Abstract
Bacterial blight is a devastating disease of rice (Oryza sativa) caused by Xanthomonas oryzae pv oryzae (Xoo). Zinc finger proteins harboring the motif with three conserved cysteine residues and one histidine residue (CCCH) belong to a large family. Although at least 67 CCCH-type zinc finger protein genes have been identified in the rice genome, their functions are poorly understood. Here, we report that one of the rice CCCH-type zinc finger proteins, C3H12, containing five typical CX8-CX5-CX3-H zinc finger motifs, is involved in the rice-Xoo interaction. Activation of C3H12 partially enhanced resistance to Xoo, accompanied by the accumulation of jasmonic acid (JA) and induced expression of JA signaling genes in rice. In contrast, knockout or suppression of C3H12 resulted in partially increased susceptibility to Xoo, accompanied by decreased levels of JA and expression of JA signaling genes in rice. C3H12 colocalized with a minor disease resistance quantitative trait locus to Xoo, and the enhanced resistance of randomly chosen plants in the quantitative trait locus mapping population correlated with an increased expression level of C3H12. The C3H12 protein localized in the nucleus and possessed nucleic acid-binding activity in vitro. These results suggest that C3H12, as a nucleic acid-binding protein, positively and quantitatively regulates rice resistance to Xoo and that its function is likely associated with the JA-dependent pathway.
A large number of genes are involved in plant resistance to pathogen invasion. These genes can be classified into two groups based on their role in defense signaling transduction: the receptor genes that include gene-for-gene disease resistance (R) genes, and pattern recognition receptor (PRR) genes and defense-responsive genes (Kou and Wang, 2010). PRRs recognize pathogen-associated molecular patterns (PAMPs), which are relatively conserved during evolution, to initiate PAMP-triggered immunity or basal resistance, whereas R proteins perceive effectors, which are more pathogen species or race specific as compared with PAMPs, to initiate effector-triggered immunity or race-specific resistance (Jones and Dangl, 2006; Thomma et al., 2011). However, there is a continuum between PAMP-triggered immunity and effector-triggered immunity, because PAMPs and effectors as well as PRRs and R proteins cannot strictly be maintained (Thomma et al., 2011). The proteins encoded by defense-responsive genes function in the pathways initiated by PRR or R proteins.
Although different types of R, PRR, and defense-responsive genes have been characterized, the roles of CCCH-type zinc finger protein genes in host-pathogen interactions are poorly understood. CCCH-type zinc finger proteins belong to a superfamily divided into nine classes (C2H2, C8, C6, C3HC4, C2HC, C2HC5, C4, C4HC3, and CCCH) according to the numbers of conserved Cys (C) and His (H) residues and the spacing between these conserved residues (Berg and Shi, 1996; