Abstract
Inducible defenses, which provide enhanced resistance after initial attack, are nearly universal in plants. This defense signaling cascade is mediated by the synthesis, movement, and perception of jasmonic acid and related plant metabolites. To characterize the long-term persistence of plant immunity, we challenged Arabidopsis (Arabidopsis thaliana) and tomato (Solanum lycopersicum) with caterpillar herbivory, application of methyl jasmonate, or mechanical damage during vegetative growth and assessed plant resistance in subsequent generations. Here, we show that induced resistance was associated with transgenerational priming of jasmonic acid-dependent defense responses in both species, caused caterpillars to grow up to 50% smaller than on control plants, and persisted for two generations in Arabidopsis. Arabidopsis mutants that are deficient in jasmonate perception (coronatine insensitive1) or in the biogenesis of small interfering RNA (dicer-like2 dicer-like3 dicer-like4 and nuclear RNA polymerase d2a nuclear RNA polymerase d2b) do not exhibit inherited resistance. The observation of inherited resistance in both the Brassicaceae and Solanaceae suggests that this trait may be more widely distributed in plants. Epigenetic resistance to herbivory thus represents a phenotypically plastic mechanism for enhanced defense across generations.
To offset their sessile life, plants have evolved diverse strategies to survive and adapt to a broad range of biotic and abiotic stresses, including insect herbivory (Howe and Jander, 2008). Resistance to herbivory is mediated by preexisting physical and chemical barriers, rapidly induced defense mechanisms (Karban and Baldwin, 1997), and priming for stronger responses to subsequent attack (van Hulten et al., 2006). Given the adaptive value of such responses, there has been tremendous interest in unraveling the mechanisms of induced defenses. The hormonal signaling cascade triggering the production of antiherbivore defenses is largely mediated by the synthesis, movement, and perception of jasmonic acid (JA) and related metabolites (Chini et al., 2007; Thines et al., 2007; Howe and Jander, 2008). Interconversion of JA with the volatile methyl jasmonate (MeJA) may allow more rapid propagation of the defense signal to other plant parts. JA-Ile conjugates are the active ligands for CORONATINE INSENSITIVE1 (COI1), the F-box component of an E3-ubiquitin ligase complex, SCFCOI1 (Chini et al., 2007; Thines et al., 2007; Sheard et al., 2010). Upon binding of JA-Ile, the SCFCOI1 protein complex targets JA ZIM domain (JAZ) proteins for degradation by the 26S proteasome. Removal of JAZ proteins, which act as repressors of JA-responsive transcription factors (e.g. MYC2), allows the transcription of these target genes. This induced gene expression leads to elevated resistance, systemic signal transmission, and defense priming (Howe and Jander, 2008).
Evidence of inherited responses to herbivory comes from research with wild radish (Raphanus raphanistrum), where insect-damaged plants produced more resistant seedlings than undamaged plants (Agrawal et al., 1999), and the demonstration of increased trichome production in yellow monkeyflower (Mimulus guttatus) that had been fed upon in the previous generation (Holeski, 2007). Transgenerational effects in plant responses to other environmental stresses have been described in several recent publications. Treatment of Arabidopsis (Arabidopsis thaliana) with UV light or bacterial flagellin caused an increased recombination frequency that persisted in subsequent generations (Molinier et al., 2006). Infection with Tobacco mosaic virus increased tobacco (Nicotiana tabacum) recombination frequency and provided increased resistance for at least two subsequent generations (Kathiria et al., 2010). In the case of abiotic stress, exposure to elevated temperature led to increased heat tolerance for at least three subsequent generations in Arabidopsis (Whittle et al., 2009), and progeny of salt-stressed plants exhibited higher salt tolerance in the next generation (Boyko et al., 2006).
Epigenetic processes, which include inherited DNA methylation and histone modifications (Molinier et al., 2006; Chinnusamy and Zhu, 2009), are a likely mechanism for retaining stress memory in subsequent generations. Small interfering RNAs (siRNAs), approximately 24-nucleotide RNAs (Hamilton and Baulcombe, 1999) that are processed from double-stranded RNA by dicer-like RNase III enzymes (Lee et al., 2004), can lead to transcriptional regulation or, in some cases, mRNA decay (Matzke et al., 2001). Since siRNAs are able to move between cells and through the plant vasculature (Chitwood and Timmermans, 2010), they represent a possible mechanism for the inheritance of acquired resistance traits.
The mechanisms of transgenerational resistance to insect herbivory remain uninvestigated. In the few published examples of this phenomenon (Agrawal et al., 1999), it was not determined whether the progeny generations are constitutively more herbivore resistant or whether they are primed to respond more vigorously if there is a subsequent attack. It is also not known whether the inheritance of acquired herbivore resistance is a maternal effect transmitted through the seed, for instance through the storage of jasmonates or defensive secondary metabolites, or a longer term effect that is inherited in an epigenetic manner. To address these questions, we designed a series of experiments to investigate the role of jasmonates and siRNA in transgenerational induced resistance to herbivory in two well-studied model species, Arabidopsis and tomato (Solanum lycopersicum).
