Abstract
This study characterized genetic interactions between the maize (Zea mays) genes dull1 (du1), encoding starch synthase III (SSIII), and isa2, encoding a noncatalytic subunit of heteromeric isoamylase-type starch-debranching enzyme (ISA1/ISA2 heteromer). Mutants lacking ISA2 still possess the ISA1 homomeric enzyme. Eight du1- mutations were characterized, and structural changes in amylopectin resulting from each were measured. In every instance, the same complex pattern of alterations in discontinuous spans of chain lengths was observed, which cannot be explained solely by a discrete range of substrates preferred by SSIII. Homozygous double mutants were constructed containing the null mutation isa2-339 and either du1-Ref, encoding a truncated SSIII protein lacking the catalytic domain, or the null allele du1-R4059. In contrast to the single mutant parents, double mutant endosperms affected in both SSIII and ISA2 were starch deficient and accumulated phytoglycogen. This phenotype was previously observed only in maize sugary1 mutants impaired for the catalytic subunit ISA1. ISA1 homomeric enzyme complexes assembled in both double mutants and were enzymatically active in vitro. Thus, SSIII is required for normal starch crystallization and the prevention of phytoglycogen accumulation when the only isoamylase-type debranching activity present is ISA1 homomer, but not in the wild-type condition, when both ISA1 homomer and ISA1/ISA2 heteromer are present. Previous genetic and biochemical analyses showed that SSIII also is required for normal glucan accumulation when the only isoamylase-type debranching enzyme activity present is ISA1/ISA heteromer. These data indicate that isoamylase-type debranching enzyme and SSIII work in a coordinated fashion to repress phytoglycogen accumulation.
Semicrystalline glucan polymers that form insoluble starch granules are found in the vast majority of organisms within the Archaeplastida lineage of primary photosynthetic eukaryotes. This group, which comprises glaucophytes, rhodophytes (red algae), and Chloroplastida (green algae and land plants), in general does not contain soluble glucan polymers of substantial size. Conversely, with a few exceptions, essentially all other eukaryotes and prokaryotes utilize the soluble polymer glycogen for the storage of Glc and lack any insoluble glucans. Starch granules and their constituent polymers are capable of storing many more Glc units than chemically similar but soluble glucans, and this is likely to have provided a selective advantage during the establishment and spread of the photosynthetic eukaryotes. Support for this suggestion comes from the facts that the primary photosynthetic eukaryotes are a monophyletic group (Rodríguez-Ezpeleta et al., 2005). In light of the important role of starch metabolism in these organisms, including the land plants, it is of interest to understand the molecular mechanisms that generate semicrystalline glucans and how these processes differ from those that generate glycogen.
Starch granules are made up of two types of glucan polymer, amylopectin and amylose. Amylopectin and glycogen are essentially identical in chemical nature but vary architecturally (Ball and Morell, 2003). Both polymers are made up of linear chains of Glc units joined by α(1→4) glycoside bonds. Such chains are attached to each other by α(1→6) glycosidic linkages to form a branched polymer. The frequency of branch points varies from approximately 5% in amylopectin to 10% in glycogen, so the average degree of polymerization (DP) of the linear chains is about 20 and 10, respectively. Furthermore, branches in glycogen are dispersed compared with their clustered locations in amylopectin. In glycogen, the chain length distribution is unimodal, whereas a polymodal distribution is observed for amylopectin linear chains (Hizukuri, 1986). These features enable the crystallization of unbranched chains within clusters, thus allowing continued growth of amylopectin molecules without generating osmotic stress within the cell. This packing efficiency increases the total number of Glc units in amylopectin compared with glycogen, which is about 107 and 106, respectively. Amylose, the other glucan polymer in starch granules, is branched at a very low frequency and so can be considered as quasilinear. Amylose is interspersed with amylopectin in insoluble granules but is not necessary for the semicrystalline nature of starch.
Starch biosynthetic enzymes in chloroplast-containing organisms include five conserved classes of starch synthase (SS), which elongate linear glucans at the nonreducing end using ADP-Glc as the monosaccharide donor, and two or three starch branching enzymes (SBEs), which generate branch linkages by cleaving an α(1→4) bond and transferring the released linear segment to a 6-hydroxyl group elsewhere in the molecule (Li et al., 2003; Deschamps et al., 2008). Four α(1→6)-glucosidases, referred to as starch debranching enzymes (DBEs), are also conserved in the green algae and land plants. These include the ISA1, ISA2, and ISA3 proteins that are classified as “isoamylase-type” DBEs based on structural and enzymatic similarities to prokaryotic isoamylases.
DBEs are hydrolases belonging to the α-amylase superfamily. Such enzymes typically are involved in glucan catabolism, and this is true of ISA3 proteins (Delatte et al., 2005; Wattebled et al., 2005). ISA1 and ISA2, however, are not necessary for starch catabolism. Rather, genetic analyses show them to be important in the generation of semicrystalline amylopectin as opposed to soluble, glycogen-like polymers. Mutations affecting ISA1, and in dicots ISA2 as well, typically cause major reductions in starch content. A soluble, glycogen-like polymer referred to as phytoglycogen accumulates in such mutants, whereas this polymer is not observed in wild-type plants. Residual starch granules in ISA1-deficient mutants are abnormal in size, shape, and number and contain structurally altered amylopectin. These observations are remarkably consistent across species and tissues, including maize (Zea mays), rice (Oryza sativa), and barley (Hordeum vulgare) endosperm, potato (Solanum tuberosum) tuber, Arabidopsis (Arabidopsis thaliana) leaf, and Chlamydomonas reinhardtii cells (James et al., 1995; Mouille et al., 1996; Zeeman et al., 1998; Kubo et al., 1999; Dauvillée et al., 2001a; Burton et al., 2002; Bustos et al., 2004; Delatte et al., 2005; Wattebled et al., 2005, 2008; Streb et al., 2008). Thus, ISA1 and ISA2 appear to have been adapted primarily for activity in starch biosynthesis rather than degradation. The role of these DBEs appears to be remodeling of a soluble glucan precursor by the removal of certain branch linkages, thus facilitating the transition to the semicrystalline state (Ball et al., 1996; Myers et al., 2000; Streb et al., 2008).
Starch decrease and phytoglycogen accumulation have been utilized as a scorable phenotype in numerous organisms in forward and reverse genetic screens, and to date, soluble glucan polymers have been observed only in mutants affected in ISA1 or ISA2 function. In Chlamydomonas, potato, or Arabidopsis, mutations in two genes condition phytoglycogen accumulation. These encode ISA1 and ISA2, which are components of a heteromeric complex. Both proteins are necessary for enzymatic function, explaining why mutation of either gene causes the same phenotype (Dauvillée et al., 2001a, 2001b; Hussain et al., 2003; Bustos et al., 2004; Delatte et al., 2005; Delvallé et al., 2005). As in other species, ISA1 is necessary for normal starch biosynthesis in cereal endosperm (James et al., 1995; Kubo et al., 1999; Burton et al., 2002); however, the lack of ISA2 does not cause phytoglycogen accumulation (Kubo et al., 2010; Utsumi et al., 2011). The explanation for the discrepancy between species is that in maize and rice endosperm, a homomeric ISA1 enzyme is active in addition to the ISA1/ISA2 heteromer (Utsumi and Nakamura, 2006; Kubo et al., 2010). The reason that ISA1 can function independently in cereal endosperm but requires ISA2 as a partner in other plant tissues is not known.
This study sought to identify additional proteins that are necessary to prevent phytoglycogen accumulation. As noted, the only proteins thus characterized in any Chloroplastida species are ISA1 and ISA2. The strategy utilized ISA2 mutations that prevent the activity of ISA1/ISA2 heteromer but do not affect ISA1 homomer. These mutants exhibit essentially normal starch biosynthesis and do not accumulate phytoglycogen in mature endosperm. Second-site mutations were then identified that conditioned phytoglycogen accumulation. The data revealed that defects in a specific class of SS, namely SSIII, result in the appearance of phytoglycogen and decreased starch content, but only in the specific condition that ISA1/ISA2 heteromer is absent.
RESULTS
Molecular Characterization of du1- Mutations
Two different alleles of the maize dull1 (du1) locus, which codes for SSIII (Gao et al., 1998), were used to test whether this enzyme can affect storage glucan crystallization. Mutations of du1 cause the “dull” kernel phenotype recognized by a tarnished surface appearance, orange color at the crown, and dimpled indentations on the abgerminal side (Fig. 1A; Cameron, 1947; Mangelsdorf, 1947). The spontaneous mutation du1-Ref first defined the locus (Mangelsdorf, 1947), and the allelic mutation du1-R4059 arose spontaneously in a genetic background with an active Mutator (Mu) transposon system (Gao et al., 1998). Endosperm tissue homozygous for du1-Ref was shown previously to contain a truncated SSIII protein reduced in amount compared with the wild type (Cao et al., 1999). SSIII was not detected in homozygous du1-R4059 endosperm, indicating a null mutation (Cao et al., 1999). These results were confirmed using an affinity-purified, specific IgG fraction in immunoblot analyses of soluble endosperm extracts from kernels harvested 20 d after pollination (DAP; Fig. 2A).
