Abstract
The Cellulose Synthase-Like D (CslD) genes have important, although still poorly defined, roles in cell wall formation. Here, we show an unexpected involvement of CslD1 from maize (Zea mays) in cell division. Both division and expansion were altered in the narrow-organ and warty phenotypes of the csld1 mutants. Leaf width was reduced by 35%, due mainly to a 47% drop in the number of cell files across the blade. Width of other organs was also proportionally reduced. In leaf epidermis, the deficiency in lateral divisions was only partially compensated by a modest, uniform increase in cell width. Localized clusters of misdivided epidermal cells also led to the formation of warty lesions, with cell clusters bulging from the epidermal layer, and some cells expanding to volumes 75-fold greater than normal. The decreased cell divisions and localized epidermal expansions were not associated with detectable changes in the cell wall composition of csld1 leaf blades or epidermal peels, yet a greater abundance of thin, dense walls was indicated by high-resolution x-ray tomography of stems. Cell-level defects leading to wart formation were traced to sites of active cell division and expansion at the bases of leaf blades, where cytokinesis and cross-wall formation were disrupted. Flow cytometry confirmed a greater frequency of polyploid cells in basal zones of leaf blades, consistent with the disruption of cytokinesis and/or the cell cycle in csld1 mutants. Collectively, these data indicate a previously unrecognized role for CSLD activity in plant cell division, especially during early phases of cross-wall formation.
The ancient, highly conserved family of Cellulose Synthase-Like D (CSLD) proteins are required for cell growth and development, yet their biochemical and cellular functions are only now emerging (Richmond and Somerville, 2000, 2001; Favery et al., 2001; Wang et al., 2001; Bernal et al., 2007, 2008; Yin et al., 2009, 2011; Park et al., 2011). CSLDs belong to one of 10 distinct subfamilies in the Cellulose Synthase superfamily, defined by amino acid sequence similarity to Cellulose Synthase (CESA; Richmond and Somerville, 2000; Hazen et al., 2002; Farrokhi et al., 2006; Fincher, 2009; Penning et al., 2009). All members of this superfamily share predicted functions based on sequence identity as membrane-bound, processive glycosyltransferases that synthesize β-linked glycan polymers, such as those of cell wall polysaccharides (Richmond and Somerville, 2000, 2001). Known products range from cellulose to hemicellulose backbones and may include additional β-linked glycan chains (Arioli et al., 1998; Dhugga et al., 2004; Liepman et al., 2005; Burton et al., 2006; Cocuron et al., 2007; Doblin et al., 2009). The CSLDs remain poorly understood despite their importance in cell development and evidence for their evolution in plant lineages extending back to nonvascular land plants and possibly before (Roberts and Roberts, 2007).
Of the cellulose synthase-like genes, CslDs are the most closely related to the CesAs themselves, leading to early suggestions that CSLDs may also function as cellulose synthases (Doblin et al., 2001). The CSLD proteins also share the greatest amino acid sequence identity (40%–50%) with CESAs and are similar or slightly larger in size (Richmond and Somerville, 2001). Other than CESAs, CSLDs are the only members of the superfamily that have the N-terminal, zinc-finger-like domain thought to function in protein-protein interactions, possibly mediating the formation of complexes or protein turnover (Richmond and Somerville, 2000; Kurek et al., 2002; Gamsjaeger et al., 2007). Also, recent evidence suggests cooperative action between CSLD proteins in Arabidopsis (Arabidopsis thaliana; Yin et al., 2011), possibly in a similar fashion to cellulose synthases (Taylor et al., 2003; Persson et al., 2007b). To date, the strongest evidence that CSLDs function as cellulose synthases comes from data for (1→ 4)-β-glucan synthase activity from a CSLD protein and the successful complementation of a csld3 mutant in Arabidopsis using a chimeric CSLD3 protein with a CESA catalytic domain (Park et al., 2011).
The suggestion that the CSLD subfamily may be ancestral to the entire Cellulose Synthase superfamily is consistent with the locations and sizes of introns in CslD genes (Richmond and Somerville, 2000, 2001; Yin et al., 2009). The CslD genes are also present in all plant genomes examined thus far, including mosses (Richmond and Somerville, 2000, 2001; Roberts and Bushoven, 2007; Yin et al., 2009). In contrast, many of the other CSL subfamilies appear only in specific taxa (Farrokhi et al., 2006; Keegstra and Walton, 2006; Vogel, 2008; Fincher, 2009; Penning et al., 2009). Of the five CSL subfamilies yet to be assigned a specific polysaccharide synthase role, only CslD and CslE subfamilies are found in both dicot and monocot genomes (unlike CslB, CslG, and CslJ). The broad distribution of the CslD genes across taxa implies a highly conserved function (Richmond and Somerville, 2000, 2001; Roberts and Bushoven, 2007; Yin et al., 2009).
Clues to the biological roles of the CSLDs have been sought by defining their biochemical activity and/or subcellular localization, but interpretation of this work has not yet been conclusive. Heterologous expression studies demonstrated that CSLA (Liepman et al., 2005), CSLF (Burton et al., 2006), CSLC (Cocuron et al., 2007), and CSLH (Doblin et al., 2009) proteins catalyze the synthesis of hemicellulose polysaccharide backbones; consequently, the CslD genes were also hypothesized to encode hemicellulose synthases (Sandhu et al., 2009). However, similar approaches have thus far been unsuccessful with CSLDs. Other important lines of evidence have led to alternative interpretations. Analysis of cell wall polysaccharides, for example, from key cell types, cell culture treatments, or genetic perturbations suggest that CSLDs could function in either the production of cellulose (Manfield et al., 2004; Li et al., 2009) or hemicellulose backbones (Bernal et al., 2007; Li et al., 2009; Yin et al., 2011). However, interpreting differences in cell wall composition is complicated by broad changes in multiple wall constituents that often occur in response to genetic perturbation (Orfila et al., 2005; Bernal et al., 2007; Persson et al., 2007a; Li et al., 2009).
Localization studies, which would indicate where CSLD functions within a cell, have also been inconclusive to date. Targeting studies show that CSLD proteins appear to localize in the Golgi, where they could aid hemicellulose biosynthesis (Favery et al., 2001; Bernal et al., 2007, 2008; Zeng and Keegstra, 2008; Li et al., 2009). However, these observations are also consistent with the transit of CSLD proteins through the Golgi en route to the plasma membrane, as has been observed for cellulose synthases (Kimura et al., 1999; Crowell et al., 2009; Gutierrez et al., 2009). Recent studies also demonstrate that CSLD proteins localize to the plasma membrane in rice (Oryza sativa) suspension culture cells (Natera et al., 2008) and in Arabidopsis root hair cells (Park et al., 2011). Polarized plasma membrane localization of CSLD3 supports a role in polysaccharide deposition at the growing tip of Arabidopsis root hair cells (Park et al., 2011).
Thus far, the majority of genetic evidence has implicated CSLDs in polarized cell expansion associated with polar tip growth typical of pollen tubes and root hairs (Favery et al., 2001; Wang et al., 2001; Kim et al., 2007; Bernal et al., 2008; Penning et al., 2009) and the intrusive tip growth typical of xylem and other sclerenchyma fibers (Samuga and Joshi, 2004). In the moss Physcomitrella patens, where tip growth predominates in caulonema cells, CslD genes constitute 46% of all ESTs from the CESA superfamily, including all CesAs and CSLs (Roberts and Bushoven, 2007). In vascular plants, root hairs and pollen tubes provide classic models for cellular tip growth (Hepler et al., 2001; Cole and Fowler, 2006), but xylem and sclerenchyma fibers also elongate by a form of intrusive tip growth (Mellerowicz et al., 2001; Samuga and Joshi, 2004). The expression of CslD2 in developing xylem of Populus is consistent with its proposed influence on xylem fiber length (Samuga and Joshi, 2004). However, the function of CSLDs in other aspects of cell growth, such as cell division, nonpolar cell expansion, or differentiation, has not been explicitly studied to date. Some mutant phenotypes cannot be readily explained by polar growth defects (Bernal et al., 2007; Li et al., 2009; Yin et al., 2011), suggesting an expanded role for CSLDs in other cellular processes.
Here, we present evidence for an unexpected, but integral, role for CSLDs in plant cell division. In csld1 mutants of maize (Zea mays), defects in cell division were identified as the underlying basis for the narrow-organ morphology as well as the characteristic epidermal warts. Using reverse genetics tools in maize, we identified several mutants whose phenotypes are caused by transposon insertion in CslD1. The csld1 phenotype was uniquely informative for dissecting the relationship between division and expansion. Through a detailed cellular analysis, we demonstrate that altered cell division in csld1 null mutants reduces cell number and is an underlying cause of the narrow-organ morphology. In-depth analysis of wall positioning, cell shape, nuclear size, cell ploidy, wall architecture, and alterations in cell number indicated that defective cell division was an early consequence of a CSLD1 deficiency. These data provide new insight into the function of CSLD proteins in plant growth, extending our understanding of their roles from tip growth to include cell division in most or all organs.
RESULTS
Phylogenetic Analyses Indicate Early Evolution and Conservation of Divergent Roles for CSLD Proteins
Phylogenetic analyses (Fig. 1A) identified three distinct clades in the CSLD subfamily and found these to represent three phenotypic classes of csld mutants. These three major phenotypic groups are, for Arabidopsis, pollen tube defects in csld1 and csld4 mutants, (Bernal et al., 2008), root hair defects in csld2 and csld3 mutants (Favery et al., 2001; Wang et al., 2001; Bernal et al., 2008), and reduced plant size in csld5 mutants (Bernal et al., 2007). Thus far, mutants in related clades of CslD genes in rice and maize have yielded phenotypes similar to those of Arabidopsis. Mutants of rice CslD1 (Kim et al., 2007), for example, and its maize homolog, CslD5 (Penning et al., 2009), result in root hair-deficient phenotypes. Additionally, alterations in rice CslD4 (the closest homolog of ZmCslD1 and AtCslD5) confer a narrow leaf and dwarf1 phenotype (Li et al., 2009; Hu et al., 2010; Wu et al., 2010). Similar functional roles are indicated by the reduced-growth phenotypes common to all three of these mutants (Fig. 1A; Bernal et al., 2007; Li et al., 2009; Hu et al., 2010; Wu et al., 2010). Collectively, these data suggest the conservation of specific developmental roles for individual CSLD proteins in plants and indicate that these arose early in plant evolution.
Figure 1.
