Abstract
The cross between Arabidopsis thaliana and the closely related species Arabidopsis arenosa results in postzygotic hybrid incompatibility, manifested as seed death. Ecotypes of A. thaliana were tested for their ability to produce live seed when crossed to A. arenosa. The identified genetic variation was used to map quantitative trait loci (QTLs) encoded by the A. thaliana genome that affect the frequency of postzygotic lethality and the phenotypes of surviving seeds. Seven QTLs affecting the A. thaliana component of this hybrid incompatibility were identified by crossing a Columbia × C24 recombinant inbred line population to diploid A. arenosa pollen donors. Additional epistatic loci were identified based on their pairwise interaction with one or several of these QTLs. Epistatic interactions were detected for all seven QTLs. The two largest additive QTLs were subjected to fine-mapping, indicating the action of at least two genes in each. The topology of this network reveals a large set of minor-effect loci from the maternal genome controlling hybrid growth and viability at different developmental stages. Our study establishes a framework that will enable the identification and characterization of genes and pathways in A. thaliana responsible for hybrid lethality in the A. thaliana × A. arenosa interspecific cross.
Hybrid inviability hinders gene flow between different populations, thus contributing to their reproductive isolation (Mayr, 1942). It can result from the incompatible interaction of diverged genes, a mechanism postulated by the Dobzhansky-Muller (D-M) model of hybrid incompatibility (Dobzhansky, 1936; Muller, 1942; Fishman and Willis, 2001; Harushima et al., 2001; Myburg et al., 2004; Moyle and Graham, 2005). According to this model, two loci produce postzygotic incompatibility due to negative interactions between alleles that have fixed changes in the separated lineages. This negative epistasis causes deleterious phenotypes, such as necrosis and hybrid sterility, reducing hybrid fitness. In plants, a few documented cases fit the basic two-locus D-M model. For example, sterility in Mimulus species hybrids is caused by the interaction of a dominant and a recessive gene (Sweigart et al., 2006, 2007). Tissue death and dwarfism are characteristic of hybrid necrotic syndromes. Functional characterization of genes identified in necrotic tomato (Solanum lycopersicum), lettuce (Lactuca sativa), rice (Oryza sativa), and intraspecific Arabidopsis thaliana hybrid populations supports the hypothesis that necrosis results from the incompatible interactions between rapidly evolving pathogen resistance genes, such as those encoding nucleotide-binding site-Leu-rich repeat proteins (Dixon et al., 1996; Krüger et al., 2002; Rooney et al., 2005; Bomblies et al., 2007; Jeuken et al., 2009; Yamamoto et al., 2010; for review, see Bomblies, 2010; Rieseberg and Blackman, 2010). Necrotic intraspecific hybrids in A. thaliana are caused by the interaction of two genes that are known to encode disease resistance factors (Bomblies et al., 2007; Bomblies and Weigel, 2007); in lettuce, interspecific hybrid incompatibility has also been attributed to a single D-M interaction between RPM1 (for resistance to Pseudomonas syringae pv maculicola 1) interacting protein 4 (RIN4) and an unknown partner, which causes a deleterious, temperature-sensitive autoimmune response (Yamamoto et al., 2010). In summary, negative epistasis can arise between intraspecific and interspecific populations, and several cases are consistent with a simple genetic base (for review, see Presgraves, 2010; Rieseberg and Blackman, 2010).
Some cases of hybrid incompatibility, such as sterility, appear more complex and are attributed to more than two loci. It has been proposed that with increased time since speciation, independently arising incompatibilities will accumulate exponentially (Orr, 1995; Orr and Turelli, 2001; Moyle and Nakazato, 2010). Evidence for this “snowball” effect has been obtained in Drosophila (Matute et al., 2010) and Solanum (Moyle and Nakazato, 2010). In Solanum interspecies hybrids, the snowball effect is supported for sterility but not for viability traits, suggesting that alternative mechanisms may play a role. Multiple smaller interactions are also found in the cross between the yeasts Saccharomyces cerevisiae and Saccharomyces paradoxus (Kao et al., 2010) and between Eucalyptus species (Myburg et al., 2004). Theoretical elaborations of the D-M model typically assume that incompatibilities arise independently (Orr, 1995; Orr and Turelli, 2001). Yet, it also seems possible that incompatibilities could be connected, such as when small-effect polymorphisms accumulate in genes that are involved in a large network (Moyle and Nakazato, 2010). Such may be the case for incompatibilities underlying hybrid postzygotic lethality. Genetic and molecular characterization of the genes responsible for incompatibility should help address the basis and complexity of this phenomenon.
Here, we examine variation at A. thaliana loci that affect incompatibility with the related species Arabidopsis arenosa. Pollination of A. thaliana by A. arenosa most often results in the death of the F1 seed. Seed survival is possible, and a prehistoric hybridization event between the two species produced the extant allotetraploid species Arabidopsis suecica (Price et al., 1994; Jakobsson et al., 2006). Failure of the F1 seed in the A. thaliana × A. arenosa crosses (the seed mother is listed first) ranges from occasional to complete, depending on the dosages of the parental genomes. Remarkably, while tetraploid A. thaliana × diploid A. arenosa crosses produce almost normal seed, crosses between parents of equal ploidy produce only 2% to 10% live seed, and diploid × tetraploid crosses lead to complete seed failure (Josefsson et al., 2006). Therefore, quantitative variation in parentally supplied factors determines this incompatibility response (Dilkes and Comai, 2004). Knockout of one such factor, PHERES1, caused a small but statistically significant improvement in seed survival, suggesting that multiple genes could be involved in the interspecies barrier (Josefsson et al., 2006). This makes Arabidopsis an appealing system in which to test whether hybrid incompatibility is controlled by a single gene pair or whether a more complex incompatibility has developed since lineages diverged over 5 million years ago. To begin the study of this hybrid incompatibility system, we chose to elucidate first the maternal components, since the genome of A. thaliana is better characterized.
We surveyed 56 A. thaliana ecotypes for their response to hybridization with A. arenosa and found quantitative variation for seed viability. Two A. thaliana ecotypes, Columbia (Col-0) and C24, displayed moderate variation, and based on preexisting genetic tools, such as the availability of a good recombinant inbred line (RIL) and near isogenic line (NIL) population, these accessions were chosen to identify genetic factors encoded by the maternal genome that are involved in hybrid incompatibility. A quantitative trait locus (QTL) mapping approach led to the identification of seven A. thaliana QTLs, all of which participated in at least one epistatic interaction. Eight additional epistatic-only loci were identified by marker-by-marker regression analysis. Most QTLs had small additive and epistatic effects, except for two QTLs, FOE and FII, which accounted for 30% and 10% of the phenotypic variance observed in the RIL population, respectively. Interestingly, fine-mapping suggested that each of the two major QTLs encompass multiple loci. Together, these observations demonstrate that the hybridization barrier between A. thaliana and A. arenosa is controlled by many genes participating in a complex genetic network.