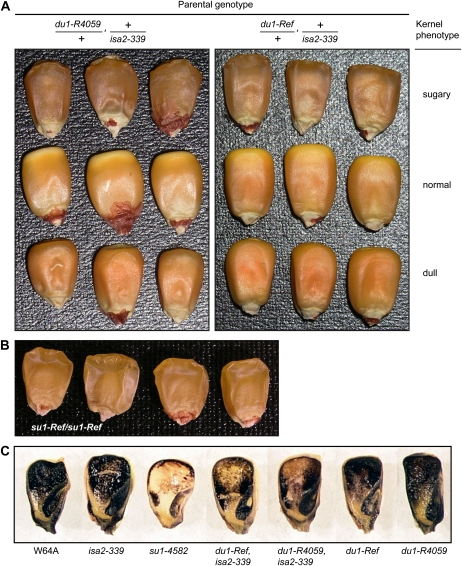
Figure 1.
Kernel phenotypes. A, Mature kernels. F1 plants of the indicated parental genotype were self-pollinated to generate segregating F2 kernel populations. Representatives of the three segregating kernel phenotypes are shown. Sugary kernels are shrunken and translucent compared with the wild type. Dull kernels have a tarnished appearance, a deeper orange color than the wild type particularly at the crown, and exhibit characteristic small indentations at the abgerminal base. B, Mature kernels from a su1-Ref/su1-Ref plant. C, Glucan stains. Kernels of the indicated genotype were harvested 20 DAP, sliced longitudinally, and dipped in I2/KI solution. Dark blue color indicates starch, and brown or tan color indicates phytoglycogen.
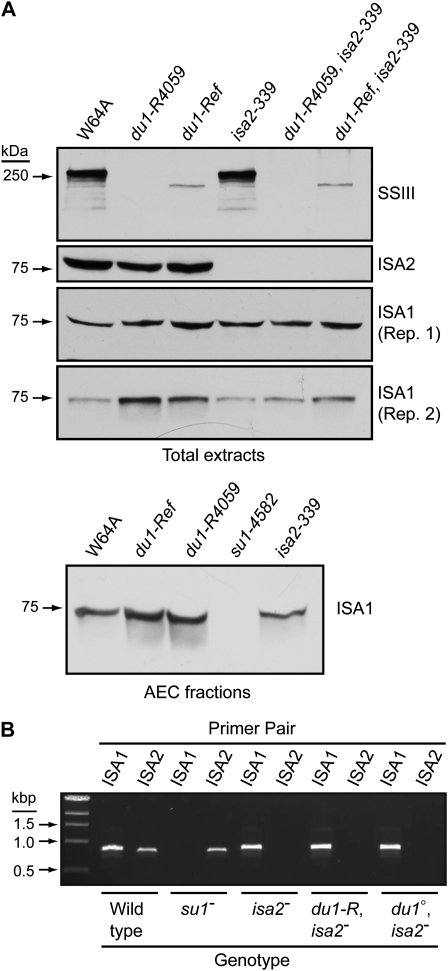
Figure 2.
Protein and transcript levels. A, Immunoblot analyses. Proteins (40 μg) from endosperm harvested 20 DAP were separated by SDS-PAGE and then probed in immunoblot analyses with antisera specific for the indicated protein. The top three panels show analyses of the same total soluble extracts. The bottom panel is an independent biological replicate analyzed for the presence of ISA1, using a short exposure of the immunoblot. In a separate analysis, AEC fractions containing ISA1 were pooled and concentrated prior to immunoblot analysis using αISA1 antibodies. Protein loads and concentration factors were identical between genotypes. B, ISA1 and ISA2 mRNA detected by RT-PCR. Kernels of the indicated genotype were harvested 20 DAP, and RNA from endosperm tissue was amplified by PCR using primer pairs specific for either transcript. Allele abbreviations are as follows: isa2-, isa2-339; du1-R, du1-Ref; du1°, du1-R4059; su1-, su1-4582.
The molecular structures of du1-Ref and du1-R4059 were determined. The wild-type allele Du1 from inbred W64A was sequenced from PCR-amplified fragments of genomic DNA, revealing a transcribed region of 11.6 kb comprising 17 exons and 16 introns (Supplemental Fig. S1A; GenBank accession no. JF273457). This sequence is 98% identical to Du1 from inbred B73 published on the maize genome database, located on chromosome 10 beginning at nucleotide 59,506,710 (http://www.plantgdb.org/ZmGDB; Ref_Gen_v2). The locus from a homozygous du1-Ref plant was also sequenced, revealing a C/T transversion at nucleotide 3,675 of the cDNA (GenBank accession no. AF023159) that creates a stop codon after residue 1,185 of the full-length protein. Premature termination would eliminate the C-terminal 489 residues of SSIII, which comprise the SS catalytic domain (Cao et al., 1999). The predicted molecular mass of the mature mutant protein is 126 kD, discounting the 69-residue plastid transit peptide predicted by the TargetP algorithm (Nielsen et al., 1997; Emanuelsson et al., 2000). The mutant protein migrates only slightly faster than the 250-kD marker, however, likely owing to anomalous behavior in SDS-PAGE (Fig. 2A). Wild-type SSIII also exhibits anomalous migration, running above the 250-kD marker even though the predicted mass of the mature protein is 181 kD (Fig. 2A).
The allele du1-R4059 is a null mutation that fails to produce transcript. DU1 mRNA was not detected by reverse transcription (RT)-PCR amplification in homozygous du1-R4059 endosperm (Supplemental Fig. S1B). The complete du1 locus including 5 kb upstream of the transcription start site was surveyed for Mu elements by PCR amplification using gene-specific primers and a degenerate primer from the Mu inverted terminal repeat. No Mu elements were found, so the molecular nature of du1-R4059 is not yet known.
Six other du1- mutations were characterized. Mu insertions were identified in five independent alleles by PCR amplification of genomic DNA with a gene-specific primer and the Mu-end primer (Supplemental Fig. S1A). Two spontaneous point mutations, du1-8801 and du1-8803, were shown to have the same molecular defect, a deletion of 27 bp (nucleotides 4,498–4,525 of the cDNA sequence) within exon 12 that would remove amino acids 1,460 to 1,468 of the full-length protein. All of these du1- alleles condition the same kernel phenotype as the null mutation (data not shown) and have the same effect on amylopectin structure (see below).
Kernel Phenotype in Mutants Affected in Both ISA2 and SSIII
Mutations at the du1 or isa2 locus do not condition phytoglycogen accumulation as single gene traits (Mangelsdorf, 1947; Gao et al., 1998; Kubo et al., 2010). Double mutant effects were tested by generating maize lines defective for both SSIII and ISA2, through combination of du1-Ref, du1-R4059, or du1-M3 with the null allele isa2-339 (Supplemental Fig. S2). Self-pollination of du1-/+, +/isa2-339 double heterozygotes yielded F2 ears that contained shrunken, translucent kernels at approximately the 1/16th frequency expected for double homozygotes (Table I). These kernels exhibited the typical “sugary” mutant phenotype caused by mutations of the sugary1 (su1) gene that eliminate ISA1 function (Fig. 1, A and B).
Table I.
Double mutant frequencies
| Parental Genotype | Progeny Phenotype | Sugary Kernels | Expected Double Homozygote Frequency | |
| Sugary | Wild Type or Dull | |||
| No. of kernels | % | |||
| +/du1-Ref × isa2-339/+ | 22 | 323 | 6.17 | 6.25 |
| +/du1-R4059 × isa2-339/+ | 30 | 456 | 6.41 | 6.25 |
The genotypes of nine sugary and 10 nonsugary progeny kernels (wild type or dull) from a du1-Ref/+, +/isa2-339 parent were determined by PCR analysis and sequencing of genomic DNA. In every instance, the sugary kernels were homozygous for both du1-Ref and isa2-339, whereas the nonsugary kernels had segregated for the wild-type and mutant alleles at both loci. The double homozygous mutant genotype was not detected in any of the nonsugary progeny. Individual progeny from the du1-R4059/+, +/isa2-339 parent were also analyzed for the isa2 genotype, and six of six sugary kernels were homozygous for the mutant allele, whereas 10 nonsugary kernels were segregating for the wild-type and mutant alleles at the isa2 locus. These data demonstrate that the sugary kernel phenotype results from the combination of du1-Ref and isa2-339, both in the homozygous state. By inference, the same conclusion is highly likely for du1-R4059, isa2-339 homozygotes. The alternative is that an unidentified mutation that conditions a sugary phenotype specifically in combination with isa2-339 was present in two independent du1- mutant lines, which is highly unlikely. Consistent with these conclusions, immunoblot analyses confirmed that endosperm from sugary kernels in the segregating F2 populations containing either du1-Ref or du1-R4059 was defective for both SSIII and ISA2 (Fig. 2A). As expected for double homozygotes, the sugary kernel phenotype bred true in successive generations.
Storage Glucan Content and Structure in Mutants Affected in Both ISA2 and SSIII
Homozygous du1-, isa2- double mutant kernels resemble su1- mutants, suggesting that they are reduced in starch content and accumulate phytoglycogen. This prediction was tested first by exposing sliced immature kernels, harvested 20 DAP, to iodine/potassium iodide (I2/KI) solution (Fig. 1C). Crystalline starch stains a dark blue color in this assay, whereas phytoglycogen stains tan or light brown. Starch was evident throughout the endosperm and embryo in wild-type kernels as well as in isa2-339, du1-Ref, or du1-R4059 single mutants, and no phytoglycogen was observed in these genotypes. The su1-4582 null mutant that lacks ISA1 contained phytoglycogen throughout the endosperm, with remnant starch detected at the periphery. The two double mutant genotypes both exhibited phytoglycogen staining in the center of the endosperm and starch at the exterior. This phenotype was intermediate between that of wild-type and su1-4582 mutant kernels.
