Abstract
An attack of plants by pathogens or treatment with certain resistance-inducing compounds can lead to the establishment of a unique primed state of defense. Primed plants show enhanced defense reactions upon further challenge with biotic or abiotic stress. Here, we report that the primed state in Arabidopsis (Arabidopsis thaliana) is still functional in the next generation without additional treatment. We compared the reactions of Arabidopsis plants that had been either primed with β-amino-butyric acid (BABA) or with an avirulent isolate of the bacteria Pseudomonas syringae pv tomato (PstavrRpt2). The descendants of primed plants showed a faster and higher accumulation of transcripts of defense-related genes in the salicylic acid signaling pathway and enhanced disease resistance upon challenge inoculation with a virulent isolate of P. syringae. In addition, the progeny of primed plants was also more resistant against the oomycete pathogen Hyaloperonospora arabidopsidis. When transgenerationally primed plants were subjected to an additional priming treatment, their descendants displayed an even stronger primed phenotype, suggesting that plants can inherit a sensitization for the priming phenomenon. Interestingly, this primed to be primed phenotype was much reduced in the Arabidopsis β-amino-butyric acid priming mutant ibs1 (induced BABA sterility1). Our results demonstrate that the primed state of plants is transferred to their progeny and confers improved protection from pathogen attack as compared to the descendants of unprimed plants.
Plants have developed sophisticated mechanisms to protect themselves against biotic and abiotic stresses (Jones and Dangl, 2006; Hirayama and Shinozaki, 2010). An impressive amount of information is available on the genes and signaling pathways involved in the plants’ reaction following exposure to a given stress situation (Feys and Parker, 2000; Thomma et al., 2001; Kunkel and Brooks, 2002; Turner et al., 2002; Fujita et al., 2006; Hirayama and Shinozaki, 2007). Triggering of the defensive state of a plant can lead to acquired resistance in the case of biotic stresses or to acclimation for abiotic stresses (van Loon, 1997).
In numerous plant species acquired resistance functions against a wide array of pathogens (Kuc, 2001). Interestingly, acquired resistance can spread systemically throughout the plant. When the induction of resistance is due to nonpathogenic rhizobacteria in the rhizosphere it is referred to as induced systemic resistance (van Loon, 1997). Induced resistance resulting from a natural induction with a pathogen or the induction with chemical inducers is referred to as systemic acquired resistance (SAR; Ryals et al., 1996; Sticher et al., 1997). The manifestation of SAR is tightly correlated with the local and systemic activation of defense-related genes such as genes coding for pathogenesis-related (PR) proteins (Ryals et al., 1996). As during pathogen stress, defense pathways can also be up-regulated by applied chemical stimuli. Resistance induced by the nonprotein amino acid β-amino-butyric acid (BABA) leads to a substantial induction of PR genes only upon challenge inoculation (Zimmerli et al., 2000). Based on northern blots it was originally described that BABA does not induce the expression of PR genes by itself (Zimmerli et al., 2000; Ton et al., 2005) but the availability of more sensitive technologies such as microarrays and PCR revealed that BABA treatment itself leads to an induction of PR genes, although to much lower levels than observed after pathogen attack (Zimmerli et al., 2008).
When a treatment puts a plant in a state of increased alertness with no or only minimal gene induction it is called sensitizing or priming (Jakab et al., 2001; Conrath et al., 2002; Prime-A-Plant Group et al., 2006). Priming of defense responses confers a fitness advantage over direct induction of resistance as observed in traditional SAR responses that infer high costs in absence of pathogens (van Hulten et al., 2006). An overview on molecular mechanism(s) possibly underlying priming for defense was recently presented by Conrath (2011). There is increasing evidence that plants have a memory of encountered stress situations that allow them to better adapt to changing conditions. In addition, several studies have shown the relevance of epigenetic mechanisms underlying plant defense responses (Alvarez-Venegas et al., 2006; March-Díaz et al., 2008; Walley et al., 2008; Jaskiewicz et al., 2011). Furthermore, the relation between PR gene expression and functionality of the SUPPRESSOR OF PR1-1 INDUCIBLE1 (SNI1) SNI1 protein has also been linked to epigenetic regulation of somatic DNA recombination. SNI1 represses PR gene transcription by modifying levels of histone H3 acetylation and histone H3K4 dimethylation (Mosher et al., 2006). Better understanding of how and in what situations plants can use this memory is of great scientific as well as applied interest.
Here, we analyze inheritance of priming. We compare the resistance of same-generation-primed plants and their descendants toward virulent Pseudomonas syringae (Pst) DC3000 and virulent isolates of the oomycete Hyaloperonospora arabidopsidis. To this end, Arabidopsis (Arabidopsis thaliana) plants either mock treated, treated with BABA, or inoculated with avirulent Pst and the progeny of these plants was analyzed for primed expression of defense-related PR genes and disease resistance. We found that when challenged with a pathogen, the progeny of primed plants showed strongly enhanced expression of defense-related genes and enhanced disease resistance compared to the progeny of nonprimed plants.
RESULTS
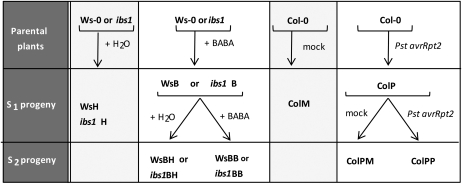
To study a possible transgenerational inheritance of priming and induced resistance, Arabidopsis lines that had been subjected to specific treatments with the goal of inducing resistance and/or priming were generated (Fig. 1). The role of BABA was tested in Arabidopsis accession Wassilewskija-0 (Ws-0) and in the priming mutant ibs1 (in the Ws-0 background; Ton et al., 2005). The parental lines Ws-0 and ibs1 were treated once with water as a control or with BABA solution in water. The plants were grown to seed set and the resulting selfed progeny (S1 progeny) was called WsH and ibs1H, respectively, in the case of water (H)-treated plants, and WsB and ibs1B, respectively, for BABA (B)-treated plants. Plants from the S1 progeny (WsB and ibs1B) were again either treated with water or with BABA, respectively. They were grown to seed set thus giving rise to the S2 progeny. S2 plants derived from S1 WsB and S1 ibs1B plants and treated with water were called WsBH and ibs1BH, respectively. S2 plants derived from S1 WsB and S1 ibs1B plants and treated with BABA were called WsBB and ibs1BB, respectively.
Figure 1.