RESULTS
Natural Variation in Hybrid Compatibility among A. thaliana Accessions
To investigate natural variation in interspecies-hybrid compatibility, multiple A. thaliana accessions were crossed as seed parents to diploid A. arenosa accession Strecno-1 and scored for hybrid seed set. F1 plants from diploid ecotypes ranged from 0% (Borky, Bor-4; Cape Verde Island, Cvi-0; Lagostera, LL-0; and Nordwijk, Nok-3) to 30%, 20%, 19%, and 17% normal seed (Sq, SQ-1; Kazakhstan, KZ-9; San Eleno, Se-0; and C24, C24, respectively; Supplemental Table S1). The high compatibility of Warschau-1, which produced more than 50% normal seed, is explained by tetraploidy (Henry et al., 2005; Josefsson et al., 2006). We have previously demonstrated that the A. thaliana × A. arenosa cross is dramatically improved by maternal genomic excess, such as in this case (Josefsson et al., 2006). The next four high-ranking accessions following Warschau-1 (SQ-1, KZ-9, Se-O, and C24) were confirmed by flow cytometric analysis to be diploid. Thus, these four are likely to carry alleles that mitigate the hybrid incompatibility present between A. thaliana and A. arenosa.
Experimental Design
An A. thaliana accession with 0% survivorship of hybrids, Col-0, and another with 17% survivorship, C24, were chosen to investigate maternally contributed incompatibility genetic factors. The experimental design is given in Supplemental Figure S1. QTLs for incompatibility were mapped using a preexisting RIL population derived from Col-0 and C24 (Törjék et al., 2006). Mature seed phenotypes were analyzed to identify loci capable of modifying F1 incompatibility and to test for genetic interaction between QTLs. Backcross populations were then used to test heritability and to validate the QTLs identified by this approach. Transmission ratio distortion (TRD) in F1 × A. arenosa crosses was used to identify which incompatibility loci, if any, were under selection during seed development.
Phenotypic Analysis of Interspecies Hybrids, and Heritability of Hybrid Seed Survival in Arabidopsis
We selected phenotypic classes suitable for testing genetic effects at specific stages of seed development: (N) normal, (G) green, (V) viviparous, and (S) shriveled/dead (Fig. 1A). The shriveled seeds were black and nearly flat, with very little volume. We infer that they fail soon after fertilization, before a discernible (when dried) amount of endosperm and embryonic tissue has formed. Inspection of developing seeds at 10 to 12 d after pollination (DAP; Fig. 1B) is consistent with this interpretation, as shrunken and dark-brown seeds were found to contain globular embryos, or no visible embryos, or cellularized endosperm, such as observed previously (Bushell et al., 2003). Green seeds contained a discernible embryo and green endosperm mass, consistent with successful early development followed by arrest at a later stage. Viviparous seeds displayed a radicle or cotyledons emerging through the seed coat, indicative of advanced embryo development, but failed maturation and dormancy. Normal seeds properly reached maturity, displaying tan seed coat color and a complete, dormant embryo that filled the seed compartment, resulting in a regular and plump seed shape (Fig. 1B). Based on phenotypic analysis of hybrid seed development, we conclude that shriveled, green, viviparous, and normal seeds are distinct traits. These seed traits are distinct to hybrids, as observations of multiple reciprocal crosses between parents (Col-0 × C24 and C24 × Col-0) did not reveal shriveled, viviparous, or green seeds. As such, it is unlikely that incompatibility results from deleterious alleles derived from A. thaliana parents. Finally, while A. thaliana and A. arenosa hybrids that form normal seeds germinate readily, the resulting adults are invariably sterile due to differences in chromosome number between parents. Therefore, phenotypic analysis of seed classes in subsequent generations was not possible.
Figure 1.
Phenotype classification scheme for hybrid seeds and influence of maternal genotype in the A. thaliana × A. arenosa cross. A, Four phenotypic classes were observed in the output from the hybrid crosses: shriveled, green, viviparous, and normal seed. The mature phenotype of these seeds corresponds to defects in major transitions in seed development. B, Visualization of a C24 × A. arenosa 10-DAP silique shows normal and shriveled seeds. The normal seed has an embryo (EM) that fills the seed compartment, whereas the shriveled seed has an embryo arrested in the heart stage. C, The maternal A. thaliana genotype used in the cross to A. arenosa is indicated on the x axis. Bar heights represent mean %NGV values, and se is displayed as black error bars. Significant differences between genotype mean values (P = 0.05) are represented by different letters above the bars (same letter, no significant difference in mean value). Mean numbers are based on data from 15 plants (approximately 1,500 seeds) for each of the Col-0, C24, and F1 crosses, 80 plants (approximately 22,900 seeds) for BC1-Col-0, and 91 plants (approximately 23,300 seeds) for BC1-C24.
To test heritability, the parental ecotypes C24 and Col-0, their F1 offspring, and two BC1 populations were grown and crossed as seed parents to diploid A. arenosa. The means and sd of seed set from these interspecies hybrids are presented in Figure 1C. The F1 mean of %NGV (normal + green + viviparous = 5.7%) fell between the two parental means but differed from the midparent value (12.0%), consistent with nonadditivity. The BC1 populations more closely resembled their respective recurrent parent, indicating that hybrid compatibility is a heritable trait. The BC1-Col-0 population mean (3.9%) was not significantly different from the Col-0 parental mean (1.3%). The BC1-C24 population mean (11.4%) differed from both the Col-0 and BC1-Col-0 means. It fell below the C24 parental mean (22.6%) and short of the predicted value for a fully additive trait, supporting the existence of nonadditive variation in this trait. Despite the general decreases in mean values within both BC1 populations, some individuals were similar to the C24 parental mean, and within the BC1-C24 population, some individuals surpassed the C24 mean. The transgressive phenotype of some BC1-C24 lines suggested the presence of Col-0 alleles with a positive effect.
Based on phenotypic variation and heritability estimates, we performed a genome-wide scan for QTLs by crossing individuals of each RIL to diploid A. arenosa and scoring five seed traits: %N, %G, %V, %NGV, and the percentage of seed that reach full maturation (%N/NV; Fig. 2). The %NGV trait was chosen to define seed that progressed through early development and produced a certain-sized embryo. The %N/NV trait defined seeds that completed normal development. Seeds in both classes achieved advanced development but varied in maturity and dormancy. Most RILs had low mean phenotypic values, similar to the Col-0 parent, while only a few lines equaled or surpassed the C24 mean. This may be due to epistatic effects, as suggested by the nonadditivity displayed in BC1 and F1 (Fig. 1C). Such a skewed distribution is not well modeled by most QTL methods. Therefore, all data were log transformed to approach normality.
Figure 2.
Histograms summarizing the frequency of seed classes in RIL × A. arenosa progeny. The means for the %N, %G, %V, %NGV, and %N/NV (x axes) were calculated from six replicates of each RIL × A. arenosa, and the number of RILs with a given mean value are tallied on the y axis. The means of the entire RIL population, the Col-0 parent, and the C24 parent are indicated by labeled arrows. The distribution of phenotypic performance depends on the trait. For %NGV, and somewhat less for %N, %G, and %V, most RILs fall close to the lower value Col-0 parent. For %N/NV, most lines perform close to the parental midpoint. For all traits, a few lines equal or surpass the mean values of C24.
Trait Correlation and Composite Interval Mapping
Correlation was used to test relationships between the five traits. Correlation values for several trait pairs determining the early and mid stages of hybrid seed development were strong (Table I), indicating that they are most likely affected by similar genes. The %N/NV trait, however, was not correlated with any other trait, suggesting that early hybrid embryo development and seed maturation are controlled by distinct genetic pathways. Likewise, the correlation between the %N and %V traits was not significant, suggesting a separate control for dormancy.
Table I.