Comparison of mutant phenotypes with CSLD protein phylogeny, and diagram of the CSLD1 protein and mutant genes in maize. A, Neighbor-joining tree of predicted protein sequences encoded by CslD genes in maize, rice, and Arabidopsis. Reported phenotypes for null alleles are shown in red (ZmCSLD5 [Penning et al., 2009], OsCSLD1 [Kim et al., 2007], AtCSLD2 [Bernal et al., 2008], AtCSLD3 [Favery et al., 2001; Wang et al., 2001], AtCSLD5 [Bernal et al., 2007], OsCSLD4 [Li et al., 2009], AtCSLD1 and AtCSLD4 [Bernal et al., 2008]). AtCslD6 and ZmCslD3 are predicted to be pseudogenes. The ZmCSLD1 protein is starred. Members of distinct clades are grouped by shapes, with dashed lines including proteins for which no genetic evidence exists. The tree was created using MEGA 4.0 (http://megasoftware.net/mega.html; Tamura et al., 2007) with 2,000 bootstrap repetitions and the pairwise deletion option. Units are amino acid substitutions per site. Supplemental Figure S1 shows the ClustalW alignments used in this phylogenetic analysis. B, Diagram of the maize CslD1 gene and protein. Sites of Mu transposon insertion (triangles) are shown on the CslD1 gene. Lengths of exons and introns are indicated below each region (in bp). Both the csld1-1 and csld1-2 mutants were obtained from the UniformMu maize population at the University of Florida (http://uniformmu.uf-genome.org [McCarty et al., 2005; Settles et al., 2007]). Smaller triangles represent Pioneer Hi-Bred International TUSC alleles (McCarty and Meeley, 2009). The predicted protein includes 1,218 amino acids, an N-terminal RING-type zinc-finger-like domain, eight transmembrane domains, and the conserved residues of an inverting, processive glycosyltransferase. The protein diagram is modeled after Richmond (2000), and protein domains were identified using SMART (http://smart.embl-heidelberg.de) and TMHMM (http://www.cbs.dtu.dk/services/TMHMM).
An Allelic Series of csld1 Mutants in Maize Enabled Functional Analysis
Seven independent loss-of-function mutants for the maize CslD1 gene were identified in reverse genetic screens, including two from the UniformMu maize population (University of Florida) and five from the Trait Utilities for Screening of Corn (TUSC) lines (Pioneer Hi-Bred International; Meeley and Briggs, 1995; McCarty and Meeley, 2009; Fig. 1B). The two UniformMu alleles, csld1-1 and csld1-2, were examined in the greatest depth because of their uniform genetic background (McCarty et al., 2005). Both of these were null for detectable expression of CslD1 mRNA by quantitative PCR (data not shown). Phenotypes of csld1-1 and csld1-2 homozygous mutants, as well as offspring from their reciprocal F1 hybrids, were indistinguishable, thus demonstrating a causal role for the dysfunctional CslD1 gene. Mutant plants showed overall reduced growth, narrow leaves, and had a rough leaf texture caused by warty protrusions from the mature leaf epidermis. Genotypic analysis of over 200 individuals from segregating families showed a 100% correspondence between this phenotype and homozygosity for the csld1-1 mutation (data not shown). Mendelian segregation ratios were typical of a recessive mutation. The five other transposon insertions in CslD1 (csld1-3–csld1-7; courtesy of Pioneer Hi-Bred International) also resulted in mutant phenotypes that included narrower organs and wart-like clusters of epidermal cells, regardless of heterologous genetic background.
Phenotype of csld1 Mutants: Narrow Organs and Wart-Like Cell Clusters
Null mutants of csld1 showed a striking phenotype that included narrow leaves, reduced stature, and highly textured leaf blades (Figs. 2 and 3). Light microscopy showed that the visibly rough texture of csld1 mutant leaves was due to irregular swelling by groups of epidermal cells that formed wart-like cell clusters (Fig. 2). Some epidermal cells expanded 75-fold in volume and were generally arranged in linear profiles along the longitudinal axis of the leaf midrib and blade (Fig. 2A) but not on leaf sheaths or stalks (data not shown). The groups of cells that formed so-called warts were interspersed with normal-appearing regions of leaf epidermis along the entire length of the leaf blade. Swollen cells remained filled with fluid until the onset of leaf senescence (Fig. 2C). Warts were present on both surfaces of mutant leaf blades but were larger and more abundant on the abaxial face (Fig. 2D). These malformed cells consistently lacked chloroplasts (Fig. 2, C and E), indicating an epidermal origin, as confirmed in serial cross-sections of mutant leaves (Fig. 2, F and G). These ballooned epidermal cells in csld1 mutants often had diameters well over 100 μm, at least 5-fold and sometimes 20-fold greater than epidermal pavement cells of nonmutant plants (Fig. 2G). Both scanning electron microscopy and optical microscopy of fresh, intact leaves showed that lesions continued to expand throughout leaf maturation and that the largest clusters included swollen cells that had collapsed. In other instances, cells remained intact, even in lesions greater than 300 μm across (Fig. 2C).
Figure 2.
Epidermal lesions on the leaf blades of csld1 mutants. A to C, Fresh, intact leaves. Epidermal warts were distributed across the entire blade and midrib of mature, fully expanded leaves. D and E, Freshly sectioned leaf blades. Swollen lesions were most abundant on the abaxial leaf blade surface. F and G, Cross-sections of fixed, embedded leaves, with F including a composite of overlapping images. Epidermal cells were more visibly affected than cells of the leaf-blade interior. Some epidermal cells had two-dimensional, cross-sectional areas up to 25-fold greater (radius = 100 μm for the swollen cell in G).
Figure 3.
The narrow-leaf phenotype of csld1 mutant plants. A, Plant height and dry weight (aboveground and belowground) were quantified for field-grown csld1 mutant and nonmutant plants. Plant height was 90.9 cm (se, 2.8; n = 20) for mutants and 102.2 cm (se, 1.7; n = 35) for nonmutants. For dry weight measurements, whole plants were sampled 3 d after ear maturity (40 d post pollination) and did not include mature ears or fine roots. Total plant dry weight was 41.9 g (se, 4.0; n = 4) for mutants and 76.5 g (se, 3.2; n = 4) for nonmutants. Belowground dry weight was 8.7 g (se, 0.2; n = 4) for mutants and 17.1 g (se, 1.1; n = 4) for nonmutants. Aboveground and belowground dry weights were reduced by similar amounts (average of 44% and 49%, respectively). se values are indicated with vertical bars and are marked by asterisks wherever values for mutant plants differed significantly from those of nonmutants (P < 0.05). B, Leaf blade length and width (at the widest point) were quantified for leaf positions 3 through 5 (as indicated) for field-grown mutant and nonmutant plants. Nonmutant plants included both wild-type and heterozygous individuals from segregating progeny after three back-crosses into the W22 inbred. Imaged blades were from leaves in position 3 from the apex. The left-most portion of each graph shows combined data from all leaf positions measured. The leaf blade width-to-length ratio was 27% less for mutant plants. Blade width and length were reduced 35% and 10%, respectively, relative to those of nonmutant plants. Data were similar for csld1-2 (data not shown) and visual appraisals of the five other csld1 mutants. Note that leaf photographs were from greenhouse-grown plants, whereas quantifications were from field-grown plants. * Significantly different from the nonmutant (P < 0.05).
Potentially analogous, wart-like epidermal swellings were described by Burton et al. (2000) in a virus-induced gene silencing experiment in tobacco (Nicotiana tabacum). Although a CesA gene was targeted, the highly similar CslD genes may also have been silenced. The transgenic tobacco had wart-like lesions on the abaxial side of leaves and an overall phenotype strikingly similar to the maize csld1 mutants (Burton et al., 2000). This commonality would be consistent with some degree of repression of the ZmCslD1 ortholog in the tobacco experiment. Alternatively, if observed lesions did result from the down-regulation of CesA genes alone, then this would support a role for CSLD proteins in cellulose biosynthesis (Doblin et al., 2001; Park et al., 2011).
Plant Dry Weight and Organ Width Are Reduced in csld1 Mutants
Total growth and organ size were reduced in homozygous csld1 mutants (Fig. 3), even though overall plant architecture, leaf number, and flowering time were similar for csld1 mutant and nonmutant siblings under field and greenhouse conditions (data not shown). At maturity, the mean height of mutant plants (to the auricle of the uppermost leaf) was only 11% less (P < 0.001), whereas dry weight decreased by a more pronounced 44% (P < 0.0025; Fig. 3A). Proportional reductions in dry weight were also evident for all organs examined, including ears, tassels, stalks, roots, and leaves (data not shown). Organ width decreased 35% (P < 0.0003) for mature leaf blades of csld1 mutant plants, but length was only 10% less (P < 0.0003; Fig. 3B). This narrow-leaf phenotype was proportional in all leaves examined, indicating a consistent defect in lateral development rather than an ontological effect at specific leaf positions (Fig. 3B).
Organ width was also reduced in stalks from csld1 mutants (Supplemental Fig. S2). The cross-sectional areas of csld1 stalks were an average of 24% less (P < 0.025). The narrow-organ phenotype extended to cobs and tassels as well (data not shown).
Cell Numbers Are Reduced in Mutants and Are More Pronounced Than Changes in Cell Size
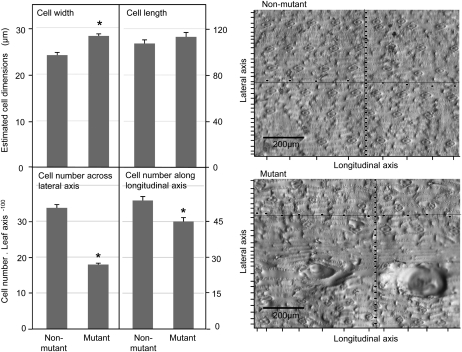
Both cell division and cell expansion contribute to the final shape of an organ; yet, it has been traditionally difficult to separate causative factors during maize leaf morphogenesis. Given the reduced leaf width in csld1 mutants, we asked whether the mutant leaf morphology was due to decreased cell number, indicating that CSLD1 acts directly or indirectly on cell division, or whether the primary cause of narrower leaves was the altered cell width. To answer this question, we quantified cell numbers and sizes in mutant and nonmutant leaf blade epidermis (Fig. 4). Examination of impressions from nonwarty areas of mutant and nonmutant leaves showed that the length of epidermal cells was not detectably different, and the width of cells from the csld1 mutant was 17% greater (P < 0.0003; Fig. 4). Interestingly, the most pronounced difference between mutant and nonmutant was in total cell numbers, and thus divisions, across the leaf blade, which was 47% less for csld1 mutants (Fig. 4) based on calculations from cell-level (Fig. 4) and leaf-level (Fig. 3) analyses.
Figure 4.
