Abstract
Systemic acquired resistance (SAR) is a plant immune response to pathogen attack. Recent evidence suggests that plant immunity involves regulation by chromatin remodeling and DNA methylation. We investigated whether SAR can be inherited epigenetically following disease pressure by Pseudomonas syringae pv tomato DC3000 (PstDC3000). Compared to progeny from control-treated Arabidopsis (Arabidopsis thaliana; C1), progeny from PstDC3000-inoculated Arabidopsis (P1) were primed to activate salicylic acid (SA)-inducible defense genes and were more resistant to the (hemi)biotrophic pathogens Hyaloperonospora arabidopsidis and PstDC3000. This transgenerational SAR was sustained over one stress-free generation, indicating an epigenetic basis of the phenomenon. Furthermore, P1 progeny displayed reduced responsiveness of jasmonic acid (JA)-inducible genes and enhanced susceptibility to the necrotrophic fungus Alternaria brassicicola. This shift in SA- and JA-dependent gene responsiveness was not associated with changes in corresponding hormone levels. Instead, chromatin immunoprecipitation analyses revealed that SA-inducible promoters of PATHOGENESIS-RELATED GENE1, WRKY6, and WRKY53 in P1 plants are enriched with acetylated histone H3 at lysine 9, a chromatin mark associated with a permissive state of transcription. Conversely, the JA-inducible promoter of PLANT DEFENSIN1.2 showed increased H3 triple methylation at lysine 27, a mark related to repressed gene transcription. P1 progeny from the defense regulatory mutant non expressor of PR1 (npr1)-1 failed to develop transgenerational defense phenotypes, demonstrating a critical role for NPR1 in expression of transgenerational SAR. Furthermore, the drm1drm2cmt3 mutant that is affected in non-CpG DNA methylation mimicked the transgenerational SAR phenotype. Since PstDC3000 induces DNA hypomethylation in Arabidopsis, our results suggest that transgenerational SAR is transmitted by hypomethylated genes that direct priming of SA-dependent defenses in the following generations.
To survive in hostile environments, plants have evolved the ability to prime their immune system against microbial pathogens. This priming results in a faster and stronger induction of defense mechanisms after pathogen attack (Conrath et al., 2006; Conrath, 2011). Although inducible defenses are often too weak to protect the host plant against disease by virulent pathogens, an augmented induction of these defenses can be highly effective, particularly when their expression precedes the delivery of susceptibility-inducing effectors by the invading pathogen (Ahmad et al., 2010).
A variety of environmental signals can trigger priming of plant defense, many of which indicate upcoming stress (Conrath et al., 2006). For example, localized pathogen attack causes systemic acquired resistance (SAR), which is associated with priming of defense (Kohler et al., 2002; Jung et al., 2009). The first systematic study of this phenomenon in tobacco (Nicotiana tabacum) revealed that SAR persists for at least 20 d (Ross, 1961). Studies over subsequent decades have mostly focused on the signaling pathways mediating SAR induction, which require endogenous accumulation of the plant hormone salicylic acid (SA) and the downstream signaling protein NON EXPRESSOR OF PR1 (NPR1; Durrant and Dong, 2004). NPR1 has also been implicated in the cross talk between SA- and jasmonic acid (JA)-dependent defense pathways, which enables plants to mount an appropriate defense reaction, depending on the nature of the attacker and the stage of infection (Spoel et al., 2003; Koornneef and Pieterse, 2008).
Recent studies have revealed that systemic accumulation of SA during the onset of SAR is preceded by a variety of metabolic signals, such jasmonates (Truman et al., 2007) and indole-derived compounds (Truman et al., 2010). The exact nature of the systemic SAR signal in Arabidopsis (Arabidopsis thaliana) after localized infection by avirulent Pseudomonas syringae remains complex and has been a matter of debate (Attaran et al., 2009). Apart from methyl salicylate (MeSA; Vlot et al., 2008a, 2008b), glycerolipids (Chaturvedi et al., 2008), azeleic acid (Jung et al., 2009), and glycerol-3-P (Chanda et al., 2011) have been implicated. As a plausible explanation, Liu et al. (2011b) recently proposed that SAR is controlled by an interaction between at least two mobile signals, MeSA and a complex formed between the lipid transfer protein DIR1 and glycerolipid or lipid derivatives. Liu et al. (2011a) also reported that the dependency of SAR on MeSA is determined by the light regime. When SAR was induced late in the day and plants received little light in subsequent hours, MeSA and its metabolizing enzymes were found to be essential for SAR. By contrast, when induction was performed in the morning and was followed by an extended light period, SAR developed in the absence of MeSA. Together, these studies illustrate that the onset of SAR in Arabidopsis is mediated by a multitude of transiently expressed signaling networks that can vary according to the environmental conditions. The mechanisms of long-lasting maintenance of SAR, on the other hand, have remained less intensely studied. Recent studies have begun to analyze how epigenetic regulatory mechanisms, such as DNA methylation and chromatin remodeling, can have long-lasting impacts on gene expression and plant immunity (Bruce et al., 2007; van den Burg and Takken, 2009; Alvarez et al., 2010).
Previously, we demonstrated that the costs of priming in Arabidopsis are outweighed by its benefits under relatively high disease pressure (van Hulten et al., 2006). This suggests that priming is a beneficial defense strategy in hostile environments. Whether priming can be inherited epigenetically from disease-exposed Arabidopsis remains unknown, even though it can be expected that transgenerational defense priming would provide benefits for short-generation plant species with limited ability to outlive disease outbreaks. The objective of this study was to examine whether disease-exposed Arabidopsis produces progeny that are primed for defense. We provide evidence for transgenerational SAR and have explored the mechanistic basis of this epigenetic immune response.
RESULTS
Plants Exposed to Fitness-Reducing Levels of Disease Produce Resistant Offspring
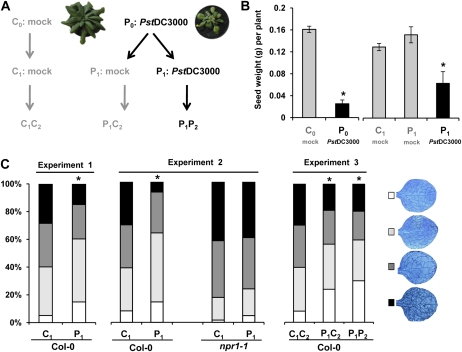
Six plants were inoculated five times over a period of 3 weeks with increasing doses of the bacterial pathogen P. syringae pv tomato DC3000 (PstDC3000), while six control plants were subjected to mock inoculations. All parental plants were allowed to set seed under similar growth conditions (Fig. 1A). Pathogen-infected parental plants (P0; P = pathogen) suffered severe fitness costs, as was evidenced by dramatically reduced growth and seed production in comparison to control-treated parental plants (C0; C = control; Fig. 1, A and 1B). Nevertheless, P1 progeny from pathogen-infected plants and C1 progeny from control-treated plants did not differ statistically in seed size, germination efficiency, or plant growth (Supplemental Fig. S1). We subsequently compared basal levels of resistance in P1 and C1 progenies against the oomycete pathogen Hyaloperonospora arabidopsidis using lactophenol trypan blue staining of infected leaves. Pathogen colonization was reduced in P1 plants compared to C1 plants (Fig. 1C), which was statistically significant for all six P1 lines from individual P0 plants (Supplemental Fig. S2A). In an independent experiment, three P1 progeny lines from the SA-insensitive npr1-1 mutant failed to develop transgenerational resistance in comparison to three corresponding C1 lines from this mutant, while three P1 wild-type lines again displayed enhanced resistance compared to the corresponding C1 wild-type lines (Fig. 1C; Supplemental Fig. S2B). Similar results were obtained after inoculation with a bioluminescent strain of PstDC3000 (PstDC3000-lux; Fan et al., 2008): Whereas P1 wild-type plants developed less bioluminescence and fewer disease symptoms than C1 wild-type plants, no such differences were observed between P1 and C1 progeny from the npr1-1 mutant (Supplemental Fig. S4). Hence, transgenerational resistance is dependent on an intact NPR1 protein and is effective against different (hemi)biotrophic pathogens.