Correlations between hybrid seed trait values from 150 Col-0/C24 RILs of A. thaliana
Significant at P < 0.0001.
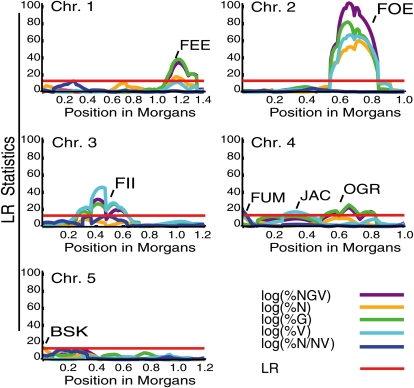
Composite interval mapping using RIL mean values was used to identify QTLs affecting each of the five traits. A plot of the likelihood ratio statistics for each trait, with significant QTLs marked, is presented in Figure 3. For five QTLs, named from the fable of Jack and the Beanstalk (Jacobs, 1902), the C24 allele had a positive effect on the trait mean, while the Col-0 allele had a positive effect on the remaining two (Supplemental Table S2, Col-0 allele-effect column), consistent with the transgressive phenotypes observed in the BC1-C24 population (Supplemental Table S3) and RIL populations (Fig. 2). The QTL additive effect on phenotype is given by Supplemental Table S2 (PVE column). Based on the observed phenotypic variance, the FOE and FII loci are responsible for major effects on the trait values. The colocalization of QTLs affecting different traits (Fig. 3), along with their trait correlations, suggest that several QTLs are pleiotropic. For example, the C24 allele at FOE on chromosome 2 affects an increase in the %N, %G, and %V values as well as in the combined %NGV measure, suggesting that FOEC24 is involved in a developmental transition common to all three traits, perhaps an early event critical to seed development, such as endosperm cellularization. Similar to what was observed in the trait correlation analyses, FUM, the QTL affecting the percentage of seed that reach full maturation (%N/NV), did not colocalize with QTLs affecting any other traits, suggesting independent controls for maturation and dormancy. On the other hand, the %N and %V traits were influenced both by trait-specific effect QTLs BSK and JAC, respectively, and common QTLs including FEE, FOE, and FII.
Figure 3.
Likelihood ratios for QTLs affecting seed set. Composite interval mapping was performed using trait mean values and marker genotypes from 150 A. thaliana RILs crossed to A. arenosa. Seed set was quantified by counting the number of seeds belonging to each of four classes, normal, green, viviparous, and shriveled, and used to calculate five traits: %NGV, %N, %G, %V, and %N/NV. Data were log transformed to improve normality. A line at a likelihood ratio (LR) of 13 is drawn to approximate statistical thresholds calculated by 1,000 permutations. The estimated thresholds for each trait were as follows: %NGV, LR ≥ 12.9; %N, LR ≥ 13.2; %G, LR ≥ 13.3; %V, LR ≥ 13.3; %N/NV, LR ≥ 13.0. All significant QTLs detected by this analysis are marked.
Confirmation of QTLs in BC1 Populations
To confirm the QTLs and heritability, we employed backcross populations to recurrent parents Col-0 and C24. The two BC1 populations were crossed to A. arenosa for phenotyping. Loci corresponding to FEE, FOE, FII, OGR, and JAC were specifically tested and found to influence hybrid seed survival in the predicted manner (Supplemental Table S2, Col-0 allele-effect column), at least for the %NGV trait (Supplemental Table S3). For example, BC1 populations that were C24 at FEE (nga280 marker) had significantly higher %NGV and %N than those with the Col-0 allele (Supplemental Table S3). The results further demonstrated the high heritability of these traits. Lack of a significant effect in this experiment might be due to a lack of statistical power associated with the small population size or to an insufficient effect on the phenotype by heterozygous alleles. QTL detection in backcross populations is based on the assumption that QTL alleles are fixed. If alleles are still segregating, analysis of multiple hybrids could lead to different results in different families, thus reducing power in QTL detection.
Epistatic Interaction Analysis
Due to the nonadditivity observed in both the RIL and earlier filial generations, we sought to determine if the hybrid compatibility traits are affected by interactions among genetic loci. We applied a pairwise regression test on markers underlying QTLs with the R statistical suite. Filtering for P < 0.01 and at least three contiguous genetic intervals, we identified 36 epistatic interactions for the five traits (Fig. 4; Supplemental Data S1; Supplemental Table S4; Supplemental Figs. S2–S9). Many of the significant epistatic interactions were detected in multiple trait analyses (Fig. 4; Supplemental Table S4; Supplemental Figs. S2–S9). For example, FEEC24 and OGRC24 are predicted to improve hybrid seed survival above the expected additive value of the genotype/locus combination for the %N trait (Supplemental Table S4).
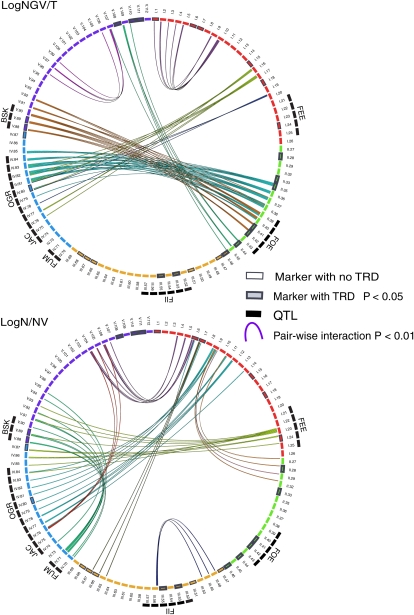
Figure 4.
Loci determining hybrid compatibility interact in a genetic network. The circles depict the A. thaliana genetic map, with intervals represented by circularly arrayed bars colored according to chromosome. A total of 15 epistatic interactions were identified for two of the traits under investigation: %NGV (top circle) and %N/NV (bottom circle). All seven additive QTLs (black outer bars) are involved in at least one epistatic interaction (colored lines). Genetic regions tested for TRD are outlined in gray, with thicker lines and gray fill for those that display TRD (P < 0.05). There are very few interactions shared by early seed traits (%NGV) and maturation/dormancy (%N/NV).
The topology of interactions indicates a complex network. As suggested by the regression test, some interactions are common to multiple traits. For example, the FOE QTL has significant interaction with BSK for %NGV, %N, and %G (Fig. 4; Supplemental Figs. S2 and S3). Interactions underlying early seed development, however, appear to be distinct from those governing maturation and dormancy, with no shared connections between %NGV and %N/NV. For most epistatic interactions, the two most beneficial alleles displayed positive epistasis (Supplemental Figs. S5–S9). Furthermore, RIL hybrids that surpass the C24 mean for both %NGV and %G traits displayed significant overrepresentation of the beneficial allele at the FOE, OGR, and BSK loci (χ2 P < 0.01). Based on additive and epistatic effects on the phenotype, FOE, OGR, and BSK are likely to be major determinants of hybrid seed survival (Supplemental Tables S2 and S3). In general, compared with the additive effect of the QTLs involved in interactions (Supplemental Table S2, PVE column), each epistatic interaction has a relatively small effect on the phenotypic variance, but the high number of interactions suggests that together they have considerable influence on the phenotype (Supplemental Table S4; Supplemental Figs. S2–S9).