Estimated dimensions of nonlesion epidermal cells, and cell numbers along lateral and longitudinal axes of leaf blades from csld1 mutant and nonmutant plants. Epidermal impressions from abaxial surfaces of fully expanded leaf blades from mature, greenhouse-grown plants were used to estimate cell dimensions and cell numbers along leaf axes. Nonmutant plants included both wild-type and heterozygous individuals from segregating progeny after three back-crosses into the W22 inbred. Cell numbers were quantified (each designated by a tick mark on impressions such as those shown to the right) along longitudinal and lateral axes of defined length. Mean cellular dimensions were determined by dividing the number of cells along a given axis by the length of that axis (mutant, n = 14; nonmutant, n = 10; longitudinal axis, 1.88 mm; lateral axis, 1.40 mm). Axes used for analyses of epidermal cell size from mutant leaves were analyzed using transects that did not cross cells in the ballooning protrusions. Approximate cell numbers across and along leaf blades were estimated by comparing cell size estimates with average leaf length and width measurements (Fig. 3). These analyses did not include the midrib (0.5 cm was subtracted from total leaf width). The mean width of epidermal cells was 17% greater in csld1 mutants, whereas mean cell numbers were 48% less across the lateral axis and 16% less along the longitudinal axis. Values are marked with asterisks where they differ significantly from those of nonmutant plants (P < 0.05).
Leaf cross-sections were examined to ascertain whether similar abnormalities were present in nonepidermal cells of csld1 mutant leaves. Leaf thickness was consistently increased in mutant leaf blades, and vascular bundles were closer together (Supplemental Fig. S3A). Fully expanded csld1 mutant leaf blades were 40% (P < 0.004) thicker than nonmutant leaf blades (Supplemental Fig. S3B). The extent of nonepidermal contributions to total leaf thickness was 43% (P < 0.002) greater for csld1 mutant leaf blades, and thus proportional to those of epidermis (Supplemental Fig. S3B). Increases in cell size were variable but most often apparent in mesophyll sheath cells. Overall organization of the mesophyll sheaths was less regular. The csd1 phenotype, therefore, was not limited to the epidermis alone. Additionally, mutant leaves showed a 12% (P < 0.03) increase in vascular bundle density (Supplemental Fig. S3C).
Levels of CslD1 mRNA Are Greatest in Regions of Active Cell Division
In order to view the phenotypes of csld1 mutants in the context of where the wild-type gene is expressed, levels of CslD1 mRNA were measured via quantitative reverse transcription (RT)-PCR across diverse tissues and stages of development (Fig. 5). The CslD1 transcript levels were greatest in young, preemergent leaves (inside the whorl) and still relatively abundant in young primary root tips and bases of more mature leaves (Fig. 5A). These mRNAs were also detected in basal zones of leaves by other recent studies of maize leaf development (Li et al., 2010). To more clearly define the pattern of transcript accumulation during leaf development, staged samples of young to mature leaves were analyzed. The CslD1 mRNA levels were highest in tissues with actively dividing cells and maximal in shoots 6 d after germination (Fig. 5B). Later in development, single-leaf analyses showed that levels of CslD1 mRNA were greatest in basal portions of blades from expanding leaves, 15 to 25 cm long. In nondividing, fully expanded leaves, CslD1 mRNA had dropped below detectable levels and remained so in the fully differentiated portions of leaves (Fig. 5B). Notably, wart formation and expansion continues in leaves through maturity, even within areas where CslD1 is no longer expressed.
Figure 5.
Levels of ZmCslD1 mRNA in diverse tissues from wild-type plants of the W22 inbred. A, Expression of ZmCslD1 in diverse organs. B, ZmCslD1 mRNA levels in leaf blades and blade regions at different stages of development. Levels of mRNA were quantified by CyberGreen quantitative real-time RT-PCR. Three biological replicates were analyzed for each tissue. Yellow areas of the leaf drawings represent immature, nongreen tissue. se values are indicated by vertical bars. DAG, Days after germination; DBP, days before pollen-shed.
Cell Wall Composition Was Similar, but Thin, Dense Walls Were More Abundant in csld1 Mutants
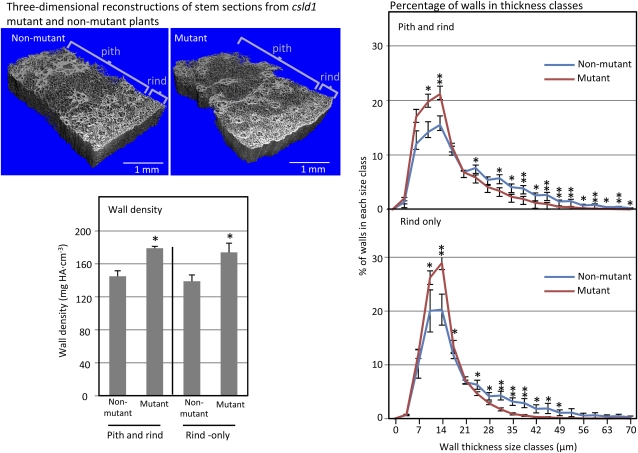
To further characterize differences between mutant and nonmutant tissues, high-resolution x-ray microcomputed tomography (micro-CT) was used to analyze hand-cut sections of stalks, since this tissue was found amenable to the x-ray approach. Three-dimensional reconstructions revealed a shift in average wall thickness toward thinner walls in the csld1 mutant (Fig. 6). Additionally, this method allowed a comparison of overall wall density, which was greater for stalks from mutant plants, regardless of whether the pith and vessel-rich rind were examined together or separately (Fig. 6).
Figure 6.
High-resolution x-ray micro-CT analysis of csld1 mutant and nonmutant stalks. Stem sections from the third internode of greenhouse-grown csld1 mutant and nonmutant plants were scanned at 3.5-μm resolution using high-resolution x-ray micro-CT. Sections for scanning (approximately 3 × 2 × 10 mm) were hand cut from the edges of stems from mutant and nonmutant plants (four each). Nonmutant plants included both wild-type and heterozygous individuals from segregating progeny after three back-crosses into the W22 inbred. Three-dimensional analyses revealed significant differences in the density of wall material (bottom left) and the distribution of wall thickness (right). Mutant stems had more dense, but generally thinner, walls, even when analyses were limited to the rind only. Volumetric analyses were calculated with an algorithm developed for trabecular bone and based on hydroxyapatite (HA) resin standards (Hildebrand and Ruegsegger, 1997). se values are indicated with vertical bars. Values from csld1 mutant plants that are significantly different from nonmutants are indicated with asterisks (** P < 0.05, * P < 0.1).
To determine whether these changes were associated with alterations to cell wall polysaccharide composition, we examined cell walls from mature leaf blades as well as isolated leaf epidermis. No significant differences in alcohol-insoluble cell wall composition were detected between csld1 mutants and nonmutants for either cellulose or sugar subunits of noncellulosic constituents (Table I). Cell walls from epidermal cells of both plant types revealed distinctive composition relative to samples from whole-leaf blades. Specifically, epidermal cell walls had less Glc, Rha, Gal, and GalUA, but more Xyl, compared with whole-leaf blades (Table I).
Table I. Cell wall sugar composition of leaf blades and epidermal peels from csld1 mutant and nonmutant plants.
Alcohol-insoluble residues were prepared from both whole-leaf blade sections and isolated epidermal strips of greenhouse-grown plants identified as mutant or wild type by PCR of a segregating family. Analysis of noncellulosic cell wall sugars was done at the Complex Carbohydrate Research Center at the University of Georgia. Values for noncellulosic sugars are given as mol %, with se in parentheses. Cellulose content was estimated based on remaining weight after the hydrolysis of hemicelluloses and lignin. Cellulose content is given as weight percentage of alcohol-insoluble cell wall fraction, with se in parentheses. None of the differences between csld1 mutant and wild-type samples were statistically significant at P < 0.05 (n = 4 for each sample). nd, Not detected; nt, not tested.
| Sample | Plant | Ara | Rha | Fuc | Xyl | Man | GalA | Gal | Glc | GlcA | Cellulose |
| Whole leaf | Wild type | 11.43 (0.33) | 0.53 (0.02) | nd | 81.40 (0.01) | nd | 1.36 (0.07) | 0.85 (0.69) | 4.43 (0.41) | nd | 26.6 (1.8) |
| csld1 | 11.84 (0.18) | 0.57 (0.05) | nd | 81.18 (1.31) | nd | 1.59 (0.016) | 0.80 (0.04) | 4.03 (1.41) | nd | 22.9 (3.1) | |
| Epidermal peels | Wild type | 10.66 (0.54) | 0.31 (0.02) | nd | 85.04 (0.69) | nd | 0.95 (0.29) | 0.65 (0.08) | 2.39 (0.34) | nd | nt |
| csld1 | 11.93 (0.63) | 0.34 (0.03) | nd | 84.37 (0.96) | nd | 0.71 (0.05) | 0.69 (0.08) | 1.97 (0.22) | nd | nt |
Multiple Cell Division Defects Are Evident in csld1 Epidermis
Dark-field images of epidermal peels from mature leaf blades showed that the normal ordering of epidermal cells was disrupted in csld1 mutants and revealed frequent anomalies in recently formed cross-walls. The latter included stubs of incomplete cell walls, particularly along the longitudinal axis of leaves (Supplemental Fig. S3). Other alterations of cell shape and misaligned cell walls were also consistent with causal defects in cell division, and these persisted in mature leaves.
To determine the developmental stage at which these cell abnormalities could first be detected, 5- to 10-cm immature leaves (sites of maximal CslD1 expression) were sampled from csld1 mutant and nonmutant plants, stained with propidium iodide, and examined by confocal microscopy (Fig. 7). Disrupted cell files and misshapen cells were evident in images of abaxial epidermal cells from the predifferentiation and postdifferentiation zones (as defined by Mitkovski and Sylvester [2003] and determined by the absence and presence of stomata, respectively; Fig. 7, A and B). Cell wall stubs were frequently observed in epidermal cells of both predifferentiation and postdifferentiation zones (Fig. 7). Again, these were typically oriented along the longitudinal axis (Fig. 7). While many epidermal cells at these stages were approximately twice the normal width (as if they had failed to undergo a single longitudinal division), nearly all cells of csld1 mutants were at least some degree wider than those of nonmutants (Fig. 7, A and B).
Figure 7.
Confocal images showing defects early in the development of csld1 mutant leaf epidermis. Propidium iodide-stained cell walls from fresh, immature csld1 mutant and nonmutant leaves are shown. A, Predifferentiation zones from the basal 3 mm of nonmutant leaf blades contrasting with the abnormal csld1 cell size, shape, and organization. Leaf blades were approximately 10 cm long. B, Postdifferentiation zones (3–10 mm above the base of the blade) showing the persistent effects of altered cell division, with the presence of large, misshapen, less ordered cells of the csld1-1 leaf epidermis. Leaf blades were approximately 10 cm long. C, A typical file of large, irregular cells bounded by individuals with incomplete cell walls protruding into them from their outer edges, consistent with the clonal inheritance of large cell size in the csld1 mutants. D, Serial optical sections revealing irregular gaps in cell walls (arrows) that occur frequently in epidermal cells of the csld1 mutant.
