Abstract
Plants exploit ubiquitination to modulate the proteome with the final aim to ensure environmental adaptation and developmental plasticity. Ubiquitination targets are specifically driven to degradation through the action of E3 ubiquitin ligases. Genetic analyses have indicated wide functions of ubiquitination in plant life; nevertheless, despite the large number of predicted E3s, only a few of them have been characterized so far, and only a few ubiquitination targets are known. In this work, we characterized durum wheat (Triticum durum) RING Finger1 (TdRF1) as a durum wheat nuclear ubiquitin ligase. Moreover, its barley (Hordeum vulgare) homolog was shown to protect cells from dehydration stress. A protein network interacting with TdRF1 has been defined. The transcription factor WHEAT BEL1-TYPE HOMEODOMAIN1 (WBLH1) was degraded in a TdRF1-dependent manner through the 26S proteasome in vivo, the mitogen-activated protein kinase TdWNK5 [for Triticum durum WITH NO LYSINE (K)5] was able to phosphorylate TdRF1 in vitro, and the RING-finger protein WHEAT VIVIPAROUS-INTERACTING PROTEIN2 (WVIP2) was shown to have a strong E3 ligase activity. The genes coding for the TdRF1 interactors were all responsive to cold and/or dehydration stress, and a negative regulative function in dehydration tolerance was observed for the barley homolog of WVIP2. A role in the control of plant development was previously known, or predictable based on homology, for wheat BEL1-type homeodomain1(WBLH1). Thus, TdRF1 E3 ligase might act regulating the response to abiotic stress and remodeling plant development in response to environmental constraints.
Plants are sessile organisms that must cope with their surrounding environments; thus, stresses such as drought, soil salinity, or extreme temperatures represent common events during their life cycle. These unfavorable conditions limit plant growth and development and strongly threat crop productivity, preventing the expression of the full genetic potential of cultivars. At the molecular level, upon plant exposition to stress, a plethora of genes are up- or down-regulated to reorganize the metabolic fluxes leading to plant adaptation as a final result. These responses provide plants with the capacity to avoid or mitigate the effects of stresses. Besides gene transcriptional changes, cells have evolved other types of molecular responses, such as posttranslational modifications of key proteins (Mazzucotelli et al., 2008). Among them, protein phosphorylation and ubiquitination are key components of the molecular response to different types of abiotic stresses. These modifications are implicated in signal transduction, positive and negative regulation of the response, and cross-connections between different signaling pathways and stress responses. The rapid and precise activation or inhibition of regulatory proteins is essential for the coordination of an array of physiological responses to cope with different stresses and to ensure a tightly controlled regulation of cellular changes induced by stress intensity and duration.
During ubiquitination, proteins are labeled with ubiquitin units through the sequential action of three enzymes: a ubiquitin-activating enzyme (E1) activates the ubiquitin moiety in an ATP-dependent reaction, a ubiquitin conjugating enzyme (E2) conjugates the ubiquitin, and finally the ubiquitin ligase enzymes (E3) specifically recruit the target proteins to covalently attach ubiquitin (Hershko and Ciechanover, 1998). Polyubiquitinated proteins are then targeted to the proteasome, where deubiquitinase activities remove the polyubiquitin chain and the 20S core proteases degrade the target protein (Smalle and Vierstra, 2004). The ubiquitination pathway is highly conserved in all eukaryotic cells (Dye and Schulman, 2007; Hunter, 2007). In plants, protein degradation mediated by ubiquitination regulates nearly every cellular process, including abiotic or biotic stress responses, circadian rhythms, cell cycle, and cell differentiation (Dreher and Callis, 2007; Trujillo and Shirasu, 2010; Lee and Kim, 2011).
In this ubiquitin-dependent labeling, the specificity of the system is ensured by E3 ligase enzymes. Different types of E3 ubiquitin ligases have been described, from E3s acting as monomers to E3 multisubunit complexes. Genomic analyses revealed that more than 1,400 Arabidopsis (Arabidopsis thaliana) genes encode E3 enzymes or subunits. Among them, the Really Interesting New Gene (RING)-finger proteins and the Skp, Cullin, F-box containing (SCF) protein-complex subunits are predominant (Smalle and Vierstra, 2004; Mazzucotelli et al., 2006). Several studies have shown that E3-mediated protein degradation plays a key role in plants, controlling various cellular processes, including hormonal responses (Zhang et al., 2007; Jurado et al., 2010), light response (del Pozo et al., 2002; Wang and Deng, 2003), circadian rhythm (Nelson et al., 2000), flowering process (Wang et al., 2003), pathogen resistance (Devoto et al., 2003; Li et al., 2011), sugar response (Huang et al., 2010), intracellular trafficking, vacuole biogenesis (Isono et al., 2010), and cell division (del Pozo et al., 2006; Jurado et al., 2008, 2010), among others.
RING E3s have been found to regulate specific molecular responses by targeting critical elements. For instance, in the response to abiotic stress, the Arabidopsis RING ubiquitin ligase DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN2A (DREB2A)-Interacting Protein1 (DRIP1) acts as negative regulator of drought response mediating the ubiquitination and the degradation of DREB2A (Qin et al., 2008). The RING-finger protein high expression of osmotically responsive gene (HOS1) is induced during cold exposure to exert a negative control of the stress response (Ishitani et al., 1998) through the ubiquitination of the key transcription factor Inducer of CRT/DRE-binding factor Expression1 (ICE1; Dong et al., 2006). In addition, there are several examples of RING E3 ligases with a pivotal role in the regulation of stress response, as well as in processes related to growth and development, thus ensuring the connections among different pathways. For instance, Delayed Seed Germination1 (OsDSG1) participates both in stress response and seed germination (Park et al., 2010). BTH-induced RING finger protein1 (OsBIRF1) is a rice (Oryza sativa) RING protein with pleiotropic effects on growth and defense response against multiple abiotic and biotic stresses (Liu et al., 2008).
The E3 ligase activities are, themselves, finely regulated through several mechanisms: changes in the expression of the corresponding mRNAs (Mazzucotelli et al., 2006), induction of multiple splice variants (Gingerich et al., 2005; Mastrangelo et al., 2005), micro-RNA-mediated gene silencing (Sunkar et al., 2007), and phosphorylation status (Pedmale and Liscum, 2007) or ubiquitin-mediated degradation (Jurado et al., 2010).
Genetic analyses have indicated the wide functions of ubiquitination in plant life; nevertheless, despite the large number of predicted E3s, only few of them have been characterized so far for their possible functions, and only few ubiquitination targets are known (Liu et al., 2010). Indeed, identification of E3 target proteins remains the most intriguing and poorly addressed aspect of the ubiquitination pathway. This work demonstrates that the durum wheat (Triticum durum ‘Ofanto’) protein Triticum durum RING Finger1 (TdRF1; formerly 6g2 in Mastrangelo et al., 2005) is an E3 ubiquitin ligase that protects cells from dehydration stress. Then, a network of proteins interacting with TdRF1 and encoded by stress-responsive genes was defined: The mitogen-activated protein (MAP) kinase TdWNK5 (for Triticum durum WITH NO LYSINE5) was able to phosphorylate TdRF1 in vitro, the transcription factor WHEAT BEL1-TYPE HOMEODOMAIN1 (WBLH1) was degraded in a TdRF1-dependent manner through the 26S proteasome in vivo, and the RING-finger protein WHEAT VIVIPAROUS-INTERACTING PROTEIN2 (WVIP2) was shown to have a strong E3 ligase activity.
RESULTS
TdRF1 Is a Functional E3 Ubiquitin Ligase
TdRF1 (formerly 6g2; Mastrangelo et al., 2005) is a 429-amino-acid protein with a predicted molecular mass of 46 kD. The corresponding mRNA (GenBank accession no. AY465427.2) was previously described as up-regulated in response to cold and drought stress (Mastrangelo et al., 2005). A predictive protein analysis through ExPASy Proteomics Server (http://www.ebi.ac.uk/Tools/InterProScan/) and SMART (http://smart.embl-heidelberg.de/) Web sites indicated that TdRF1 contains a C3H2C3-type RING-finger domain (amino acids 281–334), a putative N-myristoylation motif (http://mendel.imp.ac.at/myristate/SUPLpredictor.htm), and a putative phosphorylation site at the C terminus (http://phosphat.mpimp-golm.mpg.de/index.html; Fig. 1).
Figure 1.
Deduced amino acid sequence of TdRF1. The highlighted gray N-terminal region corresponds to the N-myristoylation motive, the underlined region is the RING-finger domain, arrows indicate the His and Cys residues coordinating Zn atoms, and the bold underlined C-terminal region corresponds to phosphorylation site.
