Abstract
Superoxide dismutases (SODs) are important antioxidant enzymes that catalyze the disproportionation of superoxide anion to oxygen and hydrogen peroxide to guard cells against superoxide toxicity. The major pathway for activation of copper/zinc SOD (CSD) involves a copper chaperone for SOD (CCS) and an additional minor CCS-independent pathway reported in mammals. We characterized the CCS-dependent and -independent activation pathways for three CSDs localized in different cellular compartments in Arabidopsis (Arabidopsis thaliana). The main activation pathway for CSD1 in the cytoplasm involved a CCS-dependent and -independent pathway, which was similar to that for human CSD. Activation of CSD2 in chloroplasts depended totally on CCS, similar to yeast (Saccharomyces cerevisiae) CSD. Peroxisome-localized CSD3 via a CCS-independent pathway was similar to nematode (Caenorhabditis elegans) CSD in retaining activity in the absence of CCS. In Arabidopsis, glutathione played a role in CCS-independent activation, as was reported in humans, but an additional factor was required. These findings reveal a highly specific and sophisticated regulation of CSD activation pathways in planta relative to other known CCS-independent activation.
Superoxide dismutases (SODs) are a group of metalloenzymes that defend against free radical species by disproportionating O2− into hydrogen peroxide and oxygen molecules (Beyer et al., 1991; Bowler et al., 1992). By a specific metal cofactor required for this superoxide-scavenging activity (McCord and Fridovich, 1969), they are classified as copper/zinc SOD (CuZnSOD), iron SOD (FeSOD), manganese SOD (MnSOD), or nickel SOD (Alscher et al., 2002; Zelko et al., 2002). Most eukaryotic cells contain two types of SODs, with CuZnSOD largely localized in the cytoplasm (Crapo et al., 1992) and MnSOD localized in mitochondria (Weisiger and Fridovich, 1973; Marres et al., 1985). Plants contain MnSOD, FeSOD, and many isoforms of CuZnSOD in different cellular compartments (Jackson et al., 1978; Kanematsu and Asada, 1989; Bowler et al., 1992; Bueno et al., 1995). Arabidopsis (Arabidopsis thaliana) has three CuZnSOD (CSD) isoforms: CSD1 in the cytoplasm, CSD2 in chloroplasts, and CSD3 in peroxisomes (Kliebenstein et al., 1998; Alscher et al., 2002).
The metal cofactors for SODs are transition metals, with the ability to readily accept or donate an electron for the superoxide dismutation process. However, these metal ions in free form via the Haber-Weiss reaction could produce harmful hydroxyl radicals (Halliwell and Gutteridge, 2007). Hence, free transition metals generally exist in a complexed form, with activated SOD required as a metallochaperone. Cu chaperones involved in Cu trafficking in yeast (Saccharomyces cerevisiae) are ATX1 (Lin and Culotta, 1995; Lin et al., 1997), COX17 (Glerum et al., 1996), and Lys-7 (Horecka et al., 1995). Lys-7 can fully restore the lys7Δ (i.e. ccsΔ) mutant function outside of Lys biosynthesis as a Cu chaperone for superoxide dismutase (CCS), which is a functional homolog of the human CCS (Horecka et al., 1995; Culotta et al., 1997). The CCS interacts with CSD, assisting in Cu incorporation and catalysis of disulfide bond formation, thereby resulting in an active CSD (Casareno et al., 1998; Lamb et al., 2001; Brown et al., 2004; Furukawa et al., 2004). In Arabidopsis, a single AtCCS gene product activated CSDs in the cytoplasm and chloroplast (Chu et al., 2005); however, the phenotype of the AtCCS-knockout mutant (Atccs) was found to be normal (Chu et al., 2005; Cohu et al., 2009). Similarly, a CCS-independent activation pathway for CSD was found in mice (Wong et al., 2000). These data suggest the existence of additional CSD-activating factors.
To date, yeast SOD1 (ySOD1) activation has been found to fully depend on yeast CCS (yCCS; Carroll et al., 2004), whereas the nematode (Caenorhabditis elegans) CSD (wSod-1) is exclusively activated independently of CCS (Jensen and Culotta, 2005). Human CSD (hSOD1) is largely activated by CCS but retains about 25% to 50% of its activity in the absence of CCS (Carroll et al., 2004). However, CCS-independent pathways for SOD activation in plants are not well understood.
It has long been suggested that reduced glutathione (GSH) may participate in intracellular Cu homeostasis by forming Cu-GSH complexes when transferring Cu to CSD (Ciriolo et al., 1990) and metallothionein (Ferreira et al., 1993) in vitro. GSH was required for CCS-independent activation of hSOD1 in yCCS mutant strains with defective GSH metabolism, and wSod-1 was inactive in the presence of CCS when GSH was depleted in yeast (Carroll et al., 2004; Jensen and Culotta, 2005). However, a direct interaction between GSH and CSD has yet to be demonstrated. Results from mutagenesis studies showed that amino acid residues 142 and 144 near the C terminus of human and yeast CSDs were important in the CCS-independent pathway (Carroll et al., 2004; Jensen and Culotta, 2005). When these residues were replaced by dual Pro residues in ySOD1, CCS-independent activities for both hSOD1 and wSod-1 were inhibited. A recent study further confirmed that the Pro at residue 144 but not 142 restricted ySOD1 disulfide formation in the absence of CCS, which played a key role in blocking CCS-independent activation (Leitch et al., 2009a). The existence of additional essential factors in this pathway remains to be determined.
In this study, we investigated the dependence of Arabidopsis CSD activity on the CCS metallochaperone in Arabidopsis and yeast knockout strains. The three CSD proteins in Arabidopsis showed unique levels of activity by CCS-dependent and -independent activation pathways. Moreover, the CSD2 lost the ability of CCS-independent activation, possibly because of an inhibitory factor(s) in chloroplasts, which has not been reported in any other species. We also confirmed in Arabidopsis, as in hSOD1, that GSH was involved in the CCS-independent pathway, with an additional factor cooperatively assisting in CSD activation.
RESULTS
CCS-Independent Activation of Arabidopsis CSD1 and CSD3 in Yeast
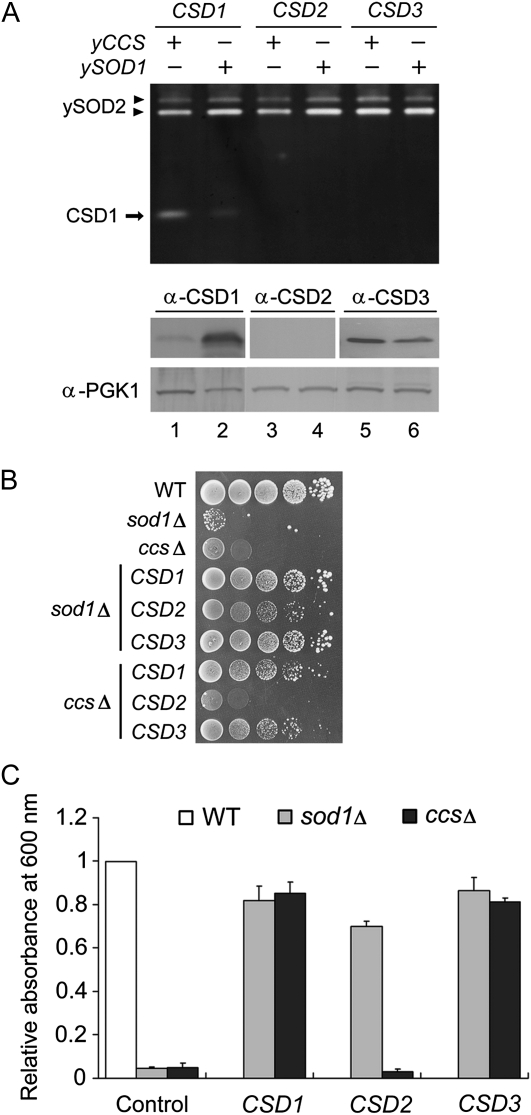
In our previous study (Chu et al., 2005), we observed residual CSD activity in Atccs (6%–30% of the wild-type levels), which indicated the existence of a CCS-independent pathway for CSD activation in Arabidopsis (Supplemental Fig. S1). Because AtCCS is a functional homolog of yCCS (Abdel-Ghany et al., 2005), the Arabidopsis CSD might be activated in the yeast system, so we expressed each of the three Arabidopsis CSD genes in yeast ySOD1- and yCCS-knockout strains (sod1Δ and ccsΔ, respectively; Fig. 1). Yeast sod1Δ and ccsΔ are auxotrophic for Lys when grown in air (Culotta et al., 1997).