Figure 1.
Transgenerational SAR in progeny from healthy and diseased Arabidopsis. A, Experimental design for the generation of progeny lines. Plants were inoculated five times at intervals of 3 to 4 d by dipping the leaves in a control solution (C0) or a solution containing PstDC3000 (P0), after which plants were allowed to set seed to provide and P1 progenies, respectively. Insets show representative growth phenotypes of C0 and P0 after mock and PstDC3000 inoculations. C1 and P1 plants were allowed to set seed under stress-free conditions, providing C1C2 and P1C2 progeny, respectively. A separate batch of P1 plants was exposed to similar PstDC3000 disease pressure as P0 plants to provide P1P2 progeny. B, Seed production by mock- and PstDC3000-inoculated parental plants. Data represent mean values (±se; n = 4–6) of grams of seed weight per plant. Asterisks indicate statistically significant differences compared to mock-inoculated C0 or C1 plants (Student’s t test; α = 0.05) C, Basal resistance against H. arabidopsidis WACO9 in C1 and P1 progenies of wild-type plants (Col-0; Experiment 1), C1 and P1 progenies of Col-0 and npr1-1 (Experiment 2), and C1C2, C1P2, and P1P2 progenies of Col-0 (Experiment 3). At 6 d after conidiospore inoculation, stained leaves were microscopically examined and assigned to different classes. Asterisks indicate statistically significant differences in class distributions in comparison to C1 or C1C2 plants (χ2 test; α = 0.05).
To examine the durability of the transgenerational resistance, four individual plants from different C1 or P1 progeny lines were allowed to set seed under stress-free conditions, providing C1C2 and P1C2 progeny lines, respectively. In addition, six individuals from different P1 lines were exposed to fitness-reducing levels of PstDC3000 disease to provide P1P2 progeny lines (Fig. 1, A and B). Compared to C1C2 plants, P1P2 and P1C2 plants were more resistant to H. arabidopsidis (Fig. 1C), which was statistically significant for independent P1P2 and P1C2 progeny lines (Supplemental Fig. S3). It can thus be concluded that transgenerational resistance is sustained over one stress-free generation.
Transgenerational Resistance Is Associated with Priming of SA-Dependent Genes
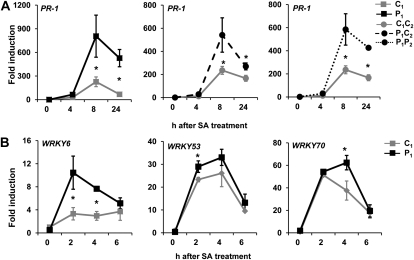
The involvement of NPR1 in transgenerational resistance resembles pathogen-induced SAR, which is based on NPR1-dependent priming of SA-inducible defense (Kohler et al., 2002; Jung et al., 2009). To examine whether transgenerational resistance is associated with similar defense priming, we quantified responsiveness of the SA-inducible PATHOGENESIS-RELATED GENE-1 (PR-1) upon treatment with SA. As is shown in Figure 2A, P1 plants displayed a faster and stronger induction of PR-1 than C1 plants, indicating that P1 progeny are primed for SA-inducible defenses. This augmented responsiveness of the PR-1 gene was also observed in plants from P1C2 and P1P2 progenies (Fig. 2A), demonstrating that the priming is maintained over one stress-free generation. We then examined whether this transgenerational priming targets regulatory genes of SA-induced defense. To this end, we profiled transcription of WRKY6, WRKY53, and WRKY70, which had previously been found to be active during priming of NPR1-dependent defense by β-amino-butyric acid (Van der Ent et al., 2009), acibenzolar S-methyl (BTH; Jaskiewicz et al., 2011), or P. syringae pv maculicola (Jaskiewicz et al., 2011). At time points preceding augmented PR-1 induction, P1 plants showed enhanced expression of WRKY6, WRKY53, and WRKY70 in comparison to C1 plants (Fig. 2B). It can thus be concluded that transgenerational priming targets multiple regulatory steps in NPR1-dependent resistance.
Figure 2.
Transgenerational priming of SA-induced defense gene expression. A, Quantitative reverse transcription PCR (qRT-PCR) analysis of SA-induced PR-1 transcription in C1 and P1 progenies (left), C1C2 and C1P2 progenies (center), and C1C2 and P1P2 progenies (right) at 4, 8, and 24 h after treatment with 0.5 mm SA. B, qRT-PCR analysis of SA-induced transcription of WRKY6, WRKY53, and WRKY70 in C1 and P1 progenies at 2, 4, and 8 h after treatment with 0.5 mm SA. Gene expression analyses were performed in 2-week-old plants. Data represent average fold induction values (±se; n = 3), relative to transcription levels before hormone treatment in C1 plants (2ΔCtPR1 = 0.00042; 2ΔCtWRKY6 = 0.0076; 2ΔCtWRKY53 = 0.00092; 2ΔCtWRKY70 = 0.0078) or in C1C2 plants (2ΔCPR1 = 0.00030). Asterisks indicate statistically significant differences in gene induction values (Student’s t test; α = 0.05).
