Abstract
The maize (Zea mays) kernel plays a critical role in feeding humans and livestock around the world and in a wide array of industrial applications. An understanding of the regulation of kernel starch, protein, and oil is needed in order to manipulate composition to meet future needs. We conducted joint-linkage quantitative trait locus mapping and genome-wide association studies (GWAS) for kernel starch, protein, and oil in the maize nested association mapping population, composed of 25 recombinant inbred line families derived from diverse inbred lines. Joint-linkage mapping revealed that the genetic architecture of kernel composition traits is controlled by 21–26 quantitative trait loci. Numerous GWAS associations were detected, including several oil and starch associations in acyl-CoA:diacylglycerol acyltransferase1-2, a gene that regulates oil composition and quantity. Results from nested association mapping were verified in a 282 inbred association panel using both GWAS and candidate gene association approaches. We identified many beneficial alleles that will be useful for improving kernel starch, protein, and oil content.
Maize (Zea mays) is the world’s most important production crop (faostat.fao.org): Its starch, protein, and oil are essential in supplying adequate food and nutrition to both humans and animals, and maize starch has recently become an important feedstock for ethanol production. Altering starch content can lead to higher yields, specialty industrial applications, and improved sweet corn varieties, while increased protein content and augmented levels of essential amino acids improve nutritional quality. Growing demand for healthy cooking oil can be met by improved oil content and composition.
Substantial effort has been spent to develop maize varieties that meet market demands for modified kernel composition. Specialty maize germplasm with unique kernel composition traits has been developed by exploiting mutations affecting kernel grain composition and quality, including opaque2 (o2), which increases Lys content (Mertz et al., 1964), amylose-free waxy1 (wx1; Lambert, 2001), sugary1 (su1), sugary enhancer (SE), and shrunken2 (sh2), which are responsible for sweet corn (Schultz and Juvik, 2004), and linoleic acid1 (ln1) with an altered fatty acid ratio (Poneleit and Alexander, 1965). Use of specialty maize germplasm with unique kernel composition has been limited, however, due to difficulties in developing agronomically superior germplasm. Future progress in kernel composition improvement will depend on understanding and exploiting quantitative trait loci (QTLs) for kernel composition traits.
The complex genetic architecture of starch, protein, and oil content has been demonstrated in the inbred line (IL) long-term selection experiment, in which more than 100 generations of recurrent selection has increased oil and protein content to approximately 20% and 27%, respectively (Moose et al., 2004). The continued phenotypic response of kernel composition provides convincing evidence that these traits are controlled by many genes. This is further demonstrated by the numerous starch, protein, and oil QTLs detected in studies involving lines derived from the IL long-term selection populations (Goldman et al., 1993, 1994; Séne et al., 2001; Laurie et al., 2004; Hill, 2005; Dudley et al., 2004, 2007; Dudley, 2008; Clark et al., 2006; Wassom et al., 2008). Little is known, however, about the causative genetic factors underlying kernel composition QTLs.
Two publically available maize genetic resources, the nested association mapping (NAM) population (McMullen et al., 2009) and the 282 IL association panel (AP; Flint-Garcia et al., 2005), were developed for high-power, high-resolution QTL analysis. The NAM population was developed by crossing 25 diverse founder ILs to the reference inbred B73 and producing 25 recombinant inbred line (RIL) families. The present NAM genetic map is based on 1,106 single nucleotide polymorphisms (SNPs) assayed on 4,699 RILs. The power and resolution of joint-linkage mapping in NAM was recently demonstrated for maize flowering time (Buckler et al., 2009). The unique structure of NAM also offers an opportunity to further dissect QTLs using genome-wide association studies (GWAS; Tian et al., 2011). Release of the first-generation maize HapMap (Gore et al., 2009) enables projection of 1.6 million SNPs and indels identified in the NAM founder lines onto the NAM RILs. Use of HapMap markers for GWAS successfully dissected leaf morphology and northern and southern leaf blight QTLs to the level of individual genes (Kump et al., 2011; Poland et al., 2011; Tian et al., 2011). The 282 IL AP exploits the rapid breakdown of linkage disequilibrium in diverse maize lines, enabling very high resolution for QTL mapping via association analysis (Flint-Garcia et al., 2005). The candidate gene association approach has been successful in identifying genes controlling various quantitative traits in maize (Thornsberry et al., 2001; Wilson et al., 2004; Harjes et al., 2008; Krill et al., 2010; Yan et al., 2010).
In this study, we evaluated the NAM population and the 282 IL APs for starch, protein, and oil content. QTLs were identified by joint-linkage analysis and further resolved with GWAS in NAM. We report kernel starch, protein, and oil composition genetic architecture is characterized primarily by additive gene action. The fine mapping resolution of NAM-enabled GWAS to resolve an oil QTL on chromosome 6 to the genic level, revealing an allelic series for acyl-coa:diacylglycerol acyltransferase1-2 (DGAT1-2), a gene involved in oil synthesis. The NAM analysis was complemented by GWAS on the 282 inbred AP using 55,000 SNPs. After multiple test correction, none of the GWAS associations in the AP were significant. However, SNPs located in specific candidate genes were significant when the candidate gene association analysis approach was used.
RESULTS
Phenotypic Assessment of NAM and AP Kernel Composition
Starch, protein, and oil content was estimated by near-infrared (NIR) spectroscopy for self-pollinated seed samples of the NAM population and 282 inbred AP grown in seven locations spanning 2 y. The Perten Ethanol Calibration Package contains over 1,700 calibration samples with the following ranges: 7.4%–37.6% for moisture, 4.9%–15.3% for protein, and 2.2%–3.5% for oil. The R2 values for the Perten calibrations are all very high (>0.94) for samples within these ranges. The proprietary Syngenta starch calibration sample set contained 814 samples ranging from 48.3% to 67.9% starch, and the R2 value was 0.94 for samples within that range. After adjusting these calibration sample composition values to a dry matter basis, the vast majority of our NAM and AP samples fell within the range of the calibration, with only 0.7%, 1.2%, and 0.9% of our values falling outside that range for starch, protein, and oil, respectively. All composition values were adjusted to a dry matter basis.
The two NAM sweet corn families (IL14H and P39) were excluded from analysis due to their extreme kernel phenotypes. Starch, protein, and oil content among the NAM founders ranged from 62.3% to 69.6%, 12.3% to 15.3%, and 3.5% to 5.5%, respectively, whereas the NAM population displayed transgressive segregation resulting in greater differences among the RILs (Table I; Supplemental Table S1). Starch, protein, and oil content among the inbreds in the 282 AP ranged from 59.6% to 70.3%, 11.5% to 17.5%, and 3.1% to 8.2%, respectively (Table I). In both the NAM population and AP, highly significant (P < 0.0001) negative phenotypic correlations were detected between starch and both protein (r = −0.66 and −0.56 for NAM and AP, respectively) and oil (r = −0.41 and −0.33 for NAM and AP, respectively), and a significant positive phenotypic correlation was detected between protein and oil (r = 0.32 and 0.29 for NAM and AP, respectively). Broad-sense heritability for these traits was high in both the NAM population and AP, ranging from 83% to 91% (Table I).
Table I. Means, ranges, difference within range, and broad-sense heritability estimates for percent starch, protein, and oil kernel composition best linear unbiased predictors on a dry matter basis in the NAM population and 282 inbred AP.