Figure 1.
The activities of three Arabidopsis CSDs in yeast sod1Δ and ccsΔ. A, Lysates of yeast expressing Arabidopsis CSD1, CSD2, and CSD3 were analyzed by in-gel SOD activity assay (top) and immunoblotting (bottom). Strains expressed on the sod1Δ or ccsΔ background are represented as yCCS+/ySOD1− or yCCS−/ySOD1+, respectively. B and C, Viability of yeast sod1Δ and ccsΔ expressing CSD1, CSD2, or CSD3 under Lys-lacking conditions. Plate (B) and liquid (C) assays are described in “Materials and Methods.” Cell density values are relative to the wild-type (WT) level. All data were from four independent tests (means ± sd).
In-gel SOD activity assay revealed a strong CSD1 activity band in sod1Δ and a weaker but visible band in ccsΔ (Fig. 1A, top, lanes 1 and 2), which demonstrated CCS-independent activation of CSD1. CSD2 and CSD3 were overexpressed in yeast, but we detected no activity (Fig. 1A, top, lanes 3–6), and the CSD2 protein was not detectable (Fig. 1A, bottom, lanes 3 and 4).
Because of the limited sensitivity of the in-gel SOD activity assay, we used a more sensitive method, the Lys-independent aerobic growth assay, to detect the relatively weak CSD activity in yeast in vivo (Wallace et al., 2004). With this assay, Lys biosynthesis ability was maintained when ySOD1 activity was just higher than 2% of the wild-type level to rescue the Lys auxotrophy of sod1Δ (Corson et al., 1998). Because ySOD1 activation depends totally on yCCS function, endogenous ySOD1 activity is lost in ccsΔ (Carroll et al., 2004), and heterologously expressed Arabidopsis CSDs could be responsible for the Lys biosynthesis ability in both sod1Δ and ccsΔ. In our experiments, neither sod1Δ nor ccsΔ was able to grow under Lys-depleted conditions, as expected (Fig. 1, B and C), which confirms the absence of ySOD1 activity in these two mutants. Lys biosynthesis ability was recovered in both sod1Δ and ccsΔ with CSD1 and CSD3 expression. With the expression of CSD2, the phenotype of sod1Δ was partially rescued, but that of ccsΔ was not restored at all, which demonstrates a weak expression and activity of CSD2 in sod1Δ. Therefore, both CSD1 and CSD3 can be activated via the CCS-independent pathway in yeast. However, the failure of CSD2 to rescue Lys biosynthesis might be related to the heterologous nature of the expression system used.
CSD2 Activity Depends on CCS Only in Chloroplasts but Is CCS Independent in Cytoplasm
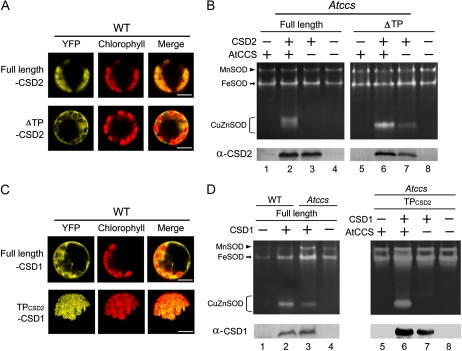
To address the question of how CSD2 is activated, CSD2 and AtCCS genes were transiently overexpressed in Atccs protoplasts (Fig. 2, A and B). Use of yellow fluorescent protein (YFP) fused to CSD2 revealed that the full-length CSD2 localized in chloroplasts, whereas the transit peptide-deleted CSD2 (ΔTP-CSD2) localized to cytoplasm (Fig. 2A). Full-length CSD2 was activated in the presence of AtCCS (Fig. 2B, lane 2) but was undetectable in the absence of AtCCS (lane 3). However, ΔTP-CSD2 showed CCS-independent activity (lane 7), which indicates that CSD2 can be activated via the CCS-independent pathway when localized in cytoplasm.
Figure 2.
Localization and SOD activity of CSD1 and CSD2 overexpressed in Atccs protoplasts. A and C, YFP fusion of full-length and chloroplast transit peptide-deleted CSD2 (ΔTP-CSD2; A) and YFP fusion of full-length CSD1 and that fused to the chloroplastic transit peptide of CSD2 (TPCSD2-CSD1; C) were expressed in Arabidopsis wild-type (WT) protoplasts for localization analysis. The chloroplasts are visualized as red autofluorescence. Bars = 10 μm. B and D, Full-length CSD1 and ΔTP-CSD2 (B) and full-length CSD1 and TPCSD2-CSD1 (D) were overexpressed with or without AtCCS coexpression in wild-type or Atccs protoplasts, then SOD activity (top) and protein level (bottom) were analyzed. Because CSD protein expression was reduced in Atccs, we loaded 3-fold more Atccs protoplasts (approximately 7.5 × 105 cells) than wild-type protoplasts (approximately 2.5 × 105 cells) to present similar protein levels in lanes 2 and 3. The CSD genes used here contained no YFP fusion.
CCS-Independent Activation of CSD1 Is Lost When Localized in Chloroplasts
To gain an understanding of CSD activation in chloroplasts, we constructed a chloroplast-directed CSD1 to observe its behavior in this compartment (Fig. 2, C and D). We used YFP fusion proteins to confirm the localization of a CSD1 fused with the chloroplast transit peptide of CSD2 (TPCSD2-CSD1) in chloroplasts (Fig. 2C). We found CCS-independent CSD1 activity in Atccs (Fig. 2D), and the efficiency of CSD1 activation (activity μg−1 protein) in Atccs was approximately 36% of that in the wild type (compare lanes 2 and 3). In contrast, chloroplast-localized CSD1 was active in the presence of AtCCS (lane 6), and its activity was undetectable in the absence of AtCCS (lane 7). This result is consistent with the observation that CCS-independent CSD activation occurs in the cytoplasm but not in the chloroplast.