RESULTS
Specificity of Transgenerational Increased Resistance
Tomato and Arabidopsis plants were challenged with caterpillar feeding, MeJA exposure, or mechanical damage. Next, progeny from control and elicited plants were grown in a common environment to identify inherited increases in herbivore resistance. All three treatments decreased the growth of Helicoverpa zea (corn earworm) on progeny of treated tomato plants by about 40% relative to controls (Fig. 1A; F3,116 = 11.269, P < 0.0001). Preliminary experiments also showed that the small amount of ethanol used to dissolve MeJA for elicitation experiments has no effect on subsequent herbivory. In the case of Arabidopsis, prior-generation feeding by Pieris rapae (white cabbage butterfly) reduced caterpillar weight gain by 40% (Fig. 1B; F1,24 = 26.848, P < 0.0001), MeJA treatment had a similar 27% effect (Fig. 1B; F1,18 = 4.108, P = 0.058), but mechanical damage alone did not affect caterpillar growth (Fig. 1B; F1,27 < 0.0001, P = 0.995). The specificity of transgenerational resistance in Arabidopsis was demonstrated by exposing progeny from a new parental population to P. rapae and three additional lepidopteran herbivores, the crucifer-specialist diamondback moth (Plutella xylostella) and two generalists, the cabbage looper (Trichoplusia ni) and the beet armyworm (Spodoptera exigua). In addition to P. rapae, only S. exigua showed reduced performance on plants that were exposed to P. rapae herbivory in the previous generation (Fig. 1C; F1,16 = 7.517, P = 0.015), likely due to the fact that there is species-specific variation in lepidopteran sensitivity to Arabidopsis defenses (Müller et al., 2010). Both P. rapae and S. exigua also grew less well on plants whose parents were fed upon by P. xylostella compared with control plants whose parents were left undamaged (Fig. 2). This indicates that the type of parental damage does not affect inherited herbivore resistance, which is consistent with observations of similar Arabidopsis transcriptional responses to different lepidopteran herbivores (Bidart-Bouzat and Kliebenstein, 2008).
Figure 1.
Transgenerational resistance in tomato and Arabidopsis. A, H. zea growth on tomato originating from parents that were either left undamaged (control) or subjected to caterpillar feeding, MeJA treatment, or mechanical damage. B, P. rapae caterpillar growth on first-generation Arabidopsis progeny originating from parents that were either left undamaged (white bars) or subjected to P. rapae feeding, MeJA elicitation, or mechanical damage (black or gray bars). C, Effect of parental generation P. rapae caterpillar feeding on P. rapae, P. xylostella, S. exigua, and T. ni caterpillar growth on progeny plants. D, P. rapae caterpillar growth on three generations of Arabidopsis after P. rapae feeding (black bars) or undamaged controls (white bars). C1, C2, and C3 = generations after control treatments; H1, H2, and H3 = generations after P. rapae herbivory. Error bars represent se. Asterisks and different letters above bars represent posthoc Student’s t tests comparing means between parental treatments (P < 0.05). DW, Dry weight.
Figure 2.
Specificity of resistance in H1 Arabidopsis plants. Shown are average dry mass ± se of P. rapae (A) and S. exigua (B) caterpillars on Arabidopsis progeny plants with parents that were undamaged (white bars), P. rapae damaged for 3 d (black bars), or P. xylostella damaged for 3 d (gray bars). Both P. rapae and P. xylostella feeding increased resistance in the next generation (for P. rapae, F2,37 = 20.949, P < 0.0001; for S. exigua, F2,22 = 4.039, P = 0.032). Letters represent posthoc Student’s t tests comparing means between parental treatments (P < 0.05).
Persistence of Transgenerational Resistance over Time
Further research to investigate the mechanisms of transgenerational insect resistance was focused primarily on the more genetically tractable Arabidopsis system. To measure the stability of the transgenerational resistance signal, we planted additional C1 (control) and H1 (herbivory in prior generation) seeds without further exposure to herbivores in the second generation. C2 and H2 seeds were harvested, and the procedure was repeated to obtain C3 and H3 seeds (third generation after control and herbivory treatments). Transgenerational resistance to P. rapae persisted in the H2 generation (Fig. 1D; F1,33 = 4.634, P = 0.039) but not in the H3 generation (Fig. 1D; F1,36 = 0.032, P = 0.858).
Seed size and provisioning of resources could affect the robustness of plants in the H1 progeny generation (Agrawal et al., 1999). Similar to what has been observed previously in using this plant-herbivore system (van Loon et al., 2003), 3 d of P. rapae feeding reduced Arabidopsis seed set by 50% (Fig. 3A; F1,18 = 49.123, P < 0.0001). However, the mass of individual seeds (Fig. 3B; F1,19 = 1.944, P = 0.180) and plant size in the progeny generation (Supplemental Fig. S1) were not altered, suggesting that there are no differences in seed provisioning. Reduced seed set could result from the smaller plant size (loss of leaf area due to herbivory) or could reflect a reallocation of resources from seed production to plant defense (Mauricio, 1998). Since Arabidopsis dies shortly after seed set, it is unlikely that plants are “saving” resources for later growth and reproduction, as might be expected from a perennial plant.
Figure 3.
Effects of P. rapae feeding on Arabidopsis seed numbers (A) and seed mass (B). Shown are averages ± se. The asterisk indicates a difference between parental treatments (P < 0.05, Student’s t test).
Mechanisms of Transgenerational Resistance
We determined whether parental generation herbivory inherently alters known defense traits in the seeds or the progeny generation. Seed content of the plant hormones JA, salicylic acid (SA), abscisic acid, and indole-3-acetic acid was not significantly affected by caterpillar feeding (Fig. 4), showing that H1 plants are not primed for insect resistance through the storage of these defense signaling molecules in the seeds. Glucosinolates, a crucifer-specific class of defensive secondary metabolites, also did not differ between seeds of control and P. rapae-treated plants (Fig. 5A). Among leaf glucosinolates in undamaged H1 plants, only 1-methoxyindol-3-ylmethylglucosinolate (1MI3M) was increased relative to C1 plants without P. rapae caterpillar feeding in the parental generation (Fig. 5B). Furthermore, the overall phenotype of the progeny plants was similar, irrespective of the parental treatment (Supplemental Fig. S1), suggesting that plant size is not growth limiting for the caterpillars. Rosette leaf trichome density, which is associated with insect resistance (Levin, 1973; Mauricio and Rausher, 1997), was not significantly altered by prior-generation herbivory (Supplemental Fig. S2).
Figure 4.
Concentrations of phytohormones in Arabidopsis seeds collected from plants with and without herbivory. Shown are averages ± se of SA, JA, abscisic acid (ABA), and indoleacetic acid (IAA) in seeds of undamaged Col-0 Arabidopsis plants (Control) or of plants that were damaged during 3 d with one caterpillar per plant of the specialist lepidopteran herbivores P. rapae or P. xylostella. No differences were found across treatments (ANOVAs for SA, F2,25 = 1.066, P = 0.359; for JA, F2,25 = 1.703, P = 0.313; for ABA, F2,25 = 0.333, P = 0.666; for IAA, F2,25 = 272, P = 0.544). FW, Fresh weight.
