner S. (2006) Chlorophyll degradation during senescence. Annu Rev Plant Biol 57: 55–77 [DOI] [PubMed] [Google Scholar]
- Hörtensteiner S. (2009) Stay-green regulates chlorophyll and chlorophyll-binding protein degradation during senescence. Trends Plant Sci 14: 155–162 [DOI] [PubMed] [Google Scholar]
- Hörtensteiner S, Kräutler B. (2011) Chlorophyll breakdown in higher plants. Biochim Biophys Acta 1807: 977–988 [DOI] [PubMed] [Google Scholar]
- Hörtensteiner S, Vicentini F, Matile P. (1995) Chlorophyll breakdown in senescent cotyledons of rape, Brassica napus L.: enzymatic cleavage of phaeophorbide a in vitro. New Phytol 129: 237–246 [DOI] [PubMed] [Google Scholar]
- Hörtensteiner S, Wüthrich KL, Matile P, Ongania K-H, Kräutler B. (1998) The key step in chlorophyll breakdown in higher plants: cleavage of pheophorbide a macrocycle by a monooxygenase. J Biol Chem 273: 15335–15339 [DOI] [PubMed] [Google Scholar]
- Joyard J, Ferro M, Masselon C, Seigneurin-Berny D, Salvi D, Garin J, Rolland N. (2009) Chloroplast proteomics and the compartmentation of plastidial isoprenoid biosynthetic pathways. Mol Plant 2: 1154–1180 [DOI] [PubMed] [Google Scholar]
- Kräutler B. (2008) Chlorophyll catabolites. Herz W, Falk H, Kirny GW, Moore RE, Tann C, eds, Progress in the Chemistry of Organic Natural Products, Vol 89. Springer, Vienna, pp 1–43 [DOI] [PubMed] [Google Scholar]
- Kräutler B, Banala S, Moser S, Vergeiner C, Müller T, Lütz C, Holzinger A. (2010) A novel blue fluorescent chlorophyll catabolite accumulates in senescent leaves of the peace lily and indicates a split path of chlorophyll breakdown. FEBS Lett 584: 4215–4221 [DOI] [PubMed] [Google Scholar]
- Kräutler B, Hörtensteiner S. (2006) Chlorophyll catabolites and the biochemistry of chlorophyll breakdown. Grimm B Porra R Rüdiger W Scheer H, eds, Chlorophylls and Bacteriochlorophylls: Biochemistry, Biophysics, Functions and Applications, Vol 25. Springer, Dordrecht, The Netherlands, pp 237–260 [Google Scholar]
- Kräutler B, Jaun B, Bortlik K-H, Schellenberg M, Matile P. (1991) On the enigma of chlorophyll degradation: the constitution of a secoporphinoid catabolite. Angew Chem Int Ed Engl 30: 1315–1318 [Google Scholar]
- Kubis SE, Lilley KS, Jarvis P. (2008) Isolation and preparation of chloroplasts from Arabidopsis thaliana plants. Methods Mol Biol 425: 171–186 [DOI] [PubMed] [Google Scholar]
- Kusaba M, Ito H, Morita R, Iida S, Sato Y, Fujimoto M, Kawasaki S, Tanaka R, Hirochika H, Nishimura M, et al. (2007) Rice NON-YELLOW COLORING1 is involved in light-harvesting complex II and grana degradation during leaf senescence. Plant Cell 19: 1362–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losey FG, Engel N. (2001) Isolation and characterization of a urobilinogenoidic chlorophyll catabolite from Hordeum vulgare L. J Biol Chem 276: 8643–8647 [DOI] [PubMed] [Google Scholar]
- Matile P, Ginsburg S, Schellenberg M, Thomas H. (1988) Catabolites of chlorophyll in senescing barley leaves are localized in the vacuoles of mesophyll cells. Proc Natl Acad Sci USA 85: 9529–9532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matile P, Schellenberg M. (1996) The cleavage of pheophorbide a is located in the envelope of barley gerontoplasts. Plant Physiol Biochem 34: 55–59 [Google Scholar]
- Matile P, Schellenberg M, Peisker C. (1992) Production and release of a chlorophyll catabolite in isolated senescent chloroplasts. Planta 187: 230–235 [DOI] [PubMed] [Google Scholar]
- Meguro M, Ito H, Takabayashi A, Tanaka R, Tanaka A. (2011) Identification of the 7-hydroxymethyl chlorophyll a reductase of the chlorophyll cycle in Arabidopsis Plant Cell 23: 3442–3453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer A, Eskandari S, Grallath S, Rentsch D. (2006) AtGAT1, a high affinity transporter for γ-aminobutyric acid in Arabidopsis thaliana. J Biol Chem 281: 7197–7204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita R, Sato Y, Masuda Y, Nishimura M, Kusaba M. (2009) Defect in non-yellow coloring 3, an α/β hydrolase-fold family protein, causes a stay-green phenotype during leaf senescence in rice. Plant J 59: 940–952 [DOI] [PubMed] [Google Scholar]
