Abstract
The duplication and divergence of heat stress (HS) response genes might help plants adapt to varied HS conditions, but little is known on the topic. Here, we examined the evolution and function of Arabidopsis (Arabidopsis thaliana) mitochondrial GrpE (Mge) proteins. GrpE acts as a nucleotide-exchange factor in the Hsp70/DnaK chaperone machinery. Genomic data show that AtMge1 and AtMge2 arose from a recent whole-genome duplication event. Phylogenetic analysis indicated that duplication and preservation of Mges occurred independently in many plant species, which suggests a common tendency in the evolution of the genes. Intron retention contributed to the divergence of the protein structure of Mge paralogs in higher plants. In both Arabidopsis and tomato (Solanum lycopersicum), Mge1 is induced by ultraviolet B light and Mge2 is induced by heat, which suggests regulatory divergence of the genes. Consistently, AtMge2 but not AtMge1 is under the control of HsfA1, the master regulator of the HS response. Heterologous expression of AtMge2 but not AtMge1 in the temperature-sensitive Escherichia coli grpE mutant restored its growth at 43°C. Arabidopsis T-DNA knockout lines under different HS regimes revealed that Mge2 is specifically required for tolerating prolonged exposure to moderately high temperature, as compared with the need of the heat shock protein 101 and the HS-associated 32-kD protein for short-term extreme heat. Therefore, with duplication and subfunctionalization, one copy of the Arabidopsis Mge genes became specialized in a distinct type of HS. We provide direct evidence supporting the connection between gene duplication and adaptation to environmental stress.
Temperature is an environmental factor that affects the growth and distribution of plants. To be successful in the survival game of nature, plants need to cope with adverse temperatures caused by daily or seasonal temperature fluctuations and by climate change. Understanding how plants adapted to temperature stress can help improve tolerance in crops (Katiyar-Agarwal et al., 2003; Shou et al., 2004).
Plants are often challenged by elevated temperature, which causes heat stress (HS). Knowledge of the plant response to HS is accumulating quickly because of the progress of genomic and functional genomic studies (Kotak et al., 2007). Many genetic components form a complex signaling transduction network and chaperone machineries. Some of these components belong to multigene families, most notably heat shock proteins (HSPs; Vierling, 1991) and heat shock transcription factors (HSFs; von Koskull-Döring et al., 2007). The HSP and HSF paralogs apparently arose from gene duplications and subsequent diversification. Gene duplication and divergence increased the capacity of organisms to adapt to environmental change (Conant and Wolfe, 2008). In addition to responding to elevated temperature, HS genes are responsive to other environmental changes. Thus, the duplication and divergence of HS genes might have allowed plants to adapt to different environmental stresses or varied HS conditions.
However, linking the adaptation and duplication of HS genes is difficult because a loss of function of one of the duplicates may not lead to an alteration in phenotype such as thermotolerance, which was often attributed to gene redundancy. Mutant phenotypes may not be revealed because the assays are not performed under the right conditions, as was shown with several Arabidopsis (Arabidopsis thaliana) HS genes (Charng et al., 2006, 2007; Meiri and Breiman, 2009; Meiri et al., 2010). Thus, determining an effective phenotyping method is critical for revealing the functions of HS genes per se and for shedding light on their evolution.
To determine the biological functions of HS response genes, we have used reverse genetic approaches by phenotyping Arabidopsis T-DNA knockout (KO) mutants under varied HS conditions (Charng et al., 2007; Liu et al., 2011). Here, we focused on the function and evolution of the mitochondrial GrpE genes. GrpE is a highly conserved and ubiquitous protein in prokaryotes and eukaryotes. It functions as a cochaperone in the DnaK/Hsp70 complex, consisting of DnaK (Hsp70), DnaJ (Hsp40), and GrpE. The complex is a chaperone machine involved in the folding of the nascent protein peptides, organellar protein translocation, and disaggregation and refolding of the denatured proteins (Mayer and Bukau, 2005). DnaK and Hsp70 homologs consist of an N-terminal ATPase domain and a C-terminal substrate domain. Current models suggest that the machine operates by propelling DnaK/Hsp70 in cycles of alternating low-affinity (ATP-bound) and high-affinity (ADP-bound) states for substrates by interaction with cochaperones, in which nucleotide dissociation catalyzed by GrpE is the rate-limiting step for substrate release (Mayer and Bukau, 2005). In Escherichia coli, both GrpE and DnaK are inducible by heat treatment. Mutants of GrpE and DnaK are sensitive to adverse high temperature (Paek and Walker, 1987; Ang and Georgopoulos, 1989).
In eukaryotes, multiple sets of the Hsp70 chaperone machineries can be found in cytosol and other organellar compartments (Sung et al., 2001). However, only those found in mitochondria and chloroplasts closely resemble their prokaryotic kin. Only one mitochondrial GrpE, Mge1, was found in yeast. Mge1 is not induced by heat shock (Ikeda et al., 1994) but is essential for yeast viability (Bolliger et al., 1994). Mge1 acts in concert with mitochondrial Hsp70s in the translocation and maturation of preproteins (Laloraya et al., 1995). Our knowledge of plant GrpEs and their biological role is limited. Two types of GrpE cDNAs were cloned from tobacco (Nicotiana tabacum) BY2 cells and found to encode mitochondrial proteins that interact with mtHsp70 when reintroduced into BY2 protoplasts (Padidam et al., 1999). In Chlamydomonas reinhardtii, the chloroplastic GrpE homolog Cge1 is strongly induced by heat shock and forms ATP-sensitive complexes with Hsp70B, the counterpart of bacterial DnaK (Schroda et al., 2001). In vitro studies showed that the Hsp70B-Cdj2-Cge1 chaperones facilitate both the assembly and disassembly of the vesicle-inducing protein in plastid oligomers (VIPP1) involved in the biogenesis of thylakoid membranes (Liu et al., 2007).
In this paper, we investigated the phylogenetic, regulatory, and functional aspects of duplicated copies of Arabidopsis Mges. Subfunctionalization at the expression and functional levels may have allowed Arabidopsis Mges to respond more efficiently to distinct HS regimes. We provide direct evidence to support a connection between gene duplication and adaptation to HS.