Number of QTLs detected in NAM by joint-linkage analysis for each trait with their respective R2 values explaining the amount of genetic variation detected by the QTLs.
| Trait | Population | Mean | Range | Difference | Broad-Sense Heritability | QTLs | R2 |
| Starch | NAM founders | 66.3 | 62.3–69.6 | 7.3 | — | ||
| NAM RILs | 67.7 | 59.7–73.0 | 13.3 | 0.85 | 21 | 59.1 | |
| AP | 66.5 | 59.6–70.3 | 10.7 | 0.88 | |||
| Protein | NAM founders | 13.7 | 12.3–15.3 | 3.0 | — | ||
| NAM RILs | 13.6 | 10.8–17.7 | 6.8 | 0.83 | 26 | 61.0 | |
| AP | 14.0 | 11.5–17.5 | 6.0 | 0.87 | |||
| Oil | NAM founders | 4.4 | 3.5–5.5 | 2.0 | — | ||
| NAM RILs | 4.2 | 2.8–6.4 | 3.6 | 0.86 | 22 | 69.7 | |
| AP | 4.4 | 3.1–8.2 | 5.2 | 0.91 |
NAM Joint QTL Linkage Analysis
Joint stepwise regression identified 21 starch, 26 protein, and 22 oil QTLs, which collectively explained 59%, 61%, and 70% of the total variation, respectively (Fig. 1; Table I). All starch, protein, and oil QTLs were shared among multiple families, with most QTLs showing significant effects among three to six families. Because the founder lines were crossed to a common reference line (B73), additive allelic effects relative to B73 can be accurately estimated. In joint-linkage mapping, we are mapping QTLs that are linked to the SNPs being tested. While the SNP markers are biallelic, each of the 23 populations was allowed to have an independent allele by fitting a population-by-marker term in the stepwise regression and final models. A total of 133 starch, 136 protein, and 114 oil alleles were significant after false discovery rate (FDR) correction (P = 0.05; Fig. 2; Supplemental Figs. S1 and S2; Supplemental Tables S5–S7). All QTL additive allelic effects were small relative to the amount of variation observed among founders, with the largest allelic effects for starch, protein, and oil QTLs being 0.65%, −0.38%, and 0.21% dry matter, respectively. Allelic series, or QTLs displaying both positive and negative additive allelic effects, were identified in 31% to 43% of the QTLs, depending on the trait.
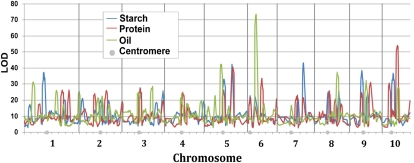
Figure 1.
Joint-linkage QTL analysis for kernel starch, protein, and oil content in NAM. Gray circles, Location of centromeres; vertical lines, chromosome boundaries; horizontal units, centiMorgans (cM); vertical units, log of odds (LOD; see also Supplemental Tables S2–S4).
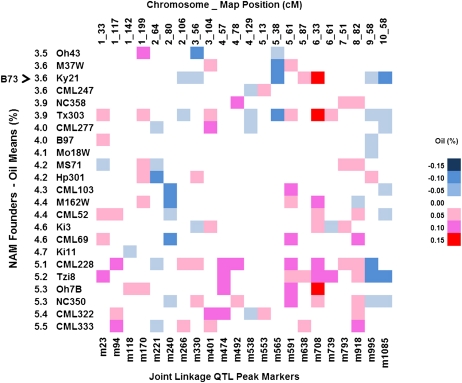
Figure 2.
Heat map displaying additive allelic effects for oil content QTLs for the 23 NAM founders relative to B73. The top horizontal axis lists the chromosome and genetic map position for each QTL peak, and the bottom axis shows the NAM map SNP selected by stepwise regression. The vertical axis displays the 23 inbred NAM founder lines sorted in increasing percent oil content on a dry matter basis. Allelic effects are color coded based on 0.05% increments.
We searched for the presence of epistatic interactions in the NAM population by testing all pairwise marker combinations. Eight significant epistatic interactions were observed for oil at the NAM level at the 5% FDR (Benjamini and Hochberg, 1995). However, none of these oil interactions remained significant when added to the full joint-linkage model. Analysis of individual families yielded only two family-specific epistatic interactions for protein that were significant after FDR correction, but these were likewise not significant in the context of the joint-linkage model.
The NAM design provides a powerful test of pleiotropy among overlapping QTL intervals from multiple traits by correlating the allelic effects across 23 families. Joint-linkage mapping with 1,106 markers produced starch, protein, and oil QTL support intervals averaging 9.1 to14.4 cM. The majority of the starch (90%), protein (85%), and oil (73%) QTL intervals overlapped a second kernel composition trait and were subsequently tested for pleiotropy. The high level of pleiotropy was expected, as starch, protein, and oil make up the bulk of the kernel’s dry matter. It is mathematically impossible to achieve a kernel with >100% dry matter, and thus as the percentage of one trait increases significantly, the percentage of the other traits must decrease. If two traits share a QTL due to pleiotropy, the allelic effects at that locus will be significantly correlated. Allelic effects were significantly correlated (P ≤ 0.001) when each pair of traits was examined (Supplemental Table S8). Each QTL was also analyzed independently, revealing 12 of 13 (92%) starch/protein, 1 of 8 (13%) starch/oil, 7 of 11 (64%) protein/oil, and 1 of 8 (13%) starch/protein/oil were pleiotropic (P ≤ 0.05; Supplemental Table S8).
GWAS in NAM and 282 Inbred AP
The NAM design, combined with the increased marker density provided by HapMap.v1 markers (Gore et al., 2009), enables further dissection of the joint-linkage mapping QTL intervals via GWAS. To perform GWAS, 1.6 million HapMap.v1 SNPs and indels identified in the 26 NAM parents were projected onto the NAM RILs (Tian et al., 2011). Two GWAS methods were tested, each run on a chromosome-by-chromosome basis accounting for the presence of QTLs on the other nine chromosomes. In the first analysis, a single forward regression model was developed for each trait based on the complete RIL data set (23 complete NAM families). The single forward regression method identified 33 starch, 31 protein, and 43 oil SNP associations (Supplemental Tables S9–S11). In order to explore a wider range of models, a second analysis was conducted based on 100 random subsamples containing 80% of the RILs from each family. The subsampling method yielded 127 starch, 118 protein, and 135 oil SNP associations with resample model inclusion probability (RMIP) ≥ 0.05 (Supplemental Tables S12–S14). More than 80% of all associations from the single regression analysis were also identified in the subsampling analysis (Supplemental Tables S9–S11).
NAM GWAS results were compared to the NAM joint-linkage QTL intervals. Between 47% and 100% of the SNPs selected by the 100 subsample method overlapped with NAM joint-linkage QTL intervals, depending on the RMIP level and trait (Supplemental Fig. S3). Between 54% and 62% of SNPs selected by both the subsampling and single forward regression GWAS methods overlapped the starch, oil, and protein NAM joint-linkage QTL intervals, respectively (Fig. 3).