Experimental design. Parent plants: Ws-0 (Ws wild-type plants), ibs1 (a priming mutant in Ws-0 background), and Col-0 (Col wild-type plants). S1 progeny: descendants from the selfed parent plants; S2 progeny: descendants from the selfed S1 plants. The plants were treated with: water = H; BABA = B; mock treated with buffer = M; or inoculated with avirulent Pst pv tomato containing avrRpt2 = P.
Arabidopsis plants accession Columbia-0 (Col-0) were inoculated with avirulent bacteria carrying the avirulence gene avrRpt2 and were tested for a possible transgenerational effect (Fig. 1). Col-0 parental plants were either mock inoculated, giving rise to a S1 progeny called ColM or inoculated with a suspension of avirulent Pst in buffer, yielding S1 progeny seeds called ColP (P standing for Pseudomonas). ColP was sown and again either mock inoculated with buffer or inoculated with a suspension of avirulent Pst in buffer. The seeds harvested from these plants (S2 progeny) were called ColPM and ColPP, respectively.
The Priming State Induced by BABA Is Transferred to the Next Generation
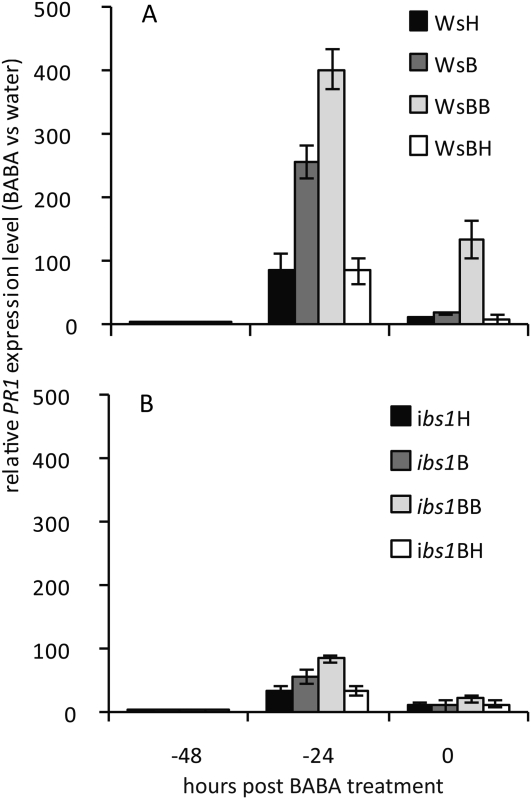
A transient accumulation of transcripts of salicylic acid (SA)-dependent marker genes is a hallmark of BABA-IR and priming (Zimmerli et al., 2000; Ton et al., 2005). Therefore, we studied PR1 expression, over a time course of 48 h, in 5-week-old plants of WsH, ibs1H, WsB, ibs1B, WsBH, ibs1BH, WsBB, and ibs1BB that were soil drenched with 25 ppm BABA or with water as a control, respectively (see Supplemental Fig. S1A for experimental design).
BABA treatment induced a transient enhancement of PR1 transcript levels in WsH plants (Fig. 2). Interestingly, transcript levels of PR1 were 3-fold higher in WsB and 4.6-fold higher in WsBB compared to WsH. In contrast, WsBH plants contained similar transcript levels as WsH (Fig. 2A), indicating that priming for gene expression was switched off after one generation. The progeny of the ibs1 mutant showed a similar pattern although the amplitude of the reaction was much reduced (Fig. 2B). Our results show that the progeny of Ws-0 as well as of ibs1 are sensitized to the priming of PR1 triggered by BABA, thus displaying a memory of the treatments to which their parents had been subjected to. Furthermore, the facts that ibs1 lines are severely affected in the induction of PR1 upon infection and that Ws-0 or ibs1 descendants do not show induction of PR1 in the absence of BABA treatments (Supplemental Fig. S2, water-treated plants) strongly indicate that this phenomenon is related to priming of defense. The results also show that the memory had to be rescued, as in the absence of further priming treatments the level of PR1 transcripts was not elevated. This is consistent with the transient nature of the priming activity.
Figure 2.
Basal PR1 expression levels in the progeny of Ws-0 plants and ibs1 plants (see Fig. 1). Four-week-old plants were soil drenched with water or a solution of BABA to a final concentration of 25 ppm in the soil. Transcript levels were analyzed with qRT-PCR. Expression was normalized to the sample treated with water at 0 h. PR1 expression in the different Ws-0 lines (A) and in the different ibs1 lines (B) is shown. The values represent means ± sd of three replicates. Similar results were obtained in three independent experiments.
Resistance against Pst DC3000 Is More Effective in the Progeny of BABA-Treated Plants
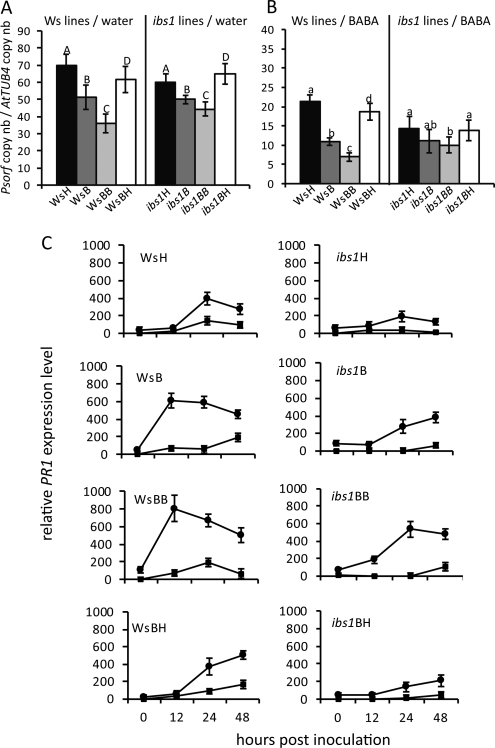
Parent Ws-0 and ibs1 plants and descendants as shown in Figure 1 were either treated by soil drench with 25 ppm BABA or water, respectively. Forty-eight hours after treatment (time 0; Supplemental Fig. S1a), plants were inoculated with virulent Pst DC3000. Bacterial growth was monitored by quantitative reverse transcription (qRT)-PCR using Pst-DC3000-specific primers over a time course of 72 h after inoculation.
Descendants of BABA-primed plants (WsB, WsBB, and WsBH) showed enhanced resistance against Pst DC3000 compared to WsH (Fig. 3; Supplemental Fig. S3). In addition, the level of resistance of the descendants was stronger according to the number of treatments accumulated through previous generations. Furthermore, a similar pattern of resistance was observed after BABA treatment prior to infection, resulting in a more pronounced resistance in the progeny of primed Arabidopsis (Fig. 3B). BABA-treated ibs1 progeny also showed transgenerational enhanced resistance against Pst DC3000 although they lacked the improved capacity to express BABA-IR observed in wild-type progeny plants.