- Moser S, Müller T, Holzinger A, Lütz C, Jockusch S, Turro NJ, Kräutler B. (2009) Fluorescent chlorophyll catabolites in bananas light up blue halos of cell death. Proc Natl Acad Sci USA 106: 15538–15543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser S, Ulrich M, Müller T, Kräutler B. (2008) A yellow chlorophyll catabolite is a pigment of the fall colours. Photochem Photobiol Sci 7: 1577–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlecker W, Kräutler B. (1996) Breakdown of chlorophyll: constitution of nonfluorescing chlorophyll-catabolites from senescent cotyledons of the dicot rape. Plant Physiol Biochem 34: 61–75 [Google Scholar]
- Mühlecker W, Ongania K-H, Kräutler B, Matile P, Hörtensteiner S. (1997) Tracking down chlorophyll breakdown in plants: elucidation of the constitution of a ‘fluorescent’ chlorophyll catabolite. Angew Chem Int Ed Engl 36: 401–404 [Google Scholar]
- Obayashi T, Hayashi S, Saeki M, Ohta H, Kinoshita K. (2009) ATTED-II provides coexpressed gene networks for Arabidopsis. Nucleic Acids Res 37: D987–D991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberhuber M, Berghold J, Breuker K, Hörtensteiner S, Kräutler B. (2003) Breakdown of chlorophyll: a nonenzymatic reaction accounts for the formation of the colorless “nonfluorescent” chlorophyll catabolites. Proc Natl Acad Sci USA 100: 6910–6915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberhuber M, Berghold J, Kräutler B. (2008) Chlorophyll breakdown by a biomimetic route. Angew Chem Int Ed Engl 47: 3057–3061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S-Y, Yu J-W, Park J-S, Li J, Yoo S-C, Lee N-Y, Lee S-K, Jeong S-W, Seo HS, Koh H-J, et al. (2007) The senescence-induced staygreen protein regulates chlorophyll degradation. Plant Cell 19: 1649–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruzinská A, Anders I, Aubry S, Schenk N, Tapernoux-Lüthi E, Müller T, Kräutler B, Hörtensteiner S. (2007) In vivo participation of red chlorophyll catabolite reductase in chlorophyll breakdown. Plant Cell 19: 369–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruzinská A, Tanner G, Anders I, Roca M, Hörtensteiner S. (2003) Chlorophyll breakdown: pheophorbide a oxygenase is a Rieske-type iron-sulfur protein, encoded by the accelerated cell death 1 gene. Proc Natl Acad Sci USA 100: 15259–15264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruzinská A, Tanner G, Aubry S, Anders I, Moser S, Müller T, Ongania K-H, Kräutler B, Youn J-Y, Liljegren SJ, et al. (2005) Chlorophyll breakdown in senescent Arabidopsis leaves: characterization of chlorophyll catabolites and of chlorophyll catabolic enzymes involved in the degreening reaction. Plant Physiol 139: 52–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren GD, Zhou Q, Wu SX, Zhang YF, Zhang LG, Huang JR, Sun ZF, Kuai BK. (2010) Reverse genetic identification of CRN1 and its distinctive role in chlorophyll degradation in Arabidopsis. J Integr Plant Biol 52: 496–504 [DOI] [PubMed] [Google Scholar]
- Rodoni S, Mühlecker W, Anderl M, Kräutler B, Moser D, Thomas H, Matile P, Hörtensteiner S. (1997) Chlorophyll breakdown in senescent chloroplasts: cleavage of pheophorbide a in two enzymic steps. Plant Physiol 115: 669–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B. (2003) An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol 53: 247–259 [DOI] [PubMed] [Google Scholar]
- Sato Y, Morita R, Katsuma S, Nishimura M, Tanaka A, Kusaba M. (2009) Two short-chain dehydrogenase/reductases, NON-YELLOW COLORING 1 and NYC1-LIKE, are required for chlorophyll b and light-harvesting complex II degradation during senescence in rice. Plant J 57: 120–131 [DOI] [PubMed] [Google Scholar]
- Schelbert S, Aubry S, Burla B, Agne B, Kessler F, Krupinska K, Hörtensteiner S. (2009) Pheophytin pheophorbide hydrolase (pheophytinase) is involved in chlorophyll breakdown during leaf senescence in Arabidopsis. Plant Cell 21: 767–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk N, Schelbert S, Kanwischer M, Goldschmidt EE, Dörmann P, Hörtensteiner S. (2007) The chlorophyllases AtCLH1 and AtCLH2 are not essential for senescence-related chlorophyll breakdown in Arabidopsis thaliana. FEBS Lett 581: 5517–5525 [DOI] [PubMed] [Google Scholar]