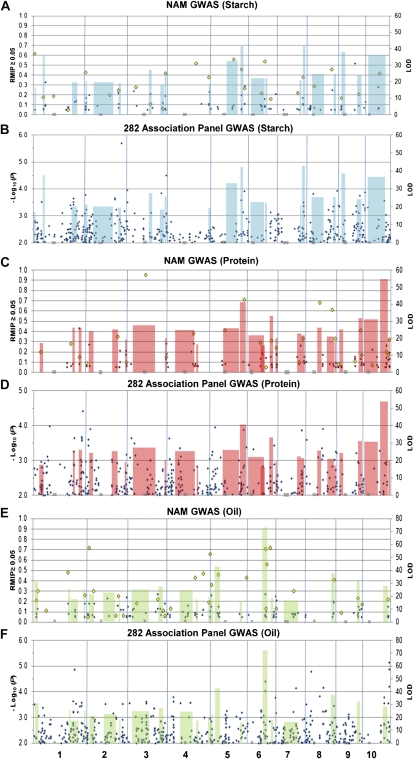
Figure 3.
Starch, protein, and oil GWAS in NAM and the 282 inbred AP compared with the NAM joint-linkage mapping analysis. The regions shaded blue (starch), red (protein), and green (oil) depict NAM joint-linkage QTL support intervals, with their height indicating log of the odds (LOD) score. Gray boxes along the horizontal axis, Centromere positions. A, C, and E, NAM, black diamonds indicate position and magnitude of associations detected by the subsampling method (RMIP ≥ 0.05; Supplemental Tables S12–S14), and yellow diamonds show the position and magnitude of associations selected by both the 100 subsample and single forward regression methods (RMIP; Supplemental Tables S9–S11). B, D, and F, 282 Inbred AP, black diamonds show the position and magnitude of GWAS SNPs selected by MLM (Q+K) analysis at P = 0.01.
Although the joint-linkage genetic QTL intervals in NAM were relatively small (average 9.1–14.4 cM), several intervals encompassed over 100 Mb of DNA sequence (Supplemental Tables S2–S4). In most cases, intervals that encompass large genomic regions correspond to low recombination regions, often representing centromeric regions (Gore et al., 2009). GWAS analysis with NAM was able to further dissect several of the QTL intervals overlapping large genomic regions into substantially smaller genomic intervals (Fig. 3).
Complementing the NAM analysis, we conducted an association analysis of kernel composition traits in an AP comprised of 282 ILs (Flint-Garcia et al., 2005) genotyped with the MaizeSNP50 BeadChip (Illumina Inc.). Removal of nonpolymorphic and low-quality SNPs resulted in a dataset of 51,741 SNPs that were used for GWAS employing the mixed linear model (MLM) method (Q+K; Yu et al., 2006) to control for population structure. None of the 51,741 genome-wide associations were significant for any of the traits after a multiple test FDR (P = 0.05) correction was applied (Benjamini and Hochberg, 1995).
Underlying Genetic Architecture
The ultimate goal of our QTL study was to identify genes underlying kernel composition traits. We identified NAM GWAS associations in several genes that are known to be important enzymes in biochemical pathways that influence starch, protein, and oil kernel content such as DGAT1-2 (RMIP 0.67), carbonic anhydrase (RMIP 0.59), Suc synthase (RMIP 0.36), pyruvate kinase (RMIP 0.23), β-amylase2 (RMIP 0.20), nitrate reductase (RMIP 0.07), and α-amylase (RMIP 0.06; Buchanan et al., 2000; Supplemental Tables S12–S14). Additionally, several significant GWAS associations were located within transcription factors, zinc finger binding proteins, kinases, and the histone H1 variant H1.2, all of which regulate complex biochemical pathways (Supplemental Tables S9–S14).
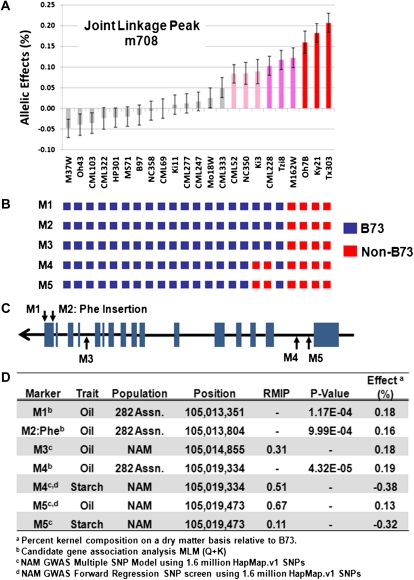
We explored the relationship between the joint-linkage oil QTLs on chromosome 6 (NAM marker m708; PZA03461.1) and a gene previously identified to affect oil content and the ratio of oleic:linoleic acids (Zheng et al., 2008). The QTL was the most significant joint-linkage QTL in our experiment and overlaps a previously identified locus, ln1, confirmed to encode a type I acyl-CoA:diacylglycerol acyltransferase located at chromosome 6: 105,013,351 to 105,020,258 (B73 RefGen_v1; (Schnable et al., 2009), which is involved in the Kennedy pathway for triacylglycerol biosynthesis (Zheng et al., 2008). The authors of the latter study identified a functional Phe insertion in the C terminus of the protein that resulted in a high oil allele of DGAT1-2 with 0.29% additive genetic effect. The NAM joint-linkage QTL on chromosome 6 overlapping DGAT1-2 showed a distinct allelic series ranging from −0.05% to 0.21% (Fig. 4A). The 23 NAM founders used in this study were genotyped for the indel conferring the Phe insertion, and the four founders (M162W, Oh7B, Ky21, and Tx303) with the highest allelic effects also have the high oil Phe insertion allele (Fig. 4B).
Figure 4.
QTL and GWAS analyses for the chromosome 6 oil QTL and candidate gene DGAT1-2. A, NAM additive percentage oil content on a dry matter basis allelic effect estimates for the m708 QTL interval overlapping the DGAT1-2 genomic position. Red bars, NAM founders possessing a significant high oil allele; blue bar, NAM founder with a significant low oil allele relative to B73. B, NAM founder genotypes for all markers displaying significant associations in DGAT1-2. C, DGAT1-2 gene model showing the position of markers with significant associations. Note that DGAT1-2 is on the negative DNA strand. D, NAM GWAS and candidate gene association analysis for DGAT1-2. M2 is the Phe:indel previously determined to be the functional polymorphism for oil content at this locus (Zheng et al., 2008).
GWAS on the NAM population further suggests that DGAT1-2 is responsible for the joint-linkage oil QTL on chromosome 6. Two biallelic SNPs (105,014,855 and 105,019,473 bp) located in DGAT1-2 were associated with oil content with RMIP scores of 0.31 and 0.67, respectively (Fig. 4, C and D). Likewise, two SNPs (105,019,334 and 105,019,473 bp) located in DGAT1-2 were associated with starch content with RMIP scores of 0.51 and 0.11, respectively (Fig. 4, C and D). The two oil SNPs had a positive estimated additive effect relative to B73 (0.13% and 0.18%), and both starch SNPs had a negative estimated additive effect relative to B73 (−0.32% and −0.38%; Fig. 4D). The negative effects for starch and positive effects for oil correspond with the significant pleiotropy (r = −0.59) detected between overlapping starch (m707) and oil (m708) joint-linkage QTLs in this region (Supplemental Table S8). The oil SNP located at 105,019,473 bp and the starch SNP located at 105,019,334 bp were also selected by the NAM GWAS single forward regression analysis.