Figure 3.
Descendants of BABA-treated Ws-0 plants are more resistant to virulent Pst. Three-week-old plants were treated with BABA (25 ppm final concentration in the soil) or water 2 d prior to inoculation with Pst DC3000 (OD600 = 0.08). A, Growth of Pst DC3000 in the water-treated Ws-0 lines (WsH, WsB, WsBB, and WsBH) and ibs1 lines (ibs1H, ibs1B, ibs1BB, and ibs1BH) at 72 hpi. B, Growth of Pst DC3000 in BABA-treated Ws-0 lines and in ibs1 lines at 72 h postinoculation. Bacterial growth was quantified by qRT-PCR as transcript levels of Psorf normalized to the transcript level of the Arabidopsis gene AtTUB4. Capital letters indicate statistically significant bacterial growth within the water-treated Ws-0 and ibs1 lines (ANOVA, Student-Newman-Keuls, n = 3, P < 0.05). Small letters indicate statistically significant bacterial growth within the BABA-treated Ws-0 or ibs1 lines. C, qRT-PCR analysis of PR1 gene expression in BABA-treated (circles) and water-treated (squares) Ws-0 and ibs1 lines. Expression was normalized to the corresponding sample treated with water at 0 h. The values represent means ± sd of three replicates. Similar results were obtained in three independent experiments.
Priming efficiency, expressed as the quotient of bacterial growth between two generations varied from 1.6 to 1.9 in BABA-treated Ws-0 lines. This quotient was between 1.1 and 1.4 for BABA-treated ibs1 lines, water-treated ibs1 lines, and for water-treated Ws-0 lines (Supplemental Table S1).
Transgenerational Priming for Defense Is Supported by SA Marker Gene Sensitization
The priming state of the plants was reflected at the molecular level in the Ws-0 lines and ibs1 lines upon inoculation with Pst DC3000. PR1 (Fig. 3C) and PR2 and PR5 transcript accumulation (Supplemental Fig. S4, A and B) remained low in all inoculated water controls. In BABA-treated plants inoculated with Pst DC3000, PR1 accumulation peaked at 24 h post inoculation (hpi) in WsH. WsB plants reacted earlier and stronger and the reaction was even more pronounced in WsBB (Fig. 3C). WsBH plants displayed similar kinetics and intensity of PR1 transcript accumulation as the WsH plants.
In the ibs1 lines the accumulation of PR1 transcripts was delayed and attenuated compared to the corresponding Ws-0 line. ibs1H plants displayed a low relative expression level peaking at 24 hpi while in ibs1B the expression level was twice as high but only at 48 hpi. ibs1BB peaked at 24 hpi with 3-fold-higher level than ibs1H. As for WsBH, in ibs1BH PR1 accumulation levels reverted to ibs1H level.
The same trend was observed for PR2 and PR5 transcript accumulation (Supplemental Fig. S4, A and B). These results show that although all plants reacted to BABA treatment with an induction of PR gene transcripts following inoculation with Pst DC3000, the reaction was much more pronounced in the priming-competent Ws-0 lines than in the priming-impaired ibs1 lines.
Transgenerational Inheritance of Priming Induced by Avirulent Pst
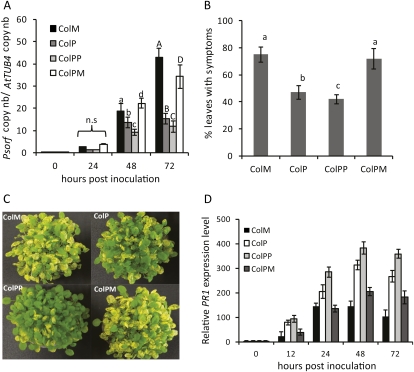
To demonstrate that transgenerational priming is not only observed following chemical induction, we performed a set of experiments using a natural priming phenomenon. Since preliminary tests had shown that S1 descendants of both Ws-0 and Col-0, respectively, showed transgenerational priming (data not shown), we decided to use Col-0 plants for this experiment to demonstrate that transgenerational priming was also not bound to a specific Arabidopsis accession. Leaves of accession Col-0 were infiltrated with avirulent Pst, leading to a localized hypersensitive reaction. An overview of treatments and generations is shown in Figure 1.
Challenge inoculation of the various Col-0 lines with virulent Pst DC3000 showed that transgeneration priming is also observed following an inducing inoculation of parent plants with avirulent Pst. At all time points post inoculation, the ColP and ColPP plants supported less bacterial growth compared to ColM plants (Fig. 4A). The restriction in bacterial growth correlated with reduced disease symptoms; ColM and ColPM plants showed more severe disease symptoms than the ColP and the ColPP plants (Fig. 4, B and C). Therefore, our data show that Arabidopsis plants exposed to same-generation priming by a necrotizing infection with avirulent bacteria transfer the experience of the encountered stress situation to their descendants. Interestingly, ColPM plants showed no significant reduction of disease symptoms and the same PR1 transcript levels compared to ColM (Fig. 4B). As it was observed with BABA-treated progeny, transgeneration priming did not last after a priming-free generation, suggesting that BABA- and PstavrRpt2-induced transgenerational resistance may be temporarily maintained via similar mechanisms.
Figure 4.
Inheritance of priming induced by avirulent Pst (PstavrRpt2). Single leaves of 4-week-old plants were infiltrated with Pst avrRpt2 or mock infiltrated with buffer. A, Col-0 lines (see Fig. 1) were challenge inoculated with Pst DC3000 and bacterial growth was quantified by qRT-PCR as described for Figure 3. Small and capital letters above error bars represent statistically significant difference in bacterial growth within Col-0 lines at 48 and 72 h postinoculation (ANOVA, Student-Newman-Keuls, n = 3, P < 0.001). Values represent means ± sd of three biological replicates. B, Quantification of disease resistance of Col lines 5 d post inoculation. Letters represent statistically significant difference in the percentage of leaves with symptoms (ANOVA, Student-Newman-Keuls, n = 30, P < 0.05). C, Disease phenotype of Col lines. D, qRT-PCR analysis of PR1 gene expression in Col lines. Expression was normalized to the sample treated with water at 0 h. The values represent means ± sd of three replicates. Similar results were obtained in three independent experiments.
The differences in basal levels of resistance among the studied lines correlated with levels of transcript accumulation of PR1 (Fig. 4D), PR2 (Supplemental Fig. S5A), and PR5 (Supplemental Fig. S5B).