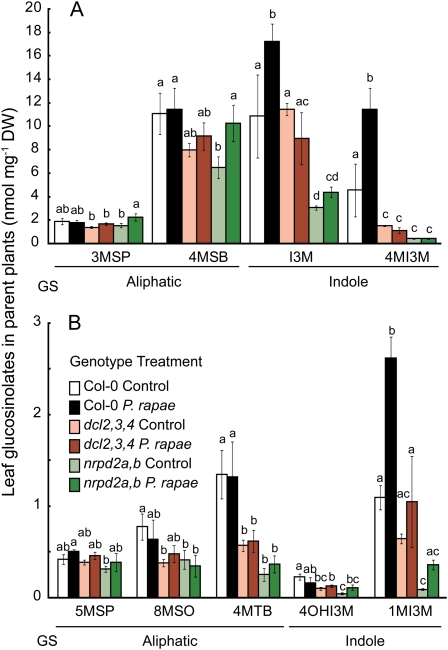
Figure 5.
Glucosinolate content of control and P. rapae-damaged Arabidopsis plants. A, Averages ± se abundance of seed aliphatic, indole, and benzoyloxy glucosinolates (GS). Seed originated from either control plants (white bars) or mature plants that were induced by 3 d of P. rapae feeding (black bars). B, Averages ± se abundance of indole and aliphatic glucosinolates in leaves from Arabidopsis plants with parents that were either left undamaged (C1; white bars) or fed upon by P. rapae caterpillars for 3 d (H1; black bars). Glucosinolates were measured on damaged leaves from P. rapae treatments and on equivalent leaf amounts in the control treatments. Plants originated from the seed batches that were used for glucosinolate assays. Wilcoxon test for 1MI3M, χ2 = 3.857, degrees of freedom = 1, P = 0.049. *P < 0.05. Glucosinolate side chain abbreviations not defined in the text are as follows: 3MSP, 3-methylsulfinylpropyl; 4MSB, 4-methylsulfinylbutyl; 5MSP, 5-methylsulfinylpentyl; 4OHI3M, 4-hydroxyindol-3-ylmethyl; 7MSH, 7-methylsulfinylheptyl; 4MTB, 4-methylthiobutyl; 8MSO, 8-methylsulfinyloctyl; I3M, indol-3-ylmethyl; 7MTH, 7-methylthioheptyl; 8MTO, 8-methylthiooctyl; 3BOP, 3-benzoyloxypropyl; 4BOB, 4-benzoyloxybutyl. DW, Dry weight.
JA Perception Is Required for Transgenerational Resistance
Since JA signaling plays a central role in the induction of plant defense responses (Howe and Jander, 2008), additional experiments were designed to determine whether inherited caterpillar resistance depends on the perception of JA via COI1 in the parental and/or progeny generation (Fig. 6A). Arabidopsis coi1-1 mutants have normal growth patterns but are unable to perceive JA-Ile conjugates and therefore fail to initiate defense-related gene expression changes in response to herbivory (Chini et al., 2007; Thines et al., 2007; Howe and Jander, 2008). Consistent with this known defense signaling function of COI1, homozygous coi1-1 mutant parent plants, which were induced by P. rapae feeding and pollinated with wild-type pollen, did not show increased resistance in the H1 generation (Fig. 6B; test for treatment effect; F1,53 = 0.023, P = 0.880). Functionally wild-type heterozygous COI1/coi1-1 plants were also subjected to P. rapae herbivory. Segregating H1 progeny from P. rapae-induced parents were more resistant to herbivory, irrespective of their genotype (Fig. 6C; for treatment effect, F1,37 = 13.143, P = 0.001; for progeny genotype effect, F1,37 = 0.656, P = 0.423; for the interaction between treatment and progeny genotypes, F1,37 = 0.025, P = 0.875). Therefore, in contrast to the parental generation (Fig. 6B), perception of JA-Ile by COI1 in the progeny generation (Fig. 6C) is not required for inherited P. rapae resistance.
Figure 6.
The role of jasmonate signaling and gene expression changes due to herbivory in the parental generation. A, Schematic of crosses used to determine when JA-Ile detection by COI1 is required. Progeny from heterozygous COI1/coi1-1 plants were raised with and without P. rapae herbivory. Homozygous coi1-1/coi1-1 plants were pollinated with wild-type (WT) Col-0. Heterozygous and wild-type plants were allowed to self-pollinate. C1 and H1 progeny were used for P. rapae growth experiments. B, P. rapae caterpillar growth on plants with jasmonate-insensitive coi1-1/coi1-1 parents. C, P. rapae caterpillar growth on jasmonate-insensitive coi1-1/coi1-1 C1 (control) and H1 (P. rapae on parents) progeny of functionally wild-type Arabidopsis (COI1/coi1-1 or COI1/COI1). D, Accumulation of JA (top) and SA (bottom) in leaves of Arabidopsis with and without P. rapae feeding in the parental and progeny generations. E, Zero- and 24-h induction of LOX2 (left) and AOS (right) gene expression in Arabidopsis with P. rapae-damaged (H1; black bars) or undamaged (C1; white bars) parents. Gene expression is in arbitrary units, with control plant expression set to 1. Error bars represent se. Asterisks and letters above bars represent differences between bars (P < 0.05, t test). DW, Dry weight; FW, fresh weight.