Transgenerational Cross-Effects on JA-Dependent Resistance
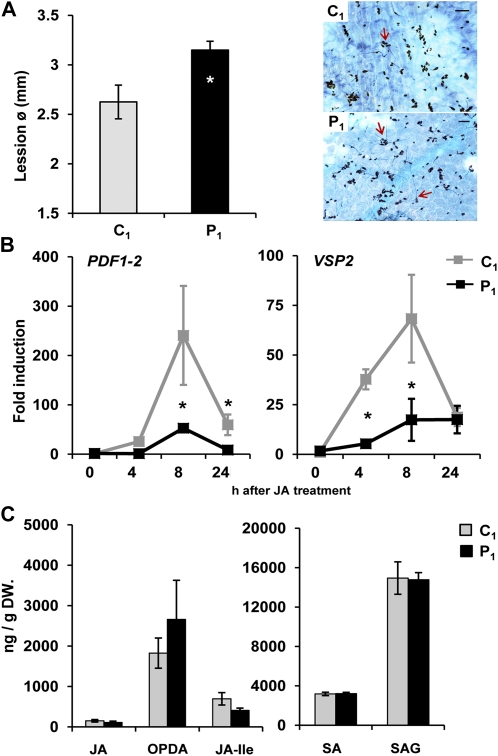
Infection with PstDC3000 activates SA-dependent defense but suppresses JA-dependent resistance against the necrotrophic fungus Alternaria brassicicola (Spoel et al., 2003, 2007). To examine whether transgenerational resistance is associated with a similar suppression of JA-dependent defense, we assessed basal resistance against the necrotrophic fungus A. brassicicola in C1 and P1 wild-type plants. At 5 d after inoculation, P1 plants developed larger chlorotic lesions and allowed increased hyphal colonization by the fungus (Fig. 3A). We subsequently investigated whether responsiveness of the JA-inducible genes PLANT DEFENSIN1.2 (PDF1.2) and VEGETATIVE STORAGE PROTEIN2 (VSP2) is affected in P1 wild-type plants. At 4 and 8 h after exogenous JA application, both marker genes showed reduced expression in P1 wild-type plants (Fig. 3B), which was absent in P1 progeny from the npr1-1 mutant (Supplemental Fig. S5). Despite these cross talk effects on JA-dependent resistance, ultraperformance liquid chromatography coupled to mass spectrometry (UPLC-MS/MS) analysis of P1 and C1 plants revealed no significant differences in endogenous levels of JA, JA-Ile, the JA precursor 12-oxo-phytodienoic acid (OPDA), SA, or SA-glucoside (SAG) between P1 and C1 plants (Fig. 3C). Hence, the shifted balance between SA- and JA-dependent defenses in P1 plants is not caused by changes in hormone levels, but rather by adjustments in the downstream response pathways.
Figure 3.
Transgenerational cross-effects on JA-dependent resistance. A, Resistance against the necrotrophic fungus A. brassicicola in 5-week-old C1 and P1 plants. Left: Average lesion diameters (±se; n = 15) at 5 d after spore inoculation. Asterisk indicates a statistically significant difference in lesion diameter between lines (Student’s t test; α = 0.05). Right: Representative photographs of fungal colonization, visualized by trypan blue staining at 4 d after inoculation. Bars = 100 μm. Red arrows indicate germinated spores. B, qRT-PCR analysis of JA-induced transcription of PDF1.2 and VSP2 in C1 and P1 progenies at 4, 8, and 24 h after treatment of the leaves with 0.1 mm JA. Gene expression analyses were based on 2-week-old plants from C1 and P1 progenies. Data represent average fold induction values (±se; n = 3) relative to transcription levels before hormone treatment in C1 plants (2ΔCtPDF1-2 = 0.0011; 2ΔCtVSP2 = 0.0060). Asterisks indicate statistically significant differences in gene induction (Student’s t test; α = 0.05). C, UPLC-MS/MS quantification of JA, JA-Ile, OPDA, SA, and SAG in mature leaves from 5-week-old plants. Shown are average values in nanograms per gram of dry weight (DW; ±se; n = 3). No statistically significant differences were detected between C1 and P1 plants (Student’s t test; α = 0.05).
Transgenerational Chromatin Modifications at Defense Gene Promoters
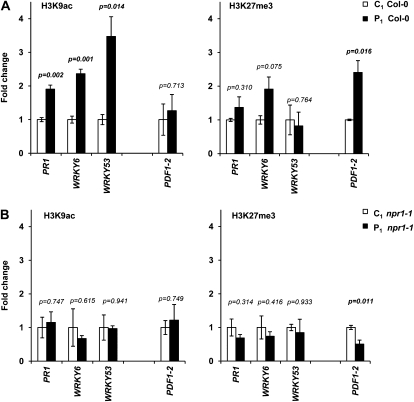
Posttranslational modifications at the N-terminal tail of histone H3 can influence defense-related gene expression (van den Burg and Takken, 2009; Alvarez et al., 2010). Because these chromatin modifications can have long-lasting impacts on plant gene expression (Vaillant and Paszkowski, 2007), we investigated whether the altered responsiveness of PR-1 and PDF1.2 in P1 plants is associated with changes in chromatin structure at the promoter regions of these genes. For this purpose, we performed chromatin immunoprecipitation (ChIP) analysis of PR-1 and PDF1-2 promoter DNA using antibodies against acetylated histone H3 at Lys-9 (H3K9ac) and triple-methylated H3 at Lys-27 (H3K27me3). Compared with C1 plants, PR-1 promoter DNA of P1 plants was associated with enhanced levels of H3K9ac (Fig. 4A), which was apparent using different primer pairs against separate regions of the promoter (Supplemental Fig. S6). By contrast, P1 progeny from the npr1-1 mutant failed to show increased levels of H3K9ac at the PR1 promoter (Figs. 4B; Supplemental Fig. S6). Acetylation of H3K9 marks an increased transcriptional capacity (Eberharter and Becker, 2002) and could therefore contribute to priming of the PR-1 gene in P1 plants. Furthermore, the PDF1-2 promoter from P1 wild-type plants was not altered in H3K9ac levels but displayed statistically significant enrichment with H3K27me3 (Fig. 4A), which was apparent with different primer pairs against this promoter (Supplemental Fig. S6). Conversely, P1 progeny from the npr1-1 mutant failed to show H3K27me3 enrichment at the PDF1.2 promoter (Fig. 4B; Supplemental Fig. S6). Since H3K27me3 is associated with transcriptional silencing (Zhang et al., 2007), this histone modification could contribute to the suppressed responsiveness of PDF1.2 in P1 plants (Fig. 3B). To further investigate the role of H3K9ac in priming of SA-dependent defense, we analyzed the promoter regions of WRKY6 and WRKY53. Both regulatory genes can be primed by pretreatment with BTH or P. syringae pv maculicola (Jaskiewicz et al., 2011) and showed augmented responsiveness to SA in P1 plants (Fig. 2B). As observed for PR-1, the promoters of WRKY6 and WRKY53 in P1 wild-type plants were enriched with H3K9ac, whereas this response was absent in P1 npr1-1 plants. Hence, transgenerational acetylation of H3K9 requires an intact NPR1 protein and targets multiple SA-inducible gene promoters.
Figure 4.
Transgenerational modifications of histone H3 at defense gene promoters in Col-0 (A) but not in npr1-1 (B). After ChIP, promoter DNA of SA-inducible PR-1, WRKY6, WRKY53, and JA-inducible PDF1.2 was quantified by qPCR relative to DNA amounts in chromatin extracts before immunoprecipitation (input) using antibodies against H3K9ac or H3K27me3. Data represent average fold change values (±se; n = 3) in P1 plants compared to C1 plants. P values indicate statistical differences between C1 and P1 progenies (Student’s t test).