Transgenerational Inheritance of Priming against H. arabidopsidis
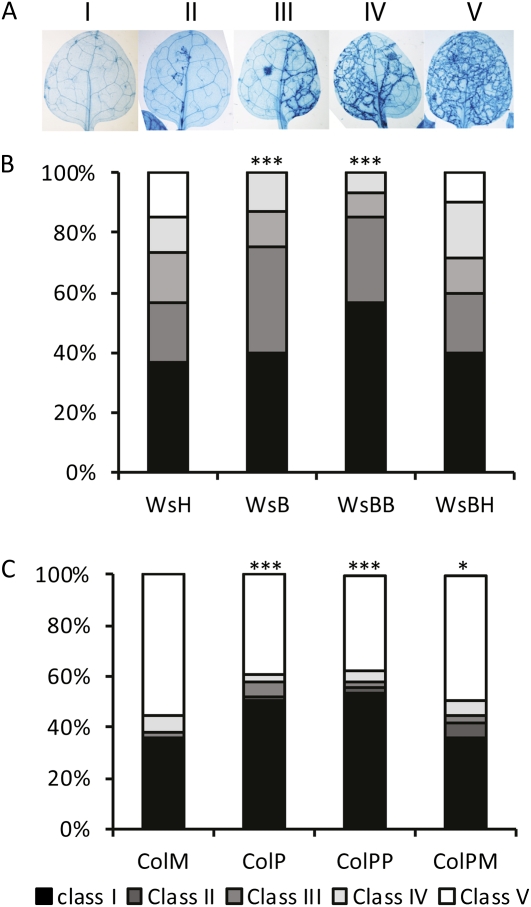
The results described above demonstrate that transgeneration priming is associated with the sensitization of the SA-dependent response. To test whether this phenomenon was also effective against other SA-sensitive pathogens, we inoculated BABA- and water-treated progeny, as well as the progeny of plants mock or avirulent Pst inoculated, with virulent strains of the obligate biotrophic oomycete H. arabidopsidis (isolate Noco for Col-0 and isolate Emwa for Ws-0). Disease severity was assessed at 7 d post inoculation in trypan-blue-stained leaves (Fig. 5). The leaves were classified into five categories according to their degree of colonization (Fig. 5A). The control lines ColM and WsH (Fig. 5) plants consistently showed a higher degree of colonization by the oomycete than the primed descendants WsB and WsBB (Fig. 5B), ColP, and ColPP (Fig. 5C). Second generation descendants whose parents were not subjected to an additional priming treatment in the S1 generation reverted to a more susceptible infection phenotype (Fig. 5, B and C). These results show that the progeny of Arabidopsis plants exposed to same-generation priming with avirulent bacteria or with BABA in the parental generation are more resistant to H. arabidopsidis.
Figure 5.
Inheritance of priming against H. arabidopsidis. A, Disease ratings were classified based on the percentage of leaf colonization by mycelium. Class I: 0%; class II: 1% to 25%; class III: 26% to 50%; class IV: 51% to 75%; and class V: 76% to 100%. Disease ratings are shown as the percentage of leaves in each resistance class. B, Disease resistance in Ws-0 lines inoculated with the Emwa strain of H. arabidopsidis. Asterisks indicate statistically significant differences in the percentage of leaves in the different classes compared to WsH (Fisher test, *** = P < 0.001). C, Disease resistance in Col lines inoculated with the Noco strain of H. arabidopsidis. Asterisks indicate statistically significant differences in the percentage of leaves in the different classes compared to ColM (Fisher test, * = P < 0.05; *** = P < 0.001).
Priming in the Progeny of BABA-Treated Plants Is Not Due to Transfer of BABA to the Next Generation through the Seeds
To assess whether the priming observed in the progeny of BABA-treated plants might be due to a transfer of BABA to the next generation through the seeds, we measured BABA concentrations in untreated plants and plants soil drenched to a final concentration of 40 ppm (Supplemental Fig. S6A) and 25 ppm BABA (Supplemental Fig. S6B), respectively. No BABA was present in untreated plants. The highest concentration of BABA was detected within days after treatment. Traces of BABA were found in the flowers of BABA-treated plants and in the derived S1 seeds (Supplemental Fig. S6A). We also analyzed BABA content in 7-d-old S1 seedlings and 5-week-old S1 rosette stage descendants of BABA-treated Col-0 plants. The traces of BABA detected (<2.2 μg/g dry weight) were considerably below the concentration threshold for resistance induction that corresponds in planta with 50 μg/g dry weight (calculated empirically from experiments performed over the years in our lab). Hence, the BABA level in the progeny of BABA-treated plants is far too low to be responsible for the observed transgenerational priming. Our conclusion is further supported by the fact that transgenerational priming is also observed in the absence of BABA in the progeny of plants inoculated with avirulent Pst.
Changes in DNA Methylation Levels in the PR1 Promotor
Since the PR1 gene has been shown to be very responsive to priming and this effect is transferred to the next generation (this article) we decided to assess the DNA methylation level in the region of the PR1 promotor where many of the elements possibly involved in such an increased induction are located. To this end we subjected plant DNA to bisulfite treatment and quantified the pattern of methylation of CGN, CHG, and CHH sites over a 503-bp-long region in the generated clones using the CyMate program (Hetzl et al., 2007). The CyMate output for methylation analysis is depicted in Supplemental Figures S7 and S8.This analysis was performed on samples of WsH, WsB, WsBB, and WsBH as well as on samples of ibs1H, ibs1B, ibs1BB, and ibs1BH. As shown in Supplemental Figure S9, only minimal changes in overall c-methylation frequency were observed in the sequenced clones. This is also true for the methylation pattern of specific motives (WRKY/W boxes; TGA binding sites; ERF binding site; PR-box and NF-κB binding site) within the PR1 promotor section tested (data not shown).
DISCUSSION
Our results show that subjecting Arabidopsis plants to a treatment with the chemical BABA or an inoculation with avirulent bacteria induces a primed state that is transmitted to the progeny. The progeny of primed plants has a higher basal level of disease resistance and an enhanced capacity to react to additional priming treatments.