Consistent with the observed increase in 1MI3M (Fig. 5), which is typically induced by insect feeding and jasmonates (Agerbirk et al., 2009), P. rapae feeding in the parental generation caused JA levels to be 2-fold higher in H1 plants compared with C1 plants in the absence of caterpillar damage (dashed lines in Fig. 6D, and parental treatment effect below). Additionally, after 72 h of feeding by neonate caterpillars, JA levels were significantly elevated in H1 plants, which had been fed upon in the previous generation, relative to C1 controls (Fig. 6D; for parent treatments, F1,58 = 5.706, P = 0.020; for induction, F1,58 = 46.117 P < 0.001; for time, F3,58 = 4.880, P = 0.004), suggesting priming for a more robust defense response. On the other hand, SA, a phytohormone that is primarily associated with pathogen defense, did not exhibit parental treatment effects. In fact, P. rapae herbivory reduced SA levels by 15% over a period of 96 h (Fig. 6D). Consistent with the elevated JA levels, the expression of LIPOXYGENASE2 (LOX2), a well-studied JA-responsive gene in Arabidopsis (Howe and Jander, 2008), was increased in plants subjected to caterpillar feeding in the prior generation (Fig. 6E; for LOX2 parental treatment effect, F1,18 = 3.135, P = 0.094; for induction effect, F1,18 = 5.389, P = 0.032; for interaction, F1,18 = 4.078, P = 0.058). ALLENE OXIDE SYNTHASE (AOS), a JA biosynthesis gene (Howe and Jander, 2008), was also more highly induced by P. rapae if the previous generation had been subjected to herbivory (Fig. 6E; for AOS parental treatment effect, F1,20 = 4.349, P = 0.050; for induction effect, F1,20 = 31.242, P < 0.0001; for interaction, F1,20 = 1.402, P = 0.250). Similar priming of defense gene expression was observed with the JA-regulated tomato PROTEASE INHIBITOR2 (PIN2) gene (Miersch and Wasternack, 2000), which was induced to a higher level by wounding if the previous generation had been exposed to MeJA (Supplemental Fig. S3).
Small RNAs Are Required for Transgenerational Resistance
The elevated caterpillar resistance (Fig. 1) and JA-mediated defenses (Fig. 6, D and E) suggest defense priming via an inherited signal from the parental plant. To test the hypothesis that this effect requires a siRNA signal, we conducted experiments with two Arabidopsis mutants that have a normal growth pattern but are deficient in small RNA biogenesis: a nuclear RNA polymerase d2a nuclear RNA polymerase d2b (nrpd2a nrpd2b) double mutant, which lacks nuclear RNA polymerases required for the synthesis of siRNAs (Pontes et al., 2006), and a dicer-like2 dicer-like3 dicer-like4 (dcl2 dcl3 dcl4) triple mutant, which is defective in siRNA processing by dicer-like enzymes (Henderson et al., 2006). Whereas feeding in the previous generation reduced the growth of P. rapae on wild-type Arabidopsis, this was not the case on the nrpd2a nrpd2b or dcl2 dcl3 dcl4 mutants (Fig. 7A; treatment effect for wild-type P. rapae-fed landrace Columbia-0 [Col-0], F1,30 = 10.198, P = 0.003; for nrpd2a nrpd2b, F1,36 = 1.405, P = 0.244; for dcl2 dcl3 dcl4, F1,26 = 0.363, P = 0.552). Similarly, application of MeJA to the parent plants was ineffective at inducing transgenerational resistance in the mutants but not on wild-type plants (Fig. 7B; treatment effect for Col-0, F1,35 = 13.034, P = 0.001; for nrpd2a nrpd2b, F1,30 = 0.001, P = 0.971; for dcl2 dcl3 dcl4, F1,31 = 0.009, P = 0.307).
Figure 7.
Transgenerational resistance to insect herbivores is absent in small RNA-deficient mutants. Shown are average mass ± se of P. rapae caterpillars on wild-type Arabidopsis compared with nrpd2a nrpd2b and dcl2 dcl3 dcl4 mutants with parent plants that were damaged by P. rapae (black bars; A), sprayed with MeJA (gray bars; B), or left as controls (white bars). Asterisks indicate differences between parental treatments (P < 0.05, t test). DW, Dry weight.
Both the nrpd2a nrpd2b and dcl2 dcl3 dcl4 mutants have reduced amounts of some glucosinolates relative to wild-type Col-0, with or without P. rapae feeding (Fig. 8; for total glucosinolate genotype effect, F2,300 = 8.772, P = 0.0002; treatment effect, F1,300 = 2.330, P = 0.128; genotype-treatment interaction, F2,300 = 1.034, P = 0.357). In particular, 4-methoxyindol-3-ylmethylglucosinolate (4MI3M), which has been associated with pathogen defense responses (Bednarek et al., 2009; Clay et al., 2009), is constitutively less abundant in the mutants. Caterpillar feeding induces the production of 4MI3M and 1MI3M in wild-type plants, but there was no significant increase in the mutant lines. Reduced caterpillar size on the nrpd2a nrpd2b and dcl2 dcl3 dcl4 mutants (Fig. 7) is correlated with a lower abundance of glucosinolates, which are P. rapae feeding stimulants (Barth and Jander, 2006; Müller et al., 2010).
Figure 8.
Constitutive and induced levels of indole and aliphatic glucosinolates (GS) in leaves of wild-type Col-0 Arabidopsis and RNA polymerase-deficient (nrpd2a nrpd2b) and dicer-like (dcl2 dcl3 dcl4) mutants. Shown are averages ± se of more abundant glucosinolates (A) and less abundant glucosinolates (B). Glucosinolates were measured in equivalent amounts of damaged (P. rapae) and undamaged (Control) leaves. Different letters above bars indicate significant differences (P < 0.05, t test) for each glucosinolate type. Glucosinolate side chain abbreviations are as in Figure 5. DW, Dry weight.
DISCUSSION
Our results demonstrate that both Arabidopsis and tomato plants that were subjected to herbivory are more resistant to subsequent attack in the next generation. In the case of Arabidopsis, this transgenerational resistance against chewing herbivores includes the priming of JA-related defense responses and requires siRNA biogenesis.