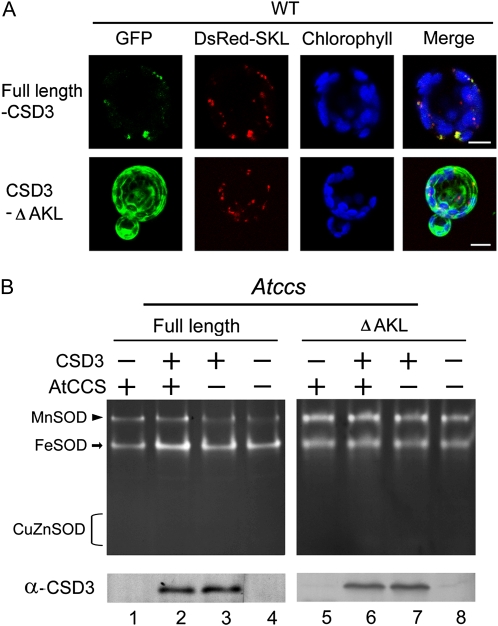
No Detection of Peroxisome-Localized CSD3 and Cytoplasm-Directed CSD3 Activities
A similar experiment was also performed with CSD3 (Fig. 3). GFP fused to full-length CSD3 showed it localized in peroxisomes and localized in cytoplasm with deletion of the C-terminal peroxisome-targeting sequence AKL (CSD3-ΔAKL; Fig. 3A). Unlike CSD1 or CSD2, CSD3 activities were undetectable even when it was overexpressed in Atccs with AtCCS coexpression (Fig. 3B) or in wild-type protoplasts (Supplemental Fig. S3).
Figure 3.
Localization and SOD activity of full-length and AKL-deleted CSD3 overexpressed in Arabidopsis protoplasts. A, GFP fusion proteins of full-length and C-terminal AKL-deleted CSD3 (CSD3-ΔAKL) were expressed in Arabidopsis wild-type (WT) protoplasts for localization analysis. DsRed-SKL is a peroxisomal marker. The autofluorescence of chlorophyll is blue. Bars = 10 μm. B, CSD3 full length and ΔAKL were overexpressed with or without AtCCS coexpression in Atccs or wild-type protoplasts, then CSD3 activity (top) and protein level (bottom) were analyzed. The CSD3 used here contained no GFP fusion.
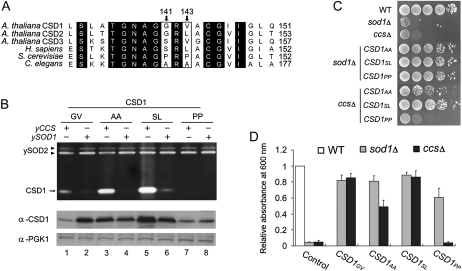
CSD1 Mutants Show Different Activity Levels in Yeast
A previous study by Carroll et al. (2004) demonstrated the importance of amino acid residues 142S and 144L of human hSOD1 for CCS-independent activation. As well, replacing these amino acids with Pro, as with dual Pro residues in yeast ySOD1, prevents activation by this pathway. We aligned CSD sequences of Arabidopsis, human, yeast, and nematode and found the corresponding residues in these three Arabidopsis CSDs to be 141G/143V in CSD1, 143G/145L in CSD2, and 147S/149V in CSD3, similar to residues 142S/144L in hSOD1 (Fig. 4A). Here, we tested only CSD1, because only CSD1 activity could be detected by in-gel SOD activity assay (Fig. 1A). We generated a series of CSD1 mutants, G141A/V143A (CSD1AA; nematode form), G141S/V143L (CSD1SL; human form), and G141P/V143P (CSD1PP; yeast form), to examine the effect on CCS-independent CSD1 activity in yeast sod1Δ and ccsΔ strains.
Figure 4.
Activity of Arabidopsis CSD1 variants in yeast sod1Δ and ccsΔ. A, Sequence alignment of CuZnSOD in Arabidopsis (CSD1, CSD2, and CSD3), human (hSOD1), S. cerevisiae (ySOD1), and C. elegans (wSod-1). Residues corresponding to 141G and 143V of CSD1 are indicated in white boxes with arrows. Identical residues are shown in black. B, Yeast sod1Δ and ccsΔ expressing the CSD1 variants were analyzed as described in Figure 1A for SOD activity (top) and CSD1 protein level (bottom). C and D, Viability of yeast sod1Δ and ccsΔ expressing the CSD1 variants under Lys-lacking conditions. Plate (C) and liquid (D) assays were as described in Figure 1, B and C. The data are from four independent tests (means ± sd). WT, Wild type.
We detected activities of wild-type CSD1 (CSD1GV) and CSD1SL in both sod1Δ and ccsΔ, but CSD1AA activity was detected only in sod1Δ (Fig. 4B, lanes 1–6). CSD1GV and CSD1SL could restore the Lys-auxotrophic phenotype of sod1Δ and ccsΔ under the Lys-depleted condition (Fig. 4, C and D). CSD1AA could also restore the viability of sod1Δ, but that of ccsΔ was only partially rescued. Although the in-gel CSD1PP activity was undetectable in both sod1Δ and ccsΔ (Fig. 4B, lanes 7 and 8), the activity of this mutant was discernible in sod1Δ but not ccsΔ by use of the Lys biosynthesis assay (Fig. 4, C and D). Therefore, the Arabidopsis CSD1GV, human form CSD1SL, and nematode form CSD1AA but not yeast form CSD1PP could be activated by the CCS-independent pathway. The CCS-independent activation in Arabidopsis and human CSDs shared the highest functional similarity.
Physiological Function of CCS-Independent CSD Activity
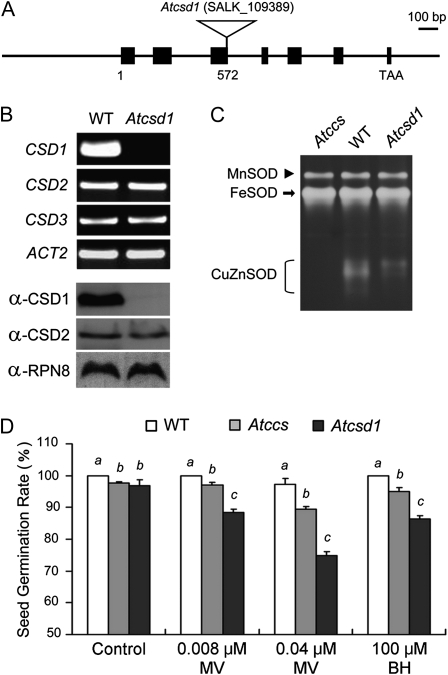
We used seed germination rate in Atccs and CSD1-knockout lines (Atcsd1) to determine whether these residual CSD activities were physiologically functional in vivo.
In the Atcsd1 mutant (Fig. 5A), CSD1 mRNA and protein were absent, but the expression levels of CSD2 and CSD3 were unchanged (Fig. 5B). The total CSD activity in Atcsd1 (with functional CSD2 and CSD3) was lower than that in the wild type but was higher than that in Atccs (Fig. 5C). When Atccs and Atcsd1 mutants were grown on half-strength Murashige and Skoog (1/2 MS) plates, their seed germination rates were only 4% lower then that of the wild type (Fig. 5D, control), which indicated that the CCS-independent activities of CSD in Atccs were sufficient for normal growth. We used treatment with reactive oxygen species (ROS)-generating reagents, methyl viologen (MV) and tert-butyl hydroperoxide (BH), to induce oxidative stress to magnify the phenotypic effects due to the decreased CSD activity. With 0.008 or 0.04 μm MV, the seed germination rate of Atccs was significantly lower than that of the wild type but higher than that of Atcsd1 (Fig. 5D, MV). Similar results were obtained with 100 μm BH treatment (Fig. 5D, BH). Hence, although the total CSD activity in Atccs (resulting from the CCS-independent CSD activity) was lower than that in Atcsd1 (resulting from CSD2 and CSD3 activities), the phenotype of inhibition of seed germination was less pronounced in Atccs than in Atcsd1.