Role of DNA Methylation in Transgenerational Resistance
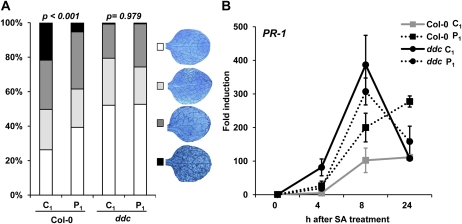
Although histone modifications can have long-lasting impacts on gene expression (Vaillant and Paszkowski, 2007), there is no convincing evidence that they can be transmitted through meiosis. By contrast, there is ample evidence for transmission of DNA methylation to following generations. It is also known that PstDC3000 triggers DNA hypomethylation in Arabidopsis (Pavet et al., 2006). To examine the role of DNA methylation in PstDC3000-induced transgenerational resistance, we compared transgenerational resistance phenotypes between wild-type plants and the drm1drm2cmt3 triple mutant (ddc), which is reduced in non-CpG DNA methylation (Chan et al., 2006). Unlike other DNA methylation mutants, the ddc mutant expressed normal growth phenotypes under our growth conditions until the onset of flowering, which would otherwise complicate the interpretation of our bioassays. Upon inoculation with H. arabidopsidis, three independent C1 and P1 progeny lines from the ddc mutant expressed similar levels of resistance, whereas the corresponding P1 lines of the wild-type displayed enhanced resistance in comparison to C1 wild-type lines. Interestingly, however, all ddc lines expressed significantly higher levels of resistance in comparison to C1 wild-type lines (Fig. 5A; Supplemental Fig. S7). The ddc mutant also expressed constitutively higher levels of resistance against PstDC3000-lux but was enhanced susceptible to the necrotrophic pathogen A. brassicicola (Supplemental Fig. S8). These results indicate that the hypomethylated DNA status in the ddc mutant mimics the transgenerational resistance phenotype of P1 wild-type plants.
Figure 5.
Role of non-CpG DNA methylation in transgenerational SAR. A, Level of resistance against H. arabidopsidis WACO9 in C1 and P1 progeny from Col-0 and the dmr1dmr2ctm3 (ddc) mutant. At 6 d after conidiospore inoculation, stained leaves were microscopically examined and assigned to different severity classes. P values indicate statistical differences in class distributions between P1 and C1 progeny of each genotype (χ2 test). B, qRT-PCR analysis of SA-induced PR-1 transcription at 4, 8, and 24 h after treatment of C1 and P1 progeny of Col-0 and ddc plants with 0.5 mm SA. Data represent average fold induction values (±se; n = 3) relative to transcription levels before hormone treatment in C1 Col-0 plants (2ΔCtPR1 = 0.0020).
To examine whether the constitutive resistance against hemibiotrophic pathogens in ddc is based on a priming of the SA response, we compared levels of PR-1 gene induction between C1 and P1 plants of wild-type and ddc plants. At 4 and 8 h after SA application, ddc plants showed significantly enhanced PR-1 transcription compared to wild-type plants (P = 0.031), which was similar in C1 and P1 progenies of the mutant (Fig. 5B). Hence, DNA hypomethylation in the ddc mutant mimics transgenerational priming of SA-dependent defense. Since infection by PstDC3000 induces DNA hypomethylation in Arabidopsis (Pavet et al., 2006), our results indicate that transgenerational resistance from PstDC3000-infected plants is transmitted by hypomethylated DNA.
DISCUSSION
Our study demonstrates that disease resistance can be carried forward to the next generation from plants exposed to fitness-reducing disease pressure. It indicates an epigenetic mechanism of disease protection, which could function as a plant memory of disease stress encountered in previous generations. A recent study by Kathiria et al. (2010) demonstrated increased homologous recombination in progeny from tobacco mosaic virus-infected tobacco, which was associated with enhanced disease resistance. Increased homologous recombination has also been reported in progeny from Arabidopsis exposed to short-wavelength radiation (ultraviolet-C) or flagellin, which persisted in subsequent, untreated generations (Molinier et al., 2006). Hence, transgenerational responses to stress are emerging as a widespread defense phenomenon in plants. Our study reveals an epigenetic mechanism for transgenerational resistance against biotic stress. Like SAR, transgenerational resistance protects against (hemi)biotrophic pathogens, requires an intact NPR1 protein, and is associated with priming of SA-dependent defense (Kohler et al., 2002; Ton et al., 2002; Durrant and Dong, 2004; Jung et al., 2009; Figs. 1 and 2). Therefore, we propose to define this phenomenon as transgenerational SAR or “next-generation SAR.”
Independently from our findings, two independent studies by Slaughter et al. (2012) and Rasmann et al. (2012) demonstrate similar transgenerational resistance phenomena in response to priming-inducing stimuli. Slaughter et al. (2012) discovered that progeny of Arabidopsis that had been treated with β-amino-butyric acid or an avirulent isolate of PstDC3000 (PstavrRpt2) are primed for SA-dependent resistance against H. arabidopsidis and PstDC3000. Furthermore, Rasmann et al. (2012) show that Arabidopsis and tomato (Solanum lycopersicum) subjected to herbivory or mechanical damage produce progeny that are primed to express JA-dependent resistance against herbivores. Together, our three studies demonstrate that transgeneration priming of defense is a robust and broadly distributed mechanism of phenotypic plasticity in plants to environmental stress. Moreover, the triplicate demonstration that the priming of defense can be transmitted to following generations indicates an epigenetic basis of the phenomenon.
Interestingly, Slaughter et al. (2012) reported that induction of transgenerational resistance induced one single inoculation with an avirulent PstDC3000 strain disappears after one stress-free generation. By contrast, our study revealed that transgenerational resistance upon repeated inoculations with plant fitness-reducing doses of the virulent PstDC3000 can be sustained over one disease-free generation (Figs. 1C and 2A). This difference indicates that the intensity of disease-related stress is proportional to the durability of transgenerational resistance in progeny. Hence, plants are capable of adjusting the stability of transgenerational defense priming according to the severity of disease pressure in their environment. Future experiments are necessary to establish the exact relationship between disease severity and epigenetic stability of transgenerational priming.