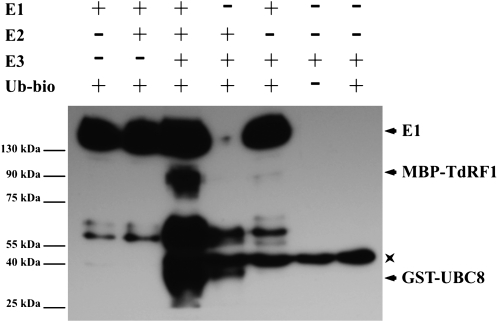
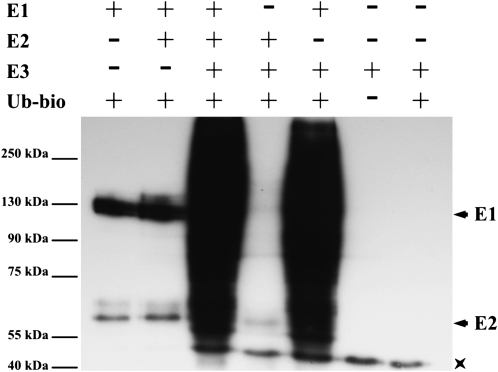
Since the RING-finger domain is a protein-protein interaction domain typical of many E3 ligase enzymes involved in protein ubiquitination (Matsuda et al., 2001), we tested whether TdRF1 has activity of ubiquitin ligase. The TdRF1 coding sequence was fused to the gene of the maltose binding protein (MBP) and expressed in Escherichia coli. The MBP-TdRF1 fusion protein was purified by affinity chromatography from the crude extract and used for in vitro ubiquitination assays with the yeast (Saccharomyces cerevisiae) E1 enzyme, the Arabidopsis E2 enzyme UBC8 (for ubiquitin conjugating) fused with glutathione S-transferase (GST), GST-UBC8, and biotinylated ubiquitin. A strong TdRF1-dependent ubiquitination signal with an apparent Mr corresponding to MBP-TdRF1 was detected as well as a signal corresponding to the ubiquitination of the GST-UBC8 (Fig. 2, lane 3). These results indicate that TdRF1 underwent autoubiquitination. Unexpectedly, TdRF1 showed a weak ubiquitin ligase activity even when mixed with the E1 enzyme only (Fig. 2, lane 5), probably due to a residual E2 activity copurified with the E1, which has a eukaryotic cell origin. A similar behavior was shown for the Arabidopsis PUB17 E3 ligase (Yang et al., 2006). Taken together, these results confirm that TdRF1 is a functional E3 ubiquitin ligase enzyme in vitro.
Figure 2.
Ubiquitination assay on TdRF1. MBP-TdRF1 fusion protein was assayed for E3 activity in the presence of the human activating enzyme (E1), the Arabidopsis conjugating enzyme UBC8 fused to GST (E2), and biotinylated ubiquitin (Ub). Samples were resolved on 10% SDS-PAGE and then blotted. Streptavidine-horseradish peroxidase was used to detect biotin-Ub. The + and − indicate the presence and the absence, respectively, of the corresponding protein in the reaction mix. The asterisk indicates a band copurified with MBP-TdRF1 and cross-reacting with streptavidine. MBP-TdRF1 and MBP-TdRF1 together with biotin-Ub were used as control.
TdRF1 Is a Nuclear-Localized E3 Ligase
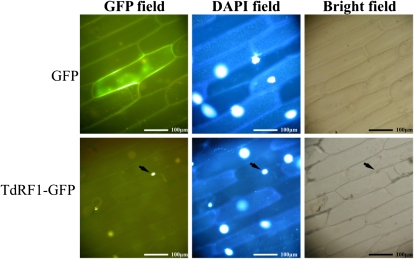
When the protein sequence of TdRF1 was analyzed with the WoLFPSORT program (a Web tool for the prediction of protein subcellular localization; http://wolfpsort.org/), a nuclear localization was suggested. To verify this prediction, TdRF1 was subcloned in frame with the gene of the GFP into an expression vector under the control of the Cauliflower mosaic virus 35S promoter. The construct was then introduced into onion (Allium cepa) epidermal cells through particle bombardment, and the subcellular localization of TdRF1-GFP was visualized by an epifluorescence microscope. Onion cells transformed with the vector expressing GFP alone, as a control, showed green fluorescence throughout the cell (Fig. 3, GFP panels). By contrast, onion cells expressing TdRF1-GFP showed the green fluorescence localized only in the nucleus, indicating the nuclear localization of TdRF1 protein (Fig. 3, TdRF1-GFP panels).
Figure 3.
Subcellular localization of TdRF1 by GFP fusion expression in onion epidermal cells. Onion epidermal cells were visualized by an epifluorescence microscope after 18 h of incubation in the dark. GFP, DAPI, and bright fields are shown for GFP alone and TdRF1-GFP fusion protein. In each of the bottom panels, the arrow indicates the nucleus of a cell transformed with TdRF1-GFP. Bar = 100 μm. [See online article for color version of this figure.]
Identification of TdRF1 Protein Interactors
The activity of the E3 ubiquitin ligases depends on their ability to interact with other proteins of the ubiquitination machinery and to recruit the target protein(s). Therefore, the identification of these components and targets is an essential step toward the understanding of the role of each E3 ubiquitin ligase. In this work, we carried out a yeast two-hybrid screening to identify TdRF1-interacting proteins.
A wheat cDNA expression library was developed using mRNA from durum wheat seedlings exposed to 3°C for 6 h, the same condition in which the TdRF1 transcript was previously isolated (Mastrangelo et al., 2005). The screening of 8 × 106 yeast clones led to the identification of three different clones. They did not show any interaction in the autoactivation assay. The putative interactors were subsequently confirmed by retransformation assay. These clones were sequenced and identified based on sequence similarity searches with BLASTN or BLASTX software at National Center for Biotechnology Information (NCBI) and The Institute for Genomic Research (TIGR) databases. The following proteins were identified: (1) WBLH1, a previously described wheat homeodomain protein (Mizumoto et al., 2011); (2) WVIP2 (Wilson et al., 2005), a protein sharing 93% amino acid similarity with the wild oat (Avena fatua) Viviparus1 Interacting Protein2 (VIP2; Jones et al., 2000); and (3) TdWNK5, a protein with 75% amino acid similarity to Arabidopsis With No Lys [K] 5 (AtWNK5), a member of the Arabidopsis WNK family of MAP kinases (Wang et al., 2008).
According to the sequence information reported in TIGR and NCBI databases, the cDNA sequences recovered from the yeast two-hybrid screening were not full length. Eight amino acids at the N terminus on a total length of 435 were missed in WVIP2; WBLH1 was 306 amino acids shorter at the N terminus on a total length of 771; TdWNK5 was both N- and C-terminal truncated with 11 and 85 amino acids missing, respectively, on a total length of 641. Nevertheless, as verified by in silico analysis (Supplemental Figs. S1–S3), the isolated WVIP2 and TdWNK5 sequences harbor all the functional and structural domains typical of the respective encoded proteins. Even the short sequence of WBLH1 harbors the complete coiled coil domain and the homeodomain typical of the encoded protein according to Müller et al. (2001). Thus, on the basis of these observations, all the subsequent experiments were carried out with the proteins encoded by the fragments recovered through the yeast two-hybrid screening and characterized by the following molecular masses: 50 kD for WBLH1, 61 kD for TdWNK5, and 47 kD for WVIP2.
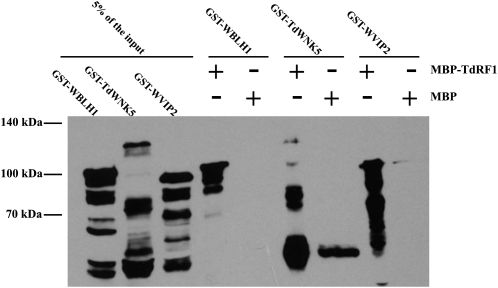
To further verify the interaction between TdRF1 and the putative interactors, pull-down experiments were performed with the following recombinant proteins: MBP-TdRF1, GST-WBLH1, GST-WVIP2, and GST-TdWNK5. MBP-TdRF1 protein was expressed in E. coli and purified with amylose beads. Then, MBP-TdRF1 beads were incubated with total protein extract from E. coli cells expressing GST-WBLH1, GST-WVIP2, or GST-TdWNK5. After several washing steps, pulled-down proteins were resolved by SDS-PAGE and immunoblotted with an anti-GST antibody. As shown in Figure 4, GST-WBLH1, GST-TdWNK5, and GST-WVIP2 were interacting with MBP-TdRF1, while no, or much less, interactions were observed when pulldown was carried out with MBP alone bound to the beads. These findings demonstrate the physical interaction between the MBP-TdRF1 E3 ligase and the three proteins identified after two-hybrid screening.
Figure 4.
In vitro pull-down assays of MBP-TdRF1 with GST-WBLH1, GST-TdWNK5, and GST-WVIP2 fusion proteins. MBP-TdRF1 fusion protein was used as a bait to pull down the putative interacting proteins expressed in E. coli cells. MBP protein was used as a bait in the negative control experiment. Samples were resolved on 10% SDS-PAGE, and then prey proteins were detected by immunoblotting with an anti-GST. An amount of protein corresponding to 5% of the total protein extract used as input to pull down the interactors was loaded as control. The + and − indicate the presence and the absence, respectively, of the corresponding protein in the reaction mix.
Expression Analysis of the Genes upon Different Stress Treatments
Since TdRF1 was originally described as up-regulated upon cold and drought treatments (Mastrangelo et al., 2005), a quantitative reverse transcription (qRT)-PCR expression analysis was carried out on genes encoding TdRF1 interactors to assess if their expression is also responsive to cold and dehydration. Moreover, given the critical role of abscisic acid (ABA) in the abiotic stress signaling, a qRT-PCR experiment upon ABA treatment was also carried out.
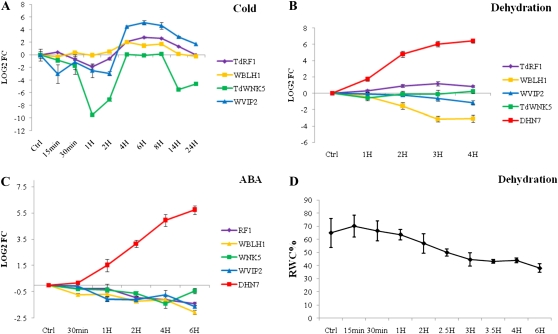
Gene expression in response to cold was assayed on leaves of durum wheat seedlings exposed to 3°C for 24 h. As shown in Figure 5, both TdRF1 and the interactor transcripts were transiently up-regulated in response to cold, but not TdWNK5, which showed a fluctuating profile (Fig. 5A). It is remarkable that the strong and transient induction of WVIP2, whose expression started after 4 h of treatment and reached its maximum level of expression after 6 h, returned to the control level after 14 h. In this expression analysis, as well as in that for ABA treatment, the constitutively expressed durum wheat gene encoding cyclophilin was used as internal reference for transcript normalization (Aprile et al., 2009).