To study the transmission of the BABA-primed state in WsB, WsBB, and WsBH plants, we analyzed levels of transient accumulation of PR gene transcripts upon BABA treatment. Basal levels of PR expression were not altered previous to BABA treatment (Fig. 2), however WsB was sensitized to additional BABA treatments, resulting in higher PR1 transcript levels compared to the progeny of water-treated plants (WsH). Additional BABA treatment of the WsB generation further increased the priming capacity in the WsBB generation (Fig. 2). Furthermore, the enhanced disease resistance in the progeny of primed lines against Pst DC3000 correlated with increased levels of SA-dependent gene transcripts of PR1, PR2, and PR5 upon infection, indicating general changes in the regulatory mechanisms of defense gene expression (Fig. 3). Interestingly, our data also show that priming induced by inoculation with avirulent Pst (PstavrRpt2) can also be inherited by the descendants. The progeny of plants treated with BABA or inoculated with avirulent Pst were significantly more resistant to virulent Pst and H. arabidopsidis, respectively, than the progeny of control-treated plants (Fig. 4). Transgeneration priming of PR1 expression was lost in the S2 generation (WsBH) in the absence of new BABA treatments, whereas the resistance phenotype was still partially detected.
Interestingly, BABA treatment of a parental generation sensitizes its descendants to be primed more effectively: The progeny is primed to be primed. In contrast, the ibs1 mutant that is defective in priming (Ton et al., 2005), was compromised in the primed to be primed response (Fig. 3). Transgenerational enhancement of basal resistance was, however, not affected in ibs1, suggesting the involvement of different underlying mechanisms in the two phenomena.
We had previously shown that BABA was able to prime plants to react faster to biotic and abiotic stress (Zimmerli et al., 2000; Jakab et al., 2005; Flors et al., 2008; Gális et al., 2009) and it is accepted that priming occurs in many plant species and with various priming agents (Agrawal, 2002; Conrath et al., 2002; Prime-A-Plant Group et al., 2006). Despite numerous reports on the same generation, examples pointing to transgenerational priming are scarce. Wild radish (Raphanus sativus) that had been fed on by Pieris rapae or treated with jasmonate, respectively, yielded progeny that displayed an increase in resistance to herbivores (Boyko et al., 2007). Enhanced virus resistance was reported in the progeny of virus-induced tobacco (Nicotiana tabacum) plants (Boyko et al., 2010; Kathiria et al., 2010). In addition, other studies describe transgeneration effects in the progeny of plants infected with the virulent Pst DC3000 or damaged by herbivory attack, which are mediated by the up-regulation of the SA- or jasmonic-acid-related signaling pathway, respectively (Luna et al., 2011; Rasmann et al., 2011). Therefore, this suggests that the molecular mechanisms underlying induced defense inheritance may depend on the stress or the priming agent to which parental lines were exposed to. The capacity for transgenerational priming on the other hand is clearly not accession specific since we observed it in both Arabidopsis accessions with Ws-0 and with Col-0 background, respectively.
In our study, for both BABA- and Pst-primed plants the primed state was only upheld in the immediate progeny. Without renewed priming treatment of the parent generation, in the progeny, the disease resistance phenotype and defense gene expression reverted to an unprimed state. In contrast, Luna et al. (2011) and Rasmann et al. (2011) demonstrated that transgenerational priming can be maintained over one stress-free generation. However, nonpersistent transgenerational adaptive effects to further generations have been described previously (Pecinka et al., 2009). Furthermore, Luna et al. (2011) exposed parental plants to recurrent pathogen stress by Pst DC3000 whereas our experiments consisted in single inoculations. The timeframe in which transgenerational priming or resistance is upheld seems to be dependent on the inducer and the severity of the disease in the parental generation.
The possibility that the transgenerational priming observed in progeny from BABA-treated parents might be due to a direct transfer of BABA through the seeds to the new generation is improbable. Our BABA measurements show that already 3 weeks after treating Arabidopsis with BABA, the remaining quantities of BABA in the plant tissues were far below the threshold needed to induce resistance (Supplemental Fig. S6). BABA was undetectable in the progeny of BABA-treated plants at the time of challenge inoculation.
For the offspring to remember a past experience from the parents, the latter have to be able to perceive the specific stress, they have to store this information, retain and transmit it to the descendants. To benefit from this information the descendants have to be able to retrieve the information and translate it into appropriate reactions. Although the molecular mechanisms of same-generation priming have recently started to be better understood (for review, see Conrath, 2011), the molecular details underlying transgenerational priming are still under study. However, recent groups have demonstrated that epigenetic mechanisms play a role in plant defense and several review articles comment on the possible involvement of epigenetic mechanisms in transgenerational phenomena (Chinnusamy and Zhu, 2009; Alvarez et al., 2010; Sano, 2010). Histone modifications are examples of epigenetic mechanisms; indeed, such modifications in the promoters of defense genes have been shown to correlate with transgenerational induced resistance in Arabidopsis against abiotic (Boyko et al., 2010) and biotic stresses (Luna et al., 2011). In addition, small RNA molecules are known to be important components of gene regulation (Vaucheret, 2006) and have been described that they play a role in transgenerational effects in plants against abiotic stress (Boyko et al., 2010). In agreement with these results, Rasmann et al. (2011) describe that mutants blocked in the enzymatic activity responsible for the biosynthesis of small RNA lacked to express transgeneration-enhanced resistance to herbivores. Finally, it has been described that different defense situations such as pathogen infection (Pavet et al., 2006) or chemical treatment with jasmonic acid and SA (Verhoeven et al., 2010) resulted in changes in DNA methylation patterns. These changes are yet the most likely signal to be transmittable through meiosis. Indeed, Luna et al. (2011) showed that reduced DNA methylation in the drm1drm2cmt3 (for domains rearranged methyltransferase1, -2, cytosine methyltransferase3) triple mutant mimics transgenerational priming of SA-dependent defenses. In combination with previous findings that Pst bacteria induce large-scale hypomethylation in Arabidopsis (Pavet et al., 2006), it is plausible that the transmission of this defense priming is mediated by hypomethylated DNA. Although DNA methylation has long been considered to be stable, spontaneous gain or loss of DNA methylation, leading to metastable heritable changes in methylated cytosines have been described recently (Schmitz et al., 2011). Such mechanisms might explain why the priming state is not transmitted to a further generation in our system or in the one described by Rasmann et al. (2011) when the priming pressure is not upheld.
Since we observed a constant and strong priming of PR1 expression in both BABA- and Pst-primed plants we tested whether demethylation in the PR1 promotor might explain the priming behavior in our Ws-0 parental and descendant lines as well as in the corresponding ibs1 lines lacking the strong priming of PR1 expression. Notable changes in DNA c-methylation were found for neither of the lines and treatments in the section of the promotor sequenced nor in the specific motives corresponding to cis-elements known to be important for PR1 expression (Supplemental Figs. S7–S9). In contrast, Pavet et al. (2006) observed a demethylation of nearly 38% following infection of Arabidopsis with Pst.