Association of the Phe:indel in DGAT1-2 was not detectable by GWAS analysis, because the indel was not present in the HapMap.v1 marker set. To verify that the Phe:indel was associated with kernel composition in our diverse inbred panel, a candidate gene association analysis approach was implemented using the MLM (Q+K) method on the 282 inbred AP. Consistent with previous results, the Phe:indel was significantly associated with oil content (P = 9.99 E-04) but was not significantly associated with either starch or protein content (Fig. 4D). In addition, there were two SNPs from the MaizeSNP50 BeadChip located in DGAT1-2 at 105,013,351 and 105,019,334 bp. While these SNPs associations were not significant after a 5% FDR correction in the context of a full genome scan, they were associated with oil content with P-values of 1.17E-04 and 4.32E-05 when a candidate gene approach was used (Fig. 4D). The additive allelic effects for these SNPs were 0.18% and 0.19%, respectively (Fig. 4D).
Comparing the NAM joint-linkage QTL allelic effects for the 23 founders to the genotypes of the significant GWAS markers in DGAT1-2 associated with oil content suggests the presence of an allelic series for DGAT1-2 (Fig. 4, A and B). The four lines with the highest estimated oil allelic effects (Tx303, Ky21, Oh7B, and M162W) have non-B73 genotypes at all five significant markers detected by GWAS and candidate gene association analysis. Two founders with intermediate oil allelic effects, CML228 and Ki3, have non-B73 genotypes at only two markers located in the N terminus of DGAT (105,019,334 and 105,019,473 bp; Fig. 4, A and B). All other founder lines have the B73 haplotype for DGAT1-2.
DISCUSSION
Joint-linkage analysis on the NAM population revealed that variation in starch, protein, and oil kernel content is controlled by at least 21 to 26 QTLs, each with relatively small effects. We compared our NAM QTL results with previous biparental QTL studies where we could determine the physical location of the markers. Previous QTL studies detected a wide range (0 to >50) in the number of kernel composition QTLs (Goldman et al., 1993, 1994; Séne et al., 2001; Dudley et al., 2004, 2007; Laurie et al., 2004; Clark et al., 2006; Dudley, 2008; Wassom et al., 2008). We found that less than one-half of these previously reported QTLs were detected in NAM (Supplemental Table S15). Several factors could be responsible for differences in position and quantity of QTLs detected in NAM versus these studies, including variation in allelic frequency, mapping resolution influenced by the magnitude of linkage disequilibrium in a population, marker density, environmental effects, and QTL analysis methods. The majority of the previous QTL studies used parental lines with extreme kernel composition phenotypes derived from the IL long-term selection program (Goldman et al., 1993, 1994; Laurie et al., 2004; Hill, 2005; Clark et al., 2006; Dudley et al., 2004, 2007; Wassom et al., 2008). The IL high- and low-oil and high- and low-protein populations were driven apart via artificial selection, and these populations likely accumulated additional variation controlled by small effect QTLs (Moose et al., 2004). This is in contrast to the NAM population, where the parents were chosen to represent overall natural variation in maize rather than variation specific to kernel composition, resulting in less extreme kernel composition variation and therefore fewer QTLs (Yu et al., 2008). While NAM was successful in capturing a representative sample of QTLs for kernel composition in naturally diverse germplasm, the 24 founders analyzed in this study do not possess all the phenotypic variation present in maize for kernel composition traits.
Epistatic additive × dominance or dominance × dominance interactions cannot be measured with the RIL structure of NAM; however, NAM has excellent power to detect additive × additive epistasis. We report that additive × additive epistatic interactions are not important for kernel composition traits in NAM; thus, the genetic architecture of starch, protein, and oil kernel content in the NAM population is characterized primarily by additive gene action. The lack of epistasis for kernel composition genetic architecture is consistent with other traits studied in NAM: flowering time, leaf morphology, and northern and southern leaf blight resistance (Buckler et al., 2009; Kump et al., 2011; Poland et al., 2011; Tian et al., 2011). In contrast, previous biparental kernel composition QTL studies reported minimal to substantial levels of epistasis (Goldman et al., 1993; Laurie et al., 2004; Dudley, 2008; Wassom et al., 2008). Variation in number of epistatic interactions among studies is not uncommon and has been observed for numerous traits and species (Barton and Keightley, 2002; Holland, 2007; Hill et al., 2008; Phillips, 2008). Interestingly, two kernel composition studies that used either RILs or S2 lines derived from the same source exhibited contrasting levels of epistasis for oil content (Laurie et al., 2004; Dudley, 2008). The study using RILs found variation in oil was predominantly explained by additive effects, leaving little variation for detection of epistatic effects (Laurie et al., 2004). In contrast, the study using S2 progeny had a higher level of oil variation described by dominant genetic effects and also detected substantial, nonadditive, epistatic interactions (Dudley, 2008).
One of the greatest challenges in developing varieties with desirable kernel quality characteristics in major crops [i.e. maize, wheat (Triticum aestivum), rice (Oryza sativa), soybeans (Glycine max), barley (Hordeum vulgare), etc.] is the strong phenotypic correlations among kernel quality traits that can be attributed to pleiotropic interactions (Simmonds, 1995; Ge et al., 2005; Panthee et al., 2005). Studies using parental lines derived from the IL long-term selection program (Goldman et al., 1994; Dudley et al., 2004, 2007; Clark et al., 2006; Wassom et al., 2008) suggest that kernel composition is regulated by a complex genetic network, resulting in strong phenotypic and pleiotropic interactions, and that it will be difficult to develop maize germplasm with high starch, protein, and oil kernel characteristics. Our analysis in NAM confirms that these traits are significantly correlated both phenotypically and genetically across diverse germplasm (Supplemental Table S8).
The NAM population was specifically constructed for high-resolution QTL dissection (Yu et al., 2008) and has proven valuable for GWAS (Kump et al., 2011; Poland et al., 2011; Tian et al., 2011). Inter- and intra-chromosomal linkage disequilibrium among SNPs in the NAM founders was reduced during NAM population development through random chromosome assortment and recombination, thereby reducing spurious unlinked associations and increasing mapping resolution. Analysis of linkage disequilibrium in NAM indicated GWAS resolution will vary but in specific cases appears sufficient to identify causal genes (Kump et al., 2011; Poland et al., 2011; Tian et al., 2011). We demonstrate the use of GWAS to identify DGAT1-2 as a strong candidate gene for a 23.5-cM oil QTL (m708; PZA03461.1) corresponding to an approximately 25-Mb genomic region on chromosome 6. GWAS identified two oil and two starch associations in DGAT1-2, a gene previously shown to influence oil content via a Phe insertion and is responsible for the ln1 mutation (Zheng et al., 2008). While DGAT1-2 was not shown to affect starch content in Zheng et al. (2008), joint-linkage QTL analysis in NAM revealed that the oil QTL (m708; PZA03461.1) overlapping DGAT1-2 was significantly pleiotropic with starch (m707; PZB01658.1) and protein (m707; PZB01658.1; Supplemental Table S8). Detection of two GWAS SNPs with positive allelic effects on oil and two GWAS SNPs with negative effects on starch in DGAT1-2 further substantiates pleiotropic effects on kernel composition.