Figure 5.
Gene expression analysis of TdRF1, WBLH1, TdWNK5, and WVIP2 in durum wheat plants exposed to cold (A), dehydration (B), or ABA (C) treatment. Leaves of treated or control untreated plants were used for RNA extraction and qRT-PCR analysis. The log(2) of fold change was calculated based on the 2−ΔΔCt method. Wheat dehydrin Dhn7 was used as control for ABA and dehydration treatment. Error bars represent se (n = 3; n = 12 for WVIP2 in C).
The effect of dehydration on the expression of TdRF1 and its interactor genes was evaluated on leaves of wheat seedlings desiccated for 4 h. As shown in Figure 5B, TdRF1 was the only up-regulated gene, WBLH1 and WVIP2 transcripts were down-regulated, while the expression of TdWNK5 remained unchanged. To assess the dehydration stress, the relative water content was calculated for each time point (Fig. 5D) and the stress responsiveness of the wheat homolog of the barley (Hordeum vulgare) dehydrin gene, Dhn7 (Choi et al., 1999), was used as treatment control. As internal reference for transcript normalization, the constitutively expressed durum wheat gene encoding polyubiquitin was used (Aprile et al., 2009).
Finally, the expression analysis carried out on leaves of wheat seedlings treated with ABA showed a moderate down-regulation for the transcripts corresponding to TdRF1 and all its interactors (Fig. 5C). The strong up-regulation of the wheat Dhn7 transcripts (Choi et al., 1999) demonstrated the effectiveness of the ABA treatment.
TdWNK5 Phosphorylates TdRF1 in Vitro
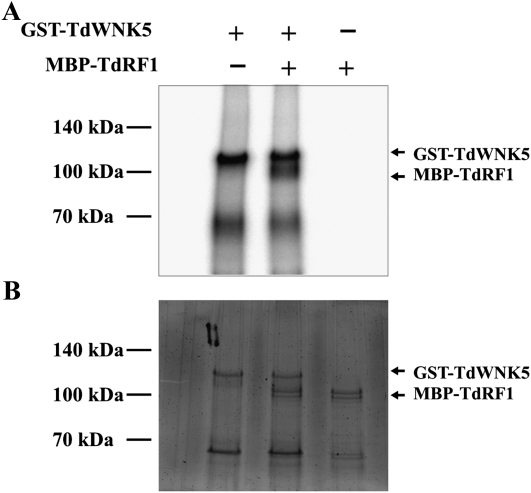
A putative phosphorylation site at the C terminus of TdRF1 sequence was identified by in silico analyses. As previously reported (Mastrangelo et al., 2005), there are four TdRF1-like genes in Arabidopsis. Based on the gene structure and on the alternative splicing features of the fourth intron, At4g39140 is likely the orthologous gene to TdRF1. Furthermore, At4g39140 was shown to be phosphorylated at the Ser in position 400, corresponding to the HFS(p)FGSR peptide (Durek et al., 2010). Although the amino acid similarity between At4g39140 and TdRF1 is only about 40%, the peptide containing the phosphosite is highly conserved, suggesting that TdRF1 could also be phosphorylated at the corresponding Ser residue. Since TdWNK5 shares a high similarity with kinase proteins and interacts with TdRF1, it is reasonable to speculate that TdWNK5 phosphorylates TdRF1. To verify this possibility, an in vitro phosphorylation assay was carried out using recombinant GST-TdWNK5 and MBP-TdRF1 proteins and [γ-32P]ATP. As shown in Figure 6A (lane 1), when GST-TdWNK5 was incubated alone in a reaction buffer containing [γ-32P]ATP, a radiolabeled protein migrated with a Mr similar to GST-TdWNK5, suggesting that TdWNK5 has a kinase activity capable of in vitro autophosphorylation. When the recombinant MBP-TdRF1 protein was added to the phophorylation reaction, besides GST-TdWNK5, an extra radiolabeled protein with the estimated Mr of MBP-TdRF1 was detected (Fig. 6A, lane 2). No signal was detected when MBP-TdRF1 only was used in the reaction (Fig. 6A, lane 3). A picture of the gel stained with SYPRO Ruby for total protein is reported in Figure 6B. These data clearly show that TdWNK5 possesses kinase activity and is able to phosphorylate TdRF1 in vitro.
Figure 6.
Phosphorylation assay of TdWNK5 and TdRF1. A, The fusion protein between GST and the TdWNK5 kinase can autophosphorylate (lane 1) and efficiently phosphorylates MBP-TdRF1 (lane 2). Samples were resolved on 10% SDS-PAGE. The + and − indicate the presence and the absence, respectively, of the corresponding proteins in the reaction mix. Arrows mark positions of the different proteins. B, Picture of the gel stained with SYPRO Ruby for visualization of total proteins.
TdRF1 Mediates the Degradation of WBLH1 in Vivo
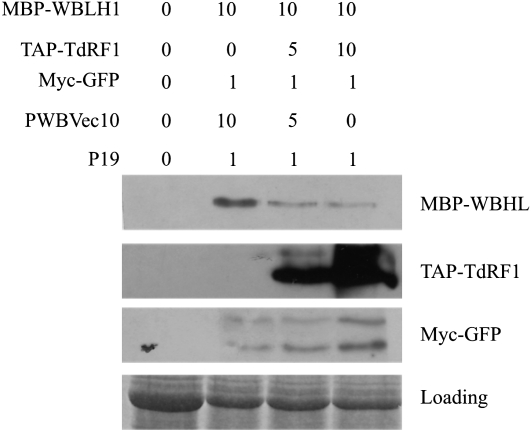
To demonstrate that TdRF1 promotes WBLH1 degradation, we performed an in vivo degradation assay as described by Liu et al. (2010). Leaves of tobacco (Nicotiana benthamiana) were coinfiltrated with Agrobacterium tumefaciens cells carrying the MBP-WBLH1 construct with increasing amounts of A. tumefaciens cells carrying the TAP-TdRF1 construct corresponding to cells ratio of 10:0, 10:5, and 10:10. The normalization of the amount of infiltrating A. tumefaciens cells was assured adding increasing amounts of pWBVec10-carrying cells, and the Myc-GFP construct was included in all infiltration mixtures as positive control of plant transformation and the construct carrying the p19 gene (Voinnet et al., 2003) as silencing suppressor. As shown in Figure 7, the levels of MBP-WBLH1 (top panel), but not of MYC-GFP, were lower when the amount of TAP-TdRF1 in the tobacco cells was higher (second panel from the top). This result suggests a causal relation between the accumulation of the E3 ligase and the degradation of its putative target.
Figure 7.
TdRF1 promotes WBLH1 degradation in vivo. Total protein extract from tobacco leaves, infiltrated with A. tumefaciens cell mixes formulated as described over the lanes, were resolved on 10% SDS. After blotting, the MBP-WBLH1, TAP-TdRF1, and Myc-GFP proteins were detected using the corresponding antibodies. pWBVec10-carrying cells were used to normalize A. tumefaciens infiltrating cells, the Myc-GFP construct was included in all infiltration mixtures as positive control of plant transformation, and the construct carrying the p19 gene was included as silencing suppressor. The Myc-GFP construct yields two proteins likely due to the presence of an additional truncated form (bottom band). As loading control, an electrophoretic gel loaded with the same amount of total proteins used in the assay and stained with Coomassie Brilliant Blue is shown.
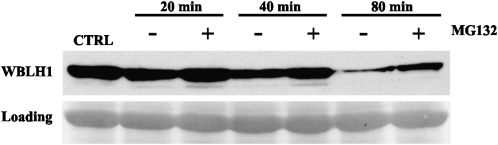
To verify the 26S-mediated proteasomal degradation of WBLH1, a protein extract from tobacco leaves infiltrated with A. tumefaciens cells carrying the MBP-WBLH1 construct was incubated with or without the proteasome inhibitor MG132. Subsequently, the amount of MBP-WBLH1 protein was analyzed over time in the presence or absence of MG132. As shown in Figure 8, WBLH1 degradation was delayed in MG132-treated samples. These findings together with the in vitro physical interaction between TdRF1 and WBLH1 strongly support that TdRF1 interacts with WBLH1 and is responsible of its degradation thorough 26S proteasome in vivo.
Figure 8.
. The 26S-mediated degradation of WBLH1. Total protein extract from tobacco leaves infiltrated with A. tumefaciens cells carrying the MBP-WBLH1 construct were incubated at 4°C for 80 min with or without MG132, with sampling at different times. Tobacco protein samples were resolved on 10% SDS and then MBP-WBLH1 was monitored using an anti-MBP antibody. The Ponceau red-stained PVDF membrane is shown as loading control.
Despite the fact that the in silico protein analysis predicts a putative N-myristoylation signal in the amino acidic sequence of TdRF1 (Fig. 1), the TdRF1 recombinant protein expressed in tobacco was easily purified in the soluble fraction; thus, TdRF1 very likely is not membrane anchored.