RESULTS
Arabidopsis Mitochondrial GrpE Genes Were Duplicated in a Recent Whole-Genome Duplication Event
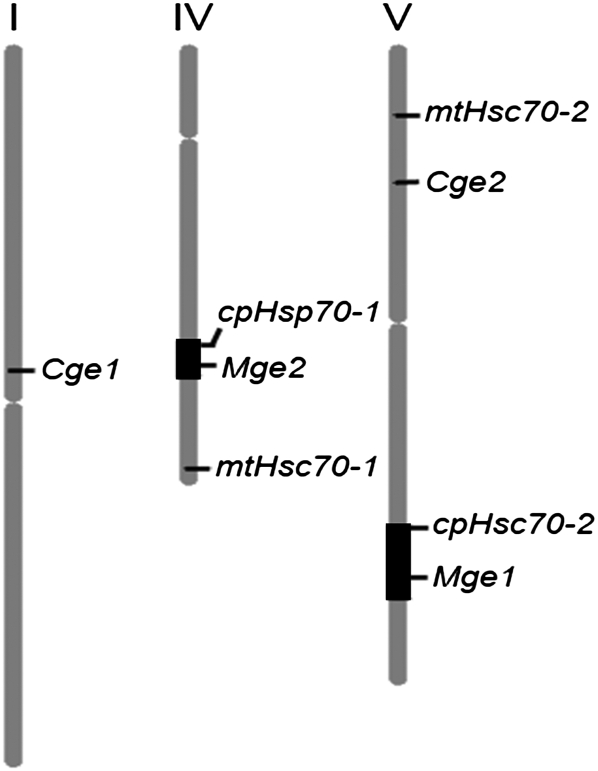
The Arabidopsis genome contains two homologous genes putatively encoding for mitochondrial GrpEs, which we named Mge1 (At5g55200) and Mge2 (At4g26780). The At4g26780 locus was previously annotated to have unknown function. Corresponding to these cochaperones are two copies of Hsp70 homologs, mtHsc70-1 (At4g37910) and mtHsc70-2 (At5g09590), encoding for mitochondrial DnaK/Hsp70 proteins. The homology between Mge1 and Mge2 suggests that they are derived from gene duplication. The Arabidopsis genome had experienced at least two rounds of whole-genome duplication (WGD) events that led to massive gene duplication (Blanc et al., 2003). To determine whether the Mges arose from either one of these events, we used the online analytic tool Paralogons to examine the locations of the Mges in the duplicated regions of the Arabidopsis genome (Blanc et al., 2003). Mge1 and Mge2 each locate in two corresponding duplicated blocks formed after the most recent WGD event (Fig. 1). However, their putatively interacting partners, mtHsc70s, came from a more ancient WGD event. Unlike the duplication time for the Mge pair, that for the two chloroplastic GrpE genes, Cge1 (At1g36390) and Cge2 (At5g17710), could not be determined, which suggests that they arose from a time before the traceable WGD events. To our surprise, the chloroplastic DnaK/Hsp70 genes, cpHsc70-1 (At5g49910) and cpHsc70-2 (At4g24280), came from the same WGD as the Mges, because they locate on the same duplicated blocks (Fig. 1). To confirm this observation, we used molecular clock theory to estimate the time of divergence of each homologous pair by using Arabidopsis genomic synonymous substitution rates (Ks) in a described formula (Lynch and Conery, 2000; Raes et al., 2003). Table I shows the Ks and time of divergence of Arabidopsis Mges, Cges, mtHsc70s, and cpHsc70s. The data are in good agreement with the chronicle of the WGD events described (Blanc et al., 2003).
Figure 1.
Location map of the mitochondrial and chloroplastic GrpE and Hsp70 duplicates in the Arabidopsis genome. The map was drawn with the use of the Chromosome Map Tool of The Arabidopsis Information Resource. The black segments on chromosomes IV and V indicate the duplicated regions derived from the recent WGD.
Table I. Approximate time of divergence of Arabidopsis GrpE and Hsp70 paralogs.
The duplication events that led to the duplication of the indicated gene pairs were determined by use of the bioinformatic program Paralogons. n/a, Undeterminable time of divergence.
| Genes | Duplication Event | Ks | Time of Divergence | |
| million years ago | ||||
| Mge1 | Mge2 | Recent | 0.7 | 57 |
| mtHsc70-1 | mtHsc70-2 | Ancient | 1.63 | 134 |
| Cge1 | Cge2 | Undefined | n/a | n/a |
| cpHsc70-1 | cpHsc70-2 | Recent | 0.71 | 59 |
Duplication of Mge Genes Occurred Independently after the Divergence of Many Distantly Related Plant Species
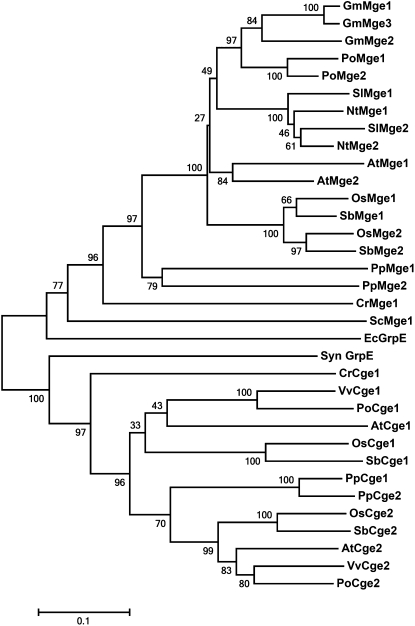
With the knowledge that Arabidopsis Mge genes arose from a recent WGD event, we investigated a phylogenetic relationship among plant Mges. With a BLAST search, we retrieved and aligned the amino acid sequences homologous to Arabidopsis Mges from various species (Supplemental Fig. S1). The aligned sequences were used to generate a phylogenetic tree based on the neighbor-joining method (Fig. 2). The tree showed two major clades of GrpE proteins, one from mitochondrion and E. coli and the other from chloroplast and cyanobacterium, which is in good agreement with the endosymbiotic theory. Two Mge homologs exist in most plants investigated in this study except soybean (Glycine max), which has three. The Mges of most plant species, except monocots or close relatives such as tobacco and tomato (Solanum lycopersicum), clustered together by species, which implies that the Mges duplicated after the divergence of the distantly related species and before the divergence of monocots and closely related species. In contrast, their chloroplastic homologs showed a different clustering pattern. Regardless of plant species, except for moss, Cge1 and Cge2 clustered together by protein type (Fig. 2), which suggests that these proteins were derived from duplication before the speciation of various plants.
Figure 2.
Phylogenetic tree of Mges and Cges of various species. The tree was generated by use of the neighbor-joining method in MEGA5. Bootstrap values (as a percentage of 1,000 replicates) are provided at the branches. The scale bar represents 10 amino acid replacements per 100 positions. At, Arabidopsis; Cr, C. reinhardtii; Ec, E. coli; Gm, soybean; Nt, tobacco; Os, Oryza sativa; Po, Populus sp.; Pp, P. patens; Sb, Sorghum bicolor; Sc, Saccharomyces cerevisiae; Sl, tomato; Syn, Synechocystis sp.; Vv, Vitis vinifera.
Retention of an In-Frame Intron Is Common in Higher Plant Mges
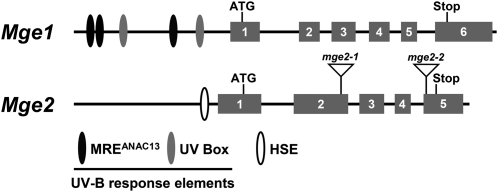
Arabidopsis Mge1 contains six exons and Mge2 has five (Fig. 3). From comparisons of the genomic DNA structures and protein sequences, exon 2 of Mge2 likely contains a retained intron that presumably corresponds to intron 2 of Mge1 (Fig. 3). This putatively retained intron in Mge2 apparently can no longer be processed by the spliceosome because of a loss of the essential features for RNA splicing and, therefore, becomes part of the coding sequence. By contrast, intron 2 of Mge1 does not contain an in-frame sequence and would disrupt the open reading frame if retained. Interestingly, we found retention of the in-frame intron 2 common in alternatively spliced (AS) transcripts for one of the two Mge genes in other higher plant species (Table II). As most of the Mge genes listed here were not annotated, we propose that the Mge genes without intron 2 retention be annotated as Mge1 (or Mge1 and Mge3 in soybean) and the Mge genes with intron 2 retention be annotated as Mge2. Notably, the sequences derived from the retained introns are conserved with a motif of four to six consecutive Arg or Lys residues (Supplemental Fig. S1). Therefore, retention of the in-frame intron 2 may be under selection pressure in higher plants. By contrast, neither Mge1 nor Mge2 of the moss Physcomitrella patens contains an in-frame intron 2.
Figure 3.