A greater cell width also remained evident later in development, even for otherwise normal-looking cells outside wart-like lesions (Fig. 4). Also, in multiple instances, files of atypically large epidermal cells were bounded at proximal and distal ends by cells that appeared to have misdivided. Cell wall stubs were visible in each and faced the large files. These files of wide cells were located at positions normally occupied by two distinct files of smaller cells (Fig. 7C). This pattern indicated either the failure of consecutive neighboring cells to divide or, more likely, a clonal file derived from a single cell that failed to complete division. In addition, three-dimensional imaging with confocal microscopy identified a large number of gaps and holes in cell walls that would otherwise have appeared to be complete when viewed by standard imaging techniques (Fig. 7D). The actual number of incomplete walls may thus be underestimated when nonconfocal approaches are used.
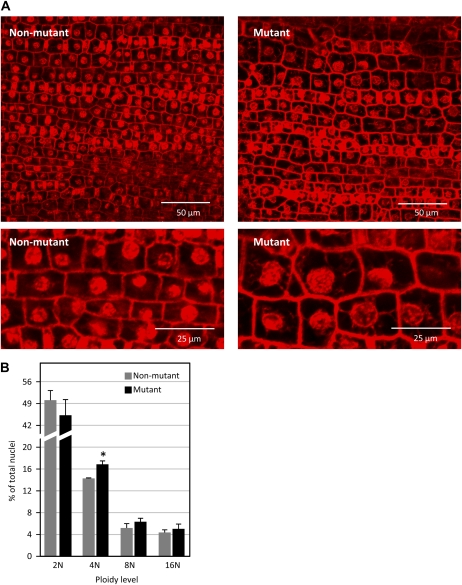
Because cells with large or multiple nuclei are commonly observed in cell division-defective mutants (Lukowitz et al., 1996; Smith et al., 1996; Spitzer et al., 2006), propidium iodide-stained nuclei were examined in basal regions of immature leaf blades from csld1 mutants (Fig. 8A). Compared with nonmutant epidermis of the same stage (predifferentiation zone of 5- to 10-cm leaves), csld1 mutant cells generally had larger nuclei (Fig. 8A). While significant variation in nuclear size was observed even in nonmutant tissue, the range in csld1 mutants was much greater, with cells containing nuclei ranging from 1-fold to approximately 4-fold the two-dimensional area of normal nuclei (Fig. 8A). Large nuclear size was consistently correlated with large cell size (Fig. 8A). Flow cytometry of nuclei from basal regions of immature leaves showed significantly more (P < 0.05) tetraploid nuclei in csld1 mutants compared with nonmutants (Fig. 8B).
Figure 8.
Nuclei of immature csld1 mutant and nonmutant epidermis. A, Confocal imaging of fixed cells stained with propidium iodide showing the abundance of large nuclei in clsd1 mutant epidermis and the correspondence between these and larger cell size. B, Percentage of total nuclei with DNA copy numbers of 2N, 4N, 8N, and 16N as determined by flow cytometry of isolated nuclei stained with propidium iodide. Nuclei were from immature tissue of basal portions of csld1 mutant and nonmutant leaves. The majority of nuclei were 2N (data not shown). se values are indicated by vertical bars. Significant difference from the nonmutant is indicated by the asterisk (P < 0.05).
DISCUSSION
CSLDs Acquired Divergent Roles Early in Plant Evolution
The integration of mutant phenotypes from maize, rice, and Arabidopsis with the CSLD phylogeny reveals ancient functional divergence and highly conserved developmental roles for subgroups within the CSLD family (Fig. 1A). Although double- and triple-mutant analyses have demonstrated some overlapping function for CslD genes in Arabidopsis (Yin et al., 2011), each single-gene mutation described thus far has a distinctive phenotype (except for the putative pseudogene AtCslD6; Favery et al., 2001; Wang et al., 2001; Bernal et al., 2007, 2008; Kim et al., 2007; Li et al., 2009; Penning et al., 2009; Hu et al., 2010; Wu et al., 2010). When we overlay data for mutant phenotypes onto a phylogentic tree, CSLD clades correspond to distinct classes of phenotypes: (1) root hair defective, (2) male transmission defective, or (3) reduced growth (Fig. 1A).
There are several notable aspects of this pattern. First, the observation that each CslD mutant has a visible phenotype indicates a limited functional redundancy within the gene family. Second, the associations shown in Figure 1A persist despite fundamental structural differences between the type I primary cell wall of Arabidopsis and the type II cell wall of rice and maize (Harris and Hartley, 1980; Carpita and Gibeaut, 1993; Carpita, 1996; Carpita et al., 2001). The roles of CslD genes thus transcend the major differences in noncellulosic cell wall constituents of diverse plant species, and the primary developmental functions of individual CslD genes appear to have been maintained. Third, previous results suggested specific contributions by CSLD proteins in tip-growing cells (Bernal et al., 2008); however, disruptions in AtCslD5, OsCslD4, and ZmCslD1 led to reduced overall plant growth without visibly altering classic tip-growing cells (Fig. 1A; Bernal et al., 2007; Li et al., 2009; Hu et al., 2010; Wu et al., 2010; Yin et al., 2011). Broader developmental functions are thus indicated for these genes. Among the reduced-growth phenotype clade, that of maize csld1 is unique in its production of wart-like lesions. This difference might lie in the greater growth rate and expansion of maize leaves. In other respects, however, commonalities between the reduced-growth phenotypes suggest a shared function for this subgroup of CSLDs.
Reduced Cell Division Is a Central Effect of the csld1 Mutation
The combination of reduced leaf width together with overly expanded cells is intriguing and points to the possibility that an analysis of the cellular effects of the mutation could provide information about the controls of cell division and expansion. We show here that cell division defects are early effects of the mutation (Figs. 2–4) and that the observed reduction in cell number contributes to the narrow-leaf phenotype (Figs. 3 and 4). Although cell size increases dramatically in epidermal cells that balloon out of the leaf plane in the csld1 mutant (Fig. 2), most epidermal cells expand by only a modest 17% and do so only along their lateral axis (Fig. 4). Collectively, decreased cell number and fewer cell divisions that affect leaf width (35% narrower) and plant dry weight (44% less; Fig. 3) are only partially compensated by enhanced cell expansion. Estimated numbers of cells across leaves are reduced by 47%. This drop in cell number can be attributed to reduced cell divisions in the lateral dimension, indicating that this specific decrease in divisions is a central effect of csld1 mutations. Although an examination of cross-sections suggested that leaf vascular bundle number and density were altered in mutant leaves, the size and shape of vascular bundles were generally unchanged. Instead, the greater thickness of csld1 leaf blades typically corresponded most closely with irregular increases in size by cells of the mesophyll sheath (Supplemental Fig. S3), indicating a broader role for CSLD1 than epidermal development alone.
A cell division role for CslD1 early in leaf development is also supported by its maximal expression in zones of wild-type leaves where cells are most actively dividing (Fig. 5) and with the occurrence of anomalous divisions within this zone in the csld1 mutant (Fig. 7). These disrupted divisions result in the formation of abnormal cell clusters implicated in later wart development and potentially affect the determination of cell file numbers across leaf blades. Expansion of the epidermal warts appears to be largely a secondary effect of the csld1 mutation, because these cells continue to expand in leaves after CslD1 mRNAs would have dropped below detectable levels (Fig. 5).
While our results are compatible with a central defect in the rate or total number of cell divisions, diverse, indirect effects may also contribute to the observed phenotype. Narrow leaves of csld1 mutants, for example (Fig. 3), could exacerbate the reductions in plant dry weight by decreasing total plant photosynthetic capacity. The smaller csld1 root system (Fig. 3) could further reduce growth. Also, the epidermis may have a prominent physical role in organ expansion and meristem geometry (Green, 1980; Moulia, 2000), providing additional potential for secondary or tertiary effects of the csld1 mutations.
Cell expansion is clearly also altered by direct and/or indirect effects of the mutation. Much of the cellular overexpansion is likely secondary, since compensatory expansion is a common response to decreases in cell number (Reynolds et al., 1998; Beemster et al., 2003; Mitkovski and Sylvester, 2003; Horiguchi and Tsukaya, 2011). However, the csld1 mutations could theoretically affect more than cross-wall deposition during division and subsequent, compensatory expansion. If the properties of mutant walls are altered, then these too could contribute to expansion-based aspects of the phenotypes (discussed further below).
Cell Wall Thickness, but Not Composition, Is Altered in the csld1 Mutant
Although the overall quantity of cell wall material is reduced by 45% in the csld1 mutant plants, cell wall composition is unaffected in either whole organs or epidermis alone (Table I). Several possibilities could account for the lack of a detectable difference. First, the CSLD1 polysaccharide product may normally be present in only small amounts, particularly if synthesis is restricted to a defined period during cytokinesis or new cell wall formation. If so, then cell wall contributions from CSLD1 could be masked by more abundant polymers. As noted above, an early developmental role for small amounts of CSLD product would be consistent with the maximal expression of CslD1 in very young leaves and basal portions of fast-growing blades (Fig. 5), since these are zones of active cell division (Sylvester et al., 1990; Freeling, 1992; Sylvester, 2000). One permutation of this suggestion is that CSLD1 might aid a specialized, directed deposition of early-arriving polymers to the growing cell plate during division. The CSLD1 product itself could thus be limited to a narrow point in time.
Another possibility is that the CSLD1 enzyme might synthesize a limiting constituent of cell walls, such that a drop in levels of the CSLD1 product would result in similar decreases for other cell wall polymers. In this way, csld1 mutants could produce less total cell wall without changing the relative proportions of individual cell wall components. This suggestion is consistent with the reduction in total dry matter. Still another, compatible scenario is that the polysaccharide product of CSLD1 may not differ in a way immediately detectable by current analyses but may nonetheless have functionally altered properties and/or capacities for interaction that affect later cell wall behavior. All three of the above possibilities remain consistent with a temporally limited contribution of a known wall component.
Cell wall analysis by high-resolution, x-ray, micro-CT (Fig. 6) provides detailed structural information on internal regions of intact tissue, including the relative density of cell wall material, based on x-ray beam attenuation (Steppe et al., 2004; Dhondt et al., 2010). Stems of csld1 mutants (tissues found most amenable to the x-ray, micro-CT approach) show slight, but consistent, increases in wall density and a greater overall abundance of thin-walled areas, regardless of position in rind or pith (Fig. 6). These findings indicate an altered cell wall or cellular architecture that is apparently independent of changes in wall composition. One, albeit speculative, possibility is that the changes in cell wall density may reflect a relative increase in cellulose crystallinity, with less amorphous cellulose being present in the cell walls of the csld1 mutant.