WVIP2 Is a Functional E3 Ubiquitin Ligase
When WVIP2 amino acidic sequence was scanned for the presence of conserved domains, a RING H2-type domain (C-X2-C-X9/39-C-X1/3-H-X2/3-H-X2-C-X1/48-C-X2-C) was identified. A subtype of E3 ubiquitin ligases contains this RING H2 domain, suggesting that WVIP2 might also act as an E3 ligase. To verify this hypothesis, an ubiquitination assay was carried out using WVIP2 as E3 ligase candidate. The GST-WVIP2 fusion protein was expressed and purified from E. coli cells and tested for E3 activity in vitro. As shown in Figure 9 (lane 3), GST-WVIP2 showed an extremely high E3 activity when mixed with the human E1, the Arabidopsis E2 (GST-UBC8), and biotinylated ubiquitin, as demonstrated by the strong signal produced by ubiquitinated GST-UBC8 and polyubiquitinated GST-WVIP2. This result indicates that WVIP2 has E3 ubiquitin ligase activity in vitro.
Figure 9.
. Ubiquitination assay on WVIP2. GST-WVIP2 fusion proteins were assayed for E3 activity in the presence of the human activating enzyme (E1), the Arabidopsis conjugating enzyme UBC8 fused to GST (E2), and biotinylated ubiquitin (Ub). Samples were resolved on 10% SDS-PAGE and then the streptavidine-horseradish peroxidase was used to detect biotin-Ub. The + and − indicate the presence and the absence, respectively, of the corresponding protein in the reaction mix. The asterisk indicates a band copurified with GST-WVIP2 and cross-reacting with streptavidine. GST-WVIP2 and GST-WVIP2 together with biotin-Ub were used as control.
TdRF1 and WVIP2 Have Opposite Effects on Cell Tolerance to Dehydration
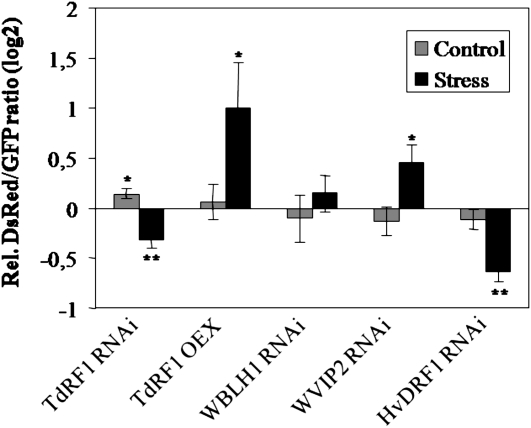
To verify if the activity of TdRF1 could have an effect on plant response to dehydration stress, transient-induced gene silencing (TIGS) and overexpression experiments were carried out in barley as previously described (Marzin et al., 2008). In this transient assay system, after a stress period of 4 d, dehydration stress tolerance is reflected by red fluorescence reporter protein DsRed that is sensitive to denaturing cellular conditions following eventual stress damage. For RNA interference (RNAi) experiments, the barley homologs to wheat TdRF1 (TC260641, 96% similarity) and to WVIP2 (TC261227, 93% similarity) as well as JuBEL1 (AF334758), representing the barley homolog of WBLH1 (94% similarity, Müller et al., 2001), were used. As shown in Figure 10, the constructs did not significantly change relative DsRed fluorescence in absence of dehydration stress, indicating absence of general non-stress-related effect by the constructs per se. The only exception was a weak but significant enhancement of DsRed fluorescence by the RNAi construct targeting TdRF1, which might suggest that resting levels of TdRF1 in unstressed leaves rather accelerated protein degradation. Under stress conditions, overexpression of the TdRF1 homolog enhanced cellular tolerance to dehydration stress, similar to the effect of TIGS of the WVIP2 homolog. Complementary to the positive TdRF1 overexpression effect on cellular tolerance to dehydration, and also complementary to the RNAi effect in the absence of stress, its RNA-mediated silencing rendered the transformed cells more sensitive to dehydration. Taken together, these results indicate a functional positive and negative role of RF1 and VIP2, respectively, on dehydration stress tolerance in barley. Assuming functional conservation of the two genes in the closely related Triticeae species barley and wheat, a similar role of TdRF1 and WVIP2 can also be proposed for durum wheat.
Figure 10.
Effect of TIGS and overexpression of TdRF1, WVIP2, and WBLH1 genes on cellular dehydration stress tolerance of bombarded barley epidermal cells. Dehydration stress tolerance was reflected by red fluorescence of the denaturation-sensitive reporter protein DsRed after a stress period of 4 d. Barley leaves were cobombarded with GFP and DsRed expression constructs in the presence of empty RNAi or overexpression vectors pIPKTA30 and pIPKTA9 or in the presence of candidate gene RNAi or overexpression constructs, as indicated below the bar chart. DsRed fluorescence was normalized to GFP fluorescence measured before onset of dehydration stress treatment, and the stress-induced change of DsRed fluorescence in the absence or presence of RNAi or overexpression constructs was determined relative to empty vector controls (black bars). As control for stress-nonrelated effects of cobombarded constructs, DsRed fluorescence normalized to empty vector controls in nonstress fully turgescent leaves is also shown (gray bars). Log(2)-transformed values below and above zero indicate enhanced sensitivity and enhanced tolerance to dehydration stress, respectively. One and two asterisks above bars indicate significant construct effects at P < 0.05 and P < 0.01, respectively (one-sample t test of log-transformed, relative values against hypothetical value 0).
DISCUSSION
Ubiquitination is a posttranslational modification of proteins playing an important role in several plant processes, such as biotic and abiotic stress responses, cell cycle progression, and hormone responses. A central function in the ubiquitination pathway is played by E3 enzymes that are responsible for docking of the activated E2-ubiquitin intermediate and the target protein.
TdRF1 was previously isolated by Mastrangelo et al. (2005) as a cold- and dehydration-regulated gene coding for a putative wheat RING-finger E3 protein; here, we attempted to assign a biological function to the isolated sequence. TdRF1 was demonstrated to be a nuclear (Fig. 3) E3 ligase able to promote protein ubiquitination in vitro (Fig. 2). Moreover, it was shown to have a protective role against cellular dehydration in an in vitro assay on barley leaves transiently transformed to obtain its silencing/overexpression (Fig. 10). Besides being overinduced by cold and dehydration in leaves of wheat plants at seedling stage (Mastrangelo et al., 2005), TdRF1 was previously shown to be induced by a controlled drought treatment very similar to environmental dry conditions of wheat cultivation (De Leonardis et al., 2007). This induction was seen in different tissues and at different developmental stages, including booting, seed development, and maturation. The general responsiveness of TdRF1 to water stress, together with its protective role against cellular dehydration in vitro, suggests that TdRF1 play a role in drought tolerance.
A Small Stress-Related Interactome around TdRF1
The main challenge in the characterization of the ubiquitination process is the identification of the E3-specific target(s) to gain knowledge about the specific cellular processes in which they are involved. Thus, using TdRF1 as a bait in a yeast two-hybrid analysis followed by confirmation through pull-down experiments (Fig. 4), we demonstrated that the RING-finger protein TdRF1 interacts with WBLH1, TdWNK5, and WVIP2. The homotranscription factor WBLH1, previously isolated by Mizumoto et al. (2011), is homologous to HvJuBEL1 (94% of amino acid similarity), which is involved in the control of leaf development (Müller et al., 2001) and is part of the BEL1-like protein family whose members are involved in various developmental aspects (Mizumoto et al., 2011). TdWNK5 is 75% similar to AtWNK5, a member of the Arabidopsis WNK family of MAP kinases (Wang et al., 2008). These kinases are mainly known to be involved in flowering time in Arabidopsis (Wang et al., 2008). WVIP2 is the homolog of AfVIP2 (Wilson et al., 2005), identified as protein interacting in vitro with AfVP1 and ABI3, respectively, the wild oat and Arabidopsis homolog to maize (Zea mays) Viviparous1 (Jones et al., 2000). Since VIVIPAROUS1 is the master regulator of embryo dormancy (McCarty et al., 1991), AfVIP2 has been proposed to be a transcription factor mediating the action of VIVIPAROUS1 (Jones et al., 2000). Our demonstration that WVIP2 has ubiquitin ligase activity suggests a new point of view on dormancy regulation. VIVIPAROUS1, which is generally expressed during embryo dormancy, could be the target of WVIP2; therefore, a mechanism based on ubiquitination could be involved in dormancy breaking.
The expression analysis of these three genes together with TdRF1 indicates a general trend of stress responsiveness. All the transcripts, except TdWNK5, were transiently up-regulated after cold treatment (Fig. 5A), with WVIP2 showing the highest induction level. This pattern is reminiscent of many early cold regulated (e-cor) genes that exert their function during the earlier response to stress (Crosatti et al., 1999; Marè et al., 2004; Mastrangelo et al., 2005). By contrast, in ABA-treated seedlings, the mRNA of all examined genes was rapidly down-regulated (Fig. 5C). An ABA-related expression was already reported for WVIP2 (Wilson et al., 2005) and AfVIP2 (Jones et al., 2000) in germinating/dormant embryos even if with contrasting behavior, thus suggesting a general regulation of this gene in response to ABA. Finally, we investigated the expression pattern of TdRF1 and its interactors upon a dehydration treatment, with experimental conditions resembling those of the barley transient assay for gene functions (Fig. 5B). Under these stress conditions, TdRF1 was the only up-regulated transcript accordingly with previous results (Mastrangelo et al., 2005), whereas WBLH1 and WVIP2 were down-regulated. The opposite regulation of TdFR1 and WVIP2 transcripts upon dehydration parallels the opposite effect detected after TIGS and transient overexpression experiments of the two closest barley homologs (Fig. 10), suggesting positive and negative roles of RF1 and VIP2, respectively, in drought tolerance.