Figure 5.
Characterization of CSD activity and seed germination rate of the wild type (WT), Atccs, and Atcsd1. A, The T-DNA insertion of the CSD1-knockout plant, Atcsd1, is in the third exon of CSD1 (nucleotide 572 of the genomic sequence). B and C, Analysis of mRNA expression of CSD1, CSD2, and CSD3 (B, top), protein levels of CSD1 and CSD2 (B, bottom), and SOD activities (C) in wild-type, Atcsd1, and Atccs 7-d-old seedlings. CSD3 protein was undetectable, so those data are not presented, and the CuZnSOD activity level is lower in Atccs young seedlings, which explains the undetectable residual activity. D, Comparison of the seed germination rates of wild-type, Atccs, and Atcsd1 plants grown on 1/2 MS plates with the indicated treatments. Data represent results from five independent experiments (means ± sd). Letters (a, b, and c) indicate difference between the wild type, Atccs, and Atcsd1 for each treatment at P < 0.05.
Glutathione Plays a Role in CCS-Independent CSD Activity
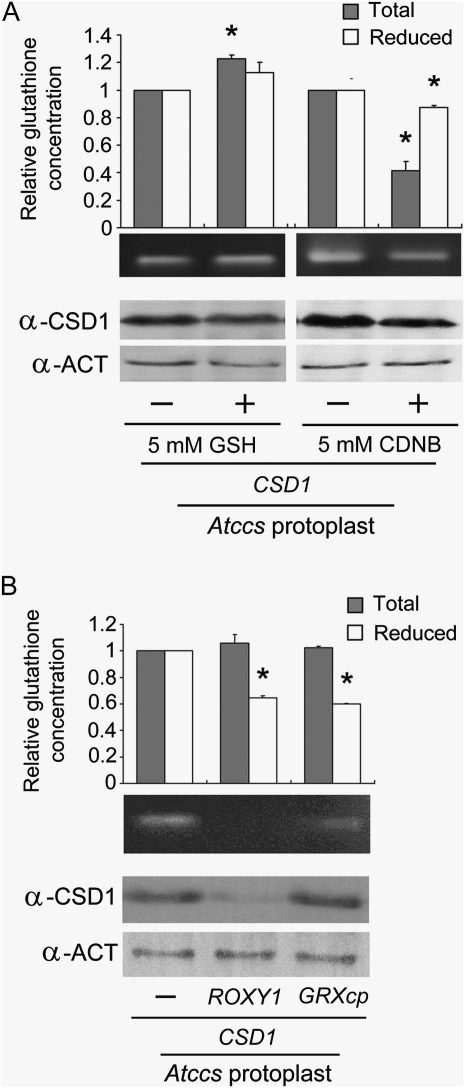
A previous study of human hSOD1 implicated a strict dependence on GSH for CCS-independent activation (Carroll et al., 2004). To investigate the possible involvement of GSH in the activation of Arabidopsis CSD, we tested the effect of GSH on Atccs protoplasts overexpressing CSD1 (Fig. 6A). The cellular concentrations of total glutathione and GSH were slightly enhanced by GSH treatment, by 1.23- and 1.13-fold, respectively, whereas the CSD1 activity did not increase noticeably. On treatment with a glutathione chelator, 1-chloro-2,4-dinitrobenzene (CDNB), both the concentrations of total glutathione and GSH decreased (to 0.42- and 0.87-fold), as did the level of CSD1 and its activity. The relatively slight change in internal GSH concentration after exogenous GSH treatments might be due to an in vivo mechanism that maintains the homeostasis of reducing power, too much of which could be potentially damaging to cells (Lockwood, 2003; Pasternak et al., 2008).
Figure 6.
Effect of glutathione concentration on the activity of CSD1 transiently expressed in Arabidopsis protoplasts. A, Atccs protoplasts overexpressing CSD1 were treated with GSH or CDNB for 2 h as indicated. B, CSD1 was transiently expressed in Atccs protoplasts together with ROXY1 or GRXcp. For both A and B, cellular extracts of the indicated protoplasts were analyzed for total glutathione and GSH level, CSD1 activity, or amount of CSD1 protein. Data represent results of three independent experiments (means ± sd). * P < 0.05.
In a further experiment, we manipulated GSH concentration by overexpressing glutaredoxin (Xing et al., 2005; Cheng et al., 2006; Rouhier, 2010), ROXY1 (a cytoplasmic glutaredoxin) or GRXcp (a chloroplastic glutaredoxin), together with CSD1, in Atccs protoplasts (Fig. 6B). GSH concentration decreased with ROXY1 or GRXcp overexpression, whereas total glutathione concentration was not significantly changed. When ROXY1 was coexpressed with CSD1, both the level and activity of CSD1 were decreased; the coexpression of GRXcp reduced CSD1 activity, with no change in protein level. Thus, we show an overall decrease in CSD1 activity with reduced intracellular GSH concentration.
Cu Ion and GSH Greatly Enhance the Activation of Apo-CSD1 in the Presence of Atccs Cellular Extracts in Vitro
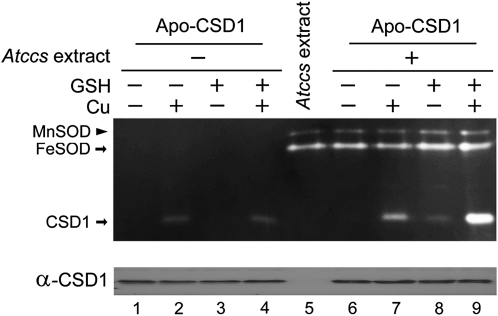
To gain a better understanding of GSH involvement in the CCS-independent pathway, we performed in vitro analyses using affinity-purified proteins. We purified the glutathione S-transferase (GST)-tagged CSD1 (GST-CSD1), then removed the GST (Holo-CSD1) by using PreScission protease (Supplemental Fig. S2, A and B). We then prepared the inactive apo-form of CSD1 (Apo-CSD1) by an acid treatment method (Lepock et al., 1981). We tested the effect of pH on this assay system and found the recovery of Apo-CSD1 activity to be maximal at pH 7 (Supplemental Fig. S2C), so we used this pH in subsequent experiments (Fig. 7). When Apo-CSD1 was treated with 0.1 μm CuSO4, CSD1 itself could spontaneously bind Cu and recover its own activity (Fig. 7, lane 2). Recovery of Apo-CSD1 activity was unsuccessful with 1 mm GSH alone (lane 3). However, CSD1 activity was not enhanced with GSH plus Cu than with Cu alone (compare lanes 2 and 4), which indicates that the Cu-GSH complex itself was not sufficient to facilitate CSD1 activation. The same experiments were performed with Atccs cellular extract (lanes 6–9). With the extract plus GSH (lane 8), the activity of CSD1 increased slightly relative to the extract alone (lane 6). However, CSD1 activity was greatly enhanced with the extract, GSH, and Cu in the reaction mixture (lane 9) as compared with only extract and Cu (lane 7). Therefore, GSH activated CSD1 efficiently under these conditions. Because the Atccs cellular extract could greatly enhance the level of CSD1 activation by GSH (compare lanes 4 and 9), the interaction between GSH and CSD1 was facilitated by an as yet unidentified cellular factor.
Figure 7.