Inheritance of alterations in DNA methylation being the most plausible explanation for the observed transgenerational inheritance of priming, it is tempting to conclude from our results that transgenerational regulation of PR1 expression in this case could be based on the activity of hypomethylated transacting elements of transcription factors that regulate the expression of PR1 and related genes. Whether this is the case in our system is currently under investigation.
CONCLUSION
The capacity of a plant to express primed resistance depends on multiple signal transduction pathways (Zimmerli et al., 2000; Jakab et al., 2005; Gális et al., 2009). The emerging picture is that priming improves a plant’s ability to cope with a given stress situation. Hence, a plant’s capacity for priming is an important survival parameter. Inheritance of the primed state as observed in transgenerational priming is expected to contribute to improved adaptation of the progeny to environmental conditions. Our findings have obvious implications for natural and agronomical ecosystems. Especially in the latter field, transgenerational priming suggests the possibility of producing disease-resistant offspring by intentionally exposing parent plants to a priming treatment.
MATERIALS AND METHODS
Biological Material
Arabidopsis (Arabidopsis thaliana) accession Col-0, Ws-0, and the ibs1 mutant were grown in Jiffy peat pellets maintained at 20°C day/18°C night temperature with 8 h light (150 μE m−2 s−1) and 70% relative humidity. About 30 seeds per pot were stratified at 4°C in the dark for 2 d and grown for 3 weeks before being used for Pst DC3000 bioassays. For the Hyaloperonospora arabidopsidis bioassays, 2-week-old plants were used. When the seedlings had reached the cotyledon stage, the number was reduced to 15/Jiffy (n = 3 in figure legends stands for n = 3 Jiffys with each 15 plants). One 4-week-old plant per Jiffy was used for stress induction with Pst avrRpt2 and for BABA-priming experiments. Bacterial strains were grown as described (Zimmerli et al., 2000) with the exception that the avirulent strain Pst avrRpt2 was grown with the addition of 25 μg mL−1 kanamycin for selection. H. arabidopsidis strains Noco and Emwa were maintained on Arabidopsis Col-0 and Ws-0, respectively.
The timing of treatments in the various bioassays is schematically presented in Supplemental Figure S1.
Generation of First and Second Progeny of BABA-Primed Lines and Pst avrRpt2 Lines
Tap water (pH 8.2) as a control treatment or an aqueous solution of BABA (pH 8.2; Sigma; final concentration of 25 ppm in the soil) were applied as a soil drench to 4-week-old Ws-0 and ibs1 plants. Each Jiffy stood in a small petri dish to guarantee that each plant received exactly the same amount of BABA. Flowering was induced by transferring plants to long-day conditions (16 h of light). S1 plants grown from the seeds of those parent plants were designated as WsB, ibs1B, WsH, and ibs1H, respectively (see Fig. 1). They were again treated with water or BABA at 4 weeks post germination. Seeds of these plants gave rise to the S2 generation WsBB, ibs1BB and WsBH, and ibs1BH lines, respectively.
Pst DC3000 carrying the avirulence gene avrRpt2 was used to generate the Pst avrRpt lines (Fig. 1). Five leaves of 4-week-old Col-0 plants were syringe infiltrated with a bacterial suspension of 108 colony-forming units mL−1 in 10 mm MgSO4. Mock-infiltrated plants were injected with 10 mm MgSO4 (pH 8.6). Plants were maintained at 100% relative humidity for 1 week. Flowering was induced by transferring plants to long-day conditions. The resulting S1 seeds (ColP and ColM, respectively) were again syringe infiltrated with bacteria or mock infiltrated to give rise to the S2 generation (ColPP and ColPM, respectively). For the generation of the transgeneration seeds we started with 15 mother plants each for every line and treatment; 15 single plants each were randomly chosen to produce the subsequent generation for each treatment.
Plant Inoculation and Sample Processing
BABA (25 ppm final concentration in the soil) or water (control) was applied as soil drench to 3-week-old plants 2 d prior to inoculation with Pst (Zimmerli et al., 2000). BABA treatment (15 ppm final concentration in the soil) or water was applied similarly on 2-week-old plants for H. arabidopsidis bioassay (Ton et al., 2005).
Plants were dip inoculated with bacteria (Zimmerli et al., 2000) and disease symptoms were scored (Jakab et al., 2005).
Seedlings were spray inoculated with H. arabidopsidis and tissue colonization was quantified (Ton et al., 2005). Colonized leaves were assigned to five different classes based on hyphal proliferation (Fig. 5A). Fisher’s exact test using R.2.11.0 software was done to show significant differences between lines and treatments.
Quantification of Pst
Quantification of Pst DC3000 growth was performed by qRT-PCR as described (Boachon et al., 2009). Genomic DNA from 3-week-old plants was extracted using the NucleoSpin plant II kit (Macherey-Nagel) and bacterial growth was quantified as transcript levels of Psorf normalized to the transcript level of the Arabidopsis gene AtTUB4. This method was used because it allows to accurately process large number of samples in a short time and to have enough material to perform gene expression analyses on the same material.
Gene Expression Analyses
Total RNA was isolated from 3-week-old plants using the NucleoSpin RNA plant kit (Macherey-Nagel) according to the manufacturer’s instructions. cDNA was obtained from 1 μg of total RNA using the PrimeScript RT kit (Takara). qRT-PCR quantification of gene expression was performed in a Rotor Gene 6000 (Qiagen). Gene-specific primer sequences were designed using Primer 3 software. Three biological replicates were collected per time point. The software program Gene-X was used to calculate the mean normalized (to the housekeeping gene SAND) expression of the genes (Vandesompele et al., 2002). Ct values ± sd for PR1, PR2, and PR5 for all the lines and treatments at time zero can be found in Supplemental Table S2.
Determination of BABA Levels
Plants were frozen in liquid nitrogen and lyophilized. Dry plant material was extracted on ice with a methanol:water mixture (10:9) containing 0.01% formic acid. After centrifugation the supernatant was recovered and filtered through a 0.22-μm cellulose filter. Aliquots were injected into an ultraperformance liquid chromatograph coupled to a triple quadruple mass spectrometer (Aquity TQD, Waters). The amount of BABA in the samples was calculated by using an external calibration curve of BABA by scoring the eluting peaks using a standard C-18 Ultra Performance Liquid Chromatography column. The mass spectrometer transition selected in positive electrospray ionization (ESI) was 104 > 44 mass-to-charge ratio. The gradient, mobile phases, and other chromatographic conditions were as described (Flors et al., 2008).