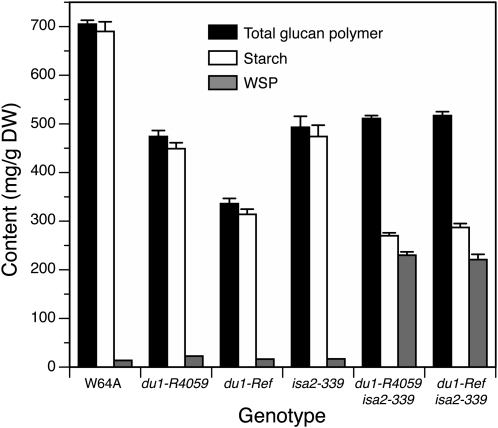
Soluble and granular glucan content in single kernels harvested 20 DAP was quantified (Fig. 3; for full data set, see Supplemental Table S1). Self-pollinated ears from at least three plants were analyzed for each genotype, with a minimum of three single kernels used as biological replicates from each ear. Because each plant provided several kernels, linear mixed-effects modeling was used for statistical analysis to take account of the correlation of the observations from the same plant. The results showed that water-soluble polysaccharide (WSP) and starch contents are significantly different between specific genotypes (P < 0.0001; Fig. 3). Distinctions in glucan quantities between different plants of the same genotype, however, or plants harvested in separate field seasons, were not statistically significant (P > 0.05). In wild-type endosperm, or du1-R4059 or du1-Ref single mutants, 93% or more of the total Glc polymer was starch. In contrast, starch accounted for only 53% or 56% of the total glucan polymer in du1-R4059, isa2-339 or du1-Ref, isa2-339 double mutants, respectively. WSP normalized to endosperm dry weight was elevated by a factor of approximately 16 in either of the double mutant endosperms compared with the wild type. Total Glc polymer normalized to dry weight was reduced to approximately 72% of the wild-type level in both double mutants. Total glucan was also reduced in the single mutants, to 48% of normal in du1-Ref and 67% in du1-R4059 endosperm.
Figure 3.
Starch and WSP contents. Soluble glucan polymer (WSP) and insoluble starch granules were isolated from endosperm tissue of single kernels harvested 20 DAP. The quantities of each polymer were determined and normalized to endosperm tissue dry weight (DW). se values are indicated. The numbers of single kernels analyzed for each genotype were as follows: W64A, parent plants (p) = 12, individual kernels (n) = 48; du1-R4059, p = 4, n = 14; du1-Ref, p = 5, n = 19; isa2-339, p = 3, n = 10; du1-R4059, isa2-339, p = 5, n = 30; du1-Ref, isa2-339, p = 3, n = 20. The full data set is shown in Supplemental Table S1. Differences from the wild type are all statistically significant (P < 0.0001) with the exception of WSP content in du1-Ref, du1-R4059, and isa2-339 single mutants.
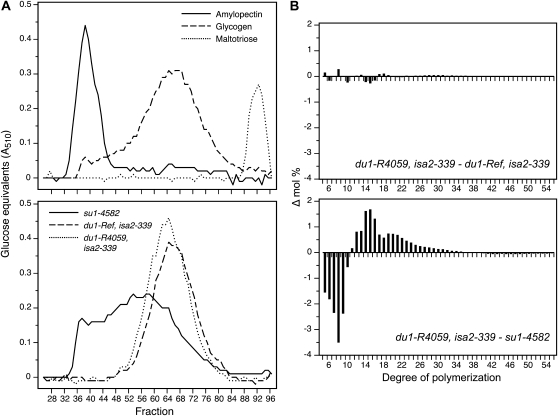
WSP present in the double mutant endosperms at 20 DAP was structurally characterized. The polymer from either double mutant eluted at approximately the same volume as rabbit liver glycogen during gel-permeation chromatography (GPC) on Sepharose CL-2B (Fig. 4A). Thus, du1-Ref, isa2-339 and du1-R4059, isa2-339 double mutant endosperm accumulates a high-molecular-mass glucan polymer designated as phytoglycogen, as opposed to maltooligosaccharides. This material was distinct from the phytoglycogen present in su1-4582 endosperm, however, because the latter exhibited a broader elution peak that eluted at an earlier volume. The distribution of linear chain lengths in the phytoglycogen of du1-, isa2- double mutants, and the ISA1-null mutant su1-4582, was determined using fluorophore-assisted carbohydrate electrophoresis (Morell et al., 1998; O’Shea et al., 1998; Supplemental Fig. S3). Difference plots of these data revealed that the two different double mutant lines had essentially identical chain length distributions (Fig. 4B). In contrast, clear differences were observed between phytoglycogen from the ISA1-null line compared with the SSIII/ISA2 double mutants. Specifically, su1-4582 phytoglycogen had a notably higher frequency of short chains of DP5 to DP9, with a corresponding decrease in abundance of DP12 to approximately DP25. Thus, the effect of the SSIII/ISA2 double mutant combination on isoamylase-type DBE function is not equivalent to the complete loss of both ISA1 homomer and ISA1/ISA2 heteromer caused by su1-4582.
Figure 4.
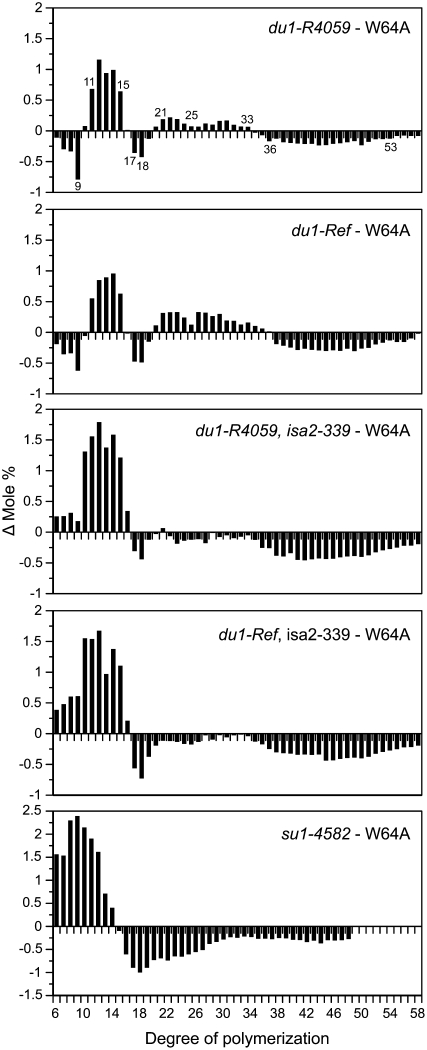
WSP characterization. A, GPC. WSP from the indicated double mutant lines and from su1-4582 single mutants was chromatographed on Sepharose-CL2B, and Glc equivalents in each fraction were quantified based on absorbance in a coupled enzyme assay. Rabbit glycogen, amylopectin from wild-type starch, and maltotriose were analyzed on the same column as approximate size standards. B, Difference plots. Chain length distributions were obtained by fluorophore-assisted carbohydrate electrophoresis of phytoglycogen isolated from su1-4582, du1-Ref, isa2-339, or du1-R4059, isa2-339 endosperms (Supplemental Fig. S3). For each DP, the frequency value for two genotypes was subtracted as indicated in the label of each panel.
Amylopectin in remnant starch of du1-, isa2- double mutants, or the nearly normal granules of du1-R4059 and du1-Ref single mutants, was also structurally characterized for chain length distribution. The frequency distributions are shown in Supplemental Figure S4, and comparisons of these data were used to reveal differences between any mutant and the wild type (Fig. 5). Changes in du1- single mutants were consistent with those reported previously in a du1- double mutant also affected in the granule-bound starch synthase (Jane et al., 1999), and the use of multiple alleles indicated the reproducibility of a complex pattern. Essentially identical results were observed in du1-Ref and du1-R4059 single mutant amylopectin. DP6 to DP9 chains were reduced in the mutants compared with the wild type, with DP9 showing the strongest defect. DP11 to DP15 chains were more abundant than in the wild type, and DP17 and DP18 were decreased in the mutants. Chains in the range of DP21 to DP33 were present at slightly elevated frequency in the mutants compared with the wild type, and DP36 to DP53 were reduced. Even the slight decrease of DP24 and DP25 chains compared with DP23 and DP26 was the same in du1-Ref and du1-R4059 (Fig. 5).
Figure 5.
Differences in amylopectin chain length distribution. The abundance of each chain length was determined by fluorophore-assisted carbohydrate electrophoresis and normalized to the total number of chains from DP6 to DP57. For each DP, the frequency value for W64A was subtracted from that of the indicated mutant. Note that the frequency difference scale is enlarged in the su1-4582 panel compared with the other genotypes.
Amylopectin chain length profiles were also obtained from du1-M1, -M2, -M3, -M4, -M6, -8801, -8802, and -8803 single mutants (Supplemental Fig. S5). The decrease in DP7 to DP9 abundance relative to the wild type, with DP9 the most extreme, was observed in all 11 independent du1- mutant starches, as was the clearly increased frequency of DP11 to DP15 chains. The decrease at DP17 and DP18 was not as reproducible, although the trend was consistent in all mutants. The elevated frequency of DP21 to DP30 was consistent, including the lesser effect on DP24 and DP25 compared with DP23 and DP26, as was the reduced frequency at DP36 to DP53. Thus, many distinct mutations affecting SSIII function all cause the same set of specific structural defects.