The Kinase TdWNK5 Regulates TdRF1 by Phosphorylation
We showed that TdWNK5 is capable to phosphorylate itself as well as TdRF1, demonstrating a functional relationship between TdRF1 and TdWNK5. It is known that phosphorylation can directly regulate the activity of RING-based E3s (Deshaies and Joazeiro, 2009). For instance, the phosphorylation of the E3 APC/C complex of budding yeast by mitotic Cdc2 kinase enhances ubiquitin ligase activity by a mechanism that appears to involve recruitment of the activator protein Cdc20 (Lahav-Baratz et al., 1995; Rudner and Murray, 2000). In a recent work, Cheng et al. (2011) demonstrated that the human Protein Kinase A-dependent phosphorylation of the E3 ligase Smad ubiquitin regulatory factor 1 (Smurf1) can switch its substrate preference between two proteins with opposite activities on axon development. The only evidence of a relationship between kinase and E3 ligase in plants concerns the LIK3 receptor-like kinase of Medicago truncatula, which phosphorylates the PUB1 E3 ligase (Mbengue et al., 2010). Two hypotheses can be advanced about the significance of the TdWNK5-TdRF1 interaction. First, the TdWNK5-mediated phosphorylation of TdRF1 could lead to its activation/inactivation, providing a fine-tuning of its activity. Alternatively, the affinity of TdRF1 for the other two interactors could be regulated by TdWNK5-mediated phosphorylation as reported for human Smurf1. Deeper analyses would be necessary to unravel the functional meaning of TdRF1 phosphorylation by TdWNK5.
TdRF1 Mediates the 26S Proteasome Degradation of the Transcription Factor WBLH1
We hypothesized that WBLH1 could be a target of the TdRF1 ubiquitin ligase activity. Despite several in vitro ubiquitination experiments, we failed to detect ubiquitinated forms of WBLH1 (data not shown), but difficulties in the detection of ligase activity on the corresponding targets in vitro are often reported (Liu et al., 2010). Nevertheless, we demonstrated that TdRF1 protein mediates the degradation of WBLH1 in vivo through the 26S proteasome (Figs. 7 and 8). The contrasting expression profile of TdRF1 and WBLH1 in response to dehydration is in agreement with the degradative action of TdRF1 on WBLH1. The homeodomain WBLH1 protein is part of the small BEL1-like (BLH1) protein family, which belongs to the three-amino acid loop extension superfamily. BLH1s are transcription factors involved in various developmental aspects like seed shattering, ovule morphogenesis, and leaf shape establishment (Mizumoto et al., 2011). Many BLH1 proteins interact with the KNOTTED1-like homeobox (KNOX) transcription factors, which also belong to the three-amino acid loop extension superfamily (Bellaoui et al., 2001) and regulate morphogenesis (Hay and Tsiantis, 2010).
In developing plant organs, the expression of transcription factors like BLH1/KNOXs in specific domains in response to positional information is responsible of differential growth and, consequently, of the morphogenesis. Thus, the degradation of WBLH1 by TdRF1 could affect the pathways of leaf morphogenesis mediated by the KNOX-BEL systems. The implication of the ubiquitin proteasome system in plant development is already well known. Indeed DELLA proteins, a small subset of transcriptional regulators involved in gibberellic acid-dependent plant growth restraining, are degraded following gibberellic acid level increase through the F-box E3 ubiquitin ligase SLEEPY1 (Achard et al., 2009).
To overcome stress conditions, plants have evolved different adaptive responses, including growth and morphology changes. For instance, drought and salt stress is known to reduce leaf growth, affecting both cell division and expansion (Fricke et al., 2006; Skirycz et al., 2010; Dosio et al., 2011), and plants overexpressing the drought-response regulator DREB2A showed a dwarf phenotype (Qin et al., 2008). Since TdRF1 is a stress-responsive gene, its degradative action on WBLH1 might mediate the effects of the stress on growth and development, thus representing a link between the stress response and leaf/plant development.
Functional Relationships between the Ubiquitin Ligases TdRF1 and WVIP2
We demonstrated that WVIP2 is a TdRF1 interactor with a RING E3 ligase activity. The corresponding transcripts are both up-regulated by cold treatment and down-regulated by ABA, whereas they showed an opposite regulation upon dehydration stress (Fig. 5) in agreement with the effects detected in transient gene silencing (TdRF1 and WVIP2) and overexpressing (TdRF1 only) experiments in barley (Fig. 10).
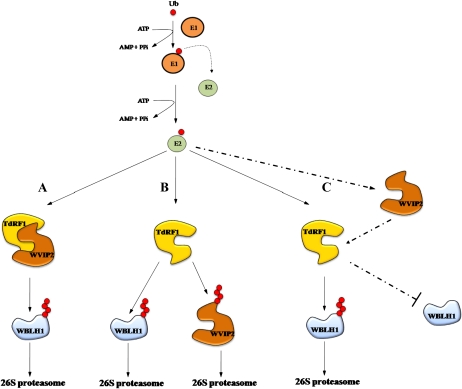
Based on the findings described in this article, we suggest three scenarios about the interaction involving TdRF1 and WVIP2. The first one hypothesizes the formation of a heterodimer between WVIP2 and TdRF1 to mediate WBLH1 degradation in response to cold, a condition where both genes are up-regulated (Fig. 11A). The formation of a heterodimer between two E3 RING ubiquitin ligases was also reported for the regulation of seed germination and growth in Arabidopsis (Hsia and Callis, 2010). Alternatively, TdRF1 can act toward both WBLH1 and WVIP2; this hypothesis meets the expression profiles of the two genes in response to drought, where the WVIP2 mRNA is down-regulated (Fig. 11B). In the third scenario, TdRF1 and WVIP2 are part of a two-step mechanism where TdRF1 is active in the first phase to propagate the stress-responsive signal and then it is removed through WVIP2, when stress conditions eventually disappear (Fig. 11C). This hypothesis can be aligned to the expression profile of the two genes at low temperature, and it results in a transient degradation of WBLH1, whose concentration is then restored by WVIP2.
Figure 11.
Representation of the three proposed scenarios for the interaction TdRF1-WVIP2. A, TdRF1 and WVIP2 form a heterodimer to degrade WBLH1 through the 26S proteasome after polyubiquitination. B, TdRF1 degrades WBLH1 as well as WVIP2. C, TdRF1 and WVIP2 are part of a two-step mechanism where TdRF1 is active in the first phase toward WBLH1 and then it is removed through WVIP2. This hypothesis results in a transient degradation of WBLH1, whose concentration is restored by WVIP2. [See online article for color version of this figure.]
CONCLUSION
The overall findings of this work together with the functions of interactors known from the literature support at least two biological meanings of the TdRF1 protein network under stress conditions. Provided the known function of BLH1-KNOX proteins, the degradation of WBLH1 mediated by TdRF1, and the expression profile of the corresponding genes in response to drought, the relationship between TdRF1 and WBLH1 is a candidate to represent a link between stress response and the regulation of leaf growth and morphology. Instead, our compelling evidence from gene function assays and expression data, as well as the known role of VIP2 in ABA signaling, demonstrate that the relationship between TdRF1 and WVIP2 regulates the response to dehydration, thus providing an increased drought tolerance phenotype when improved.
MATERIALS AND METHODS
Plant Materials and Stress Treatments
RNA for preparation of the expression library and for gene expression analysis was obtained from durum wheat (Triticum durum ‘Ofanto’); barley (Hordeum vulgare ‘Golden Promise’) was used for gene function assays and tobacco (Nicotiana benthamiana) for degradation assays.
For cold treatment, durum wheat seeds were sown in a peat pot and grown in a growth chamber for 7 d (first leaf stage) in a daily regime of 8 h light (200 μmol m−2 s−1) at 22°C/19°C and cold treated at 3°C in a growth chamber with 300 μmol m−2 s−1 up to 24 h.
Leaf dehydration was performed on 7-d-old seedlings germinated on 1% agar medium and grown under growth chamber controlled conditions at 24°C with a 16-/8-h photoperiod, 110 μmol m−2 s−1 until the first leaf stage. Plants were pulled up, air dried at room temperature, and harvested during 6 h. The relative water content was monitored during the whole experiment on four leaves for each timing (Fig. 5D).
ABA treatment of wheat seedlings was carried out as previously described (Marè et al., 2004). Briefly, 7-d-old seedlings sown in peat pot were watered and sprayed with 1 mm ABA solution and then leaves collected during 6 h.
Protein Expression and Purification
TdRF1 coding sequence was cloned in pMAL Gateway (Invitrogen) converted vector for the N-terminal fusion with MBP, and cDNAs coding for WBLH1, WVIP2, and TdWNK5 were cloned in pGEX-2KT Gateway (Invitrogen) converted vector for the N-terminal fusion with GST. The MBP or GST fusion proteins were expressed in Escherichia coli strain BL21 Rosetta in 50 mL of culture. Transformed cells were grown at 37°C to an OD600 of 0.4 to 0.5 and induced with 1 mm isopropylthio-β-galactoside for 3 h at 37°C. Cells were harvested by centrifugation and lysed by sonication in 5 mL of phosphate-buffered saline (PBS; Sigma-Aldrich), 0.1% (v/v) Triton X-100, or TAP buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 10% [v/v] glycerol, and 0.1% [v/v] Nonidet P-40). For purification, 100 μL of GST beads (Glutathione Sepharose 4B; GE Healthcare) or amylose resin (New England Biolabs) was added to the clarified lysate and incubated for 2 h at 4°C. Beads were then washed three times with 5 mL of PBS, 0.1% (v/v) Triton X-100, or TAP buffer. GST fusion proteins were eventually eluted in 100 μL of elution buffer containing 25 mm Tris-HCl, pH 7.5, 150 mm NaCl, and 0.01% (v/v) Triton X-100, supplemented with 20 mm reduced glutathione or left bound to beads if necessary. MBP fusion proteins were eventually eluted in elution buffer supplemented with 20 mm maltose or left bound to beads if requested. The UBC8 (At5g41700) cDNA was cloned into the pGEX vector (Amersham Biosciences). The recombinant GST-UBC8 was expressed in the BL21 bacterial strain in standard conditions and was purified as previously described. Recombinant protein was released from beads with 50 mm reduced glutathione in 20 mm Tris-HCl, pH 7, 20 mm NaCl, and 5% (v/v) glycerol and kept at −80°C. To assess expression and purification, purified proteins were separated by 10% SDS-PAGE and visualized by Coomassie Brilliant Blue staining or transferred to Hybond-P polyvinylidene difluoride (PVDF) membrane (GE Healthcare) and immunodetected with the anti MBP-HRP-conjugated antibody (New England Biolabs) or rabbit anti-GST antibody.