Spoel et al. (2007) reported that virulent PstDC3000 renders plants more susceptible to the necrotrophic pathogen A. brassicicola through suppression of the JA pathway. Our results suggest that this NPR1-dependent signaling cross talk can be transmitted to the following generation (Fig. 3). Interestingly, P1 plants did not accumulate different levels of endogenous SA or JA (Fig. 3C), indicating that the altered balance between SA- and JA-dependent defense is based on changes in the sensitivity of the downstream response pathways. WKRY70 has been implicated as a downstream regulator of negative cross talk between SA- and JA-dependent signaling (Li et al., 2006). Indeed, P1 plants expressed enhanced levels of WRKY70 (Fig. 2B), supporting a regulatory role of this transcription factor in transgenerational cross talk. We furthermore found that the observed change in responsiveness of SA-inducible PR-1 and JA-inducible PDF1.2 is marked by NPR1-dependent modifications of histone H3 at their gene promoters (Fig. 4). Together, our results suggest that transgenerational SAR is based on a shifted balance between SA- and JA-dependent defense, which is maintained by NPR1-dependent modifications of chromatin structure at promoters of JA- and SA-responsive defense genes.
Evidence is emerging that plant defense is regulated by changes in chromatin structure (van den Burg and Takken, 2009). Arabidopsis mutants in chromatin remodeling enzymes are affected in JA- and SA-dependent resistance (Devoto et al., 2002; March-Díaz et al., 2008; Wu et al., 2008). Furthermore, the SAR-inducing chemical BTH triggers posttranslational modifications of histone H3 in the PR-1 promoter (Mosher et al., 2006), and Jaskiewicz et al. (2011) recently demonstrated that P. syringae pv maculicola primes stress-inducible expression of WRKY genes that is associated with NPR1-dependent H3 modifications at their promoters. Although the latter two studies describe relatively short-term responses within days after treatment, they are consistent with our finding that P1 progeny are primed for NPR1-dependent defense. However, there is no evidence that histone modifications can be transmitted through meiosis. Conversely, various plant traits have been demonstrated to be transmitted by DNA methylation, which can remain stable over multiple generations (Kalisz and Purugganan, 2004). In particular DNA hypomethylation has been associated with plant defense. For instance, Arabidopsis responds to infection by PstDC3000 by hypomethylation of genomic DNA (Pavet et al., 2006). Furthermore, inbreeding of the Arabidopsis decrease in DNA methylation1 mutation, causing genome-wide DNA hypomethylation, gives rise to a heritable but metastable defense allele, called bal1, which confers NPR1-dependent resistance (Stokes et al., 2002; Yi and Richards, 2009). Hence, DNA hypomethylation can cause epigenetic inheritance of disease resistance. Our finding that non-CpG DNA hypomethylation by the ddc mutations mimics the phenotype of transgenerational SAR supports this notion (Fig. 5; Supplemental Figs. S7 and S8). Furthermore, evidence from both animal and plant systems suggests that (de)methylated DNA can direct posttranslational modifications of histone H3 (Vaillant and Paszkowski, 2007; Cedar and Bergman, 2009). Therefore, we propose that transgenerational SAR is inherited through hypomethylated regulatory genes, which direct NPR1-dependent histone H3 modifications in following generations to induce and maintain priming of SA-dependent defense genes.
Small interfering RNAs (siRNAs) can control gene transcription through changes in DNA methylation (Brodersen and Voinnet, 2006; Vaucheret, 2006). Interestingly, the complementary study by Rasmann et al. (2012) demonstrated that two Arabidopsis mutants impaired in the biogenesis of siRNAs (dcl2 dcl3 dcl4 and nrpd2a nrpd2b) failed to produce progeny with transgenerational resistance against herbivores. Since the ddc mutant is affected in RNA-directed DNA methylation (Kurihara et al., 2008), our results support a similar role by siRNAs during the onset of transgenerational SAR. PstDC3000 has recently been reported to trigger the accumulation of 27 RNA interference families in Arabidopsis (Zhang et al., 2011). Furthermore, micro-RNAs and siRNAs have both been implicated in the response of Arabidopsis to P. syringae pathogens (Li et al., 2011). However, the siRNA families eliciting transgenerational SAR and herbivore resistance must be different to prime SA- and JA-inducible defenses, respectively. Our additional observation that transgenerational SAR is associated with a suppression of JA-dependent resistance (Fig. 3) suggests that these siRNAs could act antagonistically on each other.
Priming of defense is a beneficial defense strategy in hostile environments with relatively minor costs (van Hulten et al., 2006; Ahmad et al., 2010). Epigenetic regulation of priming would allow plants to protect their progeny against recurring biotic stress without permanent genetic fixation of the trait and its associated costs. A better understanding of this phenomenon will create opportunities to improve disease resistance in agricultural crops. Food security is an important challenge in the 21st century (Baulcombe et al., 2009), and there is a pressing need to develop sustainable ways of pest and disease control to safeguard yields while minimizing pesticide use. An efficient induction of epigenetically imprinted defense priming would allow us to generate seed stocks of crops with broad-spectrum resistance that would require fewer chemicals to control disease.
CONCLUSION
Disease-exposed Arabidopsis produces progeny with enhanced disease resistance, which can be maintained over one stress-free generation. This transgenerational SAR is effective against (hemi)biotrophic pathogens, requires an intact NPR1 protein, and is associated with priming of SA-dependent genes. Furthermore, transgenerational SAR is associated with an NPR1-dependent repression of JA-dependent defense genes and enhanced susceptibility to the necrotrophic fungus A. brassicicola. This shift in the cross talk balance between SA- and JA-dependent defenses is associated with permissive and repressive histone H3 modifications at SA- and JA-inducible gene promoters, respectively. The hypomethylated ddc mutant of Arabidopsis mimics the transgenerational SAR phenotype, suggesting that SAR is inherited through pathogen-induced hypomethylation at non-CpG DNA sites.
MATERIALS AND METHODS
Plant Material
Arabidopsis (Arabidopsis thaliana) Columbia-0 (Col-0), npr1-1 (Cao et al., 1994), and drm1-2 drm2-2 cmt3-11 (Chan et al., 2006) were cultivated under standard conditions (8.5:15.5 h light:dark; 20°C photophase, 18°C scotophase; 65% relative humidity) in a sand:compost mixture (1:3). Seeds (approximately 50 per pot) were stratified at 4°C in the dark for 2 d. For experiments lasting longer than 3 weeks, plants were individually transferred at the seedling stage (approximately 10 d old) to 60-mL pots.
Imprinting of Transgenerational Defense Priming
Inoculation with Pseudomonas syringae pv tomato DC3000 (PstDC3000) was performed five times by dipping rosettes in a bacterial suspension containing 10 mm MgSO4 and 0.01% (v/v) Silwet L-77 at intervals of 3 to 5 d between treatments. To ensure constant disease pressure, the first three inoculations were performed at 108 cells/mL and the last two at 109 cells/mL. Plants were maintained at 100% relative humidity from the 1st inoculation until the 1 week after the 5th inoculation. Flowering was induced by transferring plants to long-light conditions (16:8 h light:dark) between the 3rd and 4th inoculation. Mock-inoculated plants (C0 and C1) were treated with 10 mm MgSO4 (0.01% Silwet L-77) without bacteria and maintained under similar conditions. C1 and P1 progeny lines were collected from individual C0 and P0 parental plants. To assess durability of transgenerational SAR, four individuals from different C1 and P1 lines were allowed to set seed under stress-free conditions, providing C1C2 and P1C2 progeny lines, respectively. A separate batch of four individuals from different P1 lines was exposed to fitness-reducing levels of PstDC3000 disease to provide P1P2 progeny lines.