Loss of ISA2 had a negligible effect on amylopectin chain length distribution, as demonstrated previously (Kubo et al., 2010). Amylopectin in residual starch granules of the du1-R4059, isa2-339 or du1-Ref, isa2-339 double mutant lines had chain length distributions similar to each other and distinct from any single mutant (Fig. 5). Chains of DP6 to DP9, rather than being reduced in frequency as in the du1- single mutants, were more abundant relative to the wild type in the du1- isa2- double mutants. The increased frequency of DP10 to DP15 observed in the single mutants was exaggerated in the double mutants. The reduced frequency of DP17 and DP18 observed in du1- amylopectin was maintained in the double mutants, as was the decrease at DP36 or greater. The increased frequency of DP6 to DP10 chains, as observed in du1- isa2- double mutants, is a hallmark of residual amylopectin in su1- mutants (Dinges et al., 2001; Kubo et al., 2010; Fig. 5). The magnitude of this change in du1- isa2- amylopectin, however, is not as severe as in su1- mutants. Distinct amylopectin structure compared with the ISA1-null line again indicates that the SSIII/ISA2 double mutant phenotype does not result simply from the loss of all ISA1 activity.
Expression of ISA1 in du1-, isa2- Double Mutants
A possible explanation for phytoglycogen accumulation in du1-, isa2- double mutants is that expression of the gene encoding ISA1 is abnormal. This hypothesis is consistent with the finding that ISA1 in potato tuber or Arabidopsis leaf fails to accumulate normally in ISA2 mutants (Bustos et al., 2004; Delatte et al., 2005), although this is not so in maize endosperm (Kubo et al., 2010). Accumulation of ISA1 or ISA2 mRNA in 20-DAP endosperm was characterized by RT-PCR (Fig. 2B). As expected, ISA1 mRNA was absent in endosperm of the null mutant su1-4582 and ISA2 transcript was present in that tissue. Conversely, ISA1 mRNA was detected in isa2-339 tissue and ISA2 transcript was absent. The du1-R4059, isa2-339 and du1-Ref, isa2-339 double mutants both lacked ISA2 transcript, as expected, and contained apparently wild-type levels of ISA1 mRNA (Fig. 2B). Thus, expression of ISA1 at the level of mRNA accumulation is not affected in du1-, isa2- double mutants.
Immunoblot analysis tested whether the ISA1 protein accumulates in du1-, isa2- double mutant endosperms. Antisera specific for ISA1 generated a signal in protein extracts from both double mutant endosperms that was of at least equal intensity as the wild-type band (Fig. 2A). These data demonstrate that ISA1 accumulates at least to normal levels in the absence of both SSIII and ISA2.
Assembly of ISA1 Homomeric Complexes in du1-, isa2- Double Mutants
A second possible explanation for the sugary kernel phenotype of du1-, isa2- double mutants is that ISA1 protein is unable to assemble into its functional form as a homomeric complex. The state of ISA1 assembly in du1-, isa2- double mutant endosperm was tested by anion-exchange chromatography (AEC) followed by GPC. Proteins in soluble endosperm extracts from 20-DAP kernels were first separated by AEC. All ISA1 in the extracts bound to the column, and this material was eluted in a gradient of increasing NaCl concentration. Immunoblot analysis revealed the fractions that contained ISA1. Essentially identical results were observed for the du1-Ref, isa2-339 and du1-R4059, isa2-339 double mutants compared with the wild type or the isa2-339 single mutant (Supplemental Fig. S6). Thus, simultaneous defects in SSIII and ISA2 do not affect the AEC behavior of ISA1.
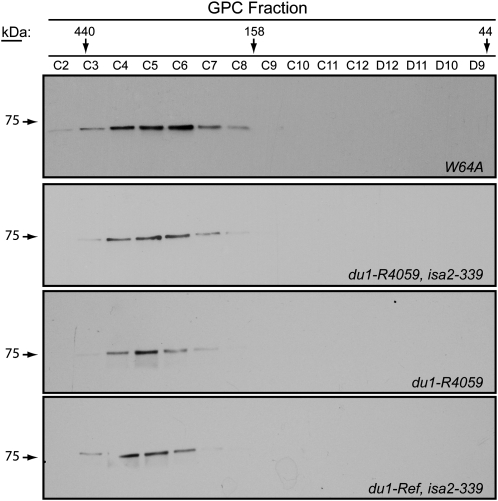
ISA1-containing fractions from the AEC elutions were pooled and concentrated. This material was then separated by GPC on Superdex 200, and fractions were tested for the presence of ISA1 by immunoblot analysis. Comparison with the elution volume of standard proteins of known molecular mass provided an estimation of the size of the ISA1-containing complexes. ISA1 in du1-R4059, isa2-339 or du1-Ref, isa2-339 double mutant endosperm assembled into a high-molecular-mass complex with an approximate molecular mass of 300 kD, similar to the wild-type ISA complexes (Fig. 6). In no instance was monomeric ISA1 detected based on GPC elution volume. Similar results were observed for du1-R4059 single mutant endosperm (Fig. 6) or du1-Ref endosperm (data not shown). Thus, the accumulation of phytoglycogen in the double mutants cannot be explained by failure of the ISA1 homomeric enzyme to assemble in the absence of both SSIII and ISA2.
Figure 6.
GPC separation of ISA complexes. ISA1 was partially purified by AEC from 20-DAP endosperm of the indicated genotype and then fractionated further by GPC. ISA1 in the eluate fractions was detected by immunoblot analysis. An equal volume of each fraction was loaded on the SDS-PAGE gel used for the immunoblot. The elution volumes of standards of known molecular mass are indicated.
ISA1 Activity in du1-, isa2- Double Mutants
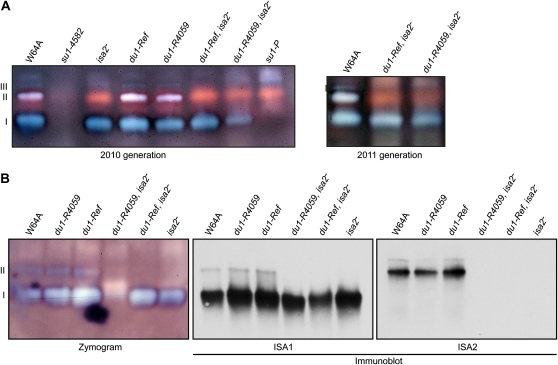
The enzymatic activity of isoamylase-type DBE complexes was compared among the wild type, du1- and isa2- single mutants, and the du1-, isa2- double mutants. Two alleles of su1 that affect ISA1 were used as controls. Proteins in total soluble endosperm extracts were separated by native-PAGE and then transferred in native conditions to a substrate gel infused with starch. ISA activities were visualized by staining the substrate gels with I2/KI solution. Endosperm from sibling plants was analyzed in independent biological replicates from two different growing seasons for each genotype. As demonstrated previously, wild-type endosperm contained three activities, an ISA1 homomeric complex and two heteromeric complexes containing both ISA1 and ISA2 (Kubo et al., 2010). The su1-4582 mutant, which lacks any ISA1, was devoid of all three activities, whereas the point mutant su1-P exhibited only an abnormal form of ISA1/ISA2 heteromer, helping to identify each of the activity bands (Kubo et al., 2010).
Single mutants homozygous for either du1-R4059 or du1-Ref exhibited both homomeric and heteromeric ISA complexes similar to the wild type (Fig. 7A). As expected, isa2-339 single mutants contained the ISA1 homomeric form but lacked ISA1/ISA2 heteromers. Both of the du1-, isa2-339 double mutant lines resembled the isa2-339 single mutant and clearly contained an enzymatically active, homomeric ISA1 complex.
Figure 7.
Isoamylase-type DBE activities. A, Total extracts. Soluble extracts from 20-DAP endosperm of the indicated genotypes were separated by native-PAGE and then electroblotted to a starch-containing gel for zymogram analysis. Activities were revealed by blue color after staining glucans on the gel with I2/KI. Band I is the ISA1 homomeric enzyme, and bands II and III are ISA1/ISA2 heteromeric enzymes. B, Concentrated fractions. ISA1 and ISA2 proteins in soluble extracts from 20-DAP endosperm were concentrated by AEC and then analyzed on activity gels as in A. In addition, the same AEC samples were subjected to immunoblot analyses using native-PAGE gels run simultaneously in a single apparatus.
Immunoblot analysis of native gels was used to confirm the proteins present in each activity complex. In order to increase the protein concentration and enable clear immunoblot signals, ISA activities were first partially purified by AEC. Duplicate native-PAGE gels were subjected to zymogram and immunoblot analyses so that the presence of ISA1 and/or ISA2 could be correlated with specific activity bands (Fig. 7B). These results confirmed the identification of the activity bands in each genotype, showing specifically that ISA1 homomeric enzyme is present in the du1-, isa2-339 double mutants. In this experiment, ISA1 homomeric enzyme activity was severely reduced in the du1-R4059, isa2-339 double mutant. This is likely explained by instability during the AEC purification steps, because active ISA1 homomer was clearly detected in total soluble extracts from the same genotype (Fig. 7A).