We complemented our studies in the NAM population with an AP of 282 ILs, using both candidate gene association and GWAS approaches to verify NAM GWAS hits. Results from GWAS using the MaizeSNP50 BeadChip produced no significant associations after performing a multiple hypothesis test correction. However, candidate gene association analysis proved effective, as we were able to detect a significant association between oil content and the Phe insertion previously identified in DGAT1-2 for increased oil (Zheng et al., 2008). Two additional SNPs on the MaizeSNP50 BeadChip located in the DGAT1-2 gene were significantly associated with oil content using the candidate gene approach.
Results from performing GWAS on both the NAM population and the AP demonstrate that NAM may be better suited for detecting associations with small effects than the AP. While NAM is genetically diverse, it captures only 80% of the diversity in the AP; thus, true associations with rare alleles present in the AP are undetected due to a lack of power. This is supported by the lower overlap between the NAM joint-linkage results and the GWAS AP hits (Supplemental Fig. S4) as compared to the NAM GWAS hits (Supplemental Fig. S3). Many associations detected by GWAS on the AP are undoubtedly real, as evident by the DGAT1-2 example. However, the need for multiple test correction requires highly significant associations, and as the number of SNPs available for GWAS approaches millions, it will become increasingly difficult to detect significant associations in an AP of the present population size using GWAS, especially for QTLs with small effects.
Other than DGAT1-2, we were surprised that we did not detect additional NAM GWAS associations with other classical kernel composition genes such as o2, pyruvate orthophosphate dikinase, amylose-free wx1 (=starch-granule-bound nucleotide diphosphate-starch glucosyl transferase), su1 (=isoamylase-type starch-debranching enzyme), prolamine box binding factor1 (pbf1), sh2 (=ADPGppase), and zein protein genes despite substantial SNP coverage within or around these genes (Mertz et al., 1964; Thompson and Larkins, 1989; Vicente-Carbajosa et al., 1997; Lambert, 2001; Schultz and Juvik, 2004; Hennen-Bierwagen et al., 2009). We did detect GWAS associations in several genes that are known to be important enzymes in biochemical pathways that influence starch, protein, and oil kernel content [i.e. carbonic anhydrase (RMIP 0.59); Suc synthase (RMIP 0.36); pyruvate kinase (RMIP 0.23); β-amylase2 (RMIP 0.20); nitrate reductase (RMIP 0.07); and α-amylase (RMIP 0.06)]. Interestingly, the majority of the significant GWAS associations located within annotated genes were elements that regulate complex molecular pathways such as transcription factors, zinc finger binding proteins, kinases, and the histone H1 variant H1.2 (Supplemental Tables S9–S14). Transcription factors and zinc finger binding proteins, such as o2, WRINKLED1 (ZmWRI1), and pbf1, have already been shown to be key regulators of kernel composition pathways, and kinases are essential for signal transduction and regulation of feedback loops (Vicente-Carbajosa et al., 1997; Manicacci et al., 2009; Pouvreau et al., 2011). Histone variants, such as the H1.2 gene that we found to be associated with oil (RMIP; 0.63), are not well characterized for kernel composition; however, chromatin remodeling has been implicated in regulation of kernel composition, and histone variants have been shown to be involved with gene-specific transcription regulation (Ascenzi and Gantt, 1997; Vicente-Carbajosa et al., 1997; Locatelli et al., 2009; Miclaus et al., 2011). We propose the prevalence of GWAS associations in regulatory elements with small effects is related to the delicate balance necessary for an inbred breeding program; breeders must manipulate multiple pleiotropic traits while simultaneously improving the overall agronomic performance of a new IL. For example, while the null mutant allele of o2 results in a dramatic increase in Lys content, it would likely be selected out of the breeding population due to its substantial negative agronomic effects (Gibbon and Larkins, 2005). Selection of subtle changes in multiple regulatory elements is a more likely mode of action in a breeding program.
Our DGAT1-2 results provide valuable confirmation that GWAS in the NAM population is capable of identifying genes influencing kernel composition QTLs. A broad inference about the accuracy of NAM GWAS for kernel composition is limited, however, by the small number of genes that have been verified to control natural variation in kernel composition. For example, we cannot rule out the possibility that the eight additional significant GWAS SNPs identified in the chromosome 6 oil QTL interval (m708; PZA03461.1) that are not located in the DGAT1-2 gene are in valid candidate genes, because their function is currently unknown. Likewise, lack of known genetic factors regulating quantitative variation in kernel composition (as opposed to the classical mutants with large effects) limits our ability to explore significant GWAS SNPs located outside the QTL intervals. Further analysis of additional significant GWAS associations will help determine if the associations are the result of the biallelic GWAS methods having more power to detect weak QTL effects versus the multi-allelic QTL methods under some conditions or if they are false positives due to linkage disequilibrium within chromosomes combined with insufficient SNP coverage in the causative gene (Gore et al., 2009; Kump et al., 2011; Tian et al., 2011). Significant SNPs should not be ignored, as they could represent real QTLs, but should be approached with caution, as they may be aberrations due to extended linkage disequilibrium. Characterization of candidate genes such as the regulatory elements previously discussed that are responsible for kernel composition QTLs will provide valuable information that can be used to “train” GWAS to detect genes associated with kernel composition traits.
We have identified many favorable alleles for improving starch, protein, and oil content in maize relative to B73. While B73 had the highest starch content of the NAM founders, it does not contain all the favorable alleles at the QTLs we identified. In fact, substituting the most favorable allele at 12 QTLs is predicted to increase the starch content of B73 from 69.6% to 79.2%. Even more striking is the potential to increase the oil content of B73 from 3.6% to 7.2% by selecting favorable alleles at 17 QTLs. The most favorable alleles are dispersed among 10 of the NAM parents in the case of starch and among 12 of the NAM parents for oil. Thus, a large, inter-mated population of the NAM parents would be required in order to bring together all these favorable alleles in a breeding program focused on kernel composition.
In conclusion, the successful resolution of kernel composition genetic architecture demonstrates the power of NAM. Analysis of the DGAT1-2 gene demonstrates NAM mapping resolution capable of identifying significant associations between traits and functional genes. Many of the significant GWAS SNP associations we detected are located in uncharacterized genes (Supplemental Tables S9–S14); hence, better gene annotation of the B73 reference genome and additional experiments will be required to determine if these genes indeed influence kernel composition. As the marker coverage on the NAM RIL population increases and the location of recombination events is improved, the ability to detect additional functional polymorphisms will also improve. Results from this study can be directly used for the development of maize germplasm with improved kernel composition traits.
MATERIALS AND METHODS
Materials and Phenotypic Analysis
Development of the NAM population has been previously described (Buckler et al., 2009; McMullen et al., 2009). The present study utilized 4,699 RILs genotyped with 1,106 SNPs. Similarly, the 282 IL AP was selected to represent the genetic diversity found in world wide collections of publically available germplasm (Flint-Garcia et al., 2005).
The NAM population and AP were planted in seven locations: five locations in 2006 (Clayton, NC; Columbia, MO; Aurora, NY; Homestead, FL; and Ponce, PR) and two locations in 2007 (Columbia, MO and Aurora, NY). Each location was arranged in an augmented lattice and consisted of a single replicate of NAM RILs, the AP, and appropriate check entries (Buckler et al., 2009). Two plants of each entry were self-pollinated to avoid zenia effects, and NIR spectroscopy analysis was performed on whole kernels with a Perten Diode Array 7200 (DA7200) instrument (Perten Instruments). The wavelength range assessed by the DA 7200 is 950 to 1650 nm. Each sample was poured into the sample cup, scanned 4 times, mixed and repacked into the sample cup, and scanned 4 more times. The two sets of scans were averaged by the Simplicity software. Starch, protein, oil, and moisture contents were predicted for each sample using a combination of the Perten Ethanol Calibration Package for moisture, protein, and oil, and the Syngenta Seeds, Inc. proprietary calibration for starch. All composition data were converted to a dry matter basis. While the raw NIR scans are not available due to the DA 7200 software and the proprietary nature of the seed industry, the raw NIR estimates are provided in Supplemental Data S1.