JA-Mediated Increased Transgenerational Resistance
Numerous studies show that jasmonates, SA, and ethylene have important functions in orchestrating plant responses to tissue damage or wounding (Howe, 2004; De Vos et al., 2005). Previous defense induction can cause plants to be primed for a more robust or rapid defense response upon subsequent attack (van Hulten et al., 2006), and treatment of seeds with JA primes plants for enhanced herbivore resistance weeks later (Worrall et al., 2011). Here, we have taken these observations a step further to show that MeJA induction in the previous generation can prime progeny plants for enhanced resistance. Additionally, using coi1 mutant plants, we demonstrate that the perception of JA is required in mother plants for increased resistance in the next generation. Further work using JA signaling-deficient mutants (e.g. jai1) will be needed to determine whether tomato also requires JA perception for increased transgenerational resistance. Unlike in the case of Arabidopsis, wounding alone primed tomato for increased caterpillar resistance in H1 plants. This could reflect inherent differences between Arabidopsis and tomato, or it could be the result of different experimental methods and perhaps insufficient mechanical wounding of the Arabidopsis plants. For instance, only frequent and controlled repetitive wounding triggered herbivore-like defense elicitation in lima bean (Phaseolus lunatus; Mithöfer et al., 2005).
Accumulating evidence demonstrates cross talk between different phytohormone signaling pathways. For example, JA-mediated defense signaling is strongly activated upon chewing herbivore attack, whereas SA and ethylene are more specific to piercing-sucking herbivores or pathogen attack (De Vos et al., 2005). Consistent with these prior reports, we observed priming for faster and higher JA induction in the H1 generation, but we did not observe increased SA accumulation in response to P. rapae attack on H1 or C1 plants. These results are consistent with a specific activation of JA signaling during chewing herbivore attack as well as with possible cross talk with the SA signaling pathway (De Vos et al., 2005).
siRNA and Transgenerational Resistance
siRNA is phloem mobile (Chitwood and Timmermans, 2010) and could provide a signal that is passed from vegetative tissue to developing seeds in response to biotic or abiotic stress. In the developing seeds and/or progeny plants, siRNA could alter gene expression through targeted mRNA degradation, regulation of translation, or DNA methylation. The inheritance of resistance over two generations (Fig. 1D) suggests that DNA methylation, which is impacted by PolIV- and DCL2-dependent siRNA production and can be inherited through meiosis, is a possible mechanism for transgenerational inheritance. Increased resistance over two generations also indicates that the signal is likely to be propagated in the embryo rather than in maternal tissue, which makes up the Arabidopsis seed coat.
Two other articles in this issue show that similar transgenerational priming of defense signaling pathways is involved in plant responses to pathogen infection. Infection with avirulent Pseudomonas syringae and treatment with β-aminobutyric acid primed Arabidopsis for the induction of SA-mediated defense responses in the subsequent generation (Slaughter et al., 2012). In another study, the descendants of primed plants showed a faster and stronger SA-mediated defense response relative to controls (Luna et al., 2012). Comparable effects in a drm1 drm2 cmt3 DNA methylation mutant suggest that hypomethylation is required to prime progeny plants for these SA-mediated defense responses.
Future Prospects
Like all forms of phenotypic plasticity, transgenerational resistance to herbivory will only be effective if the response in one generation protects seedlings in the next generation (Karban et al., 1999). A plant that employs transgenerational resistance but whose progeny are not subject to attack may suffer energetic and ecological costs without benefits (Agrawal, 2001). Many natural populations of Arabidopsis have more than one generation per growing season (Donohue, 2009; Bentsink et al., 2010), which would expose parents and progeny to similar herbivore pressures. Therefore, rapid-cycling Arabidopsis genotypes would benefit from priming of insect resistance in progeny of plants that were subjected to herbivory. A testable prediction is that defense priming should be reduced in response to longer seed dormancy, which would expose progeny plants to a likely different herbivore environment in subsequent years. Such effects have been seen in the release of gene silencing due to seed aging (Lang-Mladek et al., 2010).
Unlike most plants in natural ecosystems, the Arabidopsis and tomato lines used for these experiments had been growing in the complete absence of herbivores for several generations. Since inherited resistance in Arabidopsis attenuates over three generations (Fig. 1D), herbivore-free laboratory rearing may account for the consistent transgenerational effects that we have observed. However, even in a growth chamber setting, there are variables than cannot completely be controlled. Out of the nine independent positive control experiments testing first-generation progeny from P. rapae-treated and control plants, seven showed variable levels of increased resistance, two showed no significant differences, and none showed a negative effect (i.e. caterpillar growth was never reduced on C1 plants compared with H1 plants; Fig. 9). Similar to the protected existence of our laboratory-grown plants, some plants in agricultural settings may grow in the almost complete absence of herbivory for several generations, particularly with the heavy use of insecticides in seed production fields. Just as the application of jasmonates to seeds can promote long-term herbivore resistance (Worrall et al., 2011), defense elicitation prior to seed set could be used to prime progeny plants for increased herbivore and pathogen resistance, thereby providing a defensive benefit to tomato and other crops that are particularly vulnerable to herbivory at the seedling stage.
Figure 9.
Independent replicates of Arabidopsis transgenerational resistance to P. rapae herbivory. Shown are average P. rapae caterpillar mass after 7 d of feeding on progeny from control and P. rapae-induced Arabidopsis plants from nine independent experiments. Parent lines were either left undamaged (Control) or were induced by a P. rapae caterpillar feeding. Out of nine experiments, which include those shown in Figures 1, 2, 6, and 7, seven showed increased transgenerational resistance (solid lines) and two showed no effect (dashed lines). Paired t test between treatments across the nine experiments, t9 = −2.806, P = 0.012.
MATERIALS AND METHODS
Plants and Growth Conditions
Wild-type Arabidopsis (Arabidopsis thaliana) landrace Col-0 was obtained from the Arabidopsis Biological Resource Center (www.arabidopsis.org). Seeds of nrpd2a nrpd2b (Henderson et al., 2006) and dcl2 dcl3 dcl4 (Henderson et al., 2006) Arabidopsis mutants were obtained from Eric Richards (Boyce Thompson Institute), and coi1-1 seeds (Xie et al., 1998) were obtained from Gregg Howe (Michigan State University). Plants were grown in Conviron growth chambers in 20- × 40-cm nursery flats using Cornell Mix (by weight, 56% peat moss, 35% vermiculite, 4% lime, 4% Osmocoat slow-release fertilizer [Scotts], and 1% Unimix [Peters]) at 23°C, and 60% relative humidity, with a light intensity of 180 μmol m−2 s−1 photosynthetic photon flux density and a 16-h/8-h light/dark photoperiod. Plants were grown for 3 weeks and were used in experiments before flowering.