Effects of du1- Mutations on ISA Activity in Single Mutants
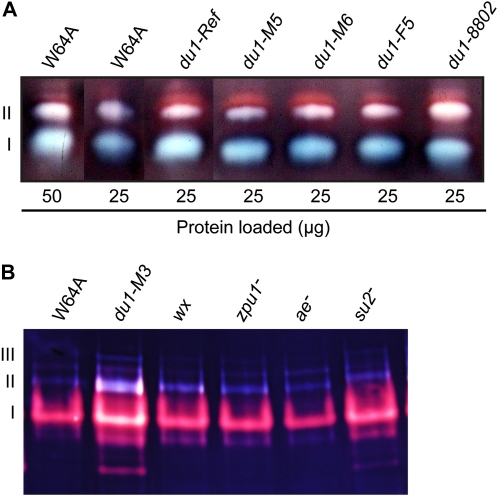
The activity of ISA1/ISA2 heteromer form II in du1-R4059 and du1-Ref single mutants appeared to be elevated compared with the wild type (Fig. 7A). This effect was tested further by examining three additional du1- mutants each carrying an independent allele. Loading equal amounts of protein on the native gel resulted in signals for ISA1 homomer and ISA1/ISA2 heteromer form II that were stronger in all five mutant lines than in wild-type extracts (Fig. 8A). This effect was also tested using amyloplast-enriched extracts of a sixth mutant, in this instance du1-M3. Increases in all three forms of ISA activity in the du1-M3 mutant were observed compared with the wild type (Fig. 8B). As controls, amyloplast extracts from mutants affected in four other enzymes involved in starch biosynthesis were examined on the same zymogram gel. These single mutations affected GBSS (wx mutant; Klösgen et al., 1986), the debranching enzyme ZPU1 (zpu1 mutant; Dinges et al., 2003), SBEIIb (ae mutant; Stinard et al., 1993), or SSIIa (su2 mutant; Zhang et al., 2004). Increased ISA activity was specific to the SSIII-deficient line.
Figure 8.
Starch-modifying enzyme activities. A, Total soluble endosperm extracts. Proteins in soluble extracts from 20-DAP endosperm tissue of the indicated genotype were subjected to zymogram analysis as in Figure 7. Band I is the ISA1 homomer, and band II is ISA1/ISA2 heteromeric enzyme. B, Purified amyloplast soluble fractions. Amyloplasts were isolated from endosperm of the indicated genotype harvested 15 to 16 DAP. Mutations affecting various starch biosynthetic enzymes were used as negative controls. The affected enzymes were GBSS (wx-), ZPU1 (zpu1-), SBEIIb (ae-), or SSIIa (su2-). In this analysis, the second form of ISA1/ISA2 heteromer is evident in band III.
An effect of du1- mutations on ISA1 was also evident at the level of total protein content. Weakly exposed immunoblots of total soluble endosperm extracts revealed a stronger ISA1 signal from du1-R4059 or du1-Ref homozygous single mutants than from the wild type (Fig. 2A). Immunoblots of pooled AEC fractions were analyzed similarly. Equal amounts of total protein were loaded onto the same HiTrap Q FF column, and for each genotype the same fractions were pooled and concentrated by the same factor. Equal volumes of the concentrated fractions were analyzed, with the result that the ISA1 signal was again stronger in the lines lacking SSIII compared with the wild type or a line lacking ISA1 (Fig. 2A). These consistent results using independent du1- alleles support the conclusion that the ISA1 protein level is elevated in SSIII single mutants. This effect was not observed in lines lacking both SSIII and ISA2 (Fig. 2A).
DISCUSSION
Effects of SSIII Deficiency in Single Mutants
Ten independent mutations of the du1 gene encoding SSIII conditioned the same set of specific changes in amylopectin structure. The abnormal structure, therefore, does not result from variation between inbred backgrounds, growth conditions, or any other extraneous factor. In addition, the mutant amylopectin structure was consistent regardless of the nature of the SSIII defect, for example, comparing the truncated protein generated from du1-Ref with the null allele du1-R4059. SSIII deficiency in rice was previously seen to alter amylopectin chain length distribution in a manner very similar to the effects shown here of maize du1- mutants (Fujita et al., 2007), so the specific chain length distribution can be taken as a general effect in cereal endosperm. In both species, loss of SSIII results in an alternating pattern of chain length abundance differences relative to the wild type in the range of DP7 to DP50. Specifically, DP7 to DP9 chains are always decreased, DP11 to DP15 chains are always increased, DP17 and DP18 are generally decreased, DP21 to DP33 are increased, and DP36 to DP50 chains are decreased. Thus, SSIII appears to affect the entire range of chains that constitute crystalline lamellae as well as those that extend through multiple crystalline clusters. In contrast, SSI and SSII are required for the normal abundance of discrete, continuous spans of chain length, which can be explained by substrate preference (Morell et al., 2003; Zhang et al., 2004, 2008; Delvallé et al., 2005; Nakamura et al., 2005; Fujita et al., 2006).
The effects of SSIII deficiency on amylopectin structure are unlikely to result from substrate specificity and/or catalytic activity based on the length of the acceptor chain, except perhaps for DP36 to DP50, which are chains that span two or three crystalline structures. Effects on these longer chains were seen in SSIII-deficient lines in numerous species (Fujita et al., 2007), and the enzyme has been shown to prefer longer chains as substrates by in vitro assay of partially purified enzymes from maize or rice (Cao et al., 2000; Fujita et al., 2006). Alternating effects on the abundance of short chains as a function of DP, if it were caused directly by substrate selection, would require that SSIII can distinguish between chain lengths differing by one or two Glc residues, for example, at the DP9 to DP11 boundary and again at the DP17 to DP19 boundary.
A more likely explanation is that SSIII recruits or aids the access of other starch biosynthetic enzymes to the growing glucan polymer, each of which has its own substrate preference relative to DP. SSIII contains a highly conserved N-terminal extension beyond the catalytic region, referred to the SSIII-homology domain (SSIIIHD; Gao et al., 1998; Li et al., 2003; Ral et al., 2004; Dian et al., 2005). This domain has been implicated in binding both to starch and to other proteins that are active in starch biosynthesis (Hennen-Bierwagen et al., 2009; Wayllace et al., 2010). Thus, beyond its catalytic role, SSIII may physically coordinate the functions of other starch biosynthetic enzymes and/or regulate their catalytic activities. According to this hypothesis, the sum of these defects would then generate the observed du1- mutant amylopectin structure.
The total glucan content in du1-Ref and du1-R4059 endosperm at 20 DAP, normalized to dry weight, was reduced to as much as half of the wild-type level. This result is generally consistent with previous reports showing moderate reduction in starch content throughout the development of du1- mutant kernels (Creech, 1965; Creech and McCardle, 1966; Singletary et al., 1997).
Effects of SSIII-ISA2 Deficiency in Double Mutants
Analyses of the du1-, isa2- double mutants described here indicate a role of SSIII in the creation of crystallization-competent structures within soluble, branched glucans that subsequently convert to amylopectin. Despite their effects on amylopectin structure, du1- mutations by themselves do not condition a change in the ratio of WSP to starch. Similarly, loss of ISA2 does not condition high levels of phytoglycogen (Kubo et al., 2010; Utsumi et al., 2011). It is striking, therefore, that endosperm lacking both ISA2 and SSIII produces nearly as much phytoglycogen as starch. This effect most likely results from altered isoamylase-type DBE function, because in all plants studied the phytoglycogen-accumulation phenotype has been observed only in mutants in which this activity is compromised.
A possible explanation for the defect in the double mutant was a requirement for either ISA2 or SSIII in order for ISA1 to be stably expressed or to assemble into functional ISA1 homomer. Several aspects of ISA1 expression were tested in the SSIII/ISA2-deficient lines. There were no differences from the wild type detected by AEC or by GPC. The latter analysis revealed that the size of the ISA1 homomer complex is approximately the same in the du1-, isa2- double mutants as endosperm lacking only ISA2. ISA1 homomer from SSIII/ISA2-deficient, ISA2-deficient, or wild-type extracts migrated at the same rate in native PAGE. Furthermore, as far as can be ascertained from zymogram analyses, the enzymatic activity of the ISA1 homomer complex was approximately the same in total extracts of the wild type, ISA2 mutants, and SSIII/ISA2 double mutants. Evidence for ISA1 homomer activity in vivo is that the specific details of the starch-decrease/phytoglycogen-accumulation phenotype differed between the du1-, isa2- double mutants and su1-4582 kernels that are devoid of both ISA1 homomer and ISA1/ISA2 heteromer. These differences included the amount of phytoglycogen present, the chain length distribution of the soluble glucan, and the chain length distribution of amylopectin in the starch that accumulates in the SSIII/ISA2-deficient lines compared with the ISA1-null line. Thus, ISA1 homomer functions to some extent in the absence of both SSIII and ISA2, but not sufficiently to prevent phytoglycogen accumulation. From these observations, it is evident that the function(s) of SSIII in preventing phytoglycogen accumulation occurs subsequent to the formation of enzymatically competent ISA1 homomer.
An apparent discrepancy from the conclusion that SSIII deficiency does not prevent ISA1 homomer catalytic activity was seen in the du1-R4059, isa2-339 total endosperm extract from harvest year 2010. This sample exhibited moderately reduced ISA1 homomer activity; however, the same line from the 2011 year had the same amount of activity as the wild type or isa2-339 single mutants. Reduced zymogram signals in some analyses of du1-R4059, isa2-339 extracts may be explained by unstable enzyme activity. Consistent with this explanation, during AEC purification, the ISA1 homomer from du1-R4059, isa2-339 endosperm remained intact but its activity was severely reduced. The total extract analyses, however, reveal clearly that ISA1 homomer is catalytically competent in the absence of both SSIII and ISA2.