Role of CslD1 in Division and Expansion
Cell-level effects of csld1 mutations highlight differences between global and local responses that lead to narrow leaves and overly expanded, warty clusters, respectively. In whole leaves, cell divisions are reduced markedly (almost 50%), while expansion is increased slightly (about 20%; Figs. 3 and 4). In clusters of abnormal cells, however, division anomalies are more frequent and expansion effects more extreme. One reason for this distinction is likely a contrast in the mechanisms by which cell expansion responds to altered cell number (Horiguchi and Tsukaya, 2011). At the whole-leaf level, classical compensatory expansion is widely recognized as a means of adjusting organ shape via cell enlargement (Reynolds et al., 1998; Beemster et al., 2003; Mitkovski and Sylvester, 2003; Tsukaya, 2008; Horiguchi and Tsukaya, 2011), whereas local responses to changes in cell number may differ and the mechanisms remain unclear (Horiguchi and Tsukaya, 2011). Primary effects of CslD1 disruption clearly include cell division, and although many aspects of expansion may be secondary, some could also be direct.
Warts Are Distinctive, Informative Features of the csld1 Mutant
In warts, swelling epidermal cells (Fig. 2) trace to cell division defects early in leaf development (Fig. 7). Assuming that CSLD1 is a wall-synthesizing enzyme, these cellular observations indicate that loss of CSLD1 activity disrupts the synthesis of early cell wall components, and this in turn alters division. The narrow organs result from a decreased number of long-axis cell divisions throughout the plant (discussed above). Wart formation also derives from disrupted divisions, but the locally extreme cell size may involve additional effects of csld1 on wall expansion. Two possible scenarios for epidermal wart formation are presented below.
Hypothesis 1 is based on a role for CSLD1 in new cell wall formation during cytokinesis and posits that epidermal warts would be initiated when rapid elongation by leaves outpaces the capacity of dividing cells to form normal cross-walls. The first set of disrupted cross-walls would lead to misplacement of the next, and the resulting clusters of irregular cells would be prone to anomalous expansion. The extent and irregularity of expansion would be exacerbated by asymmetric support from surrounding cells. Hypothesis 2 includes basic tenets of hypothesis 1 but also suggests that csld1 mutants may have broader alterations to cell wall properties that reduce the control of expansion. This in turn could contribute to both incomplete and misplaced cross-wall formation. Altered wall properties could reduce the control of cell expansion at each step in the development of irregular cell clusters and epidermal warts and could act in a slow, uniform manner and/or add a threshold component to the ballooning of epidermal cells. Under either hypothesis, the excessive expansion of cells in wart formation can be considered a secondary effect of the CslD1 deficiency, resulting from a cascade of interactions between altered cell division and expansion.
The sequence of events in hypothesis 1 begins with a central role for the CSLD1 polysaccharide product (likely cellulose) in the formation of new cross-walls. In csld1 mutants, this cross-wall formation is implicated in reducing the number of long-axis cell divisions and thus organ width. In leaf blades especially, a CSLD1-based limitation on new wall formation could have prominent effects when the demands of rapid growth outpace the compromised capacity for cross-wall production. The resulting anomalies in csld1 epidermis include instances of nondivided cells and defective cross-walls. The latter could arise from either partial formation of these walls and/or their expansion before full completion. Resulting cells have nonexistent or defective cross walls with central holes or cell wall stubs (as observed). A single cell division altered at the base of a growing leaf blade could lead to other disruptions by altering the timing and placement of the subsequent cross-walls. The abnormal divisions would then lead to the uneven and disrupted cell files as well as to large and misshapen cells (Fig. 7, A and B). These cells would lack the support otherwise afforded a normal, highly ordered, brick-like pattern of epidermal cells and would be more prone to excessive expansion during turgor-driven growth. In addition, hypothesis 1 predicts that the wartyness of csld1 phenotypes will be more extreme under conditions where growth is most rapid and cross-walls cannot form correctly. Consistent with this prediction, the phenotypes vary in plants grown under field versus greenhouse conditions (data not shown).
Another possibility is that csld1 alters cell wall properties that affect expansion and these in turn cause cell division defects that lead to wart formation (hypothesis 2). Such changes could have functional significance even if generated by trace amounts of materials contributed only during a brief period early in cell development. Although altered cell walls (missing a CSLD1 product) could be “weaker,” they might also be more responsive to effectors of cell expansion, such as endogenous signals or physical aspects of turgor pressure. Larger or irregularly shaped cells could result, and this could compromise their capacity to form accurately positioned or complete cross walls. The resulting series of anomalous, failed, or disrupted divisions would produce warty cell clusters. The suggestion that csld1 cells expand more readily would be consistent with the somewhat larger size of epidermal cells overall. Wall properties might be altered in subtle ways throughout development or impart a threshold component to ballooning cells of warts.
The CslD1 Gene Is Essential for Specific Aspects of Cell Division
Primary effects of the csld1 mutation on cell division can be clearly discerned in the narrow leaves and fine stems of the csld1 phenotype. Although the overly expanded warty lesions exhibit cascades of anomalous secondary events, the majority of cells in leaves and stems are well ordered. Reduced cell numbers and cell divisions in these organs are consistent with a global response to the csld1 mutation. In particular, long-axis divisions are decreased and proportional to reductions in leaf width, other organ diameters, and plant dry weight. Although the narrowness of leaf blades could theoretically be exacerbated by secondary effects of warts, the same cannot be said of the other narrow organs of csld1 plants. As a cell wall biosynthetic gene, CslD1 can have direct and indirect effects. The latter can include compensatory expansion (Horiguchi and Tsukaya, 2011) as well as wall-based signaling (Hématy and Höfte, 2008; Ringli, 2010; Seifert and Blaukopf, 2010; Boisson-Dernier et al., 2011). However, changes in cell number are seldom secondary consequences of altered expansion at the organ level (Wang et al., 2003; Cnops et al., 2004; Hu et al., 2006; Szécsi et al., 2006; Horiguchi et al., 2011; Horiguchi and Tsukaya, 2011), and a primary role is suggested here for CslD1 in cell division.
The deposition and integrity of new cross-walls are clearly impaired in dividing cells of csld1 mutants. Long-axis divisions are most markedly affected, with new walls having large central openings or appearing as incomplete stubs. Collective data reveal a new dimension to functions of the CslD gene subfamily, since these were previously considered largely related to tip growth (Favery et al., 2001; Wang et al., 2001; Kim et al., 2007; Bernal et al., 2008; Penning et al., 2009; Park et al., 2011). However, both tip growth and cross-wall formation during division share a common, directional aspect to the deposition of new wall material. Bednarek and Falbel (2002) previously suggested that such mechanisms could involve similar components. Our work here is consistent with that possibility as well as with contributions by CslD genes to targeted wall formation in tip-growing and dividing cells.
The csld1 mutation also disrupts cytokinesis and/or the cell cycle. Larger nuclei were observed by confocal imaging in cells of the predifferentiation zone of immature csld1 leaves (Fig. 8A). Similar, large-nucleus cells have been reported in studies of cell division-defective mutants (Lukowitz et al., 1996). Here, flow cytometry also showed a small, but significant, increase in endoreduplication (Fig. 8B). Even with 50% of epidermal cells undergoing endoreduplication, these would constitute a relatively small portion of nuclei from whole-leaf tissue. Whether the increase in endoreduplication and larger sized nuclei reflects a response to larger cell size or the arrest of the cell cycle after DNA replication, but before nuclear division, remains unclear. A more rapid entry of dividing cells into an endoreduplicative state is typically observed where cell size is increasing as a compensatory response to limited cell number in a developing organ (Beemster et al., 2005; Ferjani et al., 2007; Horiguchi and Tsukaya, 2011). The delay of cells at the G2 phase implies a disruption of the normal cell cycle due to a lack of CSLD1 activity.
CONCLUSION
The significance of our collective findings is 3-fold. (1) A previously unrecognized role in cell division is demonstrated here for a cell wall biosynthetic gene. (2) CslD1 is also found here to be central for the effective formation and integrity of new cross-walls during cell division. (3) Collective roles for the CslD gene subfamily are shown to include not only tip growth by cells but also probable contributions to other aspects of directional wall deposition. Additionally, csld1 maize mutants, besides helping our understanding of the specific functions of a clearly important cell wall-synthesizing enzyme, will provide valuable tools for dissecting the complex and interconnected processes of cell division and cell expansion. Protein localization efforts are under way and should further clarify the specific role of CSLD1 in early cell wall formation.
MATERIALS AND METHODS
Phylogenetic Analyses
Protein sequences predicted from full-length cDNAs for each of the CslD genes from rice (Oryza sativa), maize (Zea mays), and Arabidopsis (Arabidopsis thaliana) were used to create a neighbor-joining tree using MEGA 4.0 (Tamura et al., 2007; http://megasoftware.net), with 2,000 bootstrap repetitions using the pairwise deletion option. Alignments used to create the tree can be accessed in Supplemental Figure S1. The nomenclature for proteins encoded by the CslD gene family in maize was assigned as per van Erp and Walton (2009). Protein domains were identified using SMART (http://smart.embl-heidelberg.de) and TMHMM (http://www.cbs.dtu.dk/services/TMHMM).
Identification of csld1 Mutants
The UniformMu population was screened using PCR-based assays to identify Mutator (Mu) transposon inserts in ZmCslD1 as per Penning et al. (2009). Close to 15,000 UniformMu lines were screened using a series of pooled DNA samples, which were forerunners of the sequence-indexed materials currently available at MaizeGDB (http://maizegdb.org, http://uniformmu.uf-genome.org). For PCR screening, CslD1-specific primers (5′-AGTTCGTGCACTACACCGTGCACATCC-3′ and 5′-TGCTACCTGTAAGGACTGAGGATGGCCTG-3′) were used along with the Mu-specific primer TIR6 (5′-AGAGAAGCCAACGCCAWCGCCTCYATTTCGTC-3′). The resulting products were separated on 1% agarose gels, blotted onto nylon membranes, and probed with a CslD1-specific PCR product. Positive probe-binding samples were identified at X45:Y4 of the UniformMu Reverse Genetics Grid 6 (of eight total). Seeds from the UniformMu family corresponding to these coordinates (04S-1130-27) were grown and PCR genotyped to identify individuals homozygous for an insertion in CslD1. The csld1-1 allele was identified from this family, and a csld1-1 line was established after three generations of successive back-crosses to the W22 inbred.
A second mutant allele, csld1-2, was identified during a visual screen of field-grown UniformMu lines. Its phenotype was indistinguishable from that of csld1-1. PCR primers (5′-ACCAGATCCTCTTCCTCCTCGGTTTGC-3′, 5′-ACCTTGTTCCTGAGGAAGTCCCTCTTC-3′, 5′-GTGGTGATCACGCTGGCATCATTCAG-3′, and 5′-AGGAGGGCTGATGTAGACCCACAG-3′) were designed to cover nearly the entire length of the CslD1 gene and identified a Mu insertion in the third exon of CslD1. Homozygous recessive mutants of this allele were obtained from segregating progeny of this family, and the csld1-2 line was established after two successive back-crosses into the W22 inbred.