Starch, protein, and oil best linear unbiased predictors across environments were calculated for each line with ASREML version 2.0 software (Gilmor et al., 2005) and were used as phenotypic inputs for subsequent genetic analysis. A detailed description of the phenotypic data analysis conducted in ASREML has been published (Hung et al., 2011). The two sweet corn families (IL14H and P39) were excluded from all subsequent analyses.
Joint-Linkage Mapping
Genotyping and construction of the NAM map (McMullen et al., 2009) and joint-linkage mapping (Buckler et al., 2009) have been previously described. Briefly, for joint-linkage mapping, appropriate P-values (starch = 1.6 × 10−5, protein = 3.9 × 10−5, and oil = 3.3 × 10−5) were determined by 1,000 permutations and were used to conduct joint stepwise regression, where the model contained a family main effect and marker effects nested within families. The stepwise regression model was refined by a refitting procedure in order to produce a final model. Significant alleles were determined by a t test comparison of their parental means versus the B73 allele at P = 0.05. QTL support intervals were calculated by adding a single flanking marker for the QTL at a step of 0.1 cM to the full model and testing for significance at the 0.05 level (Tian et al., 2011).
To test for the presence of epistasis, all possible pairwise marker combinations were tested across the NAM panel using a modification of EPISTASY (Holland, 1998) and significance determined by false discovery rate (FDR; P = 0.05; Benjamini and Hochberg, 1995). In addition, epistasis was evaluated for each individual family.
Pleiotropy was evaluated by correlating starch, protein, and oil allele effects for QTLs with overlapping support intervals. Overlapping QTLs with a significant Pearson correlation coefficient (P < 0.05) were considered to be pleiotropic across traits.
GWAS
As described in previous reports, GWAS were conducted on the NAM population by projecting founder SNP genotypes from the maize (Zea mays) HapMap (Gore et al., 2009) onto the NAM RILs (Yu et al., 2006; Tian et al., 2011). Briefly, HapMap SNP projections were based on SNP physical position and genotype of the flanking genetic map markers. Phenotypic residuals for each RIL were calculated on a chromosome basis by fitting a model that included QTLs from the other nine chromosomes. Forward regression analyzing one chromosome at a time was used to identify significant GWAS SNPs. Significance thresholds for entry into the model were determined by 1,000 permutations for each chromosome and ranged from 9.00 × 10−7 to 1.23 × 10−7 for starch, 1.58 × 10−6 to 2.23 × 10−7 for protein, and 1.34 × 10−6 to 3.27 × 10−7 for oil.
A second GWAS method used subsampling in order to explore a wider range of multiple SNP models (Huang et al., 2009; Valdar et al., 2006, 2009) and has been successfully applied to GWAS in maize (Kump et al., 2011; Tian et al., 2011). A random subsample of 80% of the RILs from each family selected without replacement was subjected to forward regression, as described above. This procedure was repeated for 100 subsamples for each chromosome. SNPs detected as significant in at least five subsamples (RMIP ≥ 0.05) are presented. The RMIP statistic is equivalent to the BPP statistic used in previous NAM GWAS studies. The median of the additive effects and P-values across the 100 analyses was used to represent the allelic effect and P-value of the associated SNP.
The 282 inbred AP was genotyped with the Illumina MaizeSNP50 BeadChip (Ganal et al., in press), an Infinium-based assay (Peiffer et al., 2006), containing approximately 56,000 SNP markers dispersed across the maize genome. The assay was performed according to the manufacturer’s specifications (Illumina, Inc.), and alleles were called using the Illumina Genome-Studio V2010.3 software with a locally modified cluster file. Removal of nonpolymorphic and low-quality SNPs resulted in a dataset of 51,741 SNPs used to conduct GWAS in TASSEL 2.1, removing sweet corn and popcorn lines and SNPs with minor allele frequencies < 0.05 and employing the MLM (Q+K) method (Yu et al., 2006; Bradbury et al., 2007).
Candidate Gene Association Analysis
Candidate gene association analysis (Thornsberry et al., 2001) was performed on the 282 inbred AP using the MLM (Q+K) method (Yu et al., 2006) in the TASSEL 2.1 software package (Bradbury et al., 2007). Sanger sequencing was performed as previously described (Yamasaki et al., 2005) to genotype the DGAT1-2 Phe:indel. Associations were considered significant at P ≤ 0.05.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Heat map displaying additive allelic effects for starch and protein content QTLs for the 23 NAM founders relative to B73.
Supplemental Figure S2. Distribution of allele effect size derived from joint-linkage mapping in NAM for starch, protein, and oil content relative to B73.
Supplemental Figure S3. Significant SNPs from the NAM subsampling GWAS method overlapped with NAM joint-linkage QTL intervals.
Supplemental Figure S4. Significant SNP overlap from 282 inbred association panel genome scan overlapped with NAM joint-linkage QTL intervals.
Supplemental Table S1. Means and ranges for percentage starch, protein, and oil kernel composition best linear unbiased predictors in each NAM family and parental lines.
Supplemental Table S2. NAM joint-linkage mapping analysis summary for percentage starch content.
Supplemental Table S3. NAM joint-linkage mapping analysis summary for percentage protein content.
Supplemental Table S4. NAM joint-linkage mapping analysis summary for percentage oil content.
Supplemental Table S5. NAM joint-linkage mapping analysis QTL allelic effects summary for percentage starch content.
Supplemental Table S6. NAM joint-linkage mapping analysis QTL allelic effects summary for percentage protein content.
Supplemental Table S7. NAM joint-linkage mapping analysis QTL allelic effects summary for percentage oil content.
Supplemental Table S8. Pleiotropy among percentage starch, protein, and oil kernel content QTLs.
Supplemental Table S9. NAM GWAS stepwise forward regression associations overlap with NAM joint-linkage QTL intervals and NAM multiple SNP model GWAS for percentage starch.
Supplemental Table S10. NAM GWAS stepwise forward regression associations, overlap with NAM joint-linkage QTL intervals, and NAM multiple SNP model GWAS for percentage protein.
Supplemental Table S11. NAM GWAS stepwise forward regression associations, overlap with NAM joint-linkage QTL intervals, and NAM multiple SNP model GWAS for percentage oil.
Supplemental Table S12. NAM multiple SNP model GWAS associations (RMIP ≥ 0.05) and overlap with NAM joint-linkage QTL intervals for percentage starch content.
Supplemental Table S13. NAM multiple SNP model GWAS associations (RMIP ≥ 0.05) and overlap with NAM joint-linkage QTL intervals for percentage protein.
Supplemental Table S14. NAM multiple SNP model GWAS associations (RMIP ≥ 0.05) and overlap with NAM joint-linkage QTL intervals for percentage oil.