Activation of Apo-CSD1 by Cu, GSH, and Atccs cellular extract. Top, SOD activity assay. Each reaction contained 870 ng of Apo-CSD1 protein and 20 μm ZnSO4, to which 0.1 μm CuSO4, 1 mm GSH, or 15 μg Atccs cellular extract was added as indicated. Lane 5 represents 15 μg of Atccs leaf cellular extract used as a control. Bottom, a replicate of 50 ng of Apo-CSD1 was subjected to the same treatments and then analyzed for protein level by immunoblotting.
DISCUSSION
In this study, we have demonstrated the existence of both CCS-dependent and -independent activation pathways for CSD in plant cells. CSD1 can be activated by both pathways in yeast and Arabidopsis protoplasts (Figs. 1 and 2D). In contrast, CSD2 could not be activated by a CCS-independent pathway in either organism (Figs. 1 and 2B). Thus, ySOD1 and CSD2 might be activated by a similar mechanism. However, further investigation revealed significant differences between the two orthologs. The C-terminal amino acid residues 142P/144P of ySOD1 prevented its activation by the alternative pathway (Carroll et al., 2004), and this effect is mediated primarily by 144P (Leitch et al., 2009a); when 144P was mutated to 144L, this ySOD1 variant showed clear activity in the absence of CCS. For CSD2, the amino acid residues corresponding to 142P/144P of ySOD1 are 143G/145L (Fig. 4A), which are typical for a CSD protein capable of being activated by the CCS-independent pathway (Carroll et al., 2004; Leitch et al., 2009a). The CSD2 protein can be activated by this pathway because the ΔTP-CSD2 (cytoplasm-localized CSD2) was active in Atccs (Fig. 2B). Thus, the lack of CCS-independent activity of CSD2 may not result from the protein structure but rather from the presence of factor(s) in the chloroplast that inhibit the CCS-independent activation pathway. This hypothesis is supported by the results obtained with the TPCSD2-CSD1 (chloroplast-directed CSD1), which also lost CCS-independent activity (Fig. 2D). The inhibitory effect on CCS-independent CSD activity in chloroplasts may be due to the absence of chloroplastic factor(s) that are required for CCS-independent activation or to the presence of inhibitory factor(s) in the chloroplast, a novel phenomenon that has not been reported so far for any other species.
Oxygen may be a factor explaining the inhibition of the CCS-independent pathway in chloroplasts. In CCS-dependent activation, oxygen is required during the disulfide formation between the interacting CCS and CSD proteins (Brown et al., 2004; Furukawa et al., 2004): activation cannot occur under hypoxic conditions. Thus, activation by CCS in the chloroplast requires a constant supply of oxygen, which was produced by a properly functioning electron transport chain of the photosynthetic machinery. Such a mechanism can ensure the presence of CSD activity to prevent a buildup of ROS by photosynthesis. In contrast, CCS-independent activation of CSD does not require oxygen (Leitch et al., 2009a). Numerous photosynthetic enzymes require Cu as their cofactor, which could be conferred by chloroplastic CSD (Shcolnick and Keren, 2006). Abundant oxygen, produced from photosynthetic reactions, might be a signal indicating that the photosynthetic enzymes are saturated with Cu cofactors. An antioxidant system in chloroplasts regulated by oxygen could guarantee priority of the distribution of Cu to photosynthetic enzymes.
Although the CSD3 activity was undetectable by the in-gel SOD activity assay in yeast, it could complement the phenotypes of both yeast sod1Δ and ccsΔ (Fig. 1), which indicates the presence of a physiologically significant level of CSD activity. In these experiments, the activity in ccsΔ could have occurred only through the CCS-independent pathway. However, sod1Δ contains factors that can facilitate both CCS-dependent and -independent activation, so phenotype recovery by CSD3 in this system may have occurred by either or both pathways. Thus, these results demonstrate only that CCS-independent activation of CSD3 has occurred, not the level of CSD3 activity that can be conferred by CCS.
To further characterize the activation of CSD3 in a homologous plant system, we did not observe activation with overexpression of the full-length CSD3 in Arabidopsis protoplasts. Considering the possible effects of localization similar to CSD2, we deleted its peroxisome-targeting sequence but still found no activity even with AtCCS coexpression (Fig. 3; Supplemental Fig. S3). Kliebenstein et al. (1998) demonstrated similar immunoblotting signals with the same amount (12 ng) of recombinant CSD1, CSD2, and CSD3 protein when probed with α-CSD1, α-CSD2, and α-CSD3 antibodies, respectively. Thus, the amount of overexpressed CSD3 protein in protoplasts should be similar to that of overexpressed CSD1 and CSD2 (Supplemental Fig. S3) seen in Figure 2. Hence, our failure to detect CSD3 activity suggests that its level of activation by CCS is much lower than that of CSD1 and CSD2 (Supplemental Fig. S3). In addition, the level of CSD3 activation associated with both pathways (expression in Atccs with AtCCS coexpression or in wild-type protoplasts) seems not to be significantly higher than the levels associated with the CCS-independent pathway only (expression in Atccs). Therefore, CSD3 may be activated primarily by the CCS-independent pathway. Notably, the activation of nematode wSod-1 in the presence of CCS did not result in a higher activity than that by the CCS-independent pathway (Jensen and Culotta, 2005). Thus, CSD3 seems to be similar to the nematode wSod-1 in this respect.
To date, studies of CSDs in yeast, nematode, mouse, and human have shown different preferences for CCS-dependent and -independent activation (Carroll et al., 2004; Jensen and Culotta, 2005; Leitch et al., 2009b). In Arabidopsis, different types of CSDs are present in a single cell: cytoplasmic CSD1 is activated mainly depending on CCS and partially by the CCS-independent pathway (Fig. 2D); chloroplastic CSD2 activation depends completely on CCS, with inhibition of the alternative pathway in chloroplasts (Fig. 2B); and peroxisomal CSD3 may be activated mainly by the CCS-independent pathway (see above). The species studied thus far showed high variation in CSD activation. Why different organisms have evolved different preferences for CCS-dependent and -independent activation is unclear but may reflect unique lifestyle requirements for Cu and oxygen (Culotta et al., 2006). Because plants cannot move to avoid environmental stresses, the complex activation mechanisms for plant CSDs might be a solution to cope with such varied and stressful surroundings.
In investigating the physiological effects of the residual CCS-independent CSD activity (Fig. 5), we obtained a higher level of seed germination rate in Atccs than in Atcsd1 when treated with ROS-generating reagents. In Atcsd1, CSD1 activity was completely lost, whereas residual CSD1 activity was present in Atccs. Because phospholipid membranes are impermeable to O2− (Takahashi and Asada, 1983), protection of different organelles can be achieved by variable distribution of SODs in different cellular compartments. In other words, the SOD activities in one compartment cannot complement functional deficiencies in another. Therefore, the lower germination rate of Atcsd1 was possibly due to the complete absence of CSD1 activity in the cytoplasm. In addition, current studies of SODs indicate that the level of SOD activity required for normal growth is much lower than the actual activity level measured (Cohu et al., 2009; C.-H. Huang, W.-Y. Kuo, C. Weiss, and T.-L. Jinn, unpublished data). In fact, Atccs showed no obvious phenotype under normal and high-light stress conditions as compared with the wild type (Cohu et al., 2009; C.-H. Huang, W.-Y. Kuo, C. Weiss, and T.-L. Jinn, unpublished data). Therefore, loss of most of the CSD activity did not seriously compromise plant viability, so CCS-independent CSD activities are physiologically functional and sufficient to support the growth of plant cells. The high expression of CSD proteins in wild-type plants may explain their role in Cu buffering (Cohu et al., 2009).