Schematic structures of genomic DNA of Mge genes. Putative cis-elements are shown in the promoter regions of the genes in ovals. Exons are shown in rectangular boxes with numbers. Approximate T-DNA insertion sites in mge2-1 and mge2-2 are indicated by triangles.
Table II. The AS forms of Mge transcripts from algae, moss, and higher plant species.
Mges from several higher plants have AS forms in one or both of the Mges. Note that AtMges do not have any AS form. However, AtMge2 highly resembles an intron 2-retained AS form.
| Species | Gene | Exons | Description of AS Form | Accession No. |
| C. reinhardtii | CrMge1 | 6 | AS form not detected | Au9.Cre08.g370450 |
| P. patens | PpMge1 | 6 | AS form not detected | Pp1s194_146V2 |
| PpMge2 | 6 | AS form not detected | Pp1s162_160V2 | |
| Arabidopsis | AtMge1 | 6 | AS form not detected | At5g55200 |
| AtMge2 | 5 | AS form not detected; resembles intron 2 retention | At4g26780 | |
| Populus trichocarpa | PoMge1 | 6 | AS form with exon 5 spliced out | POPTR_0011s09130 |
| PoMge2 | 6 | AS form with in-frame intron 2 retention | POPTR_0015s07680 | |
| Soybean | GmMge1 | 6 | AS form not detected | Glyma18g10120 |
| GmMge2 | 6 | AS form with in-frame intron 2 retention | Glyma02g46390 | |
| GmMge3 | 6 | AS form not detected | Glyma08g43430 | |
| O. sativa | OsMge1 | 6 | AS form not detected | LOC_Os09g11250 |
| OsMge2 | 6 | AS form with in-frame intron 2 retention | LOC_Os08g25090 | |
| S. bicolor | SbMge1 | 6 | AS form not detected | Sb02g019590 |
| SbMge2 | 6 | AS form with in-frame intron 2 retention | Sb07g017190 |
Mge2 Is Localized in Mitochondria
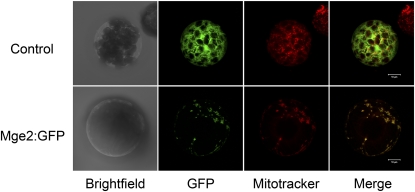
Previously, mtHsc70-1, mtHsc70-2, and Mge1 were identified in the mitochondrial proteome of Arabidopsis (Millar et al., 2001; Heazlewood et al., 2004), so these proteins are localized in mitochondria. However, the localization of Mge2 was not known. The presence of a predicted N-terminal mitochondrial targeting peptide (mTP) in Mge2 (Supplemental Fig. S1) suggests that this protein is also localized in mitochondria. This prediction was further confirmed by transient expression of the Mge2:GFP fusion protein in Arabidopsis protoplasts. We found that fusion proteins localized in the punctate foci overlapped those stained by the mitochondria-specific dye MitoTracker Orange, with the GFP control localized in the cytosol (Fig. 4).
Figure 4.
Subcellular localization of AtMge2. The plasmid containing Arabidopsis Mge2 fused to the 5′ end of GFP was introduced into Arabidopsis protoplasts to transiently express the fusion protein Mge2:GFP. Mitochondria of the transformed cells were stained with MitoTracker Orange. Transformation with plasmid containing only GFP was used as a control.
Mges Are Differentially Expressed
Duplicate genes usually undergo subfunctionalization at the regulatory and/or protein level (Zhang, 2003). Subfunctionalization at the regulatory level often leads to differential expression of homologous genes. A search of the public microarray database revealed the paralogs AtMge1 and AtMge2 to be responsive to UV-B or heat treatment. Mge1 was up-regulated by UV-B, and Mge2 was preferentially induced by heat treatment at 38°C (Supplemental Fig. S2).
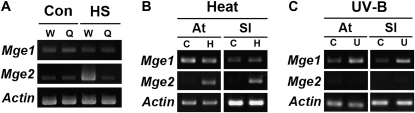
Analysis of the cis-elements in the promoters showed the Mge1 promoter containing putative UV-B-responsive elements, such as UV boxes and MREANAC13 (Safrany et al., 2008), and the Mge2 promoter containing one canonical heat shock element (HSE; Fig. 3). HSE is the conserved cis-element binding with trimerized HSF for transcription activation of HS genes in eukaryotes (Kroeger et al., 1993). We then tested whether the heat induction of Mge2 requires HsfA1s, the master regulators of the HS response in Arabidopsis (Liu et al., 2011; Yoshida et al., 2011). Semiquantitative reverse transcription (RT)-PCR showed no induction of Mge2 with heat treatment in the null mutant of HsfA1 genes, which indicates that Mge2 is under the control of HsfA1 (Fig. 5A). However, Mge1 still maintained the basal expression level regardless of HsfA1s. These results agree with the absence or presence of HSE in the promoter regions of the Mges.
Figure 5.
RT-PCR analyses of the differential expression of Mge genes. A, Mge1 and Mge2 were differentially regulated by HsfA1s in Arabidopsis. The RT-PCR products of Mge1 and Mge2 were derived from the wild type (W) and a quadruple KO mutant of HsfA1s (Q). RNA samples were purified from 5-d-old seedlings without (Con) or with (HS) heat treatment at 37°C for 1 h. B, RT-PCR products of Mge1 and Mge2 derived from Arabidopsis (At) and tomato (Sl) seedlings without (C) or with (H) heat treatment at 37°C for 1 h or 40°C for 2 h, respectively. C, RT-PCR products of Mge1 and Mge2 derived from Arabidopsis (At) and tomato (Sl) seedlings without (C) or with (U) UV-B treatment. Actin expression was used as a loading control.
To determine whether Mges are also differentially expressed in species other than Arabidopsis, we examined the transcript levels of Mge1 and Mge2 in tomato seedlings with heat and UV-B treatment. The mRNA expression of Mge1 was up-regulated in tomato by UV-B treatment, whereas that of Mge2 was induced by heat, as in Arabidopsis (Fig. 5, B and C). Therefore, the two paralogs had undergone a similar subfunctionalization process at the regulation level after the divergence of Arabidopsis and tomato.
Heterologously Expressed Arabidopsis Mge2 Confers Thermotolerance in the Heat-Sensitive E. coli grpE Mutant at Higher Temperature Than Does Mge1
Eukaryotic GrpE homologs, such as the distantly related Cge, can replace the function of E. coli GrpE (Deloche et al., 1997; Schroda et al., 2001). Because Arabidopsis Mges are differentially expressed under HS, we wondered whether Mge1 and Mge2 could replace the function of bacterial GrpE in different capacities. We performed a functional complementation assay with the E. coli grpE mutant (DA16; Ang and Georgopoulos, 1989). The mutant strain carries a point mutation that substitutes a conserved Gly at position 122 with Asp in GrpE (Harrison et al., 1997; Grimshaw et al., 2005). The mutant line exhibited a temperature-sensitive phenotype with growth abolished at 40°C (Fig. 6). We ligated Arabidopsis Mge1 and Mge2 cDNAs without the coding sequence of the predicted mTPs onto an expression vector for transformation into the mutant line to determine whether the temperature-sensitive phenotype could be rescued. The mTPs predicted as putative targeting peptides differed significantly in length in Mge1 and Mge2, with N-terminal 58 and 38 amino acids, respectively, by both TargetP (Emanuelsson et al., 2007) and MitoProt (Claros and Vincens, 1996). However, because the 20-amino acid difference is in a relatively conserved region, part of the functional region of the protein might be removed along with the stretch of peptide sequence. Therefore, we created two constructs for each gene, with one set of constructs having 38 amino acids from the N terminus removed from both Mge1 and Mge2 (designated the “long form”; Supplemental Fig. S1) and another set with 54 or 58 amino acids removed from the N terminus of both proteins (designated the “short form”).