Supplemental Table S15. Overlap between NAM joint-linkage QTLs and previously identified biparental QTLs.
Supplemental Data S1. Raw NIR kernel composition estimate data from seven grow-outs of the NAM and association panel.
Supplementary Material
Acknowledgments
We thank past and present members of the Buckler, Holland, and McMullen laboratories for self-pollinating multiple locations of the NAM experiment, and Syngenta Seeds, Inc. for conducting the NIR analysis of over 26,000 seed samples. We also thank the anonymous reviewers for their relevant suggestions. Names of products are necessary to report factually on available data; however, neither the U.S. Department of Agriculture nor any other participating institution guarantees or warrants the standard of the product and the use of the name does not imply approval of the product to the exclusion of others that may also be suitable.
References
- Ascenzi R, Gantt JS. (1997) A drought-stress-inducible histone gene in Arabidopsis thaliana is a member of a distinct class of plant linker histone variants. Plant Mol Biol 34: 629–641 [DOI] [PubMed] [Google Scholar]
- Barton NH, Keightley PD. (2002) Understanding quantitative genetic variation. Nat Rev Genet 3: 11–21 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc B Met 57: 289–300 [Google Scholar]
- Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23: 2633–2635 [DOI] [PubMed] [Google Scholar]
- Buchanan BB, Gruissem W, Jones RL. (2000) Biochemistry and Molecular Biology of Plants. American Society of Plant Biology, Rockville, MD [Google Scholar]
- Buckler ES, Holland JB, Bradbury PJ, Acharya CB, Brown PJ, Browne C, Ersoz E, Flint-Garcia S, Garcia A, Glaubitz JC, et al. (2009) The genetic architecture of maize flowering time. Science 325: 714–718 [DOI] [PubMed] [Google Scholar]
- Clark D, Dudley JW, Rocheford TR, LeDeaux JR. (2006) Genetic analysis of corn kernel chemical composition in the random mated 10 generation of the cross of generations 70 of IHO x ILO. Crop Sci 46: 807–819 [Google Scholar]
- Dudley JW. (2008) Epistatic interactions in crosses of Illinois high oil × Illinois low oil and of Illinois high protein × Illinois low protein corn strains. Crop Sci 48: 59–68 [Google Scholar]
- Dudley JW, Clark D, Rocheford TR, LeDeaux JR. (2007) Genetic analysis of corn kernel chemical composition in the random mated 7 generation of the cross of generations 70 of IHP × ILP. Crop Sci 47: 45–57 [Google Scholar]
- Dudley JW, Dijkhuizen A, Paul C, Coates ST, Rocheford TR. (2004) Effects of random mating on marker-QTL associations in the cross of the Illinois high protein x Illinois low protein maize strains. Crop Sci 44: 1419–1428 [Google Scholar]
- Flint-Garcia SA, Thuillet AC, Yu J, Pressoir G, Romero SM, Mitchell SE, Doebley J, Kresovich S, Goodman MM, Buckler ES. (2005) Maize association population: a high-resolution platform for quantitative trait locus dissection. Plant J 44: 1054–1064 [DOI] [PubMed] [Google Scholar]
- Ganal M, Durstewitz G, Polley A, Berard A, Buckler ES, Charcosset A, Clarke JD, Graner E, Hansen M, Joets J, et al. (2011) A large maize (Zea mays L.) SNP genotyping array: development and germplasm genotyping and genetic mapping to compare with the B73 reference genome. PLoS ONE 6: e28334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge XJ, Xing YZ, Xu CG, He YQ. (2005) QTL analysis of cooked rice grain elongation, volume expansion, and water absorption using a recombinant inbred population. Plant Breeding 124: 121–126 [Google Scholar]
- Gibbon BC, Larkins BA. (2005) Molecular genetic approaches to developing quality protein maize. Trends Genet 21: 227–233 [DOI] [PubMed] [Google Scholar]
- Gilmor AR, Cullis B, Gogel B, Welham SJ, Thompson R. (2005) Asreml user's guide. Release 2.0, VSN International, Hemel Hempstead, UK [Google Scholar]
- Goldman IL, Rocheford TR, Dudley JW. (1993) Quantitative trait loci influencing protein and starch concentration in the Illinois Long Term Selection maize strains. Theor Appl Genet 87: 217–224 [DOI] [PubMed] [Google Scholar]
- Goldman IL, Rocheford TR, Dudley JW. (1994) Molecular markers associated with maize kernel oil concentration in an Illinois high protein x Illinois low protein cross. Crop Sci 34: 908–915 [Google Scholar]
- Gore MA, Chia JM, Elshire RJ, Sun Q, Ersoz ES, Hurwitz BL, Peiffer JA, McMullen MD, Grills GS, Ross-Ibarra J, et al. (2009) A first-generation haplotype map of maize. Science 326: 1115–1117 [DOI] [PubMed] [Google Scholar]
- Harjes CE, Rocheford TR, Bai L, Brutnell TP, Kandianis CB, Sowinski SG, Stapleton AE, Vallabhaneni R, Williams M, Wurtzel ET, et al. (2008) Natural genetic variation in lycopene epsilon cyclase tapped for maize biofortification. Science 319: 330–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennen-Bierwagen TA, Lin Q, Grimaud F, Planchot V, Keeling PL, James MG, Myers AM. (2009) Proteins from multiple metabolic pathways associate with starch biosynthetic enzymes in high molecular weight complexes: a model for regulation of carbon allocation in maize amyloplasts. Plant Physiol 149: 1541–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill WG. (2005) Genetics. A century of corn selection. Science 307: 683–684 [DOI] [PubMed] [Google Scholar]
- Hill WG, Goddard ME, Visscher PM. (2008) Data and theory point to mainly additive genetic variance for complex traits. PLoS Genet 4: e1000008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland J. (1998) Computer note. EPISTACY: A SAS program for detecting two-locus epistatic interactions using genetic marker information. J Hered 89: 374–375 [Google Scholar]
- Holland JB. (2007) Genetic architecture of complex traits in plants. Curr Opin Plant Biol 10: 156–161 [DOI] [PubMed] [Google Scholar]
- Huang G-J, Shifman S, Valdar W, Johannesson M, Yalcin B, Taylor MS, Taylor JM, Mott R, Flint J. (2009) High resolution mapping of expression QTLs in heterogeneous stock mice in multiple tissues. Genome Res 19: 1133–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung HY, Browne C, Guill K, Coles N, Eller M, Garcia A, Lepak N, Melia-Hancock S, Oropeza-Rosas M, Salvo S, et al. (October 26, 2011) The relationship between parental genetic or phenotypic divergence and progeny variation in the maize nested association mapping population. Heredity http://dx.doi.org/10.1038/hdy.2011.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krill AM, Kirst M, Kochian LV, Buckler ES, Hoekenga OA. (2010) Association and linkage analysis of aluminum tolerance genes in maize. PLoS ONE 5: e9958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kump KL, Bradbury PJ, Wisser RJ, Buckler ES, Belcher AR, Oropeza-Rosas MA, Zwonitzer JC, Kresovich S, McMullen MD, Ware D, et al. (2011) Genome-wide association study of quantitative resistance to southern leaf blight in the maize nested association mapping population. Nat Genet 43: 163–168 [DOI] [PubMed] [Google Scholar]