Previous studies in yeast, human, and nematode have shown GSH to be involved in the alternative activation pathway (Carroll et al., 2004; Jensen and Culotta, 2005). Evidence of this involvement includes analyses of a yeast knockout line for GSH biosynthesis in which the major CCS-independent activity was abolished. We tested this hypothesis in Arabidopsis by overexpressing a glutaredoxin in Atccs protoplasts to interfere with internal GSH concentrations. However, ROXY1 expression resulted in a greatly reduced amount of CSD1, with reduced activity (Fig. 6B). Because ROXY1 colocalizes with CSD1, it can readily reduce its disulfide bond, which would lead to degradation of the disulfide-reduced CSD1 (Borchelt et al., 1994; Wang et al., 2003; Carroll et al., 2006; Rouhier, 2010). In contrast, when we overexpressed GRXcp, the GSH concentration was decreased, along with CSD1 activity, without a corresponding change in the level of CSD1 protein (Fig. 6B). GRXcp localizes in chloroplasts and cannot interact with CSD1 (Cheng et al., 2006); therefore, the effect of GRXcp on CSD1 activity is due to decreased GSH concentration only. Our result also confirmed the involvement of GSH in this pathway.
However, the results of our in vitro experiments showed that GSH was not sufficient to activate CSD1 unless it was added together with cellular extract (Fig. 7). Therefore, GSH was involved in the CCS-independent pathway in Arabidopsis in the presence of an unidentified factor. Further investigation is required to characterize the unidentified factor and elucidate the complete mechanism.
MATERIALS AND METHODS
Plants, Yeast Strains, and Growth Conditions
The Arabidopsis (Arabidopsis thaliana) AtCCS (At1g12520) and AtCSD1 (At1g08830) knockout lines, Atccs (SALK_025986) and Atcsd1 (SALK_109389), were obtained from the Arabidopsis Biological Research Center (http://abrc.osu.edu;). Seeds were incubated at 4°C for 3 d in the dark before sowing, then plants were grown in a growth chamber at 23°C with 16 h of light at an intensity of 60 to 100 μmol m−2 s−1. Saccharomyces cerevisiae BY4741 (MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0) was used as the yeast wild type, and sod1Δ (sod1::kanMX4) and ccsΔ (ccs::kanMX4) are derivatives of BY4741. Yeast was incubated at 30°C under aerobic conditions without shaking, and G418 was used at 200 μg mL−1 to maintain the ySOD1 and yCCS deletions in the yeast as required.
Lys-Independent Aerobic Growth
Yeast strains were cultured in appropriate medium overnight at 30°C without shaking. Then, cells were centrifuged, washed, and resuspended in sterile water, and the A600 was measured. For plate assays, the yeast cultures were diluted serially from optical density at 600 nm = 1 to 10−4, plated on synthetic dropout Lys medium, and incubated at 30°C under aerobic conditions for 3 d. For the liquid assay, the yeast cultures were seeded from optical density at 600 nm = 0.01 in the Lys-lacking medium, incubated at 30°C for 24 h under aerobic conditions, and then cell density was determined by measuring the A600 (Corson et al., 1998; Wallace et al., 2004).
Seed Germination Rate
Sterilized seeds of the wild type, Atccs, and Atcsd1 were plated on 1/2 MS medium with 1% (w/v) Suc, supplemented with MV or BH at the indicated concentrations, and incubated at 23°C with 16 h of light for 3 d, then seed germination rates were measured.
Constructs and Gene Cloning
For yeast expression, all cDNAs from ATG to the stop codon were inserted at the HindIII site of yeast expression vector pADNS (Colicelli et al., 1989), with the exception of CSD3 (At5g18100), which was inserted at the NotI site. The 35S promoter of pPE1000 vector (Hancock et al., 1997) was inserted between the XhoI/HindIII sites of pEYFP vectors (Clontech; http://www.clontech.com) to create p35S-EYFP. For gene overexpression in Arabidopsis protoplasts, cDNA of CSD1, CSD2 (At2g28190), and CSD3 with a stop codon was subcloned into p35S-EYFP at EcoRI/BamHI, HindIII, and EcoRI sites, respectively. For ΔTP-CSD2, the 61 amino acids at the N terminus of CSD2 were deleted and then inserted into p35S-EYFP at the HindIII site. The chloroplast transit peptides of CSD2 (TPCSD2) with EcoRI/NcoI sites and of CSD1 with NcoI/SalI sites were ligated into p35S-EYFP at EcoI/SalI sites to create a chloroplast-localized TPCSD2-CSD1. CSD3 and CSD3-ΔAKL (peroxisome-targeting sequence deleted) were inserted into the 326-GFPnt vector (Lee et al., 2001) at the SmaI/SalI sites. Both ROXY1 (At3g02000; Xing et al., 2005) and GRXcp (At3g54900; Cheng et al., 2006) cDNAs with a stop codon were inserted into p35S-EYFP at SmaI/BamHI sites. Point mutations of CSD1 were created by the megapriming method (Landt et al., 1990). For GST-tagged protein, CSD1 was inserted into pGEX-6P-1 (Amersham Pharmacia Biotech; http://www.gelifesciences.com) at the EcoRI site. All gene fragments were amplified by PCR and sequenced before making the constructs. Primers used in this study are shown in Supplemental Table S1.
RNA Extraction and Reverse Transcription-PCR
Total RNA was prepared with TRIZOL reagent (Invitrogen; http://www.invitrogen.com) and the TURBO DNA-free Kit (Applied Biosystems; http://www.appliedbiosystems.com). cDNA synthesis involved the use of high-capacity cDNA Reverse Transcription Kits (Applied Biosystems). Reverse transcription-PCR involved the use of the primers shown in Supplemental Table S1.
Protein Extraction, SOD Activity Assay, and Immunoblotting
Arabidopsis leaf cellular extracts were prepared with 150 mm Tris, pH 7.2, grinding buffer (Chu et al., 2005). Yeast crude protein was extracted by the glass bead lysis protocol (Culotta et al., 1997). Protein concentration was determined by the method of Bradford (1976). SOD activity assay and immunoblotting were performed on nondenaturing and denaturing gels, respectively (Chu et al., 2005). SOD activities and protein signals were quantified by analyzing the activity gels and immunoblotting membranes with the use of LAS-3000 (Fuji Film; http://www.fujifilm.com) and ImageQuant software (Molecular Dynamics; http://www.mdyn.com).
Protoplast Preparation and Transfection
Protoplast preparation and transfection were as described (Yoo et al., 2007). About 106 protoplasts was transfected with 100 to 300 μg of plasmid DNA for each construct, then incubated for 16 to 24 h. For protein extract, protoplasts were collected and resuspended in 150 mm Tris buffer (pH 7.2), then vortexed five times for 5 s each. Cell lysates were then subjected to the following experiments.
Glutathione Treatment and Quantification
Atccs protoplasts transfected with CSD1 were incubated at room temperature for 16 h, then 1 mm GSH or CDNB (dissolved in 95% ethanol) was added for 15 min. Total glutathione and GSH concentrations were quantified by the use of the Total Glutathione Quantification Kit (Dojindo Laboratories; http://www.dojindo.com) and the QuantiChrom Glutathione Assay Kit (BioAssay Systems; http://www.bioassaysys.com). The resulting concentrations were normalized to the protein concentration of the same sample, and the glutathione concentration values are expressed relative to those with mock treatment.