Figure 6.
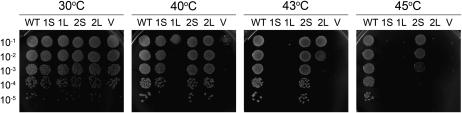
Differential complementation of the E. coli grpE mutant strain with Arabidopsis Mge1 and Mge2. Heat-sensitive E. coli mutant DA16 grpE was transformed with Arabidopsis Mges encoding long- and short-form Mge proteins with 38 and 58 putative mTPs removed from the N terminus, respectively. DA16 transformed with short Mge1, long Mge1, short Mge2, and long Mge2 are labeled 1S, 1L, 2S, and 2L, respectively. DA16 transformed with empty vector (V) was used as a negative control. The wild-type strain DA15 (WT) with an isogenic background to DA16 was transformed with the same empty vector to confer the same antibiotic resistance as the others and included as a comparison. Cell cultures were serially diluted and spotted onto LB plates and incubated overnight at 30°C, 40°C, 43°C, or 45°C.
At control temperature (30°C), all lines, whether Mge-transformed or empty vector-transformed DA16 lines, grew similar to the wild-type strain DA15. When grown at a higher temperature (40°C), DA16 with the empty vector showed the temperature-sensitive phenotype and failed to grow. However, except for the one with the long-form Mge1, the Mge-transformed lines were able to grow as normally as the wild type, which suggests that the Arabidopsis proteins and the prokaryotic homologs could function similarly. Nevertheless, at 43°C, the two Mge-transformed lines showed different behavior. DA16 transformed with Mge2, whether the short or long form, could at least in part complement the mutation, whereas Mge1 could not, and it showed abolished growth, like DA16 transformed with the empty vector (Fig. 6). The difference between Mge1 and Mge2 was not due to the level of the recombinant proteins expressed in E. coli, because the thermotolerance level was not significantly affected with or without the expression inducer anhydrotetracycline (data not shown). Therefore, Arabidopsis Mge2 could function at higher temperatures than Mge1.
Mge2 Specifically Confers Thermotolerance to Chronic HS in Arabidopsis
Because Mge2 was highly induced by elevated temperature, we wondered whether it was required for thermotolerance in Arabidopsis. We characterized two homozygous T-DNA KO lines for Mge2; the T-DNA KO line for Mge1 was not available during this work. The locations of the T-DNA insertion are shown in Figure 3 and were confirmed by PCR analysis of the mutants’ genomic DNA (Supplemental Fig. S3A). RT-PCR analysis confirmed that these mutants no longer generated transcripts like the wild type (Supplemental Fig. S3B), so the lines were null mutants. Under normal conditions, the mutants did not differ in phenotype from the wild type, which suggests that Mge2 is not essential for growth and development.
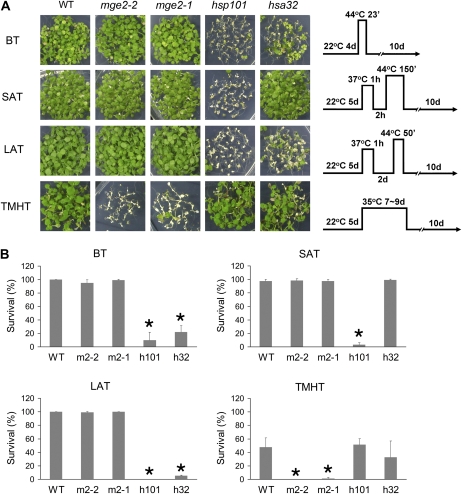
Seedlings of the mutants then underwent treatment with four different HS regimes to measure the capacity for basal thermotolerance (BT), short-term acquired thermotolerance (SAT), long-term acquired thermotolerance (LAT), and thermotolerance to moderately high temperature (TMHT), and their survival rates were measured. The first three assays involved an extremely high temperature, at 44°C, and the condition for TMHT mimicked chronic HS. The assay for SAT differed from those of LAT in recovery time between the acclimation treatment and acute HS challenge (Fig. 7A). Previously, we showed that the HS-associated 32-kD protein (Hsa32) is required for LAT but not SAT, whereas the heat shock protein 101 (Hsp101) is required for both (Charng et al., 2006, 2007). Interestingly, the two KO mutants for Mge2, mge2-1 and mge2-2, were defective in TMHT but not the other three types of thermotolerance (Fig. 7). By contrast, the hsp101 and hsa32 mutants showed defective phenotypes in at least two of the assay conditions for BT, SAT, and LAT, without substantial alteration in TMHT (Fig. 7).
Figure 7.
Thermotolerance assays of Mge2 T-DNA KO lines. A, Phenotypes of Arabidopsis seedlings of the wild type (WT) and various T-DNA KO mutants after 10 d of recovery from the assays under four different HS regimes as schematically shown on the right of each row. Seedlings in the same row were grown on the same plate. B, Survival rates of seedlings after the assay treatments shown in A. The labels for mutants are mge2-1 (m2-1), mge2-2 (m2-2), hsp101 (h101), and hsa32 (h32). The bars present means ± sd of three replicates for each treatment (n ≥ 35 each). * P < 0.01 (versus wild-type plants by Student’s t test).
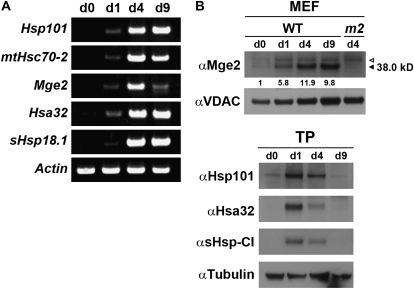
To further examine the function of Mge2 in TMHT, we assessed mRNA and protein expression patterns in seedling samples collected at different times during heat treatment. The mRNA expression of Mge2 and other HSP genes, including Hsp101 and Hsa32, was up-regulated throughout the chronic HS (Fig. 8A). To detect Arabidopsis Mge2 protein, we raised rabbit antiserum against Mge2 recombinant protein. The antibody recognized a protein band of about 38 kD in the wild type but not the mge2-2 mutant (Fig. 8B). The size determined by migration on SDS-PAGE was the same as for the short-form Mge2 expressed in E. coli described earlier, which suggests that the N-terminal 54 amino acids in the preprotein sequence of Mge2 is the mTP (Supplemental Fig. S1). With the TMHT assay, the Mge2 protein level gradually increased and remained high after 9 d (Fig. 8B). By contrast, the other HSP levels were high at day 1 but gradually decreased during treatment and were barely detectable at day 9.
Figure 8.
Transcript and protein profiles of Mge2 and other HSPs during TMHT treatment. A, Transcript levels of Mge2 and other HSPs during day (d) 0, 1, 4, and 9 of TMHT treatment detected by semiquantitative RT-PCR. Actin expression was used as a loading control. B, Total or mitochondria-enriched fraction proteins were extracted from the TMHT seedling samples in A for western-blot analyses of proteins. For Mge2, 65 μg of proteins from the mitochondria-enriched fraction (MEF) was loaded; for all other HSPs, 60 μg of total proteins (TP) was loaded. Voltage-dependent anion channel (VDAC) was used as a loading control for MEF protein. The band intensity of Mge2 was normalized to that of VDAC, with day 0 assigned as 1, and indicated below the corresponding Mge2 bands. The black arrowhead indicates the protein band of Mge2, and the white arrowhead indicates the nonspecific band. The molecular mass of Mge2 was estimated by molecular mass markers (data not shown). Tubulin was used as a loading control for total proteins. WT, Wild type; m2, mge2-2.