Fitness Assays
Growth rates and seed production were determined as described previously (van Hulten et al., 2006). Seed size was estimated on the basis of seed area (Herridge et al., 2011). Germination efficiency was determined after 3 d of cultivation under standard growth conditions following 2 d of stratification.
Basal Resistance Assays
Hyaloperonospora arabidopsidis WACO9 bioassays were performed as described before (Van der Ent et al., 2009). Infected leaves (>75) from at least 15 randomly selected plants were collected at 6 d after spray inoculation with 105 conidiospores/mL, stained with lactophenol–trypan blue (Koch and Slusarenko, 1990), and scored using light microscopy. Colonization levels were assigned to four classes: I, no pathogen growth; II, hyphal colonization without conidiophores; III, hyphal colonization with conidiophores and sporadic oospores; and IV, extensive colonization, conidiophores, and frequent oospores. Differences in class distributions between progenies were analyzed for statistical differences by χ2 contingency tests or χ2 goodness of fit tests using Genstat software (13th edition).
PstDC3000 assays were performed with a bioluminescent luxCDABE-tagged strain (PstDC3000-lux; Fan et al., 2008). Five-week-old plants were inoculated by dipping the leaves in a bacterial suspension containing 108 colony-forming units/mL in 10 mm MgSO4 and 0.01% (v/v) Silwet L-77. Symptoms were scored at 3 d after inoculation as described before (Van der Ent et al., 2009). Bacterial colonization was estimated from the intensity of bioluminescence, using a liquid nitrogen cooled CCD detector (Princeton Instruments) at maximum sensitivity. Digital photographs were taken at 3 d after inoculation under bright light (exposure time of 0.5 s) and in darkness (exposure time of 300 s) using WinView/32 software at fixed black-and-white contrast settings. Bacterial luminescence showed no major variation within colonized leaf areas and was proportional to the number of pixels on digital photographs. Bacterial titers in each plant were expressed as ratios between numbers bioluminescent pixels and total leaf pixels from bright light pictures, as described previously for digital callose quantification (Luna et al., 2011).
Alternaria brassicicola inoculation was performed when plants were 5 weeks old by applying 5-μL drops of 1 × 106 spores/mL onto four leaves of approximately similar age, as described (Ton and Mauch-Mani, 2004). Disease symptoms and pathogen colonization were evaluated at 5 d after inoculation and based on mean lesion diameters from 15 to 20 plants and microscopy examination of lactophenol-trypan blue stained leaves, respectively.
Gene Expression Assays
Hormone-induced gene expression assays were based on 2-week-old plants after spraying with water, 0.1 mm JA, or 0.5 mm SA, supplemented with 0.01% Silwet (L-77). Samples were collected at the indicated time points, consisting of pooled shoots from four to six plants per replicate. RNA extraction, cDNA synthesis, and quantitative PCR reactions were essentially performed as described before (Van der Ent et al., 2009). Briefly, two technical replicates of each sample were subjected to the quantitative PCR reaction. PCR efficiency (E) of primer pairs were estimated from multiple amplification plots using the Equation (1+E) = 10slope (Ramakers et al., 2003) and were confirmed to provide (1+E) values close to 2 (ranging between 1.9 and 2.0). Transcript levels were calculated relative to the reference gene At1g13440 (Czechowski et al., 2005) using the 2ΔCt (cycle threshold) method, where ΔCt = Ct (reference gene) − Ct (gene of interest). Primer sequences were designed against the 3′-end of the gene and have been published before (Czechowski et al., 2004; Van der Ent et al., 2009). Fold induction values of gene expression were normalized to average 2ΔCt values relative to wild-type C1 or C1C2 plants at 0 h before hormone treatments, which are indicated in the figure legends.
UPLC-Quadruple MS/MS Analysis
Mature leaf samples were collected from five plants per replicate. Extraction and quantification of SA, SAG (2-O-β-d-glucoside), JA, JA-Ile, and OPDA were analyzed by UPLC-MS/MS as described (Flors et al., 2008), with modification from (Forcat et al., 2008). Samples were analyzed by a triple quadrupole tandem mass spectrometer (TQD; Waters). Liquid chromatography separation was performed using an Acquity UPLC BEH C18 analytical column (Waters) at a flow rate of 300 mL/min. Quantifications were carried out with MassLynx 4.1 software (Waters) using internal standards as a reference for extraction recovery and the standard curves as quantifiers.
ChIP
Assays were performed according to the manufacturer’s protocol (EpiQuik Plant ChIP kit; Epigentek) using mature leaves from 5-week-old plants. Each biologically replicated sample from independent experiments (n = 3) consisted of pooled leaves from four to five plants. Chromatin samples were immunoprecipitated using antibodies against acetyl-histone H3K9 (Millipore 07-352) and trimethyl-H3K27 (Millipore 07-449). Abundance of DNA in chromatin extracts was analyzed by quantitative PCR from two independent precipitations using an ABI PRISM 7900 HT sequence detection system (Applied Biosystems). Reactions were performed in a final volume of 25 μL containing Jump Start SYBR Green (Sigma-S4438). Gene-specific primers were designed to cover gene promoter regions containing known cis-elements (Supplemental Table S1 and Supplemental Fig. S6; Koornneef et al., 2008; Jaskiewicz et al., 2011). Results were normalized to initial DNA amounts in the input control, as described (Haring et al., 2007), and standardized to levels in corresponding C1 progeny.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Fitness of C1 and P1 plants.
Supplemental Figure S2. Level of resistance against H. arabidopsidis WACO9 in independent C1 and P1 progeny lines from individual wild-type and npr1-1.
Supplemental Figure S3. Level of resistance against H. arabidopsidis WACO9 in independent C1C2, P1C1, and P1P2 progeny lines from individual wild-type C1 or P1 plants.
Supplemental Figure S4. Basal resistance against bioluminescent PstDC3000-lux in C1 and P1 wild-type and npr1-1 plants.
Supplemental Figure S5. Lack of transgenerational repression of the JA response in the npr1-1 mutant.
Supplemental Figure S6. Transgenerational modifications of histone H3 at different regions of the PR-1 and PDF1-2 gene promoters.
Supplemental Figure S7. No difference in resistance between C1 and P1 progenies of the dmr1dmr2ctm3 (ddc) mutant.
Supplemental Figure S8. Basal resistance of Col-0 and ddc plants against bioluminescent PstDC3000-lux Alternaria brassicicola.
Supplemental Table S1. Primers used for quantification of immunoprecipitated promoter DNA.
Supplementary Material
Acknowledgments
We are grateful for the technical support from Servei Central d'Instrumentació Cientıfica of the University of Jaume I. We also thank John Lucas and Maurice Moloney (Rothamsted Research) for support and useful advice throughout the project.