Fine-Mapping Analysis of Major-Effect Loci FOE and FII
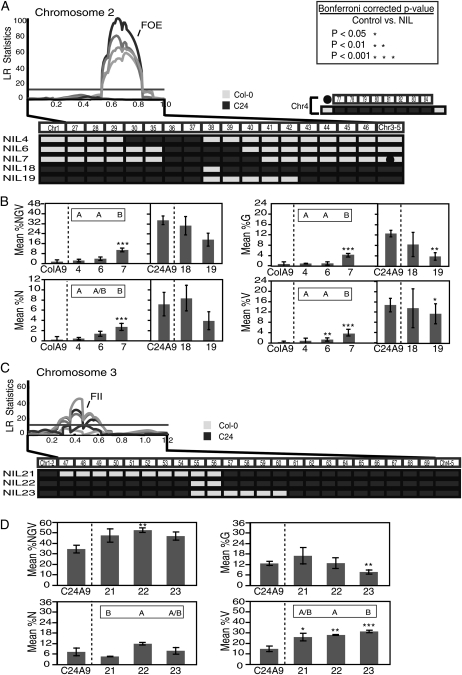
Based on their relatively large additive effects, FOE and FII were selected for further characterization. Interval-specific contrast was used to more precisely define QTL position (Fig. 5). Eight NILs, containing introgressed segments spanning either the FOE (Fig. 5A) or the FII (Fig. 5C) locus, were selected. The lines were selected from a population of 140 NILs that, by genotyping at 111 single-nucleotide polymorphisms, were confirmed to have one or two introgressions (Törjék et al., 2008). Each NIL was crossed to the background parent in order to generate NIL F1 plants (male-sterile lines of the background parents were used as females in these crosses). Even though fine-mapping with NIL F1s has the potential for reduced power, we chose to make lines heterozygous in order to test TRD as well as QTL effect in the interspecific cross (see below). Each NIL F1 was crossed to A. arenosa to assess whether the given introgression impacted hybrid seed set (Fig. 5, B and D).
Figure 5.
Fine-mapping of major-effect QTLs reveals multiple loci. A and B, Dissection of the FOE QTL. A, The QTL likelihood plot with FOE localization on chromosome 2 (Fig. 3) is displayed above the map of five NILs with introgressions spanning the FOE QTL. NIL4 to -7 have the Col-0 genomic background, while NIL18 and NIL19 have the C24 background. All contain one introgressed segment, displayed at the bottom, except for NIL7, which contains a C24 introgression on chromosome 4 (middle right). LR, Likelihood ratio. B, Histograms depicting percentage values for four seed traits under investigation (%NGV, %N, %G, and %V). The phenotype was evaluated in NIL F1 × A. arenosa crosses compared with crosses between the background parent (either Col-0 or C24) and A. arenosa (x axis). The mean for each cross is given (y axis) with sd bars. Asterisks denote significant differences in trait means from parental control values based on unpaired Student’s t test. Corresponding P values are given at top right in A. Letters above each NIL denote significant differences in NIL-versus-NIL trait mean comparisons (P < 0.01). The absence of letters above the histogram indicates that the genotypes were not significantly different. All data were derived from three independent biological replicates, and P values have been Bonferroni corrected to reflect multiple tests. C and D, Dissection of the FII QTL. C, The QTL plot displays FII localization on chromosome 3 as well as three NILs with introgressions spanning the FII QTL. All NILs are in the C24 background with one Col-0 introgression. D, Same as B.
Multiple genes within the FOE region impact seed survival. NIL7 was significantly different from its background parent for all traits (P < 0.0001). NIL6, which carries most of the introgressed fragment of NIL7, was only significantly different from its background parent for %V. This suggested a factor in the segment carried by NIL7 but not NIL6 (gene A, II.40-II.41). Since the NIL6 introgression significantly affected vivipary (%V), it may carry a gene specifically involved in this response (gene B, II.38-II.40). For example, gene A could control early events in seed development, while gene B could contribute to the induction of dormancy. The Col-0 lines carrying introgressions across the FOE QTL allow for similar observations (Fig. 5B). NIL19 is significantly different for the %G and %V traits, while NIL18 has, although not significant, decreased seed survival for the %NGV, %G, and %V traits. To further evaluate the effect of the second introgressed segment of NIL7 (chromosome 4 IV.77-IV.84; Fig. 5A), NIL7-derived F2s segregating for the C24-derived chromosomal segment were produced. Comparison of F2s confirmed a positive effect of the C24 segment on seed survival (1.81-fold increase in %NGV; χ2 P < 0.001). Because there was not a significant effect on normal seed (1.2-fold change; P = 0.32), interval IV.77-IV.84 is most important for green and viviparous seed traits. Even without the chromosome 4 introgression, NIL7 was still significantly different from the background parent for all seed traits except %G (n = 3; t test P values are as follows: %NGV, P = 0.0003; %N, P = 0.007; %G, P < 0.02; %V, P = 3.5E-05).
Like the FOE locus, the FII locus also appears to be a composite QTL. Each NIL was significantly different from its background parents for %V (Fig. 5D). This suggests a positive factor from Col-0 in the segment shared by all three NILs (gene C, III.55-II.56). Additionally, NIL23 had significantly lower green seeds than C24 (Fig. 5D, %G). Perhaps a Col-0 gene (gene D, III.57-III.60) that negatively impacts %G at the expense of %V is present in that region. Interestingly, this locus was not detected in the composite interval-mapping experiment, although it was detected in epistatic analysis (Supplemental Figs. S2 and S3). Line NIL21 had significantly lower mean %N seeds than NIL22, suggesting the presence of a third Col-0 locus (gene E, between III.47 and III.55).
In conclusion, our results suggest that the FOE QTL might be made up of at least two genes (genes A and B). Similarly, the FII QTL might comprise one subregion (gene C) in which the Col-0 allele positively impacts %V, with at least two additional flanking genes (genes D and E) that act in trans to negatively influence %N and %G. Confirmation of candidate genes could be challenging, as it appears that many genes, some with positive effects and others with negative effects on seed development, are relatively closely linked.
TRD Analysis in F1s and NILs
A TRD experiment was performed to measure whether Col-0 and C24 A. thaliana alleles were selected in the interspecific hybrid offspring of a [Col-0 × C24] F1 × A. arenosa cross and in NIL F1 × A. arenosa crosses. Distortion (deviation from the expected 1:1 ratio) indicates action before fertilization in the female gametophyte or after fertilization in the hybrid zygote. In contrast, loci acting through the maternal sporophyte cannot be detected, since all [Col-0 × C24] F1 sporophytes share the same genotype. As a control, TRD was examined in a [Col-0 × C24] F1 × Col-0 cross and NIL BC1 populations. Markers linked to FEE, FOE, FII, OGR, and BSK and epistatic-only loci OMC, MGC, HRP, OVN, MUF, and GLD were scored in offspring from the interspecific hybrid and control crosses. Markers linked to two QTLs, FOE and FII, and three epistatic-only QTLs, HRP, OVN, and GLD, were significantly distorted from the expected 1:1 transmission ratio in the interspecific hybrid offspring (Table II) while displaying no, or opposite (FII), distortion in the control cross to Col-0, with the exception of GLD (Supplemental Table S5). For each marker tested, the beneficial allele, as defined by composite interval mapping (Supplemental Table S2) or epistatic analysis (Supplemental Table S4; Supplemental Figs. S2–S9), was preferentially transmitted. The TRD in NIL F1 individuals confirmed the presence of factors acting in the FOE QTL at II.38 and in the FII QTL at III.55-III.56 (Table II). Because GLD is also distorted in control BC1 populations, this suggests that the action of GLD is not unique to the hybrid crosses.