DISCUSSION
In this study, we show that in Arabidopsis, the two mitochondrial GrpE proteins arose from a recent WGD event, estimated to occur about 24 to 75 million years ago (Lynch and Conery, 2000; Simillion et al., 2002; Blanc et al., 2003). This finding is consistent with our estimated divergence time for the Mges, about 57 million years ago (Table I). Intriguingly, our phylogenetic analysis revealed similar independent duplication events for the Mge genes in other higher plants (Fig. 2). Therefore, these Mge paralogs also likely emerged from recent WGD events, which were found to occur in many plant species around the Cretaceous-Tertiary boundary about 65 million years ago (Fawcett et al., 2009). However, we could not exclude that segmental duplication instead of WGD is the cause for some species. Nevertheless, this coincidence implies that selection acted to preserve both copies of the Mge genes in these distantly related species.
Preservation of the two Mge genes could have conferred tolerance to a wide range of environmental conditions. However, retaining both Mge copies might cause dosage imbalance, because overexpression of GrpE caused a defect in the DnaK chaperone system in E. coli (Sugimoto et al., 2008). Subfunctionalization at the regulatory level could alleviate the problem of dosage imbalance (Veitia et al., 2008). The RT-PCR and microarray data revealed Arabidopsis Mges to be differentially expressed under UV-B and HS, which is consistent with the presence of associated cis-elements in the promoters of corresponding genes (Fig. 3). We suspected that for certain eudicots, the promoter of the ancestral Mge contained all of the cis-elements required for both types of stress responses, because the tomato Mges were differentially expressed, similar to the Arabidopsis genes (Fig. 5B). As time elapsed from gene duplication, the cis-elements were differentially lost in the two promoters, probably to optimize response to HS or UV-B. This inference agrees well with the degenerative complementation model of evolution in regulatory elements after gene duplication (Papp et al., 2003; Li et al., 2005) and with the rapid expression divergence of duplicate genes in response to external changes in Arabidopsis (Ha et al., 2007). Here, we provide genetic evidence that Mge2 is involved in thermotolerance, specifically TMHT. However, the role of Mge1 in UV-B stress tolerance remains to be elucidated.
Besides expression divergence, the functions of Mge1 and Mge2 proteins were also subfunctionalized. Mge1 but not Mge2 is present in the mitochondrial proteome under a non-HS condition in cultured cells (Heazlewood et al., 2004), which suggests that Mge1 is responsible for crucial housekeeping functions such as protein import (Truscott et al., 2003). We cannot exclude the role of Mge1 in HS tolerance. Mge1 may be required for basal or acquired thermotolerance, in that it could be involved in protein folding during recovery at ambient temperature. Unfortunately, the T-DNA KO line for Mge1 was not available at the time of the study, which might be due to it being essential for viability like its counterparts in yeast and E. coli (Ang and Georgopoulos, 1989; Laloraya et al., 1994). The functional complementation assay of the E. coli grpE mutant showed the two Arabidopsis Mges with different temperature ranges required for their functions, which is consistent with results in planta. However, that Mge1 can complement the heat-sensitive phenotype at 40°C in the E. coli DA16 mutant (Fig. 6) but is not sufficient to sustain growth at 35°C in Arabidopsis Mge2 KO mutants is intriguing. One reason may be that the thermostability or function of the protein is greatly affected by the milieu; that is, Mge1 might be less stable or less efficient at 35°C in mitochondria than in bacteria cells. Alternatively, the interaction between Mge1 and bacterial DnaK might be more efficient than that between Mge1 and mitochondrial Hsp70s at 35°C. In vitro experiments may help examine the latter possibility.
A closer examination of the genomic sequences of the duplicated Mges in Arabidopsis and comparison with their homologs in other plant species strongly suggest that the two Mges in Arabidopsis might have emerged from a single ancestral Mge that could produce transcripts with two AS forms (Table II). However, following duplication, both Arabidopsis Mge genes lost the ability to generate AS transcripts. Sequence alignment (Supplemental Fig. S1) revealed that the most distinctive difference between the Arabidopsis Mge1 and Mge2 protein sequences is a 30-amino acid stretch encoded by the putatively retained intron 2 of Mge2 (Fig. 3). This stretch is located in a highly diverse region near the N-terminal end of the long α-helix (Supplemental Fig. S1) from the crystal structure of E. coli GrpE (Harrison et al., 1997). The N-terminal 33 amino acids of E. coli GrpE corresponding to this region were found to be unstructured but involved in substrate release from the DnaK (Harrison et al., 1997; Brehmer et al., 2004). Given the conservation of these intron-derived peptide sequences, the intron retention may affect the cochaperone function of Mge. Further studies of the effect of the absence or presence of the in-frame intron 2 in Arabidopsis and other plant species with use of the Arabidopsis Mge2 KO lines, TMHT assay, and E. coli complementation assay established in this study should provide better insights. Of note, temperature-dependent alternative splicing generates Cge isoforms in Chlamydomonas (Willmund et al., 2007). However, the biological function of the AS forms is also not clear.
The observation that Mge2 is involved in the thermotolerance of chronic HS but not extreme HS is, to our knowledge, the first evidence for HSPs characterized to date. The role of Mge2 in TMHT agrees well with the expression induced by heat treatment at the transcript and protein levels. Mge2 might be required for importing or folding mitochondrial proteins under moderately high temperature. Of note, one recently identified mutant of Arabidopsis, hit2, which encodes a nuclear transport receptor, was found impaired in growth at moderately high temperature (37°C for 4 d) and defective in BT but not SAT or LAT (Wu et al., 2010). However, Hit2 is not responsive to heat treatment. By contrast, Hsp101 is essential for basal and acquired thermotolerance but not TMHT, which is consistent with previous findings (Queitsch et al., 2000). These observations indicate that plants use different components for diverse HS conditions. Interestingly, despite induction at the transcription level for all HSP genes tested under the TMHT condition, several HSPs, including Hsp101, could not be sustained at the protein level, whereas the level of Mge2 substantially increased with time (Fig. 8B). This phenomenon suggests that the HSPs were under translational or posttranslational control during prolonged exposure to moderately high temperature. Thus, inferring with the function of a gene under different HS conditions based on its transcript level is difficult. Investigating the differential regulation of HSPs at the translational or posttranslational level is needed to better understand the HS response.
The differential evolution of mitochondrial and chloroplastic GrpEs and Hsp70s in Arabidopsis is of interest. Mges arose from a recent WGD and mtHsc70s from an ancient duplication event, and vice versa for their chloroplastic counterparts (Table I). With the increasing number of plant genomes deciphered, comparison of the evolution of the organellar Hsp70 complexes would help determine how common this phenomenon is in plants. Results from this study can provide basic information regarding the evolution of the chaperone machineries. Of note, two copies of Mges were found in rodents and human (Naylor et al., 1998; Oliveira et al., 2006). How and why two Mges evolved in these organisms is unknown.
Recent progress in our knowledge of chloroplast Hsp70s and cochaperones has shown that the Hsp70 chaperone system is involved in protein translocation into chloroplasts (Shi and Theg, 2010; Su and Li, 2010), development and thermotolerance (Su and Li, 2008), and the biogenesis of thylakoid membranes by regulating the assembly state of VIPP1 (Liu et al., 2007). Studies of mitochondrial Hsp70 systems are beginning to reveal the importance of the system in plants. We are interested to know whether mtHsc70-1 and mtHsc70-2 also have different roles in thermotolerance as do their cochaperones, Mges.