References
- Ahmad S, Gordon-Weeks R, Pickett J, Ton J. (2010) Natural variation in priming of basal resistance: from evolutionary origin to agricultural exploitation. Mol Plant Pathol 11: 817–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez ME, Nota F, Cambiagno DA. (2010) Epigenetic control of plant immunity. Mol Plant Pathol 11: 563–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attaran E, Zeier TE, Griebel T, Zeier J. (2009) Methyl salicylate production and jasmonate signaling are not essential for systemic acquired resistance in Arabidopsis. Plant Cell 21: 954–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe D, Crute I, Davies B, Dunwell J, Gale M, Jones J, Pretty J, Sutherland W, Toumin C. (2009) Reaping the benefits: science and the sustainable intensification of global agriculture. In Royal Society, RS Policy Document 11/09. Royal Society, London, 2009 [Google Scholar]
- Brodersen P, Voinnet O. (2006) The diversity of RNA silencing pathways in plants. Trends Genet 22: 268–280 [DOI] [PubMed] [Google Scholar]
- Bruce TJA, Matthes MC, Napier JA, Pickett JA. (2007) Stressful “memories” of plants: evidence and possible mechanisms. Plant Sci 173: 603–608 [Google Scholar]
- Cao H, Bowling SA, Gordon AS, Dong X. (1994) Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6: 1583–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedar H, Bergman Y. (2009) Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet 10: 295–304 [DOI] [PubMed] [Google Scholar]
- Chan SW, Henderson IR, Zhang X, Shah G, Chien JS, Jacobsen SE. (2006) RNAi, DRD1, and histone methylation actively target developmentally important non-CG DNA methylation in Arabidopsis. PLoS Genet 2: e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanda B, Xia Y, Mandal MK, Yu K, Sekine KT, Gao QM, Selote D, Hu Y, Stromberg A, Navarre D, et al. (2011) Glycerol-3-phosphate is a critical mobile inducer of systemic immunity in plants. Nat Genet 43: 421–427 [DOI] [PubMed] [Google Scholar]
- Chaturvedi R, Krothapalli K, Makandar R, Nandi A, Sparks AA, Roth MR, Welti R, Shah J. (2008) Plastid ω3-fatty acid desaturase-dependent accumulation of a systemic acquired resistance inducing activity in petiole exudates of Arabidopsis thaliana is independent of jasmonic acid. Plant J 54: 106–117 [DOI] [PubMed] [Google Scholar]
- Conrath U. (2011) Molecular aspects of defence priming. Trends Plant Sci 16: 524–531 [DOI] [PubMed] [Google Scholar]
- Conrath U, Beckers GJ, Flors V, García-Agustín P, Jakab G, Mauch F, Newman MA, Pieterse CM, Poinssot B, Pozo MJ, et al. ; Prime-A-Plant Group (2006) Priming: getting ready for battle. Mol Plant Microbe Interact 19: 1062–1071 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Bari RP, Stitt M, Scheible WR, Udvardi MK. (2004) Real-time RT-PCR profiling of over 1400 Arabidopsis transcription factors: unprecedented sensitivity reveals novel root- and shoot-specific genes. Plant J 38: 366–379 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible W-R. (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devoto A, Nieto-Rostro M, Xie DX, Ellis C, Harmston R, Patrick E, Davis J, Sherratt L, Coleman M, Turner JG. (2002) COI1 links jasmonate signalling and fertility to the SCF ubiquitin-ligase complex in Arabidopsis. Plant J 32: 457–466 [DOI] [PubMed] [Google Scholar]
- Durrant WE, Dong X. (2004) Systemic acquired resistance. Annu Rev Phytopathol 42: 185–209 [DOI] [PubMed] [Google Scholar]
- Eberharter A, Becker PB. (2002) Histone acetylation: a switch between repressive and permissive chromatin. Second in review series on chromatin dynamics. EMBO Rep 3: 224–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Crooks C, Lamb C. (2008) High-throughput quantitative luminescence assay of the growth in planta of Pseudomonas syringae chromosomally tagged with Photorhabdus luminescens luxCDABE. Plant J 53: 393–399 [DOI] [PubMed] [Google Scholar]
- Flors V, Ton J, van Doorn R, Jakab G, García-Agustín P, Mauch-Mani B. (2008) Interplay between JA, SA and ABA signalling during basal and induced resistance against Pseudomonas syringae and Alternaria brassicicola. Plant J 54: 81–92 [DOI] [PubMed] [Google Scholar]
- Forcat S, Bennett MH, Mansfield JW, Grant MR. (2008) A rapid and robust method for simultaneously measuring changes in the phytohormones ABA, JA and SA in plants following biotic and abiotic stress. Plant Methods 4: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haring M, Offermann S, Danker T, Horst I, Peterhansel C, Stam M. (2007) Chromatin immunoprecipitation: optimization, quantitative analysis and data normalization. Plant Methods 3: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herridge RP, Day RC, Baldwin S, Macknight RC. (2011) Rapid analysis of seed size in Arabidopsis for mutant and QTL discovery. Plant Methods 7: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaskiewicz M, Conrath U, Peterhänsel C. (2011) Chromatin modification acts as a memory for systemic acquired resistance in the plant stress response. EMBO Rep 12: 50–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung HW, Tschaplinski TJ, Wang L, Glazebrook J, Greenberg JT. (2009) Priming in systemic plant immunity. Science 324: 89–91 [DOI] [PubMed] [Google Scholar]
- Kalisz S, Purugganan MD. (2004) Epialleles via DNA methylation: consequences for plant evolution. Trends Ecol Evol 19: 309–314 [DOI] [PubMed] [Google Scholar]
- Kathiria P, Sidler C, Golubov A, Kalischuk M, Kawchuk LM, Kovalchuk I. (2010) Tobacco mosaic virus infection results in an increase in recombination frequency and resistance to viral, bacterial, and fungal pathogens in the progeny of infected tobacco plants. Plant Physiol 153: 1859–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch E, Slusarenko A. (1990) Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell 2: 437–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler A, Schwindling S, Conrath U. (2002) Benzothiadiazole-induced priming for potentiated responses to pathogen infection, wounding, and infiltration of water into leaves requires the NPR1/NIM1 gene in Arabidopsis. Plant Physiol 128: 1046–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef A, Pieterse CM. (2008) Cross talk in defense signaling. Plant Physiol 146: 839–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef A, Rindermann K, Gatz C, Pieterse CM. (2008) Histone modifications do not play a major role in salicylate-mediated suppression of jasmonate-induced PDF1.2 gene expression. Commun Integr Biol 1: 143–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara Y, Matsui A, Kawashima M, Kaminuma E, Ishida J, Morosawa T, Mochizuki Y, Kobayashi N, Toyoda T, Shinozaki K, Seki M. (2008) Identification of the candidate genes regulated by RNA-directed DNA methylation in Arabidopsis. Biochem Biophys Res Commun 376: 553–557 [DOI] [PubMed] [Google Scholar]