Tomato (Solanum lycopersicum) cv Micro-Tom was used in all tomato experiments. Seeds were originally purchased from Tomato Growers Supply. Seedlings were grown in Metromix 400 potting mix (Griffin Greenhouse & Nursery Supplies) in a greenhouse at Pennsylvania State University. The greenhouse was maintained on a 16-h/8-h light/dark photoperiod. Six-week-old plants were used for insect feeding, mechanical damage, or MeJA treatment.
Insects and Rearing Conditions
A Pieris rapae colony was established with approximately 20 adult butterflies that were collected on the Cornell University campus in 2008. Caterpillars were raised on cabbage (Brassica oleracea) var Wisconsin Golden Acre (Seedway) under the same conditions as those used for growing Arabidopsis. Adult butterflies were fed with a 20% Suc solution. Trichoplusia ni, Plutella xylostella, Spodoptera exigua, and Helicoverpa zea eggs were obtained from Benzon Research. For induction in the parental generation, individual caterpillars were allowed to feed for 3 d on each plant, and seeds were harvested 5 to 6 weeks after planting. We used about 25 parental plants for induction, and progeny were obtained from a random sampling of the mother plants. For bioassays in subsequent generations, insects were confined on the leaves of 3-week-old Arabidopsis with mesh-covered cups. A single neonate lepidopteran larva per plant was allowed to feed for 7 d before being collected and lyophilized for 1 d. Larval dry weight was determined using a precision balance.
For experiments with H. zea, neonates were fed a wheat germ- and casein-based artificial diet (Bio-Serv) for 1 d and then transferred to tomato leaves for 4 d (one larva per plant). Larval fresh weight was determined using a precision balance.
COI1 Experiments
Seeds from COI1/coi1-1 Arabidopsis were planted and grown as described above to produce a segregating population of COI1/coi1-1, COI1/COI1, and coi1-1/coi1-1 plants. Half of all plants were induced with P. rapae for 3 d as described above. Homozygous mutants (coi1-1/coi1-1), which are male sterile, were visually identified by the absence of seed pods after flowering. Homozygous coi1-1/coi1-1 mutants were pollinated with wild-type Col-0 to obtain viable heterozygous seeds. Subsequently, one neonate P. rapae larva per plant was allowed to feed for 7 d on 3-week-old, next-generation (H1 and C1) plants, which either originated from functionally wild-type parents (heterozygous COI1/coi1-1 or homozygous COI1/COI1) or from the cross between the jasmonate-insensitive mutants (coi1-1/coi1-1) and a wild-type Col-0 plant. The design of the COI1 experiments is illustrated in Figure 6A.
Phytohormone Analysis
Arabidopsis Col-0 wild-type seeds were planted as described above, and after 3 weeks of growth, half of the plants were subjected to 3 d of P. rapae feeding. Plants were allowed to self-pollinate, and seeds were collected 5 to 6 weeks after initial planting. Harvested seeds were grown as described above and divided into four treatments (with and without P. rapae feeding in the parental and progeny generations). In the progeny generation, damage was imposed by placing one first instar P. rapae caterpillar on each plant and allowing it to feed continuously for 4 d. Full-grown, visibly damaged leaves were harvested after 0, 48, 72, and 96 h. Plants from which a leaf had been harvested were discarded and not used for further experiments. Harvested leaves were weighed and placed in tubes containing 0.9 g of FastPrep matrix (BIO 101) before being flash frozen in liquid nitrogen and stored at −80°C until further use. One milliliter of extraction buffer (2:1:0.005, isopropanol:water:HCl) was added to each sample. d4-SA and d5-JA (CDN Isotopes) were added as internal standards, and samples were homogenized in a FastPrep homogenizer (MP Biomedicals) at 6 m s−1 for 45 s. Samples were dissolved in 200 μL of methanol after extraction with dichloromethane and solvent evaporation, and 15 μL was analyzed using a triple-quadrupole liquid chromatography-tandem mass spectrometry system (Quantum Access; Thermo Scientific). Analytes were separated on a C18 reverse-phase HPLC column (3 μm, 150 × 2.00 mm; Gemini-NX; Phenomenex) using a gradient of 0.1% formic acid in water (solvent A) and 0.1% formic acid in acetonitrile (solvent B) at a flow rate of 300 μL min−1. The initial condition of 10% B was kept for 2 min and increased to 100% solvent B at 20 min. Phytohormones were analyzed by negative electrospray ionization (spray voltage, 3.5 kV; sheath gas, 15; auxiliary gas, 15; capillary temperature, 350°C), collision-induced dissociation (argon gas pressure, 1.3 mTorr; energy, 16 V), and selected reaction monitoring of compound-specific parent/product ion transitions (SA, 137→93; d4-SA, 141→97; JA, 209→59; d5-JA, 214→62).
Glucosinolate Assays
Arabidopsis leaves were collected, frozen in liquid nitrogen, and lyophilized. Extraction of plant tissue and preparation of desulfoglucosinolates were done as described previously (Barth and Jander, 2006; Kim et al., 2008). Desulfoglucosinolates were separated using a Waters 2695 HPLC device and detected using a Waters 2996 photodiode array detector. For HPLC separation, the mobile phases were water (A) and 90% acetonitrile (B), with a flow rate of 1 mL min−1 at 23°C. Column linear gradients for samples were as follows: 0 to 1 min, 98% A; 1 to 6 min, 94% A; 6 to 8 min, 92% A; 8 to 16 min, 77% A; 16 to 20 min, 60% A; 20 to 25 min, 0% A; 25 to 27 min, hold at 0% A; 27 to 28 min, 98% A; 28 to 37 min, 98% A.