CONCLUSION
The duplication of Mges occurred independently in many plant species, which suggests a common tendency in the evolution of the genes. Subfunctionalization of Arabidopsis Mges at both the protein and regulatory levels allowed Mge2 to specialize in the tolerance of long-term exposure to moderately high temperature. Our data suggest that the duplication and subfunctionalization of a highly conserved HSP such as Mge constituted a critical evolutionary adjustment in plants to cope with distinct types of HS and to better adapt to a changed environment.
MATERIALS AND METHODS
Phylogenetic Analysis
Sequences of Mge and Cge homologs were found by using the Arabidopsis (Arabidopsis thaliana) Mge2 as query sequence in a TBLASTN search of the databases Dana-Farber Cancer Institute Plant Gene Indices (http://compbio.dfci.harvard.edu/tgi/plant.html), National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/), and Phytozome (http://www.phytozome.net/). The sequences were aligned by use of the ClustalW program in MEGA5 (Tamura et al., 2011). The aligned sequences (for the sections of sequences used, see Supplemental Fig. S1) were then used to generate a phylogenetic tree by the neighbor-joining method.
The determination of Arabidopsis paralogous genes and their duplication events involved the online program Paralogons (http://wolfe.gen.tcd.ie/athal/dup). To estimate the time of divergence of the two duplicate genes, DnaSP was used to first determine the Ks of each gene pair (Librado and Rozas, 2009). Then, the resulting Ks was used in the formula T = Ks/2λ, with λ = 6.1 × 10−9 as the average whole-genome Ks of Arabidopsis (Lynch and Conery, 2000; Raes et al., 2003).
In Silico Analyses of Gene Expression and cis-Elements of the Promoter
Transcript expression data for Arabidopsis Mge1 and Mge2 under abiotic stress conditions were obtained from the public microarray database maintained in the Arabidopsis eFP Browser (http://www.bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi; Winter et al., 2007). Details for the data of HS and UV-B can be found in the browser by choosing the data source “abiotic stress.” The consensus sequences for UV-B regulatory elements and HSEs were obtained from Safrany et al. (2008) and Nover et al. (2001), respectively. Sequence about 1,100 bp upstream of the ATG site was considered the promoter region. The elements were found by using the search function of Vector NTI 9.1.0 (Invitrogen).
Plant Materials and Growth Condition
The Mge2 T-DNA insertion lines SALK_075614 (mge2-1) and SALK_082197 (mge2-2) were obtained from the Arabidopsis Biological Resource Center. The mutants are derived from ecotype Columbia (Alonso et al., 2003). Homozygous lines of the mutant allele were identified by PCR analysis as described (Charng et al., 2007). The T-DNA KO mutants of Hsp101, Hsa32, and HsfA1s were described previously (Liu et al., 2011). Seeds were sown on 0.5× Murashige and Skoog (MS) medium plates containing 0.1% or 1% Suc. The sown seeds were imbibed for 3 d at 4°C in the dark before being allowed to germinate and grow at 22°C with 16 h of light (120 μmol m−2 s−1).
Determining the Subcellular Localization of Mge2
The full-length coding sequence of Mge2 (GenBank accession no. NM_118812) was amplified by PCR from a cDNA template and cloned onto the entry vector pCR8/GW/TOPO (Invitrogen), then sequenced to confirm no missense or nonsense mutation. The cDNA was cloned onto pMDC83 (Curtis and Grossniklaus, 2003) by use of LR Clonase (Invitrogen) to generate a recombinant DNA encoding a recombinant protein of GFP fused to the C terminus of Mge2. The finished construct, driven by the cauliflower mosaic virus 35S promoter, was transiently expressed in Arabidopsis mesophyll cell protoplasts as described (Wu et al., 2009). The transformed protoplasts were then stained with the mitochondria-specific probe MitoTracker Orange (Invitrogen). The GFP and MitoTracker Orange signals were observed with a Zeiss LSM 510 Meta confocal microscope.
Heat and UV-B Treatments
Five-day-old Arabidopsis seedlings were subjected to heat treatment at 37°C for 1 h as described (Charng et al., 2006). Seven-day-old tomato (Solanum lycopersicum ‘Microtom’) seedlings sown in Magenta boxes containing 0.5× MS medium and 1% Suc were subjected to heat treatment at 40°C for 2 h.
For UV-B treatment, 7-d-old Arabidopsis or tomato seedlings were exposed to 150 mJ cm−2 UV-B by using an X1000 UV-B cross-linker (Spectronics).
Semiquantitative Analysis of mRNA
Seedling samples were collected and immediately incubated in liquid nitrogen after treatment. Total RNA was harvested by the Trizol method (Invitrogen) followed by chloroform purification. RNA was reverse transcribed into cDNA by the use of oligo(dT) primers and Moloney murine leukemia virus reverse transcriptase (Promega). PCR involved Ampliqon III Taq polymerase mix with specific primers as described (Charng et al., 2007). Primer sequences are shown in Supplemental Table S1.
Functional Complementation of a Heat-Sensitive Escherichia coli Mutant Containing Defective GrpE
Primers were designed to remove the putative mTPs predicted by TargetP version 1.0 (http://www.cbs.dtu.dk/services/TargetP/) and MitoProt (http://ihg.gsf.de/ihg/mitoprot.html) from Arabidopsis Mge1 and Mge2 protein sequences. The coding sequence corresponding to the first 54 to 58 or 38 amino acids from the N terminus was removed from both Mge1 and Mge2 as mitochondrial targeting sequences. The Mge1 and Mge2 coding sequences for both species were amplified by PCR, cloned onto pASK3 plus expression vector (IBA BioTAGnology), and transformed into DA16 cells (Coli Genetic Stock Center, Yale University). The transformed DA16 lines were grown in Luria-Bertani (LB) medium containing ampicillin (100 μg mL−1) at 30°C to about an optical density at 600 nm = 0.8. The liquid cultures then were 10-fold serially diluted, and 4 μL of each dilution was dropped onto an LB plate containing ampicillin. The plates were grown in a growth chamber at 30°C or at 40°C, 43°C, or 45°C in a water bath overnight. To confirm the identities of the expressed recombinant proteins, the protein bands induced by anhydrotetracycline underwent liquid chromatography-electrospray ionization-tandem mass spectrometry analysis after trypsin digestion, performed by the Proteomics Core Laboratory of the Institute of Plant and Microbial Biology/Agricultural Biotechnology Research Center, Academia Sinica.
Thermotolerance Assays
SAT, LAT, BT, and TMHT assays were as described (Liu et al., 2011) with slight modification. For the SAT assay, 5-d-old Arabidopsis seedlings were first acclimated with 37°C heat treatment for 1 h. After a 2-h recovery at 22°C, the seedlings then were challenged with acute HS at 44°C for 150 min. For the LAT assay, 5-d-old Arabidopsis seedlings were acclimated to 37°C heat treatment for 1 h and then underwent a 2-d recovery at 22°C. The seedlings then were challenged at 44°C for 50 min. For the BT assay, 4-d-old seedlings were challenged with 44°C for 23 min without prior acclimation. All phenotypes were assessed after a 10-d recovery at 22°C post-HS. For the TMHT assay, Arabidopsis seeds were sown on 0.5× MS medium plates containing 0.1% Suc. Five-day-old seedlings were then transferred to a growth chamber with temperature of 35°C ± 0.3°C during 16 h of light (110 μmol m−2 s−1) and 33.5°C ± 0.3°C during 8 h of darkness. The seedlings then recovered at 22°C for 10 d before phenotypes were assessed.