- Li J, Brader G, Kariola T, Palva ET. (2006) WRKY70 modulates the selection of signaling pathways in plant defense. Plant J 46: 477–491 [DOI] [PubMed] [Google Scholar]
- Li Y, Wang W, Zhou J-M. (2011) Role of small RNAs in the interaction between Arabidopsis and Pseudomonas syringae. Front Biol 6: 462–467 [Google Scholar]
- Liu PP, von Dahl CC, Klessig DF. (2011a) The extent to which methyl salicylate is required for signaling systemic acquired resistance is dependent on exposure to light after infection. Plant Physiol 157: 2216–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu PP, von Dahl CC, Park SW, Klessig DF. (2011b) Interconnection between methyl salicylate and lipid-based long-distance signaling during the development of systemic acquired resistance in Arabidopsis and tobacco. Plant Physiol 155: 1762–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna E, Pastor V, Robert J, Flors V, Mauch-Mani B, Ton J. (2011) Callose deposition: a multifaceted plant defense response. Mol Plant Microbe Interact 24: 183–193 [DOI] [PubMed] [Google Scholar]
- March-Díaz R, García-Domínguez M, Lozano-Juste J, León J, Florencio FJ, Reyes JC. (2008) Histone H2A.Z and homologues of components of the SWR1 complex are required to control immunity in Arabidopsis. Plant J 53: 475–487 [DOI] [PubMed] [Google Scholar]
- Molinier J, Ries G, Zipfel C, Hohn B. (2006) Transgeneration memory of stress in plants. Nature 442: 1046–1049 [DOI] [PubMed] [Google Scholar]
- Mosher RA, Durrant WE, Wang D, Song J, Dong X. (2006) A comprehensive structure-function analysis of Arabidopsis SNI1 defines essential regions and transcriptional repressor activity. Plant Cell 18: 1750–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavet V, Quintero C, Cecchini NM, Rosa AL, Alvarez ME. (2006) Arabidopsis displays centromeric DNA hypomethylation and cytological alterations of heterochromatin upon attack by Pseudomonas syringae. Mol Plant Microbe Interact 19: 577–587 [DOI] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RH, Moorman AF. (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339: 62–66 [DOI] [PubMed] [Google Scholar]
- Rasmann S, De Vos M, Casteel CL, Tian D, Halitschke R, Sun JY, Agrawal AA, Felton GW, Jander G. (2012) Herbivory in the previous generation primes plants for enhanced insect resistance. Plant Physiol 158: 854–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross AF. (1961) Systemic acquired resistance induced by localized virus infections in plants. Virology 14: 340–358 [DOI] [PubMed] [Google Scholar]
- Slaughter A, Daniel X, Flors V, Luna E, Hohn B, Mauch-Mani B. (2012) Descendants of primed Arabidopsis plants exhibit enhanced resistance to biotic stress. Plant Physiol 158: 835–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel SH, Johnson JS, Dong X. (2007) Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proc Natl Acad Sci USA 104: 18842–18847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel SH, Koornneef A, Claessens SMC, Korzelius JP, Van Pelt JA, Mueller MJ, Buchala AJ, Métraux J-P, Brown R, Kazan K, et al. (2003) NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell 15: 760–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes TL, Kunkel BN, Richards EJ. (2002) Epigenetic variation in Arabidopsis disease resistance. Genes Dev 16: 171–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton J, Mauch-Mani B. (2004) Beta-amino-butyric acid-induced resistance against necrotrophic pathogens is based on ABA-dependent priming for callose. Plant J 38: 119–130 [DOI] [PubMed] [Google Scholar]
- Ton J, Van Pelt JA, Van Loon LC, Pieterse CMJ. (2002) Differential effectiveness of salicylate-dependent and jasmonate/ethylene-dependent induced resistance in Arabidopsis. Mol Plant Microbe Interact 15: 27–34 [DOI] [PubMed] [Google Scholar]
- Truman W, Bennett MH, Kubigsteltig I, Turnbull C, Grant M. (2007) Arabidopsis systemic immunity uses conserved defense signaling pathways and is mediated by jasmonates. Proc Natl Acad Sci USA 104: 1075–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman WM, Bennett MH, Turnbull CG, Grant MR. (2010) Arabidopsis auxin mutants are compromised in systemic acquired resistance and exhibit aberrant accumulation of various indolic compounds. Plant Physiol 152: 1562–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaillant I, Paszkowski J. (2007) Role of histone and DNA methylation in gene regulation. Curr Opin Plant Biol 10: 528–533 [DOI] [PubMed] [Google Scholar]
- van den Burg HA, Takken FL. (2009) Does chromatin remodeling mark systemic acquired resistance? Trends Plant Sci 14: 286–294 [DOI] [PubMed] [Google Scholar]
- Van der Ent S, Van Hulten M, Pozo MJ, Czechowski T, Udvardi MK, Pieterse CM, Ton J. (2009) Priming of plant innate immunity by rhizobacteria and beta-aminobutyric acid: differences and similarities in regulation. New Phytol 183: 419–431 [DOI] [PubMed] [Google Scholar]
- van Hulten M, Pelser M, van Loon LC, Pieterse CM, Ton J. (2006) Costs and benefits of priming for defense in Arabidopsis. Proc Natl Acad Sci USA 103: 5602–5607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H. (2006) Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev 20: 759–771 [DOI] [PubMed] [Google Scholar]
- Vlot AC, Klessig DF, Park S-W. (2008a) Systemic acquired resistance: the elusive signal(s). Curr Opin Plant Biol 11: 436–442 [DOI] [PubMed] [Google Scholar]
- Vlot AC, Liu P-P, Cameron RK, Park S-W, Yang Y, Kumar D, Zhou F, Padukkavidana T, Gustafsson C, Pichersky E, et al. (2008b) Identification of likely orthologs of tobacco salicylic acid-binding protein 2 and their role in systemic acquired resistance in Arabidopsis thaliana. Plant J 56: 445–456 [DOI] [PubMed] [Google Scholar]
- Wu K, Zhang L, Zhou C, Yu CW, Chaikam V. (2008) HDA6 is required for jasmonate response, senescence and flowering in Arabidopsis. J Exp Bot 59: 225–234 [DOI] [PubMed] [Google Scholar]
- Yi H, Richards EJ. (2009) Gene duplication and hypermutation of the pathogen Resistance gene SNC1 in the Arabidopsis bal variant. Genetics 183: 1227–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Gao S, Zhou X, Chellappan P, Chen Z, Zhou X, Zhang X, Fromuth N, Coutino G, Coffey M, Jin H. (2011) Bacteria-responsive microRNAs regulate plant innate immunity by modulating plant hormone networks. Plant Mol Biol 75: 93–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Clarenz O, Cokus S, Bernatavichute YV, Pellegrini M, Goodrich J, Jacobsen SE. (2007) Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol 5: e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.