Table II.
TRD tested on selected QTLs identified by composite interval mapping and epistatic analysis
| QTL (Chromosome) | Line × A. arenosa | Marker | No. of Offspring Tested | P | Allele with Higher Frequencya | Fold Overrepresentation |
| OMC (1) | F1 | MSAT1.3 | 95 | nsb | None | ns |
| MGC (1) | F1 | T27K12 | 95 | ns | None | ns |
| FEE (1) | F1 | nga692 | 95 | ns | None | ns |
| HRP (2) | F1 | MSAT2.28c | 95 | 1.7E-07 | C24 | 3.32 |
| FOE (2) | F1 | nga361 | 95 | 1.0E-10 | C24 | 4.75 |
| FOE (2) | NIL7 F1 | MSAT2.11 | 54 | 0.001 | C24 | 2.38 |
| FOE (2) | NIL18 F1 | II.38 | 60 | 0.02 | C24 | 1.86 |
| FOE (2) | F1 | nga168c | 94 | 6.1E-10 | C24 | 4.53 |
| OVN (2) | F1 | MSAT2.33c | 95 | 5.4E-08 | C24 | 3.52 |
| MUF (3) | F1 | NGA172 | 95 | ns | None | ns |
| FII (3) | F1 | GAPAbc | 94 | 0.002 | Col-0 | 1.94 |
| FII (3) | NIL21 F1 | III.56 | 231 | 1.8E-10 | Col-0 | 2.45 |
| FII (3) | NIL22 F1 | III.55 | 282 | 3.0E-18 | Col-0 | 3.48 |
| FII (3) | NIL23 F1 | III.55 | 306 | 1.5E-7 | Col-0 | 1.86 |
| OGR (4) | F1 | MSAT4.19 | 95 | ns | None | ns |
| BSK (5) | F1 | CTR1 | 95 | ns | None | ns |
| GLD (5) | F1 | MSAT5.19 | 95 | 0.006 | C24 | 1.79 |
The genotype of the preferentially transmitted allele is indicated.
ns, Nonsignificant.
See Supplemental Table S5 for controls (F1 × Col-0) for selected significantly distorted markers.
DISCUSSION
Implications of Our Experimental Design
We performed a genetic analysis of the A. thaliana genome to identify survival determinants for the A. thaliana × A. arenosa hybrid. The scope of the analysis differs from comparable studies in postzygotic incompatibility, presenting both disadvantages and advantages. By being limited to one genome, that of A. thaliana, this study did not explicitly probe for the existence of D-M loci. For example, if a single D-M pair is responsible for the incompatible response, we could detect the locus of the pair residing in the A. thaliana genome. Characterization of the epistatic nonorthologous locus in the A. arenosa genome would be required to establish conformity to the D-M model. By contrast, the study by Moyle and Nakazato (2010), which detected the snowballing increase of inviability D-M loci in diverging Solanum species, explicitly tested intergenomic epistasis between putative D-M pairs by phenotying seed formation in tomato lines with fragments of introgressed wild Solanum species. This approach, however, could not test the intragenomic epistasis of incompatibility loci, which would have required concurrent introgression of two or more chromosomal segments. A study on Eucalyptus interspecific hybridization mapped parental allele distortion in pseudo-backcrosses between the interspecific F1 generation and its parental species. In theory, it was suited to detect intragenomic epistasis, but it did not detect any (Myburg et al., 2004). Our approach (Supplemental Fig. S1) is suited to the discovery of QTLs responsible for variation in A. thaliana performance during interspecific hybridization as well as to the analysis of interactions between QTLs that contribute to incompatibility. Additionally, because viability is measured in F1 hybrid seed, chromosomal rearrangements differentiating the two species are controlled for.
A Set of Pleiotropic QTLs Influence Seed Development and Postzygotic Lethality
Composite interval mapping identified seven additive loci. Some of the hybrid seed survival traits considered in our experiment show strong correlation (Table I), and this correlation is reflected in the QTLs detected by these traits. In most cases, alleles at these QTLs affected both traits in the same direction, rather than causing the number of green seeds to go up at the expense of the number of normal seeds, for example. The pattern of correlation and shared QTL positions suggests underlying pleiotropic factors that affect several developmentally interdependent steps required for hybrid seed survival. While late-seed traits should also be correlated (Fig. 1A), we found that independent responses determine the number of seeds that will become viviparous. This is evident both by the lack of correlation between viviparous seed (%V) and normal seed (%N; Table I) and by the action of trait-specific QTLs that only participate in epistatic interactions early or late in development (i.e. FUM and JAC; Fig. 4; Supplemental Figs. S2–S9). The number of loci affecting all seed traits exceeds those found in Eucalyptus and tomato (Myburg et al., 2004; Moyle and Graham, 2005).
All but one locus were confirmed in the Col-0/C24 BC1 populations (Supplemental Table S3), despite modest population sizes and weak statistical power in this experiment. Moreover, the allelic effects at each genomic region matched that defined by the RIL experiment to predict hybrid seed survival. This result demonstrates that the mechanism responsible for incompatibility can act in early generations, and there is strong support for multiple incompatibility loci.
Nonadditive Genetic Effects on Hybridization Efficiency
The deviation of the [Col-0 × C24] F1 mean from the midparent value (Fig. 1C) suggested that nonadditive genetic mechanisms act on hybrid seed survival. Accordingly, all seven additive QTLs affected some trait through epistatic interactions that were positive and improved hybrid seed survival above the expected additive value of the genotype/locus combination; the alleles at QTLs that improved crossing success when considered in isolation provided the best combinatorial genotype in the interaction test. This gave rise to a pattern of positive synergistic interaction between alleles in which the best combination was always significantly different from the next-best genotype (Supplemental Figs. S5–S9). We plan to further test this statistical epistasis by creating NILs combining the best alleles at multiple QTLs.
Many Genes Contribute to Incompatibility
QTL identification and fine-mapping of FOE and FII were consistent with the presence of multiple small-effect incompatibility genes instead of a single gene. Together with the degree of epistasis observed in the RIL population (Fig. 4), this suggests that the observed incompatible response could involve a perturbed network of genes.
TRD analysis of selected QTLs in a [Col-0 × C24] F1 × A. arenosa cross and a NIL F1 × A. arenosa cross indicated that two loci, FOE (HRP/OVN) and FII, act on hybrid seed survival either in the female gametophyte before fertilization or in the zygote after fertilization (Table II). For two loci, the allele with higher frequency in the interspecific progeny corresponded to the allele that increased seed set in the RIL analysis: FOEC24 on chromosome 2 and FIICol-0 on chromosome 3. GLD was not identified by composite interval mapping, but it also appeared to impact seed survival, based on preferential transmission and epistatic interactions in both hybrid and control crosses. TRD in Col-0/C24 recombinant inbred line populations has been attributed to negative epistatic interactions between two loci, one on chromosome 4 and another on chromosome 5 (Törjék et al., 2006). While the distortion observed at FOEChr.2 and FIIChr.3 does not overlap with the loci identified by Törjék et al. (2006), markers linked to GLDChr.5 were also distorted in these RIL populations. Törjék et al. (2006) found that plants homozygous for Col-0 at MASC09215 (IV.81) and homozygous for C24 at MASC04350 (V.112) have low fitness due to reduced pollen fertility. In our epistatic analysis and segregation tests (Fig. 4; Table II; Supplemental Figs. S2–S9; Supplemental Table S4), the loci described by Törjék et al. (2006) did not interact, and segregation distortion indicates that hybrids that are C24 on chromosome 5 actually have higher fitness. Clearly, GLD modulates both prezygotic and postzygotic hybrid fitness, and it might be worth investigating whether it acts similarly in intraspecific and interspecific hybrid populations.