Apo-CSD1 Protein Preparation and in Vitro Treatments
GST-tagged proteins were affinity purified according to the Glutathione-Agarose user manual (Sigma; http://www.sigmaaldrich.com). GST was removed by the use of PreScission protease (Sigma), and the protein mixture was passed through a GST agarose column to remove free GST. Preparation of inactivated Apo-CSD1 was as described (Lepock et al., 1981). For the in vitro treatments, each reaction contained 870 ng of Apo-CSD1 (equals 2.9 μm) and 20 μm ZnSO4, to which 0.1 μm CuSO4, 1 mm GSH, or 15 μg of Atccs cellular extract was added as indicated. After incubation at room temperature for 30 min, 1.5 mm EDTA was added to the samples, and then native-PAGE was performed with 0.1 mm EDTA on both the nondenaturing gels and running buffer. In the treatments with both Cu and GSH, the two solutions were mixed and incubated at 4°C for 16 h before the experiment.
Statistical Analysis
All experiments were independently repeated at least three times. Statistical analysis involved the use of Student’s t test (two-tailed, unpaired). P < 0.05 was considered statistically significant.
Antibodies
α-CSD1, α-CSD2, and α-CSD3 antisera were generous gifts from D.J. Kliebenstein (Kliebenstein et al., 1998), and an Arabidopsis internal control antibody, α-RPN8, was kindly provided by H.-Y. Fu (Yang et al., 2004). We used α-ACT (Chemicon; http://www.chemicon.com) and yeast α-PGK1 (Molecular Probes; http://probes.invitrogen.com) antibodies as internal controls.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Residual CSD activities analyzed in Arabidopsis Atccs.
Supplemental Figure S2. Affinity purification of CSD1 and its activity under various pH treatments.
Supplemental Figure S3. Characterization of individual CSD activities in Arabidopsis protoplasts.
Supplemental Table S1. Primers used in this study.
Supplementary Material
Acknowledgments
We are grateful to Fang-Jen Lee for generous gifts of yeast BY4741, sod1Δ, and ccsΔ lines obtained from the Saccharomyces Genome Deletion Project; Daniel J. Kliebenstein for kindly providing the antisera of Arabidopsis CSDs; and Hong-Yong Fu for α-RPN8 antibody. We thank Chu-Yung Lin for critically reading the manuscript.
References
- Abdel-Ghany SE, Burkhead JL, Gogolin KA, Andrés-Colás N, Bodecker JR, Puig S, Peñarrubia L, Pilon M. (2005) AtCCS is a functional homolog of the yeast copper chaperone Ccs1/Lys7. FEBS Lett 579: 2307–2312 [DOI] [PubMed] [Google Scholar]
- Alscher RG, Erturk N, Heath LS. (2002) Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. J Exp Bot 53: 1331–1341 [PubMed] [Google Scholar]
- Beyer W, Imlay J, Fridovich I. (1991) Superoxide dismutases. Prog Nucleic Acid Res Mol Biol 40: 221–253 [DOI] [PubMed] [Google Scholar]
- Borchelt DR, Lee MK, Slunt HS, Guarnieri M, Xu ZS, Wong PC, Brown RH, Jr, Price DL, Sisodia SS, Cleveland DW. (1994) Superoxide dismutase 1 with mutations linked to familial amyotrophic lateral sclerosis possesses significant activity. Proc Natl Acad Sci USA 91: 8292–8296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler C, Van Montagu MV, Inzé D. (1992) Superoxide dismutase and stress tolerance. Annu Rev Plant Physiol Plant Mol Biol 43: 83–116 [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Brown NM, Torres AS, Doan PE, O’Halloran TV. (2004) Oxygen and the copper chaperone CCS regulate posttranslational activation of Cu,Zn superoxide dismutase. Proc Natl Acad Sci USA 101: 5518–5523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueno P, Varela J, Giménez-Gallego G, del Río LA. (1995) Peroxisomal copper,zinc superoxide dismutase: characterization of the isoenzyme from watermelon cotyledons. Plant Physiol 108: 1151–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll MC, Girouard JB, Ulloa JL, Subramaniam JR, Wong PC, Valentine JS, Culotta VC. (2004) Mechanisms for activating Cu- and Zn-containing superoxide dismutase in the absence of the CCS Cu chaperone. Proc Natl Acad Sci USA 101: 5964–5969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll MC, Outten CE, Proescher JB, Rosenfeld L, Watson WH, Whitson LJ, Hart PJ, Jensen LT, Culotta VC. (2006) The effects of glutaredoxin and copper activation pathways on the disulfide and stability of Cu,Zn superoxide dismutase. J Biol Chem 281: 28648–28656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casareno RL, Waggoner D, Gitlin JD. (1998) The copper chaperone CCS directly interacts with copper/zinc superoxide dismutase. J Biol Chem 273: 23625–23628 [DOI] [PubMed] [Google Scholar]
- Cheng NH, Liu JZ, Brock A, Nelson RS, Hirschi KD. (2006) AtGRXcp, an Arabidopsis chloroplastic glutaredoxin, is critical for protection against protein oxidative damage. J Biol Chem 281: 26280–26288 [DOI] [PubMed] [Google Scholar]
- Chu CC, Lee WC, Guo WY, Pan SM, Chen LJ, Li HM, Jinn TL. (2005) A copper chaperone for superoxide dismutase that confers three types of copper/zinc superoxide dismutase activity in Arabidopsis. Plant Physiol 139: 425–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciriolo MR, Desideri A, Paci M, Rotilio G. (1990) Reconstitution of Cu,Zn-superoxide dismutase by the Cu(I).glutathione complex. J Biol Chem 265: 11030–11034 [PubMed] [Google Scholar]
- Cohu CM, Abdel-Ghany SE, Gogolin Reynolds KA, Onofrio AM, Bodecker JR, Kimbrel JA, Niyogi KK, Pilon M. (2009) Copper delivery by the copper chaperone for chloroplast and cytosolic copper/zinc-superoxide dismutases: regulation and unexpected phenotypes in an Arabidopsis mutant. Mol Plant 2: 1336–1350 [DOI] [PubMed] [Google Scholar]
- Colicelli J, Birchmeier C, Michaeli T, O’Neill K, Riggs M, Wigler M. (1989) Isolation and characterization of a mammalian gene encoding a high-affinity cAMP phosphodiesterase. Proc Natl Acad Sci USA 86: 3599–3603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corson LB, Strain JJ, Culotta VC, Cleveland DW. (1998) Chaperone-facilitated copper binding is a property common to several classes of familial amyotrophic lateral sclerosis-linked superoxide dismutase mutants. Proc Natl Acad Sci USA 95: 6361–6366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crapo JD, Oury T, Rabouille C, Slot JW, Chang LY. (1992) Copper,zinc superoxide dismutase is primarily a cytosolic protein in human cells. Proc Natl Acad Sci USA 89: 10405–10409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culotta VC, Klomp LWJ, Strain J, Casareno RLB, Krems B, Gitlin JD. (1997) The copper chaperone for superoxide dismutase. J Biol Chem 272: 23469–23472 [DOI] [PubMed] [Google Scholar]