In Vitro Ubiquitination Assays
MBP-TdRF1 was expressed and purified as described above. For the E3 ligase assay, we mixed 2 μL of the MBP-TdRF1 fusion protein with 100 nm human E1 expressed and purified from insect cells (Biomol) for TdRF1 and WVIP2, 2 μL of total protein extract from E. coli expressing GST-UBC8, 0.2 mm dithiothreitol (DTT), 5 mm MgCl2, and 0.5 μg of biotinylated-ubiquitin, and 5 mm ATP in a final total volume of 20 μL. The reactions were incubated at 30°C for 45 min and then stopped by adding SDS-PAGE sample buffer with 100 mm DTT and incubating at 100°C for 10 min. The mixture was then subjected to 10% SDS-PAGE and detected with streptavidine-horseradish peroxidase. After blocking with 3% (w/v) BSA in PBS-T (PBS with 0.1% [v/v] of Tween 20) for 10 min, membranes were incubated with streptavidine-horseradish peroxidase (Sigma-Aldrich) at 1:50,000 for 45 min to detect the ubiquitin-biotin conjugated proteins. After washing, the blot was revealed with the ECL plus kit (Amersham Biosciences).
Subcellular Localization of TdRF1
To determine nuclear localization of TdRF1 protein, TdRF1 coding sequence without the STOP codon was cloned in pMDC83 (Curtis and Grossniklaus, 2003) as a C-terminal fusion with GFP and transformed in onion (Allium cepa) epidermal cells by particle bombardment. Inner epidermal peels (2 × 2 cm) of commercial white onion were directly placed on agar plates containing Murashige and Skoog medium. Gold microcarriers (1 μm diameter) were prepared essentially as described by Dal Bosco et al. (2003). Thirty-five microliters of resuspended microcarrier (60 mg/mL ethanol) were mixed with 25 μg of TdRF1-GFP plasmid or empty pMDC83 in negative control experiment. The microcarriers were delivered to the freshly transferred explants using the PDS-1000/He biolistic particle delivery system (Bio-Rad; Lemaux et al., 1996) according to the manufacturer’s instructions. A petri dish containing the plant tissue was placed 9 cm below the microcarrier launch assembly. The particles were fired using 1100-ψ rupture discs (Bio-Rad) with a partial vacuum of 28 mm Hg. After recovering the transformed onion epidermis cells at 22°C for 18 h in the dark, GFP was detected by an epifluorescence microscope. The 4′,6-diamino-phenylindole (DAPI) staining was performed before microscopy observation, leaving onion peels 15 min in 1 mm DAPI solution.
Yeast Two-Hybrid cDNA Library Construction and Screening
Total RNA was isolated with Trizol reagent (Invitrogen) from the first leaf of 7-d-old seedlings of durum wheat, cold treated as described, and harvested after 6 h of stress. Poly(A)-RNA was isolated using the FastTrack MAG Maxi mRNA isolation kit from Invitrogen according to the manufacturer’s instructions. cDNA was synthesized from 5 μg poly(A)-RNA and cloned in pEXP-AD502 using the SuperScript plasmid system kit from Invitrogen according to the manufacturer’s instructions. About 106 colony-forming units were obtained. The cDNA expression library was further amplified reaching a titer of 1011 colony-forming units.
To develop the bait construct containing the DNA BD-Gal4 fused to TdRF1, the TdRF1 coding sequence was cloned in pENTR/SD/D-TOPO (Invitrogen) according to manufacturer’s instructions and then subcloned in pDEST32 by attL × attR recombination reaction (Invitrogen). The bait construct was transformed in Saccharomyces cerevisiae strain AH109 by the LiAc/SS carrier DNA/PEG method (Gietz and Woods, 2002). The AH109 carrying bait construct strain was further transformed with the cDNA prey expression library and plated on a synthetic complete dropout (SC) medium without Leu, Trp, and His and added of 10 mm 3-amino-1,2,4-triazole to inhibit growth due to autoactivation. Several dilutions of the transformation mixture were plated on SC medium minus Leu and Trp to count the number of screened clones; they were 8 × 106. Positives clones resulted from the screening were further confirmed on SC medium minus Leu, Trp, His, and adenine and added with 10 mm 3-amino-1,2,4-triazole and with X-Gal for a colorimetric assay. To reduce clone redundancy, positive clones were analyzed by colony PCR. Half reaction was digested using a four-cutter enzyme and the other half used for Sanger sequencing reaction. Positives clones resulting from this assay were further reconfirmed by inverting bait and prey clones in a retransformation assay.
Pull-Down Assay
For pull-down assay, 50 μL of MBP-TdRF1 beads was added to 5 mL of cleared lysate, obtained from a 50 mL of E. coli cultured cells expressing GST-TdWNK5 or WVIP2 or WBLH1 recombinant proteins. The mix was incubated at 4°C for 3 h. After incubation, the beads were washed three times with TAP buffer, and SDS-PAGE buffer was added to the beads and boiled. Proteins were separated on 10% SDS-PAGE, transferred on a PVDF membrane, and immunodetected using anti-GST antibody.
qRT-PCR Analysis
To perform qRT-PCR analysis, total RNA from leaf samples was extracted using TRI reagent (Ambion) according to manufacturer’s instructions. Three micrograms of total RNA of each sample were reverse transcribed using oligo(dT)18 primer with Moloney murine leukemia virus reverse transcription reagents (M-MLV; Promega) according to the manufacturer’s instructions. Subsequently, the cDNAs were quantified using a Qubit fluorometer (Invitrogen), diluted, and used for qPCR amplifications with gene-specific primers. qRT-PCR analyses were performed using the 7300 Real-Time PCR System (Applied Biosystems) with Qiagen QuantiFast SYBR Green chemicals. Specific primers for housekeeping reference genes chosen according to Aprile et al. (2009) were used for normalization of the cDNA. They were cyclophilin for gene expression analyses in response to cold and ABA, and polyubiquitin for dehydration treatment. The wheat homolog (Ta.29352.1.S1_at) of the barley dehydrin gene Dhn7 was used as positive control in ABA and dehydration experiments.
To calculate the relative fold change of the expression of the target genes, the 2–DDCt (cycle threshold) method was used (Livak and Schmittgen, 2001), where the Ct data were expressed as average of three (or 12) experimental replicates.
In Vitro Phosphorylation Assay
The phosphorylation assay was performed essentially as previously described by Zhang et al. (2010) with some modifications. Briefly, about 1 μg of E. coli-purified MBP-TdRF1 and GST-TdWNK5 proteins were added to the kinase reaction mix, containing 1× kinase buffer (25 mm Tris-HCl, pH 7.5, 1 mm DTT, 5 mm MgCl2, and 2 mm CaCl2) and 1× ATP solution (100 μm MgCl2, 100 μm ATP-Na2, and 2 μCi [γ-32P]ATP) in a final total volume of 30 μL. Reactions were incubated at 30°C for 1 h and stopped by addition of 5 μL protein SDS-PAGE sample buffer and boiling for 10 min. Reactions were subsequently separated by 12% SDS-PAGE. After separation, acrylamide gels were washed three times for 30 min with kinase gel wash buffer (5% [w/v] trichoroacetic acid and 1% [w/v] Na2H2P2O7). After 2 d of exposure, the impressed storage phosphor screen (Amersham Biosciences Molecular Dynamics) was read by the Typhoon 9210 and the gel stained with SYPRO Ruby for total proteins (Invitrogen).
In Vivo Degradation Assay
TdRF1 coding sequence was cloned in the NTAPi binary vector (Rohila et al., 2004) as an N-terminal fusion with TAP tag, and WBLH1 cDNA was cloned in pMDC32 binary vector modified to allow the N-terminal fusion with MBP tag. In vivo degradation assay was essentially carried out as previously described by Liu et al. (2010). WBLH1 and TdRF1 constructs were transformed separately in Agrobacterium tumefaciens strain GV3101(pMP90). Leaves of 4-week-old tobacco plants grown in a growth chamber under standard conditions were coinfiltrated with Agrobacterium cells carrying MBP-WBLH1 plus Myc-GFP mixed with A. tumefaciens cells carrying TAP-TdRF1 in 10:0, 10:5, and 10:10 ratios. Moreover, A. tumefaciens cells carrying other constructs were added for specific purposes: p19 gene as silencing suppressor, Myc-GFP as internal control of transformation, and PWBVec10 for normalization of inoculating cells. Seven days after infiltration, leaves were collected, and total proteins were extracted in NB1 extraction buffer (Liu et al., 2010), separated on 12% SDS-PAGE, and transferred to a PVDF membrane. The PVDF membrane was probed using anti-MBP antibody to detect WBLH1 protein, PAP antibody to detect TdRF1, and anti-Myc antibody to detect GFP internal control. The experiment to assess the 26S proteasome-mediated degradation of WBLH1 was performed with total protein extract of tobacco overexpressing MBP-WBLH1. Protein extracts were incubated at 4°C with or without 50 μm MG132.