Preferential inheritance of incompatibility factors suggests that these loci encode factors that can modulate hybrid survival in the female gametophyte or fertilization products. Many genes with an effect on endosperm development and seed growth have been shown to be imprinted or to be required in the female gametophyte before fertilization (for review, see Makarevich et al., 2006). We previously showed that the transcriptional activation of imprinted genes occurs in the developing hybrid seed and that the degree of activation is negatively correlated with the rate of hybrid seed survival (Josefsson et al., 2006; Walia et al., 2009). An alternative hypothesis for incompatibility may be based on the duplication of a required factor followed by the inactivation of different duplicates in the parental lines. As demonstrated by Bikard et al. (2009), this leads to F2 lethality and is inconsistent with the observation of F1 lethality.
Although genetic incompatibility is often thought of as resulting from a deleterious direct interaction of gene products in the zygote, gene action in the gametophyte before fertilization may favor the survival of zygotes later on as well. For example, a larger gametophyte may provide more maternal resources and therefore may have an advantage during hybridization, similar to observations in 4× A. thaliana × 2× A. arenosa interspecies crosses (Josefsson et al., 2006). Similarly, alleles favoring the accumulation of factors advantageous during hybridization would increase in relative frequency among the hybrid progeny. This quantitative response, which may also arise due to selection favoring the allele in the zygote, is consistent with the dosage effect observed during hybridization.
What might be the molecular nature of these QTLs, and how do the underlying genes interact with the paternal A. arenosa genome to determine hybrid seed survival? Different kinds of interactions can be envisioned: temporal and physiological interplay between morphological structures of the seed, interactions between regulators and targets in a transcriptional network, or structural interactions between protein components of a regulatory complex. For example, necrotic responses caused by misregulation of defense mechanisms have been observed in hybrids (Dixon et al., 1996; Krüger et al., 2002; Rooney et al., 2005; Bomblies et al., 2007; Jeuken et al., 2009; Yamamoto et al., 2010), although not in seeds. The developing seed requires coordinated growth of at least three structures: the sporophytic seed coat, the endosperm, and the embryo. Coordination between growth of the seed coat and expansion of the endosperm is important for regulating seed growth (Garcia et al., 2005). Changes in one structure, such as the seed coat, may affect hybrid seed development and could be ameliorated or worsened by changes in another structure, such as the endosperm. The dependency of the embryo on the endosperm, and their mutual dependency on parental contributions, make this system potentially more susceptible to developmental perturbations and to the action of modifier genes. Indeed, evidence for regulatory mechanisms affecting interspecific hybridization via the seed parent (maternal sporophyte) has been described (Dilkes et al., 2008). These genes could be critical timing determinants of seed coat and endosperm growth, interacting components in a heterochromatin-silencing complex, or perhaps transcriptional master regulators affecting seed growth.
In summary, the phenotype of hybrid seed survival in the cross between A. thaliana and A. arenosa is determined by a suite of networked loci. It is possible, therefore, that the Arabidopsis network may consist of many D-M gene pairs or a single gene pair with an associated network of modifiers. The numbers and effects of the genes involved suggest that a viable strategy to elucidate the molecular basis of the postzygotic incompatibility response in Arabidopsis may involve coupling mapping information to a comparison of molecular responses in compatible and incompatible crosses through development. We have laid the foundation for future fine-mapping, epistasis tests, molecular cloning experiments, and expression profiles of hybrid seeds that will reveal the identities of these genes.
MATERIALS AND METHODS
Plant Material
The diverse ecotype set of Arabidopsis thaliana (for accession numbers, see Supplemental Table S6) is available from The Arabidopsis Information Resource (www.arabidopsis.org). A subset of 150 Col-0/C24 RILs were selected from 423 lines (Törjék et al., 2006), as described below. The diploid Arabidopsis arenosa var Strecno-1 (provided by Dr. M. Lysak) was collected from the wild in Strecno, Slovakia, by Dr. Marcus Koch. Male-sterile C24 (Paul et al., 1992) and its seventh generation backcross to Col-0 (gifts from Rod Scott and Rinke Vinkenoog), whose transgenic male sterility is encoded between markers MSAT5.22 and MSAT5.10 on chromosome 5, were used as maternal parents to create F1s of NILs and two backcross (BC1) populations: BC1-Col-0 (80 individuals) and BC1-C24 (91 individuals). NILs were selected from 140 isogenic lines (Törjék et al., 2008). Plant growth was in a 16-h light cycle and 22°C. Flow cytometric analysis of nuclear genome content (Henry et al., 2005) confirmed diploidy of highly compatible ecotypes and RIL. The reciprocal cross (A. arenosa × A. thaliana is prevented by pollen-style unilateral incompatibility (Lewis and Crowe, 1958).
Phenotyping
Temporally staggered A. thaliana plants were pollinated by a single diploid A. arenosa Strecno-1, referred to as “big pappa.” A. thaliana plants were emasculated before anthesis, pollinating the fully developed stigmas 1 d later. Crosses to male-sterile A. thaliana lines were done on open flowers having well-developed stigmas. F1 seeds were classified according to their phenotype at maturity (Fig. 1A). The mean number of seeds counted per RIL was 641 (range, 249–957; sd, 137). The mean number of seeds counted per NIL cross was 1,161 (range, 203–3,253; sd, 710). The A. thaliana (n = 5) × A. arenosa (n = 8) diploid F1 hybrids are sterile due to chromosome incompatibilities, so generational observations were not possible.
Microscopy
Hybrid siliques were dissected and visualized with a stereomicroscope at 5 to 14 DAP. Seeds were then cleared using Hoyer’s medium (chloral hydrate:distilled water:glycerol, 80:30:10 g) and imaged on a Leica DM-6000 using differential interference contrast microscopy. Photographs for Figure 1B were taken using a DFC 350 FX camera (Leica) and processed with Leica Application Suite version 3 to adjust brightness and contrast.
Selection of RILs for QTL Mapping
We used the default settings of the SAMPLEEXP function of MapPop (Vision et al., 2000) to select from the Col-0/C24 RILs 75 lines from each reciprocal cross containing the maximum number of complementary crossover sites and the smallest possible crossover intervals.
Linkage Map Construction
MapManager QTXb20 (version 0.3; Manly et al., 2001) was used to estimate the distances between the 105 markers genotyped in the RIL population (chromosome 1, 26; chromosome 2, 17; chromosome 3, 21; chromosome 4, 17; chromosome 5, 24). Marker distances were estimated based on their observed recombination in the 150 selected RILs using the Haldane mapping function of MapManager QTXb20. The total map length generated by the data set was 586 centimorgan.