- Culotta VC, Yang M, O’Halloran TV. (2006) Activation of superoxide dismutases: putting the metal to the pedal. Biochim Biophys Acta 1763: 747–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira AMD, Ciriolo MR, Marcocci L, Rotilio G. (1993) Copper(I) transfer into metallothionein mediated by glutathione. Biochem J 292: 673–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa Y, Torres AS, O’Halloran TV. (2004) Oxygen-induced maturation of SOD1: a key role for disulfide formation by the copper chaperone CCS. EMBO J 23: 2872–2881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glerum DM, Shtanko A, Tzagoloff A. (1996) Characterization of COX17, a yeast gene involved in copper metabolism and assembly of cytochrome oxidase. J Biol Chem 271: 14504–14509 [DOI] [PubMed] [Google Scholar]
- Halliwell B, Gutteridge JMC. (2007) Free Radicals in Biology and Medicine, Ed 4. Oxford University Press, Oxford
- Hancock KR, Phillips LD, White DWR, Ealing PM. (1997) pPE1000: a versatile vector for the expression of epitope-tagged foreign proteins in transgenic plants. Biotechniques 22: 861–862, 865 [DOI] [PubMed] [Google Scholar]
- Horecka J, Kinsey PT, Sprague GF., Jr (1995) Cloning and characterization of the Saccharomyces cerevisiae LYS7 gene: evidence for function outside of lysine biosynthesis. Gene 162: 87–92 [DOI] [PubMed] [Google Scholar]
- Jackson C, Dench J, Moore AL, Halliwell B, Foyer CH, Hall DO. (1978) Subcellular localisation and identification of superoxide dismutase in the leaves of higher plants. Eur J Biochem 91: 339–344 [DOI] [PubMed] [Google Scholar]
- Jensen LT, Culotta VC. (2005) Activation of CuZn superoxide dismutases from Caenorhabditis elegans does not require the copper chaperone CCS. J Biol Chem 280: 41373–41379 [DOI] [PubMed] [Google Scholar]
- Kanematsu S, Asada K. (1989) CuZn-superoxide dismutase in rice: occurrence of an active, monomeric enzyme and two types of isozyme in leaf and non-photosynthetic tissues. Plant Cell Physiol 30: 381–391 [Google Scholar]
- Kliebenstein DJ, Monde RA, Last RL. (1998) Superoxide dismutase in Arabidopsis: an eclectic enzyme family with disparate regulation and protein localization. Plant Physiol 118: 637–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb AL, Torres AS, O’Halloran TV, Rosenzweig AC. (2001) Heterodimeric structure of superoxide dismutase in complex with its metallochaperone. Nat Struct Biol 8: 751–755 [DOI] [PubMed] [Google Scholar]
- Landt O, Grunert HP, Hahn U. (1990) A general method for rapid site-directed mutagenesis using the polymerase chain reaction. Gene 96: 125–128 [DOI] [PubMed] [Google Scholar]
- Lee YJ, Kim DH, Kim YW, Hwang I. (2001) Identification of a signal that distinguishes between the chloroplast outer envelope membrane and the endomembrane system in vivo. Plant Cell 13: 2175–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch JM, Jensen LT, Bouldin SD, Outten CE, Hart PJ, Culotta VC. (2009a) Activation of Cu,Zn-superoxide dismutase in the absence of oxygen and the copper chaperone CCS. J Biol Chem 284: 21863–21871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch JM, Yick PJ, Culotta VC. (2009b) The right to choose: multiple pathways for activating copper,zinc superoxide dismutase. J Biol Chem 284: 24679–24683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepock JR, Arnold LD, Petkau A, Kelly K. (1981) Interaction of superoxide dismutase with phospholipid liposomes: an uptake, spin label and calorimetric study. Biochim Biophys Acta 649: 45–57 [DOI] [PubMed] [Google Scholar]
- Lin SJ, Culotta VC. (1995) The ATX1 gene of Saccharomyces cerevisiae encodes a small metal homeostasis factor that protects cells against reactive oxygen toxicity. Proc Natl Acad Sci USA 92: 3784–3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SJ, Pufahl RA, Dancis A, O’Halloran TV, Culotta VC. (1997) A role for the Saccharomyces cerevisiae ATX1 gene in copper trafficking and iron transport. J Biol Chem 272: 9215–9220 [PubMed] [Google Scholar]
- Lockwood TD. (2003) Redox pacing of proteome turnover: influences of glutathione and ketonemia. Arch Biochem Biophys 417: 183–193 [DOI] [PubMed] [Google Scholar]
- Marres CA, Van Loon AP, Oudshoorn P, Van Steeg H, Grivell LA, Slater EC. (1985) Nucleotide sequence analysis of the nuclear gene coding for manganese superoxide dismutase of yeast mitochondria, a gene previously assumed to code for the Rieske iron-sulphur protein. Eur J Biochem 147: 153–161 [DOI] [PubMed] [Google Scholar]
- McCord JM, Fridovich I. (1969) Superoxide dismutase: an enzymic function for erythrocuprein (hemocuprein). J Biol Chem 244: 6049–6055 [PubMed] [Google Scholar]
- Pasternak M, Lim B, Wirtz M, Hell R, Cobbett CS, Meyer AJ. (2008) Restricting glutathione biosynthesis to the cytosol is sufficient for normal plant development. Plant J 53: 999–1012 [DOI] [PubMed] [Google Scholar]
- Rouhier N. (2010) Plant glutaredoxins: pivotal players in redox biology and iron-sulphur centre assembly. New Phytol 186: 365–372 [DOI] [PubMed] [Google Scholar]
- Shcolnick S, Keren N. (2006) Metal homeostasis in cyanobacteria and chloroplasts: balancing benefits and risks to the photosynthetic apparatus. Plant Physiol 141: 805–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi MA, Asada K. (1983) Superoxide anion permeability of phospholipid membranes and chloroplast thylakoids. Arch Biochem Biophys 226: 558–566 [DOI] [PubMed] [Google Scholar]
- Wallace MA, Liou LL, Martins J, Clement MHS, Bailey S, Longo VD, Valentine JS, Gralla EB. (2004) Superoxide inhibits 4Fe-4S cluster enzymes involved in amino acid biosynthesis: cross-compartment protection by CuZn-superoxide dismutase. J Biol Chem 279: 32055–32062 [DOI] [PubMed] [Google Scholar]
- Wang J, Slunt H, Gonzales V, Fromholt D, Coonfield M, Copeland NG, Jenkins NA, Borchelt DR. (2003) Copper-binding-site-null SOD1 causes ALS in transgenic mice: aggregates of non-native SOD1 delineate a common feature. Hum Mol Genet 12: 2753–2764 [DOI] [PubMed] [Google Scholar]
- Weisiger RA, Fridovich I. (1973) Mitochondrial superoxide simutase: site of synthesis and intramitochondrial localization. J Biol Chem 248: 4793–4796 [PubMed] [Google Scholar]
- Wong PC, Waggoner D, Subramaniam JR, Tessarollo L, Bartnikas TB, Culotta VC, Price DL, Rothstein J, Gitlin JD. (2000) Copper chaperone for superoxide dismutase is essential to activate mammalian Cu/Zn superoxide dismutase. Proc Natl Acad Sci USA 97: 2886–2891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing SP, Rosso MG, Zachgo S. (2005) ROXY1, a member of the plant glutaredoxin family, is required for petal development in Arabidopsis thaliana. Development 132: 1555–1565 [DOI] [PubMed] [Google Scholar]
- Yang PZ, Fu HY, Walker J, Papa CM, Smalle J, Ju YM, Vierstra RD. (2004) Purification of the Arabidopsis 26 S proteasome: biochemical and molecular analyses revealed the presence of multiple isoforms. J Biol Chem 279: 6401–6413 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zelko IN, Mariani TJ, Folz RJ. (2002) Superoxide dismutase multigene family: a comparison of the CuZn-SOD (SOD1), Mn-SOD (SOD2), and EC-SOD (SOD3) gene structures, evolution, and expression. Free Radic Biol Med 33: 337–349 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.