DNA Extractions and Bisulphite Sequencing
Total genomic DNA was extracted from 3-week-old leaf tissue of each WsH, WsB, WsBB, WsBH, ibs1H, ibs1B, ibs1BB, and ibs1BH lines using the Qiagen DNeasy plant kit (Qiagen) according to the manufacturer’s protocol. Approximately 2 μg of genomic DNA were used for bisulphite conversion. Bisulphite modification and desulfonication of genomic DNA were performed using the MethylDetector bisulfite modification kit (Active Motif, www.activemotif.com) according to the manufacturer’s instructions.
Based on Johnson et al. (2003), the primer pair used was as follows: 5′-TCGGAGGAGTATATGTTATTGCTTAGAATCA-3′ and 5′-TTGTTTCGTATCGGTAGCTTTGCCAT-3′ (PCR conditions are available upon request). This allowed the amplification by PCR of a 672-bp fragment of the PR1 promotor containing as-1 sequences. The PCR products were inserted into pGEM-T easy vector and at least eight to 20 clones for each WsH, ibs1H, and their respective transgenerational descendant lines were selected for plasmid isolation. The clones were sequenced (www.macrogen.com) and 503-bp segments of each sequence starting at position −679 relative to transcription initiation site of the PR1 gene were then aligned using CLUSTAL version 1.83. The alignment files were saved, then submitted to CyMATE (http://www.gmi.oeaw.ac.at/research-groups/cymate/cymate) and analyzed with this program.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Experimental design of resistance tests.
Supplemental Figure S2. PR1 expression levels in Ws-0 and ibs1 lines control treated with water.
Supplemental Figure S3. Descendants of BABA-treated Ws-0 plants are more resistant to virulent Pst.
Supplemental Figure S4. qRT-PCR analysis of PR2 and PR5 transcript levels in Ws-0 and ibs1 lines upon inoculation with virulent Pst.
Supplemental Figure S5. qRT-PCR analysis of PR2 and PR5 transcript levels in Col-0 lines upon inoculation with virulent Pst.
Supplemental Figure S6. Priming of the progeny of BABA-treated lines is not due to direct transfer of BABA to the next generation.
Supplemental Figure S7. Graphical output generated by CyMate of the Ws transgenerational lines.
Supplemental Figure S8. Graphical output generated by CyMate of the ibs1 transgenerational lines.
Supplemental Figure S9. Bilsulfite sequencing data showing percentage of CGN, CHG, and CHH methylation, as computed by the CyMate program, in a 503-bp region of the PR1 promotor of Ws-0 and descendants and ibs1 and descendants, respectively.
Supplemental Table S1. Comparison of priming efficiency of different generations of Ws-0 and ibs1 lines in response to inoculation with virulent Pst.
Supplemental Table S2. Ct values for the transgenerational lines and treatments at 48 h.
Supplementary Material
Acknowledgments
We thank Felix Mauch for critical reading of the manuscript, Matthias Held for help with the statistical analysis, and the Servei Central d'Instrumentació Científica of the University Jaume I for the technical assistance.
References
- Agrawal AA. (2002) Maternal effects associated with herbivory: mechanisms and consequences of transgenerational induced plant resistance. Ecology 83: 3408–3415 [Google Scholar]
- Alvarez ME, Nota F, Cambiagno DA. (2010) Epigenetic control of plant immunity. Mol Plant Pathol 11: 563–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Venegas R, Sadder M, Hlavacka A, Baluška F, Xia Y, Lu G, Firsov A, Sarath G, Moriyama H, Dubrovsky JG, et al. (2006) The Arabidopsis homolog of trithorax, ATX1, binds phosphatidylinositol 5-phosphate, and the two regulate a common set of target genes. Proc Natl Acad Sci USA 103: 6049–6054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boachon B, Robert J, Daniel X, Mauch-Mani B. (2009) Absolute quantification of specific plant defence pathways and disease development of several microbial pathogens in Arabidopsis using real-time PCR. IOBC Meeting Proceedings of “Biological Control of Fungal and Bacterial Plant Pathogens (Phytopathogens)” Molecular Tools for Understanding and Improving Biocontrol, 43, pp 311–316 [Google Scholar]
- Boyko A, Blevins T, Yao Y, Golubov A, Bilichak A, Ilnytskyy Y, Hollunder J, Meins F, Jr, Kovalchuk I. (2010) Transgenerational adaptation of Arabidopsis to stress requires DNA methylation and the function of Dicer-like proteins. PLoS ONE 5: e9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyko A, Kathiria P, Zemp FJ, Yao Y, Pogribny I, Kovalchuk I. (2007) Transgenerational changes in the genome stability and methylation in pathogen-infected plants: (virus-induced plant genome instability). Nucleic Acids Res 35: 1714–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Zhu JK. (2009) Epigenetic regulation of stress responses in plants. Curr Opin Plant Biol 12: 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrath U. (2011) Molecular aspects of defence priming. Trends Plant Sci 16: 524–531 [DOI] [PubMed] [Google Scholar]
- Conrath U, Pieterse CM, Mauch-Mani B. (2002) Priming in plant-pathogen interactions. Trends Plant Sci 7: 210–216 [DOI] [PubMed] [Google Scholar]
- Feys BJ, Parker JE. (2000) Interplay of signaling pathways in plant disease resistance. Trends Genet 16: 449–455 [DOI] [PubMed] [Google Scholar]
- Flors V, Ton J, van Doorn R, Jakab G, García-Agustín P, Mauch-Mani B. (2008) Interplay between JA, SA and ABA signalling during basal and induced resistance against Pseudomonas syringae and Alternaria brassicicola. Plant J 54: 81–92 [DOI] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, Shinozaki K. (2006) Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr Opin Plant Biol 9: 436–442 [DOI] [PubMed] [Google Scholar]
- Gális I, Gaquerel E, Pandey SP, Baldwin IT. (2009) Molecular mechanisms underlying plant memory in JA-mediated defence responses. Plant Cell Environ 32: 617–627 [DOI] [PubMed] [Google Scholar]
- Hetzl J, Foerster AM, Raidl G, Mittelsten Scheid O. (2007) CyMATE: a new tool for methylation analysis of plant genomic DNA after bisulphite sequencing. Plant J 51: 526–536 [DOI] [PubMed] [Google Scholar]
- Hirayama T, Shinozaki K. (2007) Perception and transduction of abscisic acid signals: keys to the function of the versatile plant hormone ABA. Trends Plant Sci 12: 343–351 [DOI] [PubMed] [Google Scholar]