Gene Expression Analysis
Total RNA was extracted from frozen tissue samples using the SV Total RNA Isolation system with on-column DNase treatment (Promega). RNA integrity was verified using a 1.2% formaldehyde agarose gel. Transcript abundance of LOX2 (At3G45140), AOS (AT5G42650), and PIN2 (K03271) was analyzed by quantitative real-time reverse transcription (qRT)-PCR. Elongation Factor1-α (AT5G60390), which was used as an internal standard for Arabidopsis experiments, was identified from publicly available microarray data as stably expressed after herbivory, and this stable expression was verified across samples using qRT-PCR. Ubiquitin (X58253) was used as an internal standard for tomato qRT-PCR. After RNA extraction and DNase treatment, 1 μg of total RNA was reverse transcribed with SMART Moloney murine leukemia virus reverse transcriptase (Clontech) using an oligo(dT)12-18 primer. Gene-specific primers used for qRT-PCR were designed using Primer-Blast (http://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi) with the following criteria: melting temperature of 60°C; PCR amplicon lengths of 90 to 150 bp yielding primer sequences with lengths of 18 to 24 nucleotides, with an optimum at 21 nucleotides; and GC contents of 40% to 60%. Primer sequences can be found in Supplemental Table S1. Reactions were carried out using 5 μL of the SYBR Green PCR master mix (Applied Biosystems), with 800 nm primer, in the 7900HT instrument (Applied Biosystems). The PCRs were initiated by incubation at 95°C for 10 min to activate the enzyme. Then, the following cycle was repeated 40 times: 95°C for 15 s, 60°C for 15 s, and 72°C for 15 s. The cycle threshold values were quantified and analyzed according to the standard curve method.
Statistical Analysis
All larval growth, seed set, seed mass, glucosinolate, and trichome count data were analyzed on a parental treatment comparison (such as P. rapae induction, P. xylostella induction, MeJA induction, mechanical damage versus undamaged control plants) with one-way ANOVA. Caterpillar mass for the coi1-1 experiment was analyzed as for the parental treatments if parents were homozygous recessive coi1/coi1-1 but with two-way ANOVA for the heterozygous parent side of the experiment to test interaction between parental treatments and segregating genotypes in the progeny generation. Phytohormone induction data were analyzed with one-way ANOVA with induction (control plants versus P. rapae-fed plants), parental treatment (control versus P. rapae induction), and time as main factors. Gene expression data were analyzed with two-way ANOVA, with parental treatment and timing of induction as main effects. Because of the small sample size, leaf and seed glucosinolates of Col-0 plants were individually analyzed with nonparametric tests. Data were checked for normality and log transformed where needed. In addition, as needed, data for caterpillar growth were blocked for plant position or batch to control for position effect in growth chamber experiments. All analyses were performed with JMP 8 software (SAS Institute).
Supplemental Data
The following materials are available in the online version of this article
Supplemental Figure S1. Herbivory in the parental generation does not have a significant effect on the overall size and morphology of progeny plants.
Supplemental Figure S2. Trichome production on C1 and H1 Arabidopsis rosette leaves.
Supplemental Figure S3. Expression of PIN2 in tomato with and without MeJA treatment in the previous generation.
Supplemental Table S1. Primers used for qRT-PCR in this study.
Supplementary Material
Acknowledgments
We thank E. Richards, G. Howe, and B. Meyers for Arabidopsis seeds and experimental advice.
References
- Agerbirk N, De Vos M, Kim JH, Jander G. (2009) Indole glucosinolate breakdown and its biological effects. Phytochem Rev 8: 101–120 [Google Scholar]
- Agrawal AA. (2001) Transgenerational consequences of plant responses to herbivory: an adaptive maternal effect? Am Nat 157: 555–569 [DOI] [PubMed] [Google Scholar]
- Agrawal AA, Laforsch C, Tollrian R. (1999) Transgenerational induction of defences in animals and plants. Nature 401: 60–63 [Google Scholar]
- Barth C, Jander G. (2006) Arabidopsis myrosinases TGG1 and TGG2 have redundant function in glucosinolate breakdown and insect defense. Plant J 46: 549–562 [DOI] [PubMed] [Google Scholar]
- Bednarek P, Piślewska-Bednarek M, Svatoš A, Schneider B, Doubský J, Mansurova M, Humphry M, Consonni C, Panstruga R, Sanchez-Vallet A, et al. (2009) A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323: 101–106 [DOI] [PubMed] [Google Scholar]
- Bentsink L, Hanson J, Hanhart CJ, Blankestijn-de Vries H, Coltrane C, Keizer P, El-Lithy M, Alonso-Blanco C, de Andrés MT, Reymond M, et al. (2010) Natural variation for seed dormancy in Arabidopsis is regulated by additive genetic and molecular pathways. Proc Natl Acad Sci USA 107: 4264–4269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidart-Bouzat MG, Kliebenstein DJ. (2008) Differential levels of insect herbivory in the field associated with genotypic variation in glucosinolates in Arabidopsis thaliana. J Chem Ecol 34: 1026–1037 [DOI] [PubMed] [Google Scholar]
- Boyko A, Hudson D, Bhomkar P, Kathiria P, Kovalchuk I. (2006) Increase of homologous recombination frequency in vascular tissue of Arabidopsis plants exposed to salt stress. Plant Cell Physiol 47: 736–742 [DOI] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Fernández G, Adie B, Chico JM, Lorenzo O, García-Casado G, López-Vidriero I, Lozano FM, Ponce MR, et al. (2007) The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- Chinnusamy V, Zhu JK. (2009) Epigenetic regulation of stress responses in plants. Curr Opin Plant Biol 12: 133–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood DH, Timmermans MCP. (2010) Small RNAs are on the move. Nature 467: 415–419 [DOI] [PubMed] [Google Scholar]