- Schoch S, Vielwerth FX. (1983) Chlorophyll degradation in senescent tobacco cell culture (Nicotiana tabacum var. “Samsun”). Z Pflanzenphysiol 110: 309–317 [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al. (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimokawa K, Hashizume A, Shioi Y. (1990) Pyropheophorbide a, a catabolite of ethylene-induced chlorophyll a degradation. Phytochemistry 29: 2105–2106 [Google Scholar]
- Shioi Y, Tatsumi Y, Shimokawa K. (1991) Enzymatic degradation of chlorophyll in Chenopodium album. Plant Cell Physiol 32: 87–93 [Google Scholar]
- Shioi Y, Watanabe K, Takamiya K. (1996) Enzymatic conversion of pheophorbide a to a precursor of pyropheophorbide a in leaves of Chenopodium album. Plant Cell Physiol 37: 1143–1149 [Google Scholar]
- Smith D, Schnell D, Fitzpatrick L, Keegstra K. (2002) In vitro analysis of chloroplast protein import. Curr Protoc Cell Biol 14: 11.16.11–11.16.21 [DOI] [PubMed] [Google Scholar]
- Strain HH, Cope BT, Svec WA. (1971) Analytical procedures for the isolation, identification, estimation and investigation of the chlorophylls. Methods Enzymol 23: 452–476 [Google Scholar]
- Suzuki T, Kunieda T, Murai F, Morioka S, Shioi Y. (2005) Mg-dechelation activity in radish cotyledons with artificial and native substrates, Mg-chlorophyllin a and chlorophyllide a. Plant Physiol Biochem 43: 459–464 [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Amano T, Shioi Y. (2006) Characterization and cloning of the chlorophyll-degrading enzyme pheophorbidase from cotyledons of radish. Plant Physiol 140: 716–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Doi M, Shioi Y. (2002) Two enzymatic reaction pathways in the formation of pyropheophorbide a. Photosynth Res 74: 225–233 [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Shioi Y. (1999) Detection of chlorophyll breakdown products in the senescent leaves of higher plants. Plant Cell Physiol 40: 909–915 [Google Scholar]
- Suzuki Y, Soga K, Yoshimatsu K, Shioi Y. (2008) Expression and purification of pheophorbidase, an enzyme catalyzing the formation of pyropheophorbide during chlorophyll degradation: comparison with the native enzyme. Photochem Photobiol Sci 7: 1260–1266 [DOI] [PubMed] [Google Scholar]
- Takamiya KI, Tsuchiya T, Ohta H. (2000) Degradation pathway(s) of chlorophyll: what has gene cloning revealed? Trends Plant Sci 5: 426–431 [DOI] [PubMed] [Google Scholar]
- Tanaka R, Hirashima M, Satoh S, Tanaka A. (2003) The Arabidopsis-accelerated cell death gene ACD1 is involved in oxygenation of pheophorbide a: inhibition of the pheophorbide a oxygenase activity does not lead to the “stay-green” phenotype in Arabidopsis. Plant Cell Physiol 44: 1266–1274 [DOI] [PubMed] [Google Scholar]
- Vlot AC, Liu PP, Cameron RK, Park SW, Yang Y, Kumar D, Zhou FS, Padukkavidana T, Gustafsson C, Pichersky E, et al. (2008) Identification of likely orthologs of tobacco salicylic acid-binding protein 2 and their role in systemic acquired resistance in Arabidopsis thaliana. Plant J 56: 445–456 [DOI] [PubMed] [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, Wang MB, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, et al. (2001) Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J 27: 581–590 [DOI] [PubMed] [Google Scholar]
- Wessel D, Flügge UI. (1984) A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem 138: 141–143 [DOI] [PubMed] [Google Scholar]
- Wüthrich KL, Bovet L, Hunziker PE, Donnison IS, Hörtensteiner S. (2000) Molecular cloning, functional expression and characterisation of RCC reductase involved in chlorophyll catabolism. Plant J 21: 189–198 [DOI] [PubMed] [Google Scholar]
- Yang Y, Xu R, Ma CJ, Vlot AC, Klessig DF, Pichersky E. (2008) Inactive methyl indole-3-acetic acid ester can be hydrolyzed and activated by several esterases belonging to the AtMES esterase family of Arabidopsis. Plant Physiol 147: 1034–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler R, Blaheta A, Guha N, Schönegge B. (1988) Enzymatic formation of pheophorbide and pyropheophorbide during chlorophyll degradation in a mutant of Chlorella fusca Shiria et Kraus. J Plant Physiol 132: 327–332 [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.