QTL Analyses
Phenotypic data were log transformed to account for nonnormality in the data and to restore residual variation to normality. Five hybrid seed traits were analyzed: %NGV (i.e. seed that advanced substantially through development; embryo 20% of seed volume), %N , %G, %V, and %N/NV (i.e. viable, dormant embryos). Since trait values of zero interfered with the log transformation, we used pseudocounts (addition of 1 to all phenotypic counts; Laplace’s rule of succession [Laplace, 1995]) in the analysis.
Zmapqtl in QTL Cartographer (version 1.17) was used to calculate likelihood ratio statistics (Basten et al., 1994). Mapping parameters used were model 6, a 15-centimorgan window, and a maximum of 10 background markers. A statistical threshold value of α = 0.05 was determined by 1,000 permutations for each trait (Churchill and Doerge, 1994). The criterion used to define QTL intervals was 2 log of the odds (=9.22 likelihood ratio units) away from each side of the peak QTL marker.
Fine-Mapping
A subset of the QTLs identified in the RIL population were further tested. Individuals in two BC1 populations, BC1-Col-0 (n = 80) and BC1-C24 (n = 91), three replicates of NIL F1 spanning FOE and FII, as well as parental controls ColA9 and C24A9 were pollinated with A. arenosa, phenotyped for hybrid seed set, and genotyped at markers linked to the QTL. Unpaired Student’s t test (NIL versus control and NIL versus NIL) with Bonferroni-corrected P value (Curtin and Schulz, 1998) were used. The effect of two introgression NILs (i.e. NIL7) was assayed in F2 by phenotyping lines with and without a second introgression and calculating fold change in seed set for six biological replicates (mean n = 458 seeds; sd = 167). Reciprocal crosses were performed to minimize maternal effects. A χ2 test was performed (Bonferroni-corrected P < 0.001).
Segregation Distortion
A TRD analysis was performed on surviving progeny from [Col-0 × C24] F1 × A. arenosa and NIL F1 × A. arenosa hybrid crosses. Marker genotypes were determined by gel electrophoresis of the PCR product according to a standard procedure. Markers can be found at The Arabidopsis Information Resource (www.arabidopsis.org); new markers are listed in Supplemental Table S7. Marker deviations from the expected 1:1 transmission ratio of Col-0:C24 alleles to the offspring were detected via χ2 tests.
Epistatic Analysis
Pairwise marker combinations were evaluated for epistatic interactions by regression. The significance of interaction effects for markers at QTL peak positions was determined using an ANOVA type III F statistic in R with the model T = μ + m1 + m2 + m1 × m2, where T corresponds to mean seed class frequency and m1 and m2 are marker genotypes. This derives a P value for the interaction between markers underlying QTL (m1 × m2). Whole-genome interaction plots (Supplemental Figs. S2–S4) were generated using Circos (Krzywinski et al., 2009), filtering for marker pairs where P < 0.01 and supported by at least three adjacent interactions. The most significant pair for each set of linked pairs was evaluated in more detail (Supplemental Figs. S5–S9) by regression using a fit model (least squares, means) in JMP 6.0 (SAS Institute). The interaction terms were calculated by dividing the explained sum of squares for the regression coefficient in the multiple regression by the total sum of squares for the trait.
Supplemental Data
The following materials are available in the online version of this article
Supplemental Figure S1. Experimental design.
Supplemental Figure S2. Graphical representation of pairwise interactions for %N trait.
Supplemental Figure S3. Graphical representation of pairwise interactions for %G trait.
Supplemental Figure S4. Graphical representation of pairwise interactions for %V trait.
Supplemental Figure S5. Least squares means plots for the %NGV trait.
Supplemental Figure S6. Least squares means plots for the %N trait.
Supplemental Figure S7. Least squares means plots for the %G trait.
Supplemental Figure S8. Least squares means plots for the %V trait.
Supplemental Figure S9. Least squares means plots for the %N/NV trait.
Supplemental Table S1. Range of %normal seed produced in hybrid crosses.
Supplemental Table S2. QTL interval and likelihood ratio statistics.
Supplemental Table S3. Seed phenotypes of BC1 populations.
Supplemental Table S4. Epistatic interactions by trait as identified by marker-by-marker regression test.
Supplemental Table S5. Controls for transmission ratio distortion (TRD) analysis.
Supplemental Table S6. A. thaliana ecotype seed stock ID.
Supplemental Data S1. Summary of marker-by-marker regression test, M1*M2 interaction P values by trait.
Supplementary Material
Acknowledgments
We thank Rebecca Doerge and Alexander Lipka for verifying our QTL analysis. We thank Julin Maloof, Dan Kliebenstein, Diana Zwerling, Sam Hwang, Brian Watson, Kathie Ngo, and Sameer Thadani for technical assistance or suggestions, and we thank Douglas Ewing and the staff of the New Botany Greenhouse at the University of Washington for assistance in plant growth.
References
- Basten C, Weir B, Zeng Z. (1994) Zmap-a QTL cartographer. Proceedings of the 5th World Congress on Genetics Applied to Livestock Production: Computing Strategies and Software, Vol 22. pp 65–66. Published by the Organizing Committee, 5th World Congress on Genetics Applied to Livestock Production, Guelph, Ontario, Canada [Google Scholar]
- Bikard D, Patel D, Le Metté C, Giorgi V, Camilleri C, Bennett MJ, Loudet O. (2009) Divergent evolution of duplicate genes leads to genetic incompatibilities within A. thaliana. Science 323: 623–626 [DOI] [PubMed] [Google Scholar]
- Bomblies K. (2010) Doomed lovers: mechanisms of isolation and incompatibility in plants. Annu Rev Plant Biol 61: 109–124 [DOI] [PubMed] [Google Scholar]
- Bomblies K, Lempe J, Epple P, Warthmann N, Lanz C, Dangl JL, Weigel D. (2007) Autoimmune response as a mechanism for a Dobzhansky-Muller-type incompatibility syndrome in plants. PLoS Biol 5: 1962–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies K, Weigel D. (2007) Hybrid necrosis: autoimmunity as a potential gene-flow barrier in plant species. Nat Rev Genet 8: 382–393 [DOI] [PubMed] [Google Scholar]
- Bushell C, Spielman M, Scott RJ. (2003) The basis of natural and artificial postzygotic hybridization barriers in Arabidopsis species. Plant Cell 15: 1430–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill GA, Doerge RW. (1994) Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin F, Schulz P. (1998) Multiple correlations and Bonferroni’s correction. Biol Psychiatry 44: 775–777 [DOI] [PubMed] [Google Scholar]
- Dilkes BP, Comai L. (2004) A differential dosage hypothesis for parental effects in seed development. Plant Cell 16: 3174–3180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilkes BP, Spielman M, Weizbauer R, Watson B, Burkart-Waco D, Scott RJ, Comai L. (2008) The maternally expressed WRKY transcription factor TTG2 controls lethality in interploidy crosses of Arabidopsis. PLoS Biol 6: 2707–2720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon MS, Jones DA, Keddie JS, Thomas CM, Harrison K, Jones JD. (1996) The tomato Cf-2 disease resistance locus comprises two functional genes encoding leucine-rich repeat proteins. Cell 84: 451–459 [DOI] [PubMed] [Google Scholar]