Additional Mu-insert alleles of csld1 were identified from the TUSC population of Pioneer Hi-Bred International as per McCarty and Meeley (2009). Primers used were 5′-ACCAGATCCTCTTCCTCCTCGGTTTGC-3′, 5′-ACCTTGTTCCTGAGGAAGTCCCTCTTC-3′, 5′-GTGGTGATCACGCTGGCATCATTCAG-3′, and 5′-AGGAGGGCTGATGTAGACCCACAG-3′, which identified five additional mutant alleles designated csld1-3 through csld1-7.
Overall Phenotypic Analyses and Size Measurements
Plant height was measured from the soil level to the auricles at the base of the uppermost leaf blades for 55 field-grown, mature plants (25 mutant, 30 nonmutant). These same 55 plants were used to measure width (at the widest point) and length of leaves at positions 3, 4, and 5 (relative to the apex). For dry weight measurements, whole-plant samples (including released root mass) were collected 3 d after ear harvest and did not include mature ears. Samples were weighed after drying for 4 weeks at 38°C. Belowground and aboveground dry weights were determined by separating root masses from the aerial portions of these plants.
Tissue Fixation and Sectioning
One-centimeter squares were excised from leaves of greenhouse-grown plants and fixed in 10% formaldehyde (Fisher lot no. 992720), 5% acetic acid, and 50% ethanol. Samples were vacuum infiltrated overnight at 4°C and then shaken at 4°C during a dehydration series using ethanol in phosphate-buffered saline (60 min each; progressing from 1× phosphate-buffered saline with 30% ethanol to 40%, 50%, 60%, 70%, 85%, and finally 95% ethanol). Samples were stained overnight with eosin in 95% ethanol, followed by four 1-h incubations in 100% ethanol and eosin at 25°C. Wax embedding was initiated by introducing CitriSolv (Fisher catalog no. 22-143975) into samples using a series of 1-h incubations (while shaking) in ethanol with increasing CitriSolv:ethanol content (25:75, 50:50, 75:25, and 100:0). Paraplast wax chips (Fisher catalog no. 23-021-399; 1 g wax mL−1 CitriSolv) were added to the 100% CitriSolv and incubated overnight at 25°C. Additional wax was added, followed by a 2-h incubation at 42°C. Samples were transferred to 60°C, where wax was poured off and replaced eight times over 4 d before samples were allowed to harden in molds. Sections (10 μm; cut with a Leitz 1512 microtome) were dewaxed with three 5-min incubations in xylene (Fisher lot no. 083423) and then washed twice in 100% ethanol (5 min each) and once in 95% ethanol (3 min). Slides were dried and examined with an Olympus BH2 light microscope.
Cell Volume Estimates
The extent of maximal expansion was estimated for ballooning epidermal cells of the csld1 mutant by comparing their volume with standard epidermal pavement cells of nonmutant plants. Cells were considered to be roughly cylindrical (V = πr2 × m), where V is volume and m is length, with nonmutant pavement cell dimensions approximately 40 μm (diameter) × 200 μm (length; Figs. 4 and 5). Ballooned epidermal cells of blades from csld1 mutant plants were often as large as 200 μm (diameter) × 600 μm (length; Figs. 4 and 5).
Epidermal Impressions and Nonwarty Cell Size Determination
Fresh samples from mature leaves of greenhouse-grown plants were cut into 1-cm2 pieces and firmly pressed into Superglue on glass slides. Glue was allowed to dry completely before leaf tissue was removed, leaving detailed epidermal impressions. These were imaged with a light microscope (Olympus BH2) with an RT SPOT camera (Diagnostic Instruments). Average cell length and width were determined by quantifying the total number of cells in a given area (1.88 × 1.40 mm). Longitudinal and lateral transects were used that did not include warty clusters.
Real-Time Quantitative RT-PCR
For each sample, RNA was extracted from approximately 200 mg of tissue, initially frozen in liquid nitrogen, and then homogenized in 1.0 mL of Trizol (Invitrogen catalog no. 15596-018) using a Q-BIOgene FastPrep 120 with Lysing Matrix D (MP Biomedicals catalog no. 116913). Samples were incubated for 5 min at 25°C with frequent vortexing. Chloroform (200 μL) was added, and samples were vortexed 15 s before, and after, a 1-min incubation at 25°C. Phases were separated by centrifuging for 10 min at 15,000g, and 200 μL of the aqueous layers was transferred to 700 μL of Qiagen RLT buffer (from the RNeasy Plant Mini kit; Qiagen catalog no. 74904). Ethanol was added (500 μL, 100% ethanol), and samples were vortexed. Half of this volume was used to clean and elute total RNA as per the RNeasy Plant Mini kit (Qiagen catalog no. 74904). The resulting RNA was treated with DNase-1 (Ambion catalog no. AM1906) and quantified using a Bio-Rad SmartSpec 3000. The cDNA was synthesized using the SuperScript One-Step kit and protocol (Invitrogen catalog no. 10928-042).
Levels of CslD1 mRNA were quantified in diverse maize tissues and in leaf blades at a range of developmental stages via real-time RT-PCR using a Step One Plus Real-Time PCR System (ABI). At least three biological replicates were analyzed for each tissue or time point, and for each of these replicates, reactions were performed in duplicate. A given reaction included 10 μL of Fast SYBR Green Master Mix (ABI lot no. 1003024), 5.0 μL of cDNA sample (diluted 10× from cDNA reaction), and 100 nm of each gene-specific primer (forward, 5′-GCCGCTCACGTCAATGG-3′; reverse, 5′-CTGGGCATCTTCATGGAGTGT-3′) in a final volume of 20 μL. The relative abundance of transcripts was normalized with 18S rRNA controls (Taqman rRNA Control Reagents; ABI lot no. 0804133) as per Eveland et al. (2008). Primer pairs for CslD1 were designed using Primer Express 3.0 (ABI).
High-Resolution X-Ray CT Analysis
Field-grown plants were sampled 3 d after harvesting ears and dried at 38°C for 3 weeks. Sections (approximately 0.5 cm) from the middle of the second internode of the conditioned stems (approximately 9% moisture content) were cut using a small band saw and scanned using a Scanco Medical Ag uCT35 instrument. Initial measurements were taken on whole-stem sections at 10-μm resolution. Regions including pith and rind (3 × 4 mm) were hand cut from the edge of these sections and scanned at 3.5-μm resolution over a 0.88-mm-high region to quantify cell wall and air space sizes. The 232 slices from each scan were reconstructed into three-dimensional images and contoured over whole stems for volumetric analyses. All scans were conducted with integration times of 600 μs and averaging two times. Both a fixed, common threshold and an adaptive threshold were used to segment cell wall from air space, and volumetric analyses were calculated with an algorithm developed for trabecular bone (Hildebrand and Ruegsegger, 1997). For rind-only analyses, hand-drawn contours were used to isolate the vascular bundle-rich region along the edge of the stem prior to three-dimensional reconstruction.
Cell Wall Composition Analysis
Samples from leaves and epidermal peels were ground in liquid nitrogen along with 200 μL of extraction buffer (50 mm Tris-Cl with 1% SDS at pH 7.2). Homogenate was transferred to 14-mL round-bottom polypropylene tubes (Falcon product no. 352059) along with 9 mL of extraction buffer, incubated for 15 min at 80°C, and centrifuged at 3,500 rpm for 5 min (approximately 2,000g) in a swinging-bucket rotor centrifuge (ThermoForma 1LGP). Supernatant was removed with an aspirator, and pellets (water-insoluble cell wall fraction) were washed, resuspended, and repelleted three times in about 10 mL of 80°C water. The same process was repeated three times with 50% ethanol at 80°C, followed by three washes with 80°C water. Samples were transferred to 1.5-mL Eppendorf tubes, alcohol-insoluble cell wall fractions were pelleted and dried, and noncellulosic sugar composition was analyzed by the Complex Carbohydrate Research Center at the University of Georgia.
For cellulose content, the alcohol-insoluble cell wall fractions from whole-leaf samples were isolated in the same way, dried for 30 h at 60°C, transferred to 14-mL round-bottom polypropylene tubes, and weighed. For each sample, approximately 50 mg of cell wall isolate was used, to which 3 mL of 80% aqueous acetic acid and 300 μL of 70% nitric acid were added. Tubes were incubated in an oil bath at 110°C and 120°C for 20 min each to hydrolyze hemicellulose and lignin (Sun et al., 2004). Samples were cooled, 1.8 mL of distilled water was added, tubes were centrifuged for 5 min (approximately 2,000g), and supernatant was removed with an aspirator. Cellulose was rinsed thoroughly with water (three times) and 95% ethanol (three times) and dried for 30 h at 60°C. Samples were weighed and compared for cellulose content as a fraction of alcohol-insoluble cell wall isolate.
Propidium Iodide Staining
Immature leaves (10–15 cm) were dissected from whorls of csld1 mutant and nonmutant plants. The basal portions (2 cm) of these leaves were immediately submerged in a solution of 0.1 mg mL−1 propidium iodide and allowed to absorb the dye for 5 min at 25°C. Samples were then rinsed thoroughly in water to remove excess stain and flattened on a glass slide. The abaxial epidermis was imaged using a Zeiss confocal microscope. For the visualization of nuclei, the same process was followed, but leaf samples were first fixed in 10% formaldehyde (Fisher lot no. 992720), 5% acetic acid, and 50% ethanol before staining with propidium iodide.
Flow Cytometry
The basal 1 cm of immature leaves (2–3 cm) was dissected and finely sliced (approximately 0.5 mm) with a razor blade in ice-cold chopping buffer (4% MOPS [0.5 m; pH 7.2], 9% MgCl2 [0.5 m], 6% Na3 citrate [0.5 m], 0.1% Triton X-100 [Sigma lot no. MKBD6639V], and 1 mg of RNase [Thermo Scientific; catalog no. AB-0549] in water). Homogenate was filtered through 50-μm nylon mesh followed by 20-μm nylon mesh and then transferred to a 1.5-mL microcentrifuge tube. Nuclei were pelleted at 1,000 rpm for 3 min, and the supernatant was discarded. Pellets were resuspended in staining buffer (chopping buffer plus 1% propidium iodide [5 mg mL−1]) and incubated at room temperature for 5 min. Nuclei were repelleted at 1,000 rpm for 3 min, and the supernatant was discarded. Pellets were resuspended in 300 μL of staining buffer and analyzed on an LSR-II cytometer (BD Biosciences). Nuclei were excited using a solid-state laser emitting 100 mW at 488 nm. Forward light scatter and orange fluorescence (575 ± 13 nm) were collected on up to 5,000 particles per sample. Small particles of debris were gated out using a fluorescence versus forward light scatter dot plot. Peaks were identified on a fluorescence histogram plotted on a logarithmic scale, and the geometric and median fluorescence values for each peak were calculated. The software used was Diva 6.1.2 (BD Biosciences).