Immunoblotting
The full-length Arabidopsis Mge2 recombinant protein with a C-terminal tag of six His residues was produced and purified from E. coli cells as described (Charng et al., 2006). Immunization and serum collection were performed by a commercial service (LTK Biotechnology). The antibodies against Hsp101, Hsa32, sHsp-CI, and tubulin were as described (Charng et al., 2006; Chi et al., 2009). The total protein of plant samples was extracted as described (Charng et al., 2006). The mitochondria-enriched protein fraction was extracted with 30 mm MOPS (pH 7.5) buffer with 0.35 m mannitol and 2 mm EDTA. The lysate was first centrifuged at 4,500g for 5 min at 4°C to pellet the undesired crude fraction containing chloroplasts and other cell debris, then the supernatant was centrifuged at 16,000g for 15 min at 4°C. The resulting mitochondria-enriched protein fraction pellet was resuspended with Tris-HCl extraction buffer as for the extraction of total protein. The protein amount was measured by use of DC Protein Assay reagents (Bio-Rad) with bovine serum albumin used as a standard. Immunoblot analysis was performed as described (Charng et al., 2006; Chi et al., 2009).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Protein sequence alignment of Mge homologs.
Supplemental Figure S2. In silico analysis of the expression of Arabidopsis Mges.
Supplemental Figure S3. Molecular characterization of mge2-1 and mge2-2 T-DNA insertion lines.
Supplemental Table S1. List of primers used in this study.
Supplementary Material
Acknowledgments
We thank the Scientific Instrument Center of Academia Sinica for the use of the LSM 510 Meta NLO Duo Scan inverted confocal microscope and the assistance of Ms. Shu-Chen Shen; the DNA Sequencing Core Facility of the Institute of Biomedical Sciences of Academia Sinica for providing DNA sequencing services; Dr. Duan-nan Wen for protein identification by liquid chromatography-electrospray ionization-tandem mass spectrometry; the Plant Tech Core Facility of the Agricultural Biotechnology Research Center of Academia Sinica for performing the transient expression technique; Dr. Daryi Wang for instruction and assistance in the calculations pertaining to the estimation of gene divergence time; Ms. Hsiu-ting Liao for assistance in experiments; Drs. Tzyy-jen Chiou and Kuo-chen Yeh for assisting in molecular cloning with pMDC83 and pASK3 plus, respectively; Drs. Kuo-chen Yeh and Daryi Wang for critically reading the manuscript and comments; Dr. Hong-Yong Fu for lending a UV-B cross-linker; and Ms. Laura Smales for English editing.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Ang D, Georgopoulos C. (1989) The heat-shock-regulated grpE gene of Escherichia coli is required for bacterial growth at all temperatures but is dispensable in certain mutant backgrounds. J Bacteriol 171: 2748–2755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Hokamp K, Wolfe KH. (2003) A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res 13: 137–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolliger L, Deloche O, Glick BS, Georgopoulos C, Jenö P, Kronidou N, Horst M, Morishima N, Schatz G. (1994) A mitochondrial homolog of bacterial GrpE interacts with mitochondrial hsp70 and is essential for viability. EMBO J 13: 1998–2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehmer D, Gässler C, Rist W, Mayer MP, Bukau B. (2004) Influence of GrpE on DnaK-substrate interactions. J Biol Chem 279: 27957–27964 [DOI] [PubMed] [Google Scholar]
- Charng YY, Liu HC, Liu NY, Chi WT, Wang CN, Chang SH, Wang TT. (2007) A heat-inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in Arabidopsis. Plant Physiol 143: 251–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charng YY, Liu HC, Liu NY, Hsu FC, Ko SS. (2006) Arabidopsis Hsa32, a novel heat shock protein, is essential for acquired thermotolerance during long recovery after acclimation. Plant Physiol 140: 1297–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi WT, Fung RWM, Liu HC, Hsu CC, Charng YY. (2009) Temperature-induced lipocalin is required for basal and acquired thermotolerance in Arabidopsis. Plant Cell Environ 32: 917–927 [DOI] [PubMed] [Google Scholar]
- Claros MG, Vincens P. (1996) Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem 241: 779–786 [DOI] [PubMed] [Google Scholar]
- Conant GC, Wolfe KH. (2008) Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet 9: 938–950 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. (2003) A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deloche O, Kelley WL, Georgopoulos C. (1997) Structure-function analyses of the Ssc1p, Mdj1p, and Mge1p Saccharomyces cerevisiae mitochondrial proteins in Escherichia coli. J Bacteriol 179: 6066–6075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H. (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc 2: 953–971 [DOI] [PubMed] [Google Scholar]
- Fawcett JA, Maere S, Van de Peer Y. (2009) Plants with double genomes might have had a better chance to survive the Cretaceous-Tertiary extinction event. Proc Natl Acad Sci USA 106: 5737–5742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimshaw JPA, Siegenthaler RK, Züger S, Schönfeld H-J, Z’graggen BR, Christen P. (2005) The heat-sensitive Escherichia coli grpE280 phenotype: impaired interaction of GrpE(G122D) with DnaK. J Mol Biol 353: 888–896 [DOI] [PubMed] [Google Scholar]
- Ha M, Li W-H, Chen ZJ. (2007) External factors accelerate expression divergence between duplicate genes. Trends Genet 23: 162–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison CJ, Hayer-Hartl M, Di Liberto M, Hartl F-U, Kuriyan J. (1997) Crystal structure of the nucleotide exchange factor GrpE bound to the ATPase domain of the molecular chaperone DnaK. Science 276: 431–435 [DOI] [PubMed] [Google Scholar]
- Heazlewood JL, Tonti-Filippini JS, Gout AM, Day DA, Whelan J, Millar AH. (2004) Experimental analysis of the Arabidopsis mitochondrial proteome highlights signaling and regulatory components, provides assessment of targeting prediction programs, and indicates plant-specific mitochondrial proteins. Plant Cell 16: 241–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda E, Yoshida S, Mitsuzawa H, Uno I, Toh-e A. (1994) YGE1 is a yeast homologue of Escherichia coli grpE and is required for maintenance of mitochondrial functions. FEBS Lett 339: 265–268 [DOI] [PubMed] [Google Scholar]
- Katiyar-Agarwal S, Agarwal M, Grover A. (2003) Heat-tolerant basmati rice engineered by over-expression of hsp101. Plant Mol Biol 51: 677–686 [DOI] [PubMed] [Google Scholar]
- Kotak S, Larkindale J, Lee U, von Koskull-Döring P, Vierling E, Scharf K-D. (2007) Complexity of the heat stress response in plants. Curr Opin Plant Biol 10: 310–316 [DOI] [PubMed] [Google Scholar]
- Kroeger PE, Sarge KD, Morimoto RI. (1993) Mouse heat shock transcription factors 1 and 2 prefer a trimeric binding site but interact differently with the HSP70 heat shock element. Mol Cell Biol 13: 3370–3383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloraya S, Dekker PJ, Voos W, Craig EA, Pfanner N. (1995) Mitochondrial GrpE modulates the function of matrix Hsp70 in translocation and maturation of preproteins. Mol Cell Biol 15: 7098–7105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloraya S, Gambill BD, Craig EA. (1994) A role for a eukaryotic GrpE-related protein, Mge1p, in protein translocation. Proc Natl Acad Sci USA 91: 6481–6485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W-H, Yang J, Gu X. (2005) Expression divergence between duplicate genes. Trends Genet 21: 602–607 [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J. (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452 [DOI] [PubMed] [Google Scholar]