- Clay NK, Adio AM, Denoux C, Jander G, Ausubel FM. (2009) Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323: 95–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos M, Van Oosten VR, Van Poecke RM, Van Pelt JA, Pozo MJ, Mueller MJ, Buchala AJ, Métraux JP, Van Loon LC, Dicke M, et al. (2005) Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol Plant Microbe Interact 18: 923–937 [DOI] [PubMed] [Google Scholar]
- Donohue K. (2009) Completing the cycle: maternal effects as the missing link in plant life histories. Philos Trans R Soc Lond B Biol Sci 364: 1059–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC. (1999) A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286: 950–952 [DOI] [PubMed] [Google Scholar]
- Henderson IR, Zhang X, Lu C, Johnson L, Meyers BC, Green PJ, Jacobsen SE. (2006) Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat Genet 38: 721–725 [DOI] [PubMed] [Google Scholar]
- Holeski LM. (2007) Within and between generation phenotypic plasticity in trichome density of Mimulus guttatus. J Evol Biol 20: 2092–2100 [DOI] [PubMed] [Google Scholar]
- Howe GA. (2004) Jasmonates as signals in the wound response. J Plant Growth Regul 23: 223–237 [Google Scholar]
- Howe GA, Jander G. (2008) Plant immunity to insect herbivores. Annu Rev Plant Biol 59: 41–66 [DOI] [PubMed] [Google Scholar]
- Karban R, Agrawal AA, Thaler JS, Adler LS. (1999) Induced plant responses and information content about risk of herbivory. Trends Ecol Evol 14: 443–447 [DOI] [PubMed] [Google Scholar]
- Karban R, Baldwin I. (1997) Induced Responses to Herbivory. University of Chicago Press, Chicago [Google Scholar]
- Kathiria P, Sidler C, Golubov A, Kalischuk M, Kawchuk LM, Kovalchuk I. (2010) Tobacco mosaic virus infection results in an increase in recombination frequency and resistance to viral, bacterial, and fungal pathogens in the progeny of infected tobacco plants. Plant Physiol 153: 1859–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Lee BW, Schroeder FC, Jander G. (2008) Identification of indole glucosinolate breakdown products with antifeedant effects on Myzus persicae (green peach aphid). Plant J 54: 1015–1026 [DOI] [PubMed] [Google Scholar]
- Lang-Mladek C, Popova O, Kiok K, Berlinger M, Rakic B, Aufsatz W, Jonak C, Hauser M-T, Luschnig C. (2010) Transgenerational inheritance and resetting of stress-induced loss of epigenetic gene silencing in Arabidopsis. Mol Plant 3: 594–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Nakahara K, Pham JW, Kim K, He ZY, Sontheimer EJ, Carthew RW. (2004) Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117: 69–81 [DOI] [PubMed] [Google Scholar]
- Levin DA. (1973) The role of trichomes in plant defense. Q Rev Biol 48: 3–15 [Google Scholar]
- Luna E, Bruce TJA, Roberts MR, Flors V, Ton J. (2012) Next generation systemic acquired resistance. Plant Physiol 158: 844–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke MA, Matzke AJM, Pruss GJ, Vance VB. (2001) RNA-based silencing strategies in plants. Curr Opin Genet Dev 11: 221–227 [DOI] [PubMed] [Google Scholar]
- Mauricio R. (1998) Costs of resistance to natural enemies in field populations of the annual plant Arabidopsis thaliana. Am Nat 151: 20–28 [DOI] [PubMed] [Google Scholar]
- Mauricio R, Rausher MD. (1997) Experimental manipulation of putative selective agents provides evidence for the role of natural enemies in the evolution of plant defense. Evolution 51: 1435–1444 [DOI] [PubMed] [Google Scholar]
- Miersch O, Wasternack C. (2000) Octadecanoid and jasmonate signaling in tomato (Lycopersicon esculentum Mill.) leaves: endogenous jasmonates do not induce jasmonate biosynthesis. Biol Chem 381: 715–722 [DOI] [PubMed] [Google Scholar]
- Mithöfer A, Wanner G, Boland W. (2005) Effects of feeding Spodoptera littoralis on lima bean leaves. II. Continuous mechanical wounding resembling insect feeding is sufficient to elicit herbivory-related volatile emission. Plant Physiol 137: 1160–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinier J, Ries G, Zipfel C, Hohn B. (2006) Transgeneration memory of stress in plants. Nature 442: 1046–1049 [DOI] [PubMed] [Google Scholar]
- Müller R, de Vos M, Sun JY, Sønderby IE, Halkier BA, Wittstock U, Jander G. (2010) Differential effects of indole and aliphatic glucosinolates on lepidopteran herbivores. J Chem Ecol 36: 905–913 [DOI] [PubMed] [Google Scholar]
- Pontes O, Li CF, Costa Nunes P, Haag J, Ream T, Vitins A, Jacobsen SE, Pikaard CS. (2006) The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell 126: 79–92 [DOI] [PubMed] [Google Scholar]
- Sheard LB, Tan X, Mao H, Withers J, Ben-Nissan G, Hinds TR, Kobayashi Y, Hsu F-F, Sharon M, Browse J, et al. (2010) Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468: 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaughter A, Daniela X, Flors V, Luna E, Hohn B, Mauch-Mani B. (2012) Descendants of primed Arabidopsis plants exhibit resistance to biotic stress. Plant Physiol 158: 835–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu GH, Nomura K, He SY, Howe GA, Browse J. (2007) JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665 [DOI] [PubMed] [Google Scholar]
- van Hulten M, Pelser M, van Loon LC, Pieterse CM, Ton J. (2006) Costs and benefits of priming for defense in Arabidopsis. Proc Natl Acad Sci USA 103: 5602–5607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon JAA, de Boer JG, Dicke M. (2003) Parasitoid-plant mutualism: parasitoid attack of herbivore increases plant reproduction. Entomol Exp Appl 97: 219–227 [Google Scholar]
- Whittle CA, Otto SP, Johnston MO, Krochko JE. (2009) Adaptive epigenetic memory of ancestral temperature regime in Arabidopsis thaliana. Botany-Botanique 87: 650–657 [Google Scholar]
- Worrall D, Holroyd GH, Moore JP, Glowacz M, Croft P, Taylor JE, Paul ND, Roberts MR. (December 5, 2011) Treating seeds with activators of plant defence generates long-lasting priming of resistance to pests and pathogens. New Phytol http://dx.doi.org/10.1111/j.1469-8137.2011.03987.x [DOI] [PubMed] [Google Scholar]
- Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG. (1998) COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.