- Hirayama T, Shinozaki K. (2010) Research on plant abiotic stress responses in the post-genome era: past, present and future. Plant J 61: 1041–1052 [DOI] [PubMed] [Google Scholar]
- Jakab G, Cottier V, Toquin V, Rigoli G, Zimmerli L, Métraux JP, Mauch-Mani B. (2001) β-Aminobutyric acid-induced resistance in plants. Eur J Plant Pathol 107: 29–37 [Google Scholar]
- Jakab G, Ton J, Flors V, Zimmerli L, Métraux JP, Mauch-Mani B. (2005) Enhancing Arabidopsis salt and drought stress tolerance by chemical priming for its abscisic acid responses. Plant Physiol 139: 267–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaskiewicz M, Conrath U, Peterhänsel C. (2011) Chromatin modification acts as a memory for systemic acquired resistance in the plant stress response. EMBO Rep 12: 50–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C, Boden E, Arias J. (2003) Salicylic acid and NPR1 induce the recruitment of trans-activating TGA factors to a defense gene promoter in Arabidopsis. Plant Cell 15: 1846–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL. (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kathiria P, Sidler C, Golubov A, Kalischuk M, Kawchuk LM, Kovalchuk I. (2010) Tobacco mosaic virus infection results in an increase in recombination frequency and resistance to viral, bacterial, and fungal pathogens in the progeny of infected tobacco plants. Plant Physiol 153: 1859–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuc J. (2001) Concepts and direction of induced systemic resistance in plants and its application. Eur J Plant Pathol 107: 7–12 [Google Scholar]
- Kunkel BN, Brooks DM. (2002) Cross talk between signaling pathways in pathogen defense. Curr Opin Plant Biol 5: 325–331 [DOI] [PubMed] [Google Scholar]
- Luna E, Bruce TJA, Roberts MR, Flors V, Ton J. (2011) Next generation systemic acquired resistance. Plant Physiol 158: 844–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- March-Díaz R, García-Domínguez M, Lozano-Juste J, León J, Florencio FJ, Reyes JC. (2008) Histone H2A.Z and homologues of components of the SWR1 complex are required to control immunity in Arabidopsis. Plant J 53: 475–487 [DOI] [PubMed] [Google Scholar]
- Mosher RA, Durrant WE, Wang D, Song J, Dong X. (2006) A comprehensive structure-function analysis of Arabidopsis SNI1 defines essential regions and transcriptional repressor activity. Plant Cell 18: 1750–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavet V, Quintero C, Cecchini NM, Rosa AL, Alvarez ME. (2006) Arabidopsis displays centromeric DNA hypomethylation and cytological alterations of heterochromatin upon attack by pseudomonas syringae. Mol Plant Microbe Interact 19: 577–587 [DOI] [PubMed] [Google Scholar]
- Pecinka A, Rosa M, Schikora A, Berlinger M, Hirt H, Luschnig C, Mittelsten Scheid O. (2009) Transgenerational stress memory is not a general response in Arabidopsis. PLoS ONE 4: e5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prime-A-Plant Group, Conrath U, Beckers GJ, Flors V, García-Agustín P, Jakab G, Mauch F, Newman MA, Pieterse CM, Poinssot B, et al. (2006) Priming: getting ready for battle. Mol Plant Microbe Interact 19: 1062–1071 [DOI] [PubMed] [Google Scholar]
- Rasmann S, De Vos M, Casteel CL, Tian D, Halitschke R, Sun JY, Agrawal AA, Felton GW, Jander G. (2011) Herbivory in the previous generation primes Arabidopsis and tomato for enhanced insect resistance. Plant Physiol 158: 854–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals JA, Neuenschwander UH, Willits MG, Molina A, Steiner HY, Hunt MD. (1996) Systemic acquired resistance. Plant Cell 8: 1809–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano H. (2010) Inheritance of acquired traits in plants: reinstatement of Lamarck. Plant Signal Behav 5: 346–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz RJ, Schultz MD, Lewsey MG, O’Malley RC, Urich MA, Libiger O, Schork NJ, Ecker JR. (2011) Transgenerational epigenetic instability is a source of novel methylation variants. Science 334: 369–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sticher L, Mauch-Mani B, Métraux JP. (1997) Systemic acquired resistance. Annu Rev Phytopathol 35: 235–270 [DOI] [PubMed] [Google Scholar]
- Thomma BP, Penninckx IA, Broekaert WF, Cammue BP. (2001) The complexity of disease signaling in Arabidopsis. Curr Opin Immunol 13: 63–68 [DOI] [PubMed] [Google Scholar]
- Ton J, Jakab G, Toquin V, Flors V, Iavicoli A, Maeder MN, Métraux JP, Mauch-Mani B. (2005) Dissecting the beta-aminobutyric acid-induced priming phenomenon in Arabidopsis. Plant Cell 17: 987–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JG, Ellis C, Devoto A. (2002) The jasmonate signal pathway. Plant Cell (Suppl) 14: S153–S164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hulten M, Pelser M, van Loon LC, Pieterse CMJ, Ton J. (2006) Costs and benefits of priming for defense in Arabidopsis. Proc Natl Acad Sci USA 103: 5602–5607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon LC. (1997) Induced resistance in plants and the role of pathogenesis-related proteins. Eur J Plant Pathol 103: 753–765 [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, van Roy N, De Paepe A, Speleman F. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: RESEARCH0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H. (2006) Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev 20: 759–771 [DOI] [PubMed] [Google Scholar]
- Verhoeven KJF, Jansen JJ, van Dijk PJ, Biere A. (2010) Stress-induced DNA methylation changes and their heritability in asexual dandelions. New Phytol 185: 1108–1118 [DOI] [PubMed] [Google Scholar]
- Walley JW, Rowe HC, Xiao Y, Chehab EW, Kliebenstein DJ. (2008) The chromatin remodeler SPLAYED regulates specific stress signaling pathways. PLoS Pathog 4: e1000237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerli L, Hou BH, Tsai CH, Jakab G, Mauch-Mani B, Somerville S. (2008) The xenobiotic β-aminobutyric acid enhances Arabidopsis thermotolerance. Plant J 53: 144–156 [DOI] [PubMed] [Google Scholar]
- Zimmerli L, Jakab G, Metraux JP, Mauch-Mani B. (2000) Potentiation of pathogen-specific defense mechanisms in Arabidopsis by beta-aminobutyric acid. Proc Natl Acad Sci USA 97: 12920–12925 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.