Accession numbers for each of the gene sequences referred to in this work are as follows: AtCslD1, AT2G33100.1; AtCslD2, AT5G16910.1; AtCslD3, AT3G03050.1; AtCslD4, AT4G38190.1; AtCslD5, AT1G02730; AtCslD6, AT1G32180.1; OsCslD1, AC027037.6; OsCslD2, Os06g0111800; OsCslD3, AC091687.1; OsCslD4, AK242601.1; OsCslD5, Os06g0336500; ZmCslD1, GRMZM2G015886; ZmCslD2, GRMZM2G052149; ZmCslD3, GRMZM2G061764; ZmCslD4, GRMZM2G044269; and ZmCslD5, GRMZM2G436299.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. ClustalW alignments of CSLD proteins.
Supplemental Figure S2. Analysis of mutant and nonmutant stalks.
Supplemental Figure S3. Analysis of internal leaf structure.
Supplementary Material
References
- Arioli T, Peng L, Betzner AS, Burn J, Wittke W, Herth W, Camilleri C, Höfte H, Plazinski J, Birch R, et al. (1998) Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279: 717–720 [DOI] [PubMed] [Google Scholar]
- Bednarek SY, Falbel TG. (2002) Membrane trafficking during plant cytokinesis. Traffic 3: 621–629 [DOI] [PubMed] [Google Scholar]
- Beemster GT, De Veylder L, Vercruysse S, West G, Rombaut D, Van Hummelen P, Galichet A, Gruissem W, Inzé D, Vuylsteke M. (2005) Genome-wide analysis of gene expression profiles associated with cell cycle transitions in growing organs of Arabidopsis. Plant Physiol 138: 734–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beemster GT, Fiorani F, Inzé D. (2003) Cell cycle: the key to plant growth control? Trends Plant Sci 8: 154–158 [DOI] [PubMed] [Google Scholar]
- Bernal AJ, Jensen JK, Harholt J, Sørensen S, Moller I, Blaukopf C, Johansen B, de Lotto R, Pauly M, Scheller HV, et al. (2007) Disruption of ATCSLD5 results in reduced growth, reduced xylan and homogalacturonan synthase activity and altered xylan occurrence in Arabidopsis. Plant J 52: 791–802 [DOI] [PubMed] [Google Scholar]
- Bernal AJ, Yoo C-M, Mutwil M, Jensen JK, Hou G, Blaukopf C, Sørensen I, Blancaflor EB, Scheller HV, Willats WGT. (2008) Functional analysis of the cellulose synthase-like genes CSLD1, CSLD2, and CSLD4 in tip-growing Arabidopsis cells. Plant Physiol 148: 1238–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson-Dernier A, Kessler SA, Grossniklaus U. (2011) The walls have ears: the role of plant CrRLK1Ls in sensing and transducing extracellular signals. J Exp Bot 62: 1581–1591 [DOI] [PubMed] [Google Scholar]
- Burton RA, Gibeaut DM, Bacic A, Findlay K, Roberts K, Hamilton A, Baulcombe DC, Fincher GB. (2000) Virus-induced silencing of a plant cellulose synthase gene. Plant Cell 12: 691–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton RA, Wilson SM, Hrmova M, Harvey AJ, Shirley NJ, Medhurst A, Stone BA, Newbigin EJ, Bacic A, Fincher GB. (2006) Cellulose synthase-like CslF genes mediate the synthesis of cell wall (1,3;1,4)-beta-D-glucans. Science 311: 1940–1942 [DOI] [PubMed] [Google Scholar]
- Carpita NC. (1996) Structure and biogenesis of the cell walls of grasses. Annu Rev Plant Physiol Plant Mol Biol 47: 445–476 [DOI] [PubMed] [Google Scholar]
- Carpita NC, Defernez M, Findlay K, Wells B, Shoue DA, Catchpole G, Wilson RH, McCann MC. (2001) Cell wall architecture of the elongating maize coleoptile. Plant Physiol 127: 551–565 [PMC free article] [PubMed] [Google Scholar]
- Carpita NC, Gibeaut DM. (1993) Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J 3: 1–30 [DOI] [PubMed] [Google Scholar]
- Cnops G, Jover-Gil S, Peters JL, Neyt P, De Block S, Robles P, Ponce MR, Gerats T, Micol JL, Van Lijsebettens M. (2004) The rotunda2 mutants identify a role for the LEUNIG gene in vegetative leaf morphogenesis. J Exp Bot 55: 1529–1539 [DOI] [PubMed] [Google Scholar]
- Cocuron JC, Lerouxel O, Drakakaki G, Alonso AP, Liepman AH, Keegstra K, Raikhel N, Wilkerson CG. (2007) A gene from the cellulose synthase-like C family encodes a beta-1,4 glucan synthase. Proc Natl Acad Sci USA 104: 8550–8555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole RA, Fowler JE. (2006) Polarized growth: maintaining focus on the tip. Curr Opin Plant Biol 9: 579–588 [DOI] [PubMed] [Google Scholar]
- Crowell EF, Bischoff V, Desprez T, Rolland A, Stierhof Y-D, Schumacher K, Gonneau M, Höfte H, Vernhettes S. (2009) Pausing of Golgi bodies on microtubules regulates secretion of cellulose synthase complexes in Arabidopsis. Plant Cell 21: 1141–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhondt S, Vanhaeren H, Van Loo D, Cnudde V, Inzé D. (2010) Plant structure visualization by high-resolution x-ray computed tomography. Trends Plant Sci 15: 419–422 [DOI] [PubMed] [Google Scholar]
- Dhugga KS, Barreiro R, Whitten B, Stecca K, Hazebroek J, Randhawa GS, Dolan M, Kinney AJ, Tomes D, Nichols S, et al. (2004) Guar seed beta-mannan synthase is a member of the cellulose synthase super gene family. Science 303: 363–366 [DOI] [PubMed] [Google Scholar]
- Doblin MS, De Melis L, Newbigin E, Bacic A, Read SM. (2001) Pollen tubes of Nicotiana alata express two genes from different β-glucan synthase families. Plant Physiol 125: 2040–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doblin MS, Pettolino FA, Wilson SM, Campbell R, Burton RA, Fincher GB, Newbigin E, Bacic A. (2009) A barley cellulose synthase-like CSLH gene mediates (1,3;1,4)-beta-D-glucan synthesis in transgenic Arabidopsis. Proc Natl Acad Sci USA 106: 5996–6001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eveland AL, McCarty DR, Koch KE. (2008) Transcript profiling by 3′-untranslated region sequencing resolves expression of gene families. Plant Physiol 146: 32–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrokhi N, Burton RA, Brownfield L, Hrmova M, Wilson SM, Bacic A, Fincher GB. (2006) Plant cell wall biosynthesis: genetic, biochemical and functional genomics approaches to the identification of key genes. Plant Biotechnol J 4: 145–167 [DOI] [PubMed] [Google Scholar]
- Favery B, Ryan E, Foreman J, Linstead P, Boudonck K, Steer M, Shaw P, Dolan L. (2001) KOJAK encodes a cellulose synthase-like protein required for root hair cell morphogenesis in Arabidopsis. Genes Dev 15: 79–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferjani A, Horiguchi G, Yano S, Tsukaya H. (2007) Analysis of leaf development in fugu mutants of Arabidopsis reveals three compensation modes that modulate cell expansion in determinate organs. Plant Physiol 144: 988–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fincher GB. (2009) Revolutionary times in our understanding of cell wall biosynthesis and remodeling in the grasses. Plant Physiol 149: 27–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M. (1992) A conceptual framework for maize leaf development. Dev Biol 153: 44–58 [DOI] [PubMed] [Google Scholar]
- Gamsjaeger R, Liew CK, Loughlin FE, Crossley M, Mackay JP. (2007) Sticky fingers: zinc-fingers as protein-recognition motifs. Trends Biochem Sci 32: 63–70 [DOI] [PubMed] [Google Scholar]
- Green PB. (1980) Organogenesis: a biophysical view. Annu Rev Plant Physiol 31: 51–82 [Google Scholar]
- Gutierrez R, Lindeboom JJ, Paredez AR, Emons AMC, Ehrhardt DW. (2009) Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat Cell Biol 11: 797–806 [DOI] [PubMed] [Google Scholar]
- Harris PJ, Hartley RD. (1980) Phenolic constituents of the cell walls of monocotyledons. Biochem Syst Ecol 8: 153–160 [Google Scholar]
- Hazen SP, Scott-Craig JS, Walton JD. (2002) Cellulose synthase-like genes of rice. Plant Physiol 128: 336–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hématy K, Höfte H. (2008) Novel receptor kinases involved in growth regulation. Curr Opin Plant Biol 11: 321–328 [DOI] [PubMed] [Google Scholar]
- Hepler PK, Vidali L, Cheung AY. (2001) Polarized cell growth in higher plants. Annu Rev Cell Dev Biol 17: 159–187 [DOI] [PubMed] [Google Scholar]
- Hildebrand T, Ruegsegger P. (1997) A new method for the model-independent assessment of thickness in three-dimensional images. J Microsc 185: 67–75 [Google Scholar]
- Horiguchi G, Mollá-Morales A, Pérez-Pérez JM, Kojima K, Robles P, Ponce MR, Micol JL, Tsukaya H. (2011) Differential contributions of ribosomal protein genes to Arabidopsis thaliana leaf development. Plant J 65: 724–736 [DOI] [PubMed] [Google Scholar]
- Horiguchi G, Tsukaya H. (2011) Organ size regulation in plants: insights from compensation. Front Plant Biol 2: 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Zhu L, Zeng D, Gao Z, Guo L, Fang Y, Zhang G, Dong G, Yan M, Liu J, et al. (2010) Identification and characterization of NARROW AND ROLLED LEAF 1, a novel gene regulating leaf morphology and plant architecture in rice. Plant Mol Biol 73: 283–292 [DOI] [PubMed] [Google Scholar]
- Hu Y, Poh HM, Chua N-H. (2006) The Arabidopsis ARGOS-LIKE gene regulates cell expansion during organ growth. Plant J 47: 1–9 [DOI] [PubMed] [Google Scholar]
- Keegstra K, Walton J. (2006) Plant science. β-Glucans: brewer’s bane, dietician’s delight. Science 311: 1872–1873 [DOI] [PubMed] [Google Scholar]
- Kim CM, Park SH, Je BI, Park SH, Park SJ, Piao HL, Eun MY, Dolan L, Han C-D. (2007) OsCSLD1, a cellulose synthase-like D1 gene, is required for root hair morphogenesis in rice. Plant Physiol 143: 1220–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Laosinchai W, Itoh T, Cui X, Linder CR, Brown RM., Jr (1999) Immunogold labeling of rosette terminal cellulose-synthesizing complexes in the vascular plant Vigna angularis. Plant Cell 11: 2075–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurek I, Kawagoe Y, Jacob-Wilk D, Doblin M, Delmer D. (2002) Dimerization of cotton fiber cellulose synthase catalytic subunits occurs via oxidation of the zinc-binding domains. Proc Natl Acad Sci USA 99: 11109–11114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Xiong G, Li R, Cui J, Tang D, Zhang B, Pauly M, Cheng Z, Zhou Y. (2009) Rice cellulose synthase-like D4 is essential for normal cell-wall biosynthesis and plant growth. Plant J 60: 1055–1069 [DOI] [PubMed] [Google Scholar]