TIGS and Overexpression Experiments
TIGS and overexpression constructs were generated in plasmid vectors pIPKTA9 and pIPKTA30, and the transient assay for gene function during dehydration stress was performed as described (Marzin et al., 2008). Briefly, barley leaf segments of ‘Golden Promise’ were bombarded with gold particles (diameter of 1 μm) that had been coated with a mixture of plasmid DNA consisting of three different vectors: pGFP (Schweizer et al., 1999) for normalization, pUbi-DsRed-nos for assessment of RNAi effects (Panstruga et al., 2003), and pIPKTA30_Target (RNAi constructs) and pIPKTA9_Candidate (overexpression constructs) as described (Douchkov et al., 2005). Bombarded leaf segments were incubated in closed petri dishes on phytoagar-containing benzimidazole as senescence inhibitor. At 24 h after bombardment, GFP-expressing epidermal cells were counted by fluorescence microscopy. For the subsequent exposure to dehydration stress, the leaf segments were placed on filter paper and air-dried to a fresh weight of 60%, relative to the initial fresh weight measured immediately before the dehydration treatment (RFW). The dehydration treatment was carried out at 20°C, 40% ± 5% relative air humidity, natural daylight conditions, and lasted 90 ± 30 min. Control leaf segments remained on water-phytoagar. After the dehydration-stressed leaves had reached the desired RFW, the filter paper was moisturized with 0.35 mL water, and petri dishes of 9-cm diameter were closed and sealed with Parafilm (Pechiney). This resulted in constant dehydration conditions during the entire stress treatment of 4 d. At the end of the stress treatment, RFW was measured again and DsRed-expressing epidermal cells were counted in control and dehydration-stressed leaves under the fluorescence microscope. The DsRed/GFP ratio was finally calculated in control and stressed leaf segments. Putative TIGS or overexpression effects of a given candidate gene were determined by first calculating the stress-induced reduction of the denaturation-sensitive DsRed reporter in the presence or absence of RNAi or overexpression construct, followed by dividing the stress-induced reduction in the presence of test construct by the one observed in the presence of the empty vector control. Values below 1 [corresponding to values below zero after log(2) transformation] indicate enhanced stress sensitivity induced by the test constructs, whereas values above 1 (zero if log-transformed) indicate enhanced stress tolerance by the test construct. Statistical significance of the relative values were tested against to the theoretical value “zero” for the null hypothesis [after log(2) transformation] by a one-sample t test with P < 0.05 as significance threshold. All data are based on a total number of five independent bombardment/stress experiments per construct.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AY465427.2 (TdRF1) and AB546643.1 (WBLH1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Alignment between amino acidic sequences of HvJUBEL1 and WBLH1.
Supplemental Figure S2. Alignment between amino acidic sequences of AfVIP2 and WVIP2.
Supplemental Figure S3. Alignment between amino acidic sequences of AtWNK5 and TdWNK5.
Supplementary Material
Acknowledgments
We thank Dr. S. Masiero from University of Milan, M.D. Curtis from the University of Zurich, and Dr. J.S. Rohila from South Dakota State University for kindly providing the yeast strains and the vectors used in this work. The technical support of Gabi Brantin of Leibniz-Institute of Plant Genetics and Crop Plant Research is also acknowledged.
References
- Achard P, Gusti A, Cheminant S, Alioua M, Dhondt S, Coppens F, Beemster GT, Genschik P. (2009) Gibberellin signaling controls cell proliferation rate in Arabidopsis. Curr Biol 19: 1188–1193 [DOI] [PubMed] [Google Scholar]
- Aprile A, Mastrangelo AM, De Leonardis AM, Galiba G, Roncaglia E, Ferrari F, De Bellis L, Turchi L, Giuliano G, Cattivelli L. (2009) Transcriptional profiling in response to terminal drought stress reveals differential responses along the wheat genome. BMC Genomics 10: 279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellaoui M, Pidkowich MS, Samach A, Kushalappa K, Kohalmi SE, Modrusan Z, Crosby WL, Haughn GW. (2001) The Arabidopsis BELL1 and KNOX TALE homeodomain proteins interact through a domain conserved between plants and animals. Plant Cell 13: 2455–2470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng PL, Lu H, Shelly M, Gao H, Poo MM. (2011) Phosphorylation of E3 ligase Smurf1 switches its substrate preference in support of axon development. Neuron 69: 231–243 [DOI] [PubMed] [Google Scholar]
- Choi DW, Zhu B, Close TJ. (1999) The barley (Hordeum vulgare L.) dehydrin multigene family: sequences, allele types, chromosome assignments, and expression characteristics of 11 Dhn genes of cv Dicktoo. Theor Appl Genet 98: 1234–1247 [Google Scholar]
- Crosatti C, Polverino de Laureto P, Bassi R, Cattivelli L. (1999) The interaction between cold and light controls the expression of the cold-regulated barley gene cor14b and the accumulation of the corresponding protein. Plant Physiol 119: 671–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Bosco C, Busconi M, Govoni C, Baldi P, Stanca AM, Crosatti C, Bassi R, Cattivelli L. (2003) cor Gene expression in barley mutants affected in chloroplast development and photosynthetic electron transport. Plant Physiol 131: 793–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Leonardis AM, Marone D, Mazzucotelli E, Neffar F, Rizza F, Di Fonzo N, Cattivelli L, Mastrangelo AM. (2007) Durum wheat genes up-regulated in the early phase of cold stress are modulated by drought in a developmental and genotype dependent manner. Plant Sci 172: 1005–1016 [Google Scholar]
- del Pozo JC, Boniotti MB, Gutierrez C. (2002) Arabidopsis E2Fc functions in cell division and is degraded by the ubiquitin-SCF(AtSKP2) pathway in response to light. Plant Cell 14: 3057–3071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo JC, Diaz-Trivino S, Cisneros N, Gutierrez C. (2006) The balance between cell division and endoreplication depends on E2FC-DPB, transcription factors regulated by the ubiquitin-SCFSKP2A pathway in Arabidopsis. Plant Cell 18: 2224–2235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA. (2009) RING domain E3 ubiquitin ligases. Annu Rev Biochem 78: 399–434 [DOI] [PubMed] [Google Scholar]
- Devoto A, Muskett PR, Shirasu K. (2003) Role of ubiquitination in the regulation of plant defence against pathogens. Curr Opin Plant Biol 6: 307–311 [DOI] [PubMed] [Google Scholar]
- Dong CH, Agarwal M, Zhang Y, Xie Q, Zhu JK. (2006) The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc Natl Acad Sci USA 103: 8281–8286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosio GA, Tardieu F, Turc O. (2011) Floret initiation, tissue expansion and carbon availability at the meristem of the sunflower capitulum as affected by water or light deficits. New Phytol 189: 94–105 [DOI] [PubMed] [Google Scholar]
- Douchkov D, Nowara D, Zierold U, Schweizer P. (2005) A high-throughput gene-silencing system for the functional assessment of defense-related genes in barley epidermal cells. Mol Plant Microbe Interact 18: 755–761 [DOI] [PubMed] [Google Scholar]
- Dreher K, Callis J. (2007) Ubiquitin, hormones and biotic stress in plants. Ann Bot (Lond) 99: 787–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durek P, Schmidt R, Heazlewood JL, Jones A, MacLean D, Nagel A, Kersten B, Schulze WX. (2010) PhosPhAt: the Arabidopsis thaliana phosphorylation site database. An update. Nucleic Acids Res 38(Database issue): D828–D834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye BT, Schulman BA. (2007) Structural mechanisms underlying posttranslational modification by ubiquitin-like proteins. Annu Rev Biophys Biomol Struct 36: 131–150 [DOI] [PubMed] [Google Scholar]
- Fricke W, Akhiyarova G, Wei W, Alexandersson E, Miller A, Kjellbom PO, Richardson A, Wojciechowski T, Schreiber L, Veselov D, et al. (2006) The short-term growth response to salt of the developing barley leaf. J Exp Bot 57: 1079–1095 [DOI] [PubMed] [Google Scholar]
- Gietz RD, Woods RA. (2002) Transformation of yeast by the Liac/SS carrier DNA/PEG method. Methods Enzymol 350: 87–96 [DOI] [PubMed] [Google Scholar]
- Gingerich DJ, Gagne JM, Salter DW, Hellmann H, Estelle M, Ma L, Vierstra RD. (2005) Cullins 3a and 3b assemble with members of the broad complex/tramtrack/bric-a-brac (BTB) protein family to form essential ubiquitin-protein ligases (E3s) in Arabidopsis. J Biol Chem 280: 18810–18821 [DOI] [PubMed] [Google Scholar]
- Hay A, Tsiantis M. (2010) KNOX genes: versatile regulators of plant development and diversity. Development 137: 3153–3165 [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. (1998) The ubiquitin system. Annu Rev Biochem 67: 425–479 [DOI] [PubMed] [Google Scholar]
- Hsia MM, Callis J. (2010) BRIZ1 and BRIZ2 proteins form a heteromeric E3 ligase complex required for seed germination and post-germination growth in Arabidopsis thaliana. J Biol Chem 285: 37070–37081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Li CY, Pattison DL, Gray WM, Park S, Gibson SI. (2010) SUGAR-INSENSITIVE3, a RING E3 ligase, is a new player in plant sugar response. Plant Physiol 152: 1889–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T. (2007) The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol Cell 28: 730–738 [DOI] [PubMed] [Google Scholar]