Hypotheses involving either direct or indirect mechanisms can be proposed to explain why SSIII is needed to prevent phytoglycogen accumulation when ISA1/ISA2 heteromer is absent. Considering direct mechanisms, SSIII potentially could be a regulator of ISA1 homomer activity in vivo, or contact with SSIII could affect the positioning of ISA1 homomer upon its substrate. In either instance, SSIII may be directly responsible or some other protein(s) that associates with SSIII could mediate the regulation. Direct interaction between SSIII and ISA1 has not been demonstrated in coimmunoprecipitation or other affinity-based experiments, despite the fact that several proteins known to bind SSIII, including SBEIIa, SBEIIb, and PPDK (Hennen-Bierwagen et al., 2009), were detected as positive controls in the same experiments (data not shown). However, the absence of affinity-based evidence does not preclude an interaction between SSIII and the ISA complexes in vivo, because such an association may be transient and/or may require a physiological condition such as plastid integrity.
Considering indirect mechanisms, SSIII activity may affect the structure of the branched glucans that become substrates of ISA1 homomer. In the SSIII/ISA2 double mutants, therefore, the proper substrate would not appear and ISA1 homomer would be unable to modify branched glucans normally destined to become amylopectin. Another potential indirect mechanism is that the ISA1 homomer and SSIII function completely independently, but both activities are necessary to affect the balance of chain length, branch frequency, and branch positioning in the precursor soluble glucans so as to facilitate subsequent crystallization.
The question remains why du1- single mutants do not accumulate phytoglycogen (i.e. when both ISA1 homomer and ISA1/ISA2 heteromer are present). One possible explanation is that the heteromeric complex functions differently from the homomer such that SSIII function is not necessary in order to prevent phytoglycogen accumulation. Alternatively, the critical factor may be the total level of isoamylase-type DBE activity. Compromise of that enzyme activity below a certain level may cause SSIII to become required in order to maintain nearly normal starch production.
Another instance of combining SSIII deficiency with compromised isoamylase-type DBE function was reported previously. These studies showed that du1-Ref in combination with the ISA1 point mutation su1-P (also denoted su1-am) conditions a moderate sugary kernel phenotype including starch reduction, accumulation of phytoglycogen, and increased Suc content relative to the wild type (Cameron, 1947; Mangelsdorf, 1947). Analogous to our study, single mutants defective only for su1-P exhibit normal kernel appearance and starch composition (Cameron, 1947; Mangelsdorf, 1947; Kubo et al., 2010). The molecular defect of ISA1 in su1-P mutants prevents the assembly of ISA1 homomer but allows the formation of an abnormal but active ISA1/ISA2 heteromeric complex (Kubo et al., 2010). Thus, compromise of isoamylase-type DBE owing to an alteration of either ISA1 homomer or ISA1/ISA2 heteromer can necessitate a requirement for SSIII in order for phytoglycogen accumulation to be blocked.
Elevated ISA1 Protein Levels and Activity in SSIII-Deficient Mutants
Additional connections between SSIII and isoamylase-type DBE were observed by comparing protein and activity levels between du1- single mutants and the wild type. The amount of ISA1 protein as judged by immunoblot analysis was elevated in both du1-R4059 and du1-Ref single mutants (Fig. 2A). The apparent enzymatic activity of ISA1 homomer and at least one of the ISA1/ISA2 heteromer bands in zymograms of total extracts, normalized to total protein, appeared to be elevated in comparison with the wild type (Figs. 7A and 8A). Although such assays are not quantitative, the qualitative estimation of activity was consistently elevated in five different mutants, each carrying a distinct allele of du1-. This effect was also observed in amylopast-enriched extracts from a line carrying a sixth du1- mutant allele, and in this instance, the apparent activities of ISA1 homomer and both forms of ISA1/ISA2 heteromer were markedly enhanced (Fig. 8B). Taken together, these data provide convincing evidence that ISA1 is activated above normal levels in the absence of SSIII.
The molecular explanation for these results remains obscure in the absence of demonstrated direct interactions between SSIII and ISA1 or ISA2. The effects of SSIII deficiency on ISA1 protein level and ISA1 homomer enzyme activity were not observed in du1-, isa2- double mutants, indicating that ISA2 protein is required. This is consistent with the suggestion that ISA1 protein production is elevated as a result of SSIII deficiency, and assembly into a heteromeric enzyme is necessary to allow the stability of that protein (i.e. to prevent degradative turnover). Although further analyses are necessary to understand the mechanism of this interaction, the data lend further support to the overall conclusion that the SSIII and isoamylase-type DBE function in an interrelated manner to accomplish the overall function of preventing the formation of phytoglycogen during the biosynthesis of amylopectin.
CONCLUSION
The effects of SSIII deficiency by itself on starch structure cannot be explained by catalytic activity alone, providing strong evidence that this protein coordinates the functions of multiple starch biosynthetic enzymes. SSIII and isoamylase-type DBE(s) function together in a concerted manner such that glucan polymer crystallization occurs normally. SSIII, therefore, is a determinant of the ability to form crystalline branched glucans in the sense that, in some circumstances, it is required to prevent the accumulation of phytoglycogen.
MATERIALS AND METHODS
Nomenclature and Plant Materials
Gene loci are designated in italic lowercase letters (e.g. du1). Wild-type alleles are designated with an uppercase letter (e.g. Du1) or by the “+” symbol. Specific mutations are designated by the locus name followed by a dash and a unique allele name (e.g. du1-Ref), and unspecified nonfunctional mutations are designated by the allele name with a superscript dash (e.g. du1-). mRNA or cDNA produced from a locus is indicated by italic uppercase letters (e.g. DU1 mRNA). Proteins are indicated by uppercase letters without italics (e.g. SSIII).
Maize (Zea mays) plants were field grown in summer nurseries at Iowa State University. Isolation of the du1 mutations in a Mu-active population was described previously (Gao et al., 1998). Correlation with allele names used in the previous work is as follows: M1, du1-R2197; M2, du1-R2339; M3, du1-R2649; M4, du1-R2370; M6, du1-R1178. Lines bearing du1-8801, du1-8802, and du1-8803 were provided by the Maize Genetics Cooperation Stock Center (http://maizecoop.cropsci.uiuc.edu/). These spontaneous mutations were identified by the appearance of the dull phenotype on segregating ears of self-pollinated F1 plants. Crosses of heterozygotes to du1-Ref homozygotes yielded ears with 50% dull kernels, thus confirming the new mutations as du1 alleles.
All maize lines used in the study were in the W64A inbred genetic background. Each mutation was backcrossed to wild-type W64A for at least five generations, with self-pollination of the outcrossed plants used to identify the presence of each du1- mutation. Self-pollination of the final heterozygote in the outcross series yielded homozygous seed that generated F0 plants used to initiate this study. The crossing scheme employed to generate the lines analyzed here is shown in Supplemental Figure S2.
Developing kernels from self-pollinated ears were collected at specified DAP, frozen in liquid N2, and stored at −80°C. These kernels provided endosperm tissue for biochemical analyses. Amyloplasts were purified from whole fresh kernels harvested 15 or 16 DAP, as described previously (Hennen-Bierwagen et al., 2008).
RT-PCR and Genotype Characterization by PCR
Genomic DNA was isolated from seedling leaves as described previously (Saghai-Maroof et al., 1984; Steiner et al., 1995). Overlapping fragments of the du1 locus were amplified from genomic DNA by PCR using forward- and reverse-strand oligonucleotide primers from the coding region, specified in Supplemental Table S2. PCR amplification used Platinum Taq Polymerase (Invitrogen; catalog no. 11304-029) in 5% dimethyl sulfoxide (DMSO), 1.5 mm MgCl2, and other buffer conditions specified by the manufacturer. Temperature cycling conditions were 95°C for 3 min; 35 cycles of 95°C for 1 min, 50°C for 1 min, and 72°C for 2 min; and 72°C for 10 min. Analysis of the amplified fragments revealed the complete genomic sequence of the transcribed region of du1 (GenBank accession no. JF273457). The same primers were also used in combination with the Mu terminal inverted repeat end primer 9242 (5′-AGAGAAGCCAACGCCAWCGCCTCYA-3′) to amplify junction fragments containing the transposon and its genomic flanking sequences. The sequences of those PCR fragments were determined to reveal the insertion site of each Mu-induced allele. Primers DURef-F and DURef-R (Supplemental Table S2) were used to amplify the region of du1 that contained the nucleotide altered in du1-Ref. The isa2 genotype was determined by PCR amplification of genomic DNA using Mu end primer 9242 and gene-specific primers as described (Kubo et al., 2010).
Total RNA isolation from endosperm tissue and RT-PCR amplification of ISA1 and ISA2 mRNA were described previously (Kubo et al., 2010). DU1 mRNA was detected by RT-PCR using primers du1-F1 and du-L1 (Supplemental Table S2). Thermocycling conditions for the detection of DU1 mRNA were 95°C for 2 min; 35 cycles of 95°C for 30 s, 60.5°C for 30 s, and 72°C for 1 min; and 72°C for 5 min. GoTaq Flexi DNA Polymerase (Promega; catalog no. M8298) was used in reaction buffer containing 1.5 mm MgCl2 and 5% DMSO.
Protein Isolation and Fractionation, Zymogram Analysis, and Immunological Methods
Preparation of total soluble endosperm extracts, AEC fractionation using HiTrap Q HP columns, and GPC fractionation using a Superdex 200 10/300 column were described previously. Standard procedures were used for the separation of proteins by SDS-PAGE, electroblotting to nitrocellulose filters, probing with affinity-purified IgG fractions, and detection of antibody binding (Kubo et al., 2010). The purified IgG fractions used were αISA1, αISA2, αSSIIIHD, and αDU1N (Hennen-Bierwagen et al., 2008; Kubo et al., 2010). Fractionation of total soluble extracts, amyloplast-enriched fractions, or partially purified AEC fractions by native-PAGE, native transfer to starch-containing gels, and detection of carbohydrate-modifying enzyme activities by staining with I2/KI solution followed previously described procedures (Kubo et al., 2010). In some analyses, duplicate gels run in the same apparatus were transferred to nitrocellulose and analyzed by immunoblot to allow the identification of specific proteins comigrating with each ISA activity. All zymogram analyses were repeated at least twice starting from independent samples of biological material.
Carbohydrate Characterization
Starch, WSP, and free Glc contents as a function of endosperm dry weight were determined by modification of a previously published method (Dinges et al., 2001). Ears were collected 20 DAP from multiple plants of each genotype during the 2009, 2010, and 2011 field seasons. From each ear, three to six individual kernels were characterized as follows. Pericarp and embryo were removed, and the single endosperm was ground to a fine slurry in a total of 4 mL of water using a mortar and pestle. From the crude lysate, 1 mL was dried at 80°C to determine dry weight for standardization. Two other 1-mL aliquots were analyzed as A and B samples for each kernel. Starch and WSP were separated by centrifugation at 10,000g for 10 min at 4°C. The soluble phase was collected and boiled for 20 min to inactivate potential degradative enzymes. The pellet containing starch was washed with 1 mL of cold 80% ethanol and then collected by centrifugation for 5 min. That pellet was dispersed in a total volume of 1.5 mL of 100% DMSO, then boiled for 30 min. Glucan polymer in both the DMSO-dissolved starch and the soluble phase was hydrolyzed to Glc in a volume of 500 μL of 100 mm sodium acetate, pH 5.0, 5 mm CaCl2, containing approximately 33 units of amyloglucosidase (Megazyme E-AMGDF100), during incubation for 45 min at 50°C. The maximum DMSO content in these reactions was 10%. Control reactions lacking amyloglucosidase were performed to quantify free Glc. Glc in the amyloglucosidase digestions was quantified using a colorimetric Glc oxidase-peroxidase (GOPOD) assay (Megazyme K-GLUC). These reactions were performed on a microtiter plate with each well containing 225 μL of the GOPOD assay reagent and 15 μL of each Glc sample. GOPOD reactions were incubated for 20 min at 50°C, then A490 was determined. Glc standard curves were included on each plate to allow calculation of the Glc equivalents in the starch, WSP, and free-Glc fractions of each individual endosperm, normalized to total dry weight.
WSP to be analyzed by GPC was collected by a similar procedure. Three dissected endosperms were ground in ice-cold water in a total volume of 5 mL. After centrifugation for 10 min at 10,000g at 4°C, the top two-thirds of the supernatant was collected and transferred to a fresh tube. Centrifugation and collection were repeated two additional times, to ensure that no starch granules contaminated the soluble glucan fraction. WSP was quantified by GOPOD assay of Glc equivalents, and 5 mg was precipitated with 5 volumes of 100% ethanol overnight at −20°C. The precipitate was dissolved in 1 mL of 10 mm NaOH, clarified by centrifugation at top speed in a microfuge for 2 min, and applied to a Sepharose CL-2B column (1.5 cm diameter × 75 cm length) run in 10 mm NaOH. Glucans were eluted at a flow rate of 0.5 mL min−1 while collecting 1.3-mL fractions. Glc equivalents in each fraction were determined by amyloglucosidase digestion and GOPOD assay as described.
For the determination of linear chain length distributions, whole starch was first solubilized by boiling in 100% DMSO. After appropriate dilutions, the material was then debranched with commercial isoamylase (Megazyme E-ISAMY), derivatized, and analyzed as described previously (O’Shea et al., 1998; Dinges et al., 2003) using a Beckman P/ACE capillary system. Each genotype was analyzed at least twice from biological replicates of sibling plants with essentially identical results.
Statistical Analyses
Statistical Analysis Software package SAS 9.2 from the SAS Institute was used for all analyses. Mean and se values were calculated from all kernels within the same genotype. The differences between WSP and starch were compared among different genotypes using linear mixed-effects modeling. Holm’s procedure was used to adjust for multiple comparisons. Values of P < 0.05 after adjustment for multiple comparisons are considered significant.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number JF273457.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Map of the du1 locus and du1- mutations.
Supplemental Figure S2. Crossing diagram.
Supplemental Figure S3. Phytoglycogen chain length distribution plots.
Supplemental Figure S4. Amylopectin chain length distribution plots.
Supplemental Figure S5. Additional amylopectin chain length distribution difference data.
Supplemental Figure S6. Anion-exchange fractionation of ISA1.
Supplemental Table S1. Full data set for starch and WSP quantification.
Supplemental Table S2. Primers for the amplification of du1 genomic DNA or cDNA.
Supplementary Material
Acknowledgments
DNA sequence analysis and the synthesis of oligonucleotides were performed by the Iowa State University DNA Facility. Capillary electrophoresis utilized instrumentation provided by the W.M. Keck Metabolomics Research Laboratory of Iowa State University. We acknowledge the contributions of Yongjie Miao for statistical analysis and Amanda Gebauer for technical assistance.
References
- Ball S, Guan H-P, James M, Myers A, Keeling P, Mouille G, Buléon A, Colonna P, Preiss J. (1996) From glycogen to amylopectin: a model for the biogenesis of the plant starch granule. Cell 86: 349–352 [DOI] [PubMed] [Google Scholar]
- Ball SG, Morell MK. (2003) From bacterial glycogen to starch: understanding the biogenesis of the plant starch granule. Annu Rev Plant Biol 54: 207–233 [DOI] [PubMed] [Google Scholar]
- Burton RA, Jenner H, Carrangis L, Fahy B, Fincher GB, Hylton C, Laurie DA, Parker M, Waite D, van Wegen S, et al. (2002) Starch granule initiation and growth are altered in barley mutants that lack isoamylase activity. Plant J 31: 97–112 [DOI] [PubMed] [Google Scholar]
- Bustos R, Fahy B, Hylton CM, Seale R, Nebane NM, Edwards A, Martin C, Smith AM. (2004) Starch granule initiation is controlled by a heteromultimeric isoamylase in potato tubers. Proc Natl Acad Sci USA 101: 2215–2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron JW. (1947) Chemico-genetic bases for the reserve carbohydrates in maize endosperm. Genetics 32: 459–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Imparl-Radosevich J, Guan H, Keeling PL, James MG, Myers AM. (1999) Identification of the soluble starch synthase activities of maize endosperm. Plant Physiol 120: 205–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, James MG, Myers AM. (2000) Purification and characterization of soluble starch synthases from maize endosperm. Arch Biochem Biophys 373: 135–146 [DOI] [PubMed] [Google Scholar]
- Creech RG. (1965) Genetic control of carbohydrate synthesis in maize endosperm. Genetics 52: 1175–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creech RG, McCardle FJ. (1966) Gene interaction for quantitative changes in carbohydrates in maize kernels. Crop Sci 6: 192–194 [Google Scholar]
- Dauvillée D, Colleoni C, Mouille G, Buléon A, Gallant DJ, Bouchet B, Morell MK, d’Hulst C, Myers AM, Ball SG. (2001a) Two loci control phytoglycogen production in the monocellular green alga Chlamydomonas reinhardtii. Plant Physiol 125: 1710–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauvillée D, Colleoni C, Mouille G, Morell MK, d’Hulst C, Wattebled F, Liénard L, Delvallé D, Ral JP, Myers AM.et al (2001b) Biochemical characterization of wild-type and mutant isoamylases of Chlamydomonas reinhardtii supports a function of the multimeric enzyme organization in amylopectin maturation. Plant Physiol 125: 1723–1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delatte T, Trevisan M, Parker ML, Zeeman SC. (2005) Arabidopsis mutants Atisa1 and Atisa2 have identical phenotypes and lack the same multimeric isoamylase, which influences the branch point distribution of amylopectin during starch synthesis. Plant J 41: 815–830 [DOI] [PubMed] [Google Scholar]
- Delvallé D, Dumez S, Wattebled F, Roldán I, Planchot V, Berbezy P, Colonna P, Vyas D, Chatterjee M, Ball S, et al. (2005) Soluble starch synthase I: a major determinant for the synthesis of amylopectin in Arabidopsis thaliana leaves. Plant J 43: 398–412 [DOI] [PubMed] [Google Scholar]
- Deschamps P, Moreau H, Worden AZ, Dauvillée D, Ball SG. (2008) Early gene duplication within chloroplastida and its correspondence with relocation of starch metabolism to chloroplasts. Genetics 178: 2373–2387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dian W, Jiang H, Wu P. (2005) Evolution and expression analysis of starch synthase III and IV in rice. J Exp Bot 56: 623–632 [DOI] [PubMed] [Google Scholar]
- Dinges JR, Colleoni C, James MG, Myers AM. (2003) Mutational analysis of the pullulanase-type debranching enzyme of maize indicates multiple functions in starch metabolism. Plant Cell 15: 666–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinges JR, Colleoni C, Myers AM, James MG. (2001) Molecular structure of three mutations at the maize sugary1 locus and their allele-specific phenotypic effects. Plant Physiol 125: 1406–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Fujita N, Yoshida M, Asakura N, Ohdan T, Miyao A, Hirochika H, Nakamura Y. (2006) Function and characterization of starch synthase I using mutants in rice. Plant Physiol 140: 1070–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Yoshida M, Kondo T, Saito K, Utsumi Y, Tokunaga T, Nishi A, Satoh H, Park JH, Jane JL, et al. (2007) Characterization of SSIIIa-deficient mutants of rice: the function of SSIIIa and pleiotropic effects by SSIIIa deficiency in the rice endosperm. Plant Physiol 144: 2009–2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Wanat J, Stinard PS, James MG, Myers AM. (1998) Characterization of dull1, a maize gene coding for a novel starch synthase. Plant Cell 10: 399–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennen-Bierwagen TA, Lin Q, Grimaud F, Planchot V, Keeling PL, James MG, Myers AM. (2009) Proteins from multiple metabolic pathways associate with starch biosynthetic enzymes in high molecular weight complexes: a model for regulation of carbon allocation in maize amyloplasts. Plant Physiol 149: 1541–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennen-Bierwagen TA, Liu F, Marsh RS, Kim S, Gan Q, Tetlow IJ, Emes MJ, James MG, Myers AM. (2008) Starch biosynthetic enzymes from developing maize endosperm associate in multisubunit complexes. Plant Physiol 146: 1892–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hizukuri S. (1986) Polymodal distribution of the chain lengths of amylopectin, and its significance. Carbohydr Res 147: 342–347 [Google Scholar]
- Hussain H, Mant A, Seale R, Zeeman S, Hinchliffe E, Edwards A, Hylton C, Bornemann S, Smith AM, Martin C, et al. (2003) Three isoforms of isoamylase contribute different catalytic properties for the debranching of potato glucans. Plant Cell 15: 133–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James MG, Robertson DS, Myers AM. (1995) Characterization of the maize gene sugary1, a determinant of starch composition in kernels. Plant Cell 7: 417–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jane J, Chen YY, Lee LF, McPherson AE, Wong KS, Radosavljevic M, Kasemsuwan T. (1999) Effects of amylopectin branch chain length and amylose content on the gelatinization and pasting properties of starch. Cereal Chem 76: 629–637 [Google Scholar]
- Klösgen RB, Gierl A, Schwarz-Sommer Z, Saedler H. (1986) Molecular analysis of the waxy locus of Zea mays. Mol Gen Genet 203: 237–244 [Google Scholar]
- Kubo A, Colleoni C, Dinges JR, Lin Q, Lappe RR, Rivenbark JG, Meyer AJ, Ball SG, James MG, Hennen-Bierwagen TA, et al. (2010) Functions of heteromeric and homomeric isoamylase-type starch-debranching enzymes in developing maize endosperm. Plant Physiol 153: 956–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo A, Fujita N, Harada K, Matsuda T, Satoh H, Nakamura Y. (1999) The starch-debranching enzymes isoamylase and pullulanase are both involved in amylopectin biosynthesis in rice endosperm. Plant Physiol 121: 399–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Sun F, Xu S, Chu X, Mukai Y, Yamamoto M, Ali S, Rampling L, Kosar-Hashemi B, Rahman S, et al. (2003) The structural organisation of the gene encoding class II starch synthase of wheat and barley and the evolution of the genes encoding starch synthases in plants. Funct Integr Genomics 3: 76–85 [DOI] [PubMed] [Google Scholar]
- Mangelsdorf PC. (1947) The inheritance of amylaceous sugary endosperm and its derivatives in maize. Genetics 32: 448–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morell MK, Kosar-Hashemi B, Cmiel M, Samuel MS, Chandler P, Rahman S, Buleon A, Batey IL, Li Z. (2003) Barley sex6 mutants lack starch synthase IIa activity and contain a starch with novel properties. Plant J 34: 173–185 [DOI] [PubMed] [Google Scholar]
- Morell MK, Samuel MS, O’Shea MG. (1998) Analysis of starch structure using fluorophore-assisted carbohydrate electrophoresis. Electrophoresis 19: 2603–2611 [DOI] [PubMed] [Google Scholar]
- Mouille G, Maddelein ML, Libessart N, Talaga P, Decq A, Delrue B, Ball S. (1996) Preamylopectin processing: a mandatory step for starch biosynthesis in plants. Plant Cell 8: 1353–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers AM, Morell MK, James MG, Ball SG. (2000) Recent progress toward understanding biosynthesis of the amylopectin crystal. Plant Physiol 122: 989–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Francisco PB, Jr, Hosaka Y, Sato A, Sawada T, Kubo A, Fujita N. (2005) Essential amino acids of starch synthase IIa differentiate amylopectin structure and starch quality between japonica and indica rice varieties. Plant Mol Biol 58: 213–227 [DOI] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng 10: 1–6 [DOI] [PubMed] [Google Scholar]
- O’Shea MG, Samuel MS, Konik CM, Morell MK. (1998) Fluorophore-assisted carbohydrate electrophoresis (FACE) of oligosaccharides: efficiency of labelling and high-resolution separation. Carbohydr Res 307: 1–12 [Google Scholar]
- Ral JP, Derelle E, Ferraz C, Wattebled F, Farinas B, Corellou F, Buléon A, Slomianny MC, Delvalle D, d’Hulst C, et al. (2004) Starch division and partitioning: a mechanism for granule propagation and maintenance in the picophytoplanktonic green alga Ostreococcus tauri. Plant Physiol 136: 3333–3340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Ezpeleta N, Brinkmann H, Burey SC, Roure B, Burger G, Löffelhardt W, Bohnert HJ, Philippe H, Lang BF. (2005) Monophyly of primary photosynthetic eukaryotes: green plants, red algae, and glaucophytes. Curr Biol 15: 1325–1330 [DOI] [PubMed] [Google Scholar]
- Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW. (1984) Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci USA 81: 8014–8018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singletary GW, Banisadr R, Keeling PL. (1997) Influence of gene dosage on carbohydrate synthesis and enzymatic activities in endosperm of starch-deficient mutants of maize. Plant Physiol 113: 293–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner JJ, Poklemba CJ, Fjellstrom RG, Elliott LF. (1995) A rapid one-tube genomic DNA extraction process for PCR and RAPD analyses. Nucleic Acids Res 23: 2569–2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinard PS, Robertson DS, Schnable PS. (1993) Genetic isolation, cloning, and analysis of a Mutator-induced, dominant antimorph of the maize amylose extender1 locus. Plant Cell 5: 1555–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streb S, Delatte T, Umhang M, Eicke S, Schorderet M, Reinhardt D, Zeeman SC. (2008) Starch granule biosynthesis in Arabidopsis is abolished by removal of all debranching enzymes but restored by the subsequent removal of an endoamylase. Plant Cell 20: 3448–3466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utsumi Y, Nakamura Y. (2006) Structural and enzymatic characterization of the isoamylase1 homo-oligomer and the isoamylase1-isoamylase2 hetero-oligomer from rice endosperm. Planta 225: 75–87 [DOI] [PubMed] [Google Scholar]
- Utsumi Y, Utsumi C, Sawada T, Fujita N, Nakamura Y. (2011) Functional diversity of isoamylase oligomers: the ISA1 homo-oligomer is essential for amylopectin biosynthesis in rice endosperm. Plant Physiol 156: 61–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattebled F, Dong Y, Dumez S, Delvallé D, Planchot V, Berbezy P, Vyas D, Colonna P, Chatterjee M, Ball S, et al. (2005) Mutants of Arabidopsis lacking a chloroplastic isoamylase accumulate phytoglycogen and an abnormal form of amylopectin. Plant Physiol 138: 184–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattebled F, Planchot V, Dong Y, Szydlowski N, Pontoire B, Devin A, Ball S, D’Hulst C. (2008) Further evidence for the mandatory nature of polysaccharide debranching for the aggregation of semicrystalline starch and for overlapping functions of debranching enzymes in Arabidopsis leaves. Plant Physiol 148: 1309–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayllace NZ, Valdez HA, Ugalde RA, Busi MV, Gomez-Casati DF. (2010) The starch-binding capacity of the noncatalytic SBD2 region and the interaction between the N- and C-terminal domains are involved in the modulation of the activity of starch synthase III from Arabidopsis thaliana. FEBS J 277: 428–440 [DOI] [PubMed] [Google Scholar]
- Zeeman SC, Umemoto T, Lue W-L, Au-Yeung P, Martin C, Smith AM, Chen J. (1998) A mutant of Arabidopsis lacking a chloroplastic isoamylase accumulates both starch and phytoglycogen. Plant Cell 10: 1699–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Colleoni C, Ratushna V, Sirghie-Colleoni M, James MG, Myers AM. (2004) Molecular characterization demonstrates that the Zea mays gene sugary2 codes for the starch synthase isoform SSIIa. Plant Mol Biol 54: 865–879 [DOI] [PubMed] [Google Scholar]
- Zhang X, Szydlowski N, Delvallé D, D’Hulst C, James MG, Myers AM. (2008) Overlapping functions of the starch synthases SSII and SSIII in amylopectin biosynthesis in Arabidopsis. BMC Plant Biol 8: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.