- Lambert RJ. (2001) High-oil corn hybrids. In AR Hallauer, ed. In Specialty Corns. CRC Press, Boca Raton, FL. pp 131–154 [Google Scholar]
- Laurie CC, Chasalow SD, LeDeaux JR, McCarroll R, Bush D, Hauge B, Lai C, Clark D, Rocheford TR, Dudley JW. (2004) The genetic architecture of response to long-term artificial selection for oil concentration in the maize kernel. Genetics 168: 2141–2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locatelli S, Piatti P, Motto M, Rossi V. (2009) Chromatin and DNA modifications in the Opaque2-mediated regulation of gene transcription during maize endosperm development. Plant Cell 21: 1410–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manicacci D, Camus-Kulandaivelu L, Fourmann M, Arar C, Barrault S, Rousselet A, Feminias N, Consoli L, Francès L, Méchin V, et al. (2009) Epistatic interactions between Opaque2 transcriptional activator and its target gene CyPPDK1 control kernel trait variation in maize. Plant Physiol 150: 506–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullen MD, Kresovich S, Villeda HS, Bradbury P, Li H, Sun Q, Flint-Garcia S, Thornsberry J, Acharya C, Bottoms C, et al. (2009) Genetic properties of the maize nested association mapping population. Science 325: 737–740 [DOI] [PubMed] [Google Scholar]
- Mertz ET, Bates LS, Nelson OE. (1964) Mutant gene that changes protein composition and increases lysine content of maize endosperm. Science 145: 279–280 [DOI] [PubMed] [Google Scholar]
- Miclaus M, Xu J-H, Messing J. (2011) Differential gene expression and epiregulation of alpha zein gene copies in maize haplotypes. PLoS Genet 7: e1002131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moose SP, Dudley JW, Rocheford TR. (2004) Maize selection passes the century mark: a unique resource for 21st century genomics. Trends Plant Sci 9: 358–364 [DOI] [PubMed] [Google Scholar]
- Panthee DR, Pantalone VR, West DR, Saxton AM, Sams CE. (2005) Quantitative trait loci for seed protein and oil concentration, and seed size in soybean. Crop Sci 45: 2015–2022 [Google Scholar]
- Peiffer DA, Le JM, Steemers FJ, Chang W, Jenniges T, Garcia F, Haden K, Li J, Shaw CA, Belmont J, et al. (2006) High-resolution genomic profiling of chromosomal aberrations using Infinium whole-genome genotyping. Genome Res 16: 1136–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips PC. (2008) Epistasis: the essential role of gene interactions in the structure and evolution of genetic systems. Nat Rev Genet 9: 855–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland JA, Bradbury PJ, Buckler ES, Nelson RJ. (2011) Genome-wide nested association mapping of quantitative resistance to northern leaf blight in maize. Proc Natl Acad Sci USA 108: 6893–6898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poneleit CG, Alexander DE. (1965) Inheritance of linoleic and oleic acids in maize. Science 147: 1585–1586 [DOI] [PubMed] [Google Scholar]
- Pouvreau B, Baud S, Vernoud V, Morin V, Gendrot G, Py C, Pichon J-P, Rouster J, Paul W, Rogowsky PM. (2011) Duplicate maize Wrinkled1 transcription factors activate target genes involved in seed oil biosynthesis. Plant Physiol. 156: 674–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al. (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Schultz JA, Juvik JA. (2004) Current models for starch synthesis and the sugary enhancer1 (se1) mutation in Zea mays. Plant Physiol Biochem 42: 457–464 [DOI] [PubMed] [Google Scholar]
- Séne M, Thévenot C, Hoffmann D, Bénétrix F, Causse M, Prioul JL. (2001) QTLs for grain dry milling properties, composition and vitreousness in maize recombinant inbred lines. Theor Appl Genet 102: 591–599 [Google Scholar]
- Simmonds NW. (1995) The relation between yield and protein in cereal grain. J Sci Food Agric 67: 309–315 [Google Scholar]
- Thompson GA, Larkins BA. (1989) Structural elements regulating zein gene expression. Bioessays 10: 108–113 [DOI] [PubMed] [Google Scholar]
- Thornsberry JM, Goodman MM, Doebley J, Kresovich S, Nielsen D, Buckler ES., IV (2001) Dwarf8 polymorphisms associate with variation in flowering time. Nat Genet 28: 286–289 [DOI] [PubMed] [Google Scholar]
- Tian F, Bradbury PJ, Brown PJ, Hung H, Sun Q, Flint-Garcia S, Rocheford TR, McMullen MD, Holland JB, Buckler ES. (2011) Genome-wide association study of leaf architecture in the maize nested association mapping population. Nat Genet 43: 159–162 [DOI] [PubMed] [Google Scholar]
- Valdar W, Holmes CC, Mott R, Flint J. (2009) Mapping in structured populations by resample model averaging. Genetics 182: 1263–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdar W, Solberg LC, Gauguier D, Burnett S, Klenerman P, Cookson WO, Taylor MS, Rawlins JNP, Mott R, Flint J. (2006) Genome-wide genetic association of complex traits in heterogeneous stock mice. Nat Genet 38: 879–887 [DOI] [PubMed] [Google Scholar]
- Vicente-Carbajosa J, Moose SP, Parsons RL, Schmidt RJ. (1997) A maize zinc-finger protein binds the prolamin box in zein gene promoters and interacts with the basic leucine zipper transcriptional activator Opaque2. Proc Natl Acad Sci USA 94: 7685–7690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassom JJ, Wong JC, Martinez E, King JJ, DeBaene J, Hotchkiss JR, Mikkilineni V, Bohn MO, Rocheford TR. (2008) QTL associated with maize kernel oil, protein, and starch concentrations; kernel mass; and grain yield in Illinois high oil x B73 backcross-derived lines. Crop Sci 48: 243–252 [Google Scholar]
- Wilson LM, Whitt SR, Ibáñez AM, Rocheford TR, Goodman MM, Buckler ES., IV (2004) Dissection of maize kernel composition and starch production by candidate gene association. Plant Cell 16: 2719–2733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki M, Tenaillon MI, Bi IV, Schroeder SG, Sanchez-Villeda H, Doebley JF, Gaut BS, McMullen MD. (2005) A large-scale screen for artificial selection in maize identifies candidate agronomic loci for domestication and crop improvement. Plant Cell 17: 2859–2872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Kandianis CB, Harjes CE, Bai L, Kim E-H, Yang X, Skinner DJ, Fu Z, Mitchell S, Li Q, et al. (2010) Rare genetic variation at Zea mays crtRB1 increases beta-carotene in maize grain. Nat Genet 42: 322–327 [DOI] [PubMed] [Google Scholar]
- Yu J, Holland JB, McMullen MD, Buckler ES. (2008) Genetic design and statistical power of nested association mapping in maize. Genetics 178: 539–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Pressoir G, Briggs WH, Vroh Bi I, Yamasaki M, Doebley JF, McMullen MD, Gaut BS, Nielsen DM, Holland JB, et al. (2006) A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet 38: 203–208 [DOI] [PubMed] [Google Scholar]
- Zheng P, Allen WB, Roesler K, Williams ME, Zhang S, Li J, Glassman K, Ranch J, Nubel D, Solawetz W, et al. (2008) A phenylalanine in DGAT is a key determinant of oil content and composition in maize. Nat Genet 40: 367–372 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.