- Ishitani M, Xiong L, Lee H, Stevenson B, Zhu JK. (1998) HOS1, a genetic locus involved in cold-responsive gene expression in Arabidopsis. Plant Cell 10: 1151–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isono E, Katsiarimpa A, Müller IK, Anzenberger F, Stierhof YD, Geldner N, Chory J, Schwechheimer C. (2010) The deubiquitinating enzyme AMSH3 is required for intracellular trafficking and vacuole biogenesis in Arabidopsis thaliana. Plant Cell 22: 1826–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones HD, Kurup S, Peters NC, Holdsworth MJ. (2000) Identification and analysis of proteins that interact with the Avena fatua homologue of the maize transcription factor VIVIPAROUS 1. Plant J 21: 133–142 [DOI] [PubMed] [Google Scholar]
- Jurado S, Abraham Z, Manzano C, López-Torrejón G, Pacios LF, Del Pozo JC. (2010) The Arabidopsis cell cycle F-box protein SKP2A binds to auxin. Plant Cell 22: 3891–3904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurado S, Díaz-Triviño S, Abraham Z, Manzano C, Gutierrez C, del Pozo C. (2008) SKP2A, an F-box protein that regulates cell division, is degraded via the ubiquitin pathway. Plant J 53: 828–841 [DOI] [PubMed] [Google Scholar]
- Lahav-Baratz S, Sudakin V, Ruderman JV, Hershko A. (1995) Reversible phosphorylation controls the activity of cyclosome-associated cyclin-ubiquitin ligase. Proc Natl Acad Sci USA 92: 9303–9307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Kim WT. (2011) Regulation of abiotic stress signal transduction by E3 ubiquitin ligases in Arabidopsis. Mol Cells 31: 201–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaux PG, Cho MJ, Louwerse JD, Williams R, Wan Y. (1996) Bombardment mediated transformation methods for barley. BioRad US/EG Bull 2007: 1–6 [Google Scholar]
- Li W, Zhong S, Li G, Li Q, Mao B, Deng Y, Zhang H, Zeng L, Song F, He Z. (2011) Rice RING protein OsBBI1 with E3 ligase activity confers broad-spectrum resistance against Magnaporthe oryzae by modifying the cell wall defence. Cell Res 21: 835–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Zhang H, Yang Y, Li G, Yang Y, Wang X, Basnayake BM, Li D, Song F. (2008) Functional analysis reveals pleiotropic effects of rice RING-H2 finger protein gene OsBIRF1 on regulation of growth and defense responses against abiotic and biotic stresses. Plant Mol Biol 68: 17–30 [DOI] [PubMed] [Google Scholar]
- Liu L, Zhang Y, Tang S, Zhao Q, Zhang Z, Zhang H, Dong L, Guo H, Xie Q. (2010) An efficient system to detect protein ubiquitination by agroinfiltration in Nicotiana benthamiana. Plant J 61: 893–903 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Marè C, Mazzucotelli E, Crosatti C, Francia E, Stanca AM, Cattivelli L. (2004) Hv-WRKY38: a new transcription factor involved in cold- and drought-response in barley. Plant Mol Biol 55: 399–416 [DOI] [PubMed] [Google Scholar]
- Marzin S, Mihaly R, Pauk J, Schweizer P. (2008) A transient assay system for the assessment of cell-autonomous gene function in dehydration-stressed barley. J Exp Bot 59: 3359–3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastrangelo AM, Belloni S, Barilli S, Ruperti B, Di Fonzo N, Stanca AM, Cattivelli L. (2005) Low temperature promotes intron retention in two e-cor genes of durum wheat. Planta 221: 705–715 [DOI] [PubMed] [Google Scholar]
- Matsuda N, Suzuki T, Tanaka K, Nakano A. (2001) Rma1, a novel type of RING finger protein conserved from Arabidopsis to human, is a membrane-bound ubiquitin ligase. J Cell Sci 114: 1949–1957 [DOI] [PubMed] [Google Scholar]
- Mazzucotelli E, Belloni S, Marone D, De Leonardis AM, Guerra D, Di Fonzo N, Cattivelli L, Mastrangelo AM. (2006) The e3 ubiquitin ligase gene family in plants: regulation by degradation. Curr Genomics 7: 509–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzucotelli E, Mastrangelo AM, Crosatti C, Guerra D, Stanca AM, Cattivelli L. (2008) Abiotic stress response in plants: when post-transcriptional and post-translational regulations control transcription. Plant Sci 174: 420–431 [Google Scholar]
- Mbengue M, Camut S, de Carvalho-Niebel F, Deslandes L, Froidure S, Klaus-Heisen D, Moreau S, Rivas S, Timmers T, Hervé C, et al. (2010) The Medicago truncatula E3 ubiquitin ligase PUB1 interacts with the LYK3 symbiotic receptor and negatively regulates infection and nodulation. Plant Cell 22: 3474–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty DR, Hattori T, Carson CB, Vasil V, Lazar M, Vasil IK. (1991) The Viviparous-1 developmental gene of maize encodes a novel transcriptional activator. Cell 66: 895–905 [DOI] [PubMed] [Google Scholar]
- Mizumoto K, Hatano H, Hirabayashi C, Murai K, Takumi S. (2011) Characterization of wheat Bell1-type homeobox genes in floral organs of alloplasmic lines with Aegilops crassa cytoplasm. BMC Plant Biol 11: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J, Wang Y, Franzen R, Santi L, Salamini F, Rohde W. (2001) In vitro interactions between barley TALE homeodomain proteins suggest a role for protein-protein associations in the regulation of Knox gene function. Plant J 27: 13–23 [DOI] [PubMed] [Google Scholar]
- Nelson DC, Lasswell J, Rogg LE, Cohen MA, Bartel B. (2000) FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis. Cell 101: 331–340 [DOI] [PubMed] [Google Scholar]
- Panstruga R, Kim MC, Cho MJ, Schulze-Lefert P. (2003) Testing the efficiency of dsRNAi constructs in vivo: a transient expression assay based on two fluorescent proteins. Mol Biol Rep 30: 135–140 [DOI] [PubMed] [Google Scholar]
- Park GG, Park JJ, Yoon J, Yu SN, An G. (2010) A RING finger E3 ligase gene, Oryza sativa Delayed Seed Germination 1 (OsDSG1), controls seed germination and stress responses in rice. Plant Mol Biol 74: 467–478 [DOI] [PubMed] [Google Scholar]
- Pedmale UV, Liscum E. (2007) Regulation of phototropic signaling in Arabidopsis via phosphorylation state changes in the phototropin 1-interacting protein NPH3. J Biol Chem 282: 19992–20001 [DOI] [PubMed] [Google Scholar]
- Qin F, Sakuma Y, Tran LS, Maruyama K, Kidokoro S, Fujita Y, Fujita M, Umezawa T, Sawano Y, Miyazono K, et al. (2008) Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell 20: 1693–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohila JS, Chen M, Cerny R, Fromm ME. (2004) Improved tandem affinity purification tag and methods for isolation of protein heterocomplexes from plants. Plant J 38: 172–181 [DOI] [PubMed] [Google Scholar]
- Rudner AD, Murray AW. (2000) Phosphorylation by Cdc28 activates the Cdc20-dependent activity of the anaphase-promoting complex. J Cell Biol 149: 1377–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer P, Pokorny J, Abderhalden O, Dudler R. (1999) A transient assay system for the functional assessment of defense-related genes in wheat. Mol Plant Microbe Interact 12: 647–654 [Google Scholar]
- Skirycz A, De Bodt S, Obata T, De Clercq I, Claeys H, De Rycke R, Andriankaja M, Van Aken O, Van Breusegem F, Fernie AR, et al. (2010) Developmental stage specificity and the role of mitochondrial metabolism in the response of Arabidopsis leaves to prolonged mild osmotic stress. Plant Physiol 152: 226–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J, Vierstra RD. (2004) The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol 55: 555–590 [DOI] [PubMed] [Google Scholar]
- Sunkar R, Chinnusamy V, Zhu J, Zhu JK. (2007) Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci 12: 301–309 [DOI] [PubMed] [Google Scholar]
- Trujillo M, Shirasu K. (2010) Ubiquitination in plant immunity. Curr Opin Plant Biol 13: 402–408 [DOI] [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D. (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Wang H, Deng XW. (2003) Dissecting the phytochrome A-dependent signaling network in higher plants. Trends Plant Sci 8: 172–178 [DOI] [PubMed] [Google Scholar]
- Wang X, Feng S, Nakayama N, Crosby WL, Irish V, Deng XW, Wei N. (2003) The COP9 signalosome interacts with SCF UFO and participates in Arabidopsis flower development. Plant Cell 15: 1071–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Liu K, Liao H, Zhuang C, Ma H, Yan X. (2008) The plant WNK gene family and regulation of flowering time in Arabidopsis. Plant Biol (Stuttg) 10: 548–562 [DOI] [PubMed] [Google Scholar]
- Wilson ID, Barker GL, Lu C, Coghill JA, Beswick RW, Lenton JR, Edwards KJ. (2005) Alteration of the embryo transcriptome of hexaploid winter wheat (Triticum aestivum cv. Mercia) during maturation and germination. Funct Integr Genomics 5: 144–154 [DOI] [PubMed] [Google Scholar]
- Yang CW, González-Lamothe R, Ewan RA, Rowland O, Yoshioka H, Shenton M, Ye H, O’Donnell E, Jones JD, Sadanandom A. (2006) The E3 ubiquitin ligase activity of Arabidopsis PLANT U-BOX17 and its functional tobacco homolog ACRE276 are required for cell death and defense. Plant Cell 18: 1084–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Nodzynski T, Pencík A, Rolcík J, Friml J. (2010) PIN phosphorylation is sufficient to mediate PIN polarity and direct auxin transport. Proc Natl Acad Sci USA 107: 918–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yang C, Li Y, Zheng N, Chen H, Zhao Q, Gao T, Guo H, Xie Q. (2007) SDIR1 is a RING finger E3 ligase that positively regulates stress-responsive abscisic acid signaling in Arabidopsis. Plant Cell 19: 1912–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.