- Liu C, Willmund F, Golecki JR, Cacace S, Hess B, Markert C, Schroda M. (2007) The chloroplast HSP70B-CDJ2-CGE1 chaperones catalyse assembly and disassembly of VIPP1 oligomers in Chlamydomonas. Plant J 50: 265–277 [DOI] [PubMed] [Google Scholar]
- Liu HC, Liao HT, Charng YY. (2011) The role of class A1 heat shock factors (HSFA1s) in response to heat and other stresses in Arabidopsis. Plant Cell Environ 34: 738–751 [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. (2000) The evolutionary fate and consequences of duplicate genes. Science 290: 1151–1155 [DOI] [PubMed] [Google Scholar]
- Mayer MP, Bukau B. (2005) Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci 62: 670–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiri D, Breiman A. (2009) Arabidopsis ROF1 (FKBP62) modulates thermotolerance by interacting with HSP90.1 and affecting the accumulation of HsfA2-regulated sHSPs. Plant J 59: 387–399 [DOI] [PubMed] [Google Scholar]
- Meiri D, Tazat K, Cohen-Peer R, Farchi-Pisanty O, Aviezer-Hagai K, Avni A, Breiman A. (2010) Involvement of Arabidopsis ROF2 (FKBP65) in thermotolerance. Plant Mol Biol 72: 191–203 [DOI] [PubMed] [Google Scholar]
- Millar AH, Sweetlove LJ, Giegé P, Leaver CJ. (2001) Analysis of the Arabidopsis mitochondrial proteome. Plant Physiol 127: 1711–1727 [PMC free article] [PubMed] [Google Scholar]
- Naylor DJ, Stines AP, Hoogenraad NJ, Høj PB. (1998) Evidence for the existence of distinct mammalian cytosolic, microsomal, and two mitochondrial GrpE-like proteins, the co-chaperones of specific Hsp70 members. J Biol Chem 273: 21169–21177 [DOI] [PubMed] [Google Scholar]
- Nover L, Bharti K, Döring P, Mishra SK, Ganguli A, Scharf KD. (2001) Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell Stress Chaperones 6: 177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira CL, Borges JC, Torriani IL, Ramos CH. (2006) Low resolution structure and stability studies of human GrpE#2, a mitochondrial nucleotide exchange factor. Arch Biochem Biophys 449: 77–86 [DOI] [PubMed] [Google Scholar]
- Padidam M, Reddy VS, Beachy RN, Fauquet CM. (1999) Molecular characterization of a plant mitochondrial chaperone GrpE. Plant Mol Biol 39: 871–881 [DOI] [PubMed] [Google Scholar]
- Paek KH, Walker GC. (1987) Escherichia coli dnaK null mutants are inviable at high temperature. J Bacteriol 169: 283–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp B, Pál C, Hurst LD. (2003) Evolution of cis-regulatory elements in duplicated genes of yeast. Trends Genet 19: 417–422 [DOI] [PubMed] [Google Scholar]
- Queitsch C, Hong SW, Vierling E, Lindquist S. (2000) Heat shock protein 101 plays a crucial role in thermotolerance in Arabidopsis. Plant Cell 12: 479–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raes J, Vandepoele K, Simillion C, Saeys Y, Van de Peer Y. (2003) Investigating ancient duplication events in the Arabidopsis genome. J Struct Funct Genomics 3: 117–129 [PubMed] [Google Scholar]
- Safrany J, Haasz V, Mate Z, Ciolfi A, Feher B, Oravecz A, Stec A, Dallmann G, Morelli G, Ulm R, et al. (2008) Identification of a novel cis-regulatory element for UV-B-induced transcription in Arabidopsis. Plant J 54: 402–414 [DOI] [PubMed] [Google Scholar]
- Schroda M, Vallon O, Whitelegge JP, Beck CF, Wollman FA. (2001) The chloroplastic GrpE homolog of Chlamydomonas: two isoforms generated by differential splicing. Plant Cell 13: 2823–2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L-X, Theg SM. (2010) A stromal heat shock protein 70 system functions in protein import into chloroplasts in the moss Physcomitrella patens. Plant Cell 22: 205–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou H, Bordallo P, Fan J-B, Yeakley JM, Bibikova M, Sheen J, Wang K. (2004) Expression of an active tobacco mitogen-activated protein kinase kinase kinase enhances freezing tolerance in transgenic maize. Proc Natl Acad Sci USA 101: 3298–3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simillion C, Vandepoele K, Van Montagu MCE, Zabeau M, Van de Peer Y. (2002) The hidden duplication past of Arabidopsis thaliana. Proc Natl Acad Sci USA 99: 13627–13632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su P-H, Li HM. (2008) Arabidopsis stromal 70-kD heat shock proteins are essential for plant development and important for thermotolerance of germinating seeds. Plant Physiol 146: 1231–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su P-H, Li HM. (2010) Stromal Hsp70 is important for protein translocation into pea and Arabidopsis chloroplasts. Plant Cell 22: 1516–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto S, Saruwatari K, Higashi C, Sonomoto K. (2008) The proper ratio of GrpE to DnaK is important for protein quality control by the DnaK-DnaJ-GrpE chaperone system and for cell division. Microbiology 154: 1876–1885 [DOI] [PubMed] [Google Scholar]
- Sung DY, Vierling E, Guy CL. (2001) Comprehensive expression profile analysis of the Arabidopsis Hsp70 gene family. Plant Physiol 126: 789–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truscott KN, Brandner K, Pfanner N. (2003) Mechanisms of protein import into mitochondria. Curr Biol 13: R326–R337 [DOI] [PubMed] [Google Scholar]
- Veitia RA, Bottani S, Birchler JA. (2008) Cellular reactions to gene dosage imbalance: genomic, transcriptomic and proteomic effects. Trends Genet 24: 390–397 [DOI] [PubMed] [Google Scholar]
- Vierling E. (1991) The roles of heat shock proteins in plants. Annu Rev Plant Physiol Plant Mol Biol 42: 579–620 [Google Scholar]
- von Koskull-Döring P, Scharf K-D, Nover L. (2007) The diversity of plant heat stress transcription factors. Trends Plant Sci 12: 452–457 [DOI] [PubMed] [Google Scholar]
- Willmund F, Mühlhaus T, Wojciechowska M, Schroda M. (2007) The NH2-terminal domain of the chloroplast GrpE homolog CGE1 is required for dimerization and cochaperone function in vivo. J Biol Chem 282: 11317–11328 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F-H, Shen S-C, Lee L-Y, Lee S-H, Chan M-T, Lin C-S. (2009) Tape-Arabidopsis Sandwich: a simpler Arabidopsis protoplast isolation method. Plant Methods 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S-J, Wang L-C, Yeh C-H, Lu C-A, Wu S-J. (2010) Isolation and characterization of the Arabidopsis heat-intolerant 2 (hit2) mutant reveal the essential role of the nuclear export receptor EXPORTIN1A (XPO1A) in plant heat tolerance. New Phytol 186: 833–842 [DOI] [PubMed] [Google Scholar]
- Yoshida T, Ohama N, Nakajima J, Kidokoro S, Mizoi J, Nakashima K, Maruyama K, Kim J-M, Seki M, Todaka D, et al. (2011) Arabidopsis HsfA1 transcription factors function as the main positive regulators in heat shock-responsive gene expression. Mol Genet Genomics 286: 321–332 [DOI] [PubMed] [Google Scholar]
- Zhang J. (2003) Evolution by gene duplication: an update. Trends Ecol Evol 18: 292–298 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.