- Dobzhansky T. (1936) Studies on hybrid sterility. II. Localization of sterility factors in Drosophila pseudoobscura hybrids. Genetics 21: 113–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman L, Willis JH. (2001) Evidence for Dobzhansky-Muller incompatibilites contributing to the sterility of hybrids between Mimulus guttatus and M. nasutus. Evolution 55: 1932–1942 [DOI] [PubMed] [Google Scholar]
- Garcia D, Fitz Gerald JN, Berger F. (2005) Maternal control of integument cell elongation and zygotic control of endosperm growth are coordinated to determine seed size in Arabidopsis. Plant Cell 17: 52–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harushima Y, Nakagahra M, Yano M, Sasaki T, Kurata N. (2001) A genome-wide survey of reproductive barriers in an intraspecific hybrid. Genetics 159: 883–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry IM, Dilkes BP, Young K, Watson B, Wu H, Comai L. (2005) Aneuploidy and genetic variation in the Arabidopsis thaliana triploid response. Genetics 170: 1979–1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs J. (1902) English Fairy Tales, Ed 3. GP Putnam’s Sons, New York [Google Scholar]
- Jakobsson M, Hagenblad J, Tavaré S, Säll T, Halldén C, Lind-Halldén C, Nordborg M. (2006) A unique recent origin of the allotetraploid species Arabidopsis suecica: evidence from nuclear DNA markers. Mol Biol Evol 23: 1217–1231 [DOI] [PubMed] [Google Scholar]
- Jeuken MJ, Zhang NW, McHale LK, Pelgrom K, den Boer E, Lindhout P, Michelmore RW, Visser RG, Niks RE. (2009) Rin4 causes hybrid necrosis and race-specific resistance in an interspecific lettuce hybrid. Plant Cell 21: 3368–3378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefsson C, Dilkes B, Comai L. (2006) Parent-dependent loss of gene silencing during interspecies hybridization. Curr Biol 16: 1322–1328 [DOI] [PubMed] [Google Scholar]
- Kao KC, Schwartz K, Sherlock G. (2010) A genome-wide analysis reveals no nuclear Dobzhansky-Muller pairs of determinants of speciation between S. cerevisiae and S. paradoxus, but suggests more complex incompatibilities. PLoS Genet 6: e1001038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger J, Thomas CM, Golstein C, Dixon MS, Smoker M, Tang S, Mulder L, Jones JD. (2002) A tomato cysteine protease required for Cf-2-dependent disease resistance and suppression of autonecrosis. Science 296: 744–747 [DOI] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. (2009) Circos: an information aesthetic for comparative genomics. Genome Res 19: 1639–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplace PS. (1995) A Philosophical Essay on Probabilities. Dover Publications, New York [Google Scholar]
- Lewis D, Crowe LK. (1958) Unilateral interspecific incompatibility in flowering plants. Heredity 12: 233–256 [Google Scholar]
- Makarevich G, Leroy O, Akinci U, Schubert D, Clarenz O, Goodrich J, Grossniklaus U, Köhler C. (2006) Different Polycomb group complexes regulate common target genes in Arabidopsis. EMBO Rep 7: 947–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manly KF, Cudmore RH, Jr, Meer JM. (2001) Map Manager QTX, cross-platform software for genetic mapping. Mamm Genome 12: 930–932 [DOI] [PubMed] [Google Scholar]
- Matute DR, Butler IA, Turissini DA, Coyne JA. (2010) A test of the snowball theory for the rate of evolution of hybrid incompatibilities. Science 329: 1518–1521 [DOI] [PubMed] [Google Scholar]
- Mayr E. (1942) Systematics and the Origin of Species. Columbia University Press, New York [Google Scholar]
- Moyle LC, Graham EB. (2005) Genetics of hybrid incompatibility between Lycopersicon esculentum and L. hirsutum. Genetics 169: 355–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyle LC, Nakazato T. (2010) Hybrid incompatibility “snowballs” between Solanum species. Science 329: 1521–1523 [DOI] [PubMed] [Google Scholar]
- Muller H. (1942) Isolating mechanisms, evolution and temperature. Biol Symp 6: 71–125 [Google Scholar]
- Myburg AA, Vogl C, Griffin AR, Sederoff RR, Whetten RW. (2004) Genetics of postzygotic isolation in Eucalyptus: whole-genome analysis of barriers to introgression in a wide interspecific cross of Eucalyptus grandis and E. globulus. Genetics 166: 1405–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA. (1995) The population genetics of speciation: the evolution of hybrid incompatibilities. Genetics 139: 1805–1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA, Turelli M. (2001) The evolution of postzygotic isolation: accumulating Dobzhansky-Muller incompatibilities. Evolution 55: 1085–1094 [DOI] [PubMed] [Google Scholar]
- Paul W, Hodge R, Smartt S, Draper J, Scott R. (1992) The isolation and characterisation of the tapetum-specific Arabidopsis thaliana A9 gene. Plant Mol Biol 19: 611–622 [DOI] [PubMed] [Google Scholar]
- Presgraves DC. (2010) The molecular evolutionary basis of species formation. Nat Rev Genet 11: 175–180 [DOI] [PubMed] [Google Scholar]
- Price RA, Palmer JD, Al-Shehbaz IA. (1994) Systematic relationships of Arabidopsis: a molecular and morphological perspective. Meyerowitz EM, Somerville CR, , Arabidopsis. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 7–20 [Google Scholar]
- Rieseberg LH, Blackman BK. (2010) Speciation genes in plants. Ann Bot (Lond) 106: 439–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney HC, Van’t Klooster JW, van der Hoorn RA, Joosten MH, Jones JD, de Wit PJ. (2005) Cladosporium Avr2 inhibits tomato Rcr3 protease required for Cf-2-dependent disease resistance. Science 308: 1783–1786 [DOI] [PubMed] [Google Scholar]
- Sweigart AL, Fishman L, Willis JH. (2006) A simple genetic incompatibility causes hybrid male sterility in Mimulus. Genetics 172: 2465–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweigart AL, Mason AR, Willis JH. (2007) Natural variation for a hybrid incompatibility between two species of Mimulus. Evolution 61: 141–151 [DOI] [PubMed] [Google Scholar]
- Törjék O, Meyer RC, Zehnsdorf M, Teltow M, Strompen G, Witucka-Wall H, Blacha A, Altmann T. (2008) Construction and analysis of 2 reciprocal Arabidopsis introgression line populations. J Hered 99: 396–406 [DOI] [PubMed] [Google Scholar]
- Törjék O, Witucka-Wall H, Meyer RC, von Korff M, Kusterer B, Rautengarten C, Altmann T. (2006) Segregation distortion in Arabidopsis C24/Col-0 and Col-0/C24 recombinant inbred line populations is due to reduced fertility caused by epistatic interaction of two loci. Theor Appl Genet 113: 1551–1561 [DOI] [PubMed] [Google Scholar]
- Vision TJ, Brown DG, Shmoys DB, Durrett RT, Tanksley SD. (2000) Selective mapping: a strategy for optimizing the construction of high-density linkage maps. Genetics 155: 407–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walia H, Josefsson C, Dilkes B, Kirkbride R, Harada J, Comai L. (2009) Dosage-dependent deregulation of an AGAMOUS-LIKE gene cluster contributes to interspecific incompatibility. Curr Biol 19: 1128–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto E, Takashi T, Morinaka Y, Lin S, Wu J, Matsumoto T, Kitano H, Matsuoka M, Ashikari M. (2010) Gain of deleterious function causes an autoimmune response and Bateson-Dobzhansky-Muller incompatibility in rice. Mol Genet Genomics 283: 305–315 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.