- Li P, Ponnala L, Gandotra N, Wang L, Si Y, Tausta SL, Kebrom TH, Provart N, Patel R, Myers CR, et al. (2010) The developmental dynamics of the maize leaf transcriptome. Nat Genet 42: 1060–1067 [DOI] [PubMed] [Google Scholar]
- Liepman AH, Wilkerson CG, Keegstra K. (2005) Expression of cellulose synthase-like (Csl) genes in insect cells reveals that CslA family members encode mannan synthases. Proc Natl Acad Sci USA 102: 2221–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz W, Mayer U, Jürgens G. (1996) Cytokinesis in the Arabidopsis embryo involves the syntaxin-related KNOLLE gene product. Cell 84: 61–71 [DOI] [PubMed] [Google Scholar]
- Manfield IW, Orfila C, McCartney L, Harholt J, Bernal AJ, Scheller HV, Gilmartin PM, Mikkelsen JD, Paul Knox J, Willats WGT. (2004) Novel cell wall architecture of isoxaben-habituated Arabidopsis suspension-cultured cells: global transcript profiling and cellular analysis. Plant J 40: 260–275 [DOI] [PubMed] [Google Scholar]
- McCarty DR, Meeley RB. (2009) Transposon resources for forward and reverse genetics in maize. In JL Bennetzen, S Hake, eds, Handbook of Maize. Springer, New York, pp 561–584 [Google Scholar]
- McCarty DR, Settles AM, Suzuki M, Tan BC, Latshaw S, Porch T, Robin K, Baier J, Avigne W, Lai J, et al. (2005) Steady-state transposon mutagenesis in inbred maize. Plant J 44: 52–61 [DOI] [PubMed] [Google Scholar]
- Meeley RB, Briggs SP. (1995) Reverse genetics for maize. Maize Genet Coop Newsl 69: 67–82 [Google Scholar]
- Mellerowicz EJ, Baucher M, Sundberg B, Boerjan W. (2001) Unravelling cell wall formation in the woody dicot stem. Plant Mol Biol 47: 239–274 [PubMed] [Google Scholar]
- Mitkovski M, Sylvester AW. (2003) Analysis of cell patterns in developing maize leaves: dark-induced cell expansion restores normal division orientation in the mutant tangled. Int J Plant Sci 164: 113–124 [Google Scholar]
- Moulia B. (2000) Leaves as shell structures: double curvature, auto-stresses, and minimal mechanical energy constraints on leaf rolling in grasses. J Plant Growth Regul 19: 19–30 [DOI] [PubMed] [Google Scholar]
- Natera SHA, Ford KL, Cassin AM, Patterson JH, Newbigin EJ, Bacic A. (2008) Analysis of the Oryza sativa plasma membrane proteome using combined protein and peptide fractionation approaches in conjunction with mass spectrometry. J Proteome Res 7: 1159–1187 [DOI] [PubMed] [Google Scholar]
- Orfila C, Sørensen SO, Harholt J, Geshi N, Crombie H, Truong HN, Reid JS, Knox JP, Scheller HV. (2005) QUASIMODO1 is expressed in vascular tissue of Arabidopsis thaliana inflorescence stems, and affects homogalacturonan and xylan biosynthesis. Planta 222: 613–622 [DOI] [PubMed] [Google Scholar]
- Park S, Szumlanski AL, Gu F, Guo F, Nielsen E. (2011) A role for CSLD3 during cell-wall synthesis in apical plasma membranes of tip-growing root-hair cells. Nat Cell Biol 13: 973–980 [DOI] [PubMed] [Google Scholar]
- Penning BW, Hunter CT, III, Tayengwa R, Eveland AL, Dugard CK, Olek AT, Vermerris W, Koch KE, McCarty DR, Davis MF, et al. (2009) Genetic resources for maize cell wall biology. Plant Physiol 151: 1703–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S, Caffall KH, Freshour G, Hilley MT, Bauer S, Poindexter P, Hahn MG, Mohnen D, Somerville C. (2007a) The Arabidopsis irregular xylem8 mutant is deficient in glucuronoxylan and homogalacturonan, which are essential for secondary cell wall integrity. Plant Cell 19: 237–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S, Paredez A, Carroll A, Palsdottir H, Doblin M, Poindexter P, Khitrov N, Auer M, Somerville CR. (2007b) Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proc Natl Acad Sci USA 104: 15566–15571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds JO, Eisses JF, Sylvester AW. (1998) Balancing division and expansion during maize leaf morphogenesis: analysis of the mutant, warty-1. Development 125: 259–268 [DOI] [PubMed] [Google Scholar]
- Richmond T. (2000) Higher plant cellulose synthases. Genome Biol 1: reviews3001.1–reviews3001.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond TA, Somerville CR. (2000) The cellulose synthase superfamily. Plant Physiol 124: 495–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond TA, Somerville CR. (2001) Integrative approaches to determining Csl function. Plant Mol Biol 47: 131–143 [PubMed] [Google Scholar]
- Ringli C. (2010) Monitoring the outside: cell wall-sensing mechanisms. Plant Physiol 153: 1445–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AW, Bushoven JT. (2007) The cellulose synthase (CESA) gene superfamily of the moss Physcomitrella patens. Plant Mol Biol 63: 207–219 [DOI] [PubMed] [Google Scholar]
- Roberts AW, Roberts E. (2007) Evolution of the cellulose synthase (CesA) gene family: insights from green algae and seedless plants. In RM Brown, IM Saxena, eds, Cellulose: Molecular and Structural Biology. Springer, Dordrecht, The Netherlands, pp 17–34 [Google Scholar]
- Samuga A, Joshi CP. (2004) Cloning and characterization of cellulose synthase-like gene, PtCSLD2 from developing xylem of aspen trees. Plant Physiol 120: 631–641 [DOI] [PubMed] [Google Scholar]
- Sandhu AP, Randhawa GS, Dhugga KS. (2009) Plant cell wall matrix polysaccharide biosynthesis. Mol Plant 2: 840–850 [DOI] [PubMed] [Google Scholar]
- Seifert GJ, Blaukopf C. (2010) Irritable walls: the plant extracellular matrix and signaling. Plant Physiol 153: 467–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settles AM, Holding DR, Tan BC, Latshaw SP, Liu J, Suzuki M, Li L, O’Brien BA, Fajardo DS, Wroclawska E, et al. (2007) Sequence-indexed mutations in maize using the UniformMu transposon-tagging population. BMC Genomics 8: 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LG, Hake S, Sylvester AW. (1996) The tangled-1 mutation alters cell division orientations throughout maize leaf development without altering leaf shape. Development 122: 481–489 [DOI] [PubMed] [Google Scholar]
- Spitzer C, Schellmann S, Sabovljevic A, Shahriari M, Keshavaiah C, Bechtold N, Herzog M, Müller S, Hanisch F-G, Hülskamp M. (2006) The Arabidopsis elch mutant reveals functions of an ESCRT component in cytokinesis. Development 133: 4679–4689 [DOI] [PubMed] [Google Scholar]
- Steppe K, Cnudde V, Girard C, Lemeur R, Cnudde J-P, Jacobs P. (2004) Use of x-ray computed microtomography for non-invasive determination of wood anatomical characteristics. J Struct Biol 148: 11–21 [DOI] [PubMed] [Google Scholar]
- Sun JX, Sun XF, Zhao H, Sun RC. (2004) Isolation and characterization of cellulose from sugarcane bagasse. Polym Degrad Stabil 84: 331–339 [Google Scholar]
- Sylvester AW. (2000) Division decisions and the spatial regulation of cytokinesis. Curr Opin Plant Biol 3: 58–68 [DOI] [PubMed] [Google Scholar]
- Sylvester AW, Cande WZ, Freeling M. (1990) Division and differentiation during normal and liguleless-1 maize leaf development. Development 110: 985–1000 [DOI] [PubMed] [Google Scholar]
- Szécsi J, Joly C, Bordji K, Varaud E, Cock JM, Dumas C, Bendahmane M. (2006) BIGPETALp, a bHLH transcription factor is involved in the control of Arabidopsis petal size. EMBO J 25: 3912–3920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Taylor NG, Howells RM, Huttly AK, Vickers K, Turner SR. (2003) Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc Natl Acad Sci USA 100: 1450–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukaya H. (2008) Controlling size in multicellular organs: focus on the leaf. PLoS Biol 6: e174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Erp H, Walton JD. (2009) Regulation of the cellulose synthase-like gene family by light in the maize mesocotyl. Planta 229: 885–897 [DOI] [PubMed] [Google Scholar]
- Vogel J. (2008) Unique aspects of the grass cell wall. Curr Opin Plant Biol 11: 301–307 [DOI] [PubMed] [Google Scholar]
- Wang X, Cnops G, Vanderhaeghen R, De Block S, Van Montagu M, Van Lijsebettens M. (2001) AtCSLD3, a cellulose synthase-like gene important for root hair growth in Arabidopsis. Plant Physiol 126: 575–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Henriksson E, Söderman E, Henriksson KN, Sundberg E, Engström P. (2003) The Arabidopsis homeobox gene, ATHB16, regulates leaf development and the sensitivity to photoperiod in Arabidopsis. Dev Biol 264: 228–239 [DOI] [PubMed] [Google Scholar]
- Wu C, Fu Y, Hu G, Si H, Cheng S, Liu W. (2010) Isolation and characterization of a rice mutant with narrow and rolled leaves. Planta 232: 313–324 [DOI] [PubMed] [Google Scholar]
- Yin L, Verhertbruggen Y, Oikawa A, Manisseri C, Knierim B, Prak L, Jensen JK, Knox JP, Auer M, Willats WGT, et al. (2011) The cooperative activities of CSLD2, CSLD3, and CSLD5 are required for normal Arabidopsis development. Mol Plant 4: 1024–1037 [DOI] [PubMed] [Google Scholar]
- Yin Y, Huang J, Xu Y. (2009) The cellulose synthase superfamily in fully sequenced plants and algae. BMC Plant Biol 9: 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, Keegstra K. (2008) AtCSLD2 is an integral Golgi membrane protein with its N-terminus facing the cytosol. Planta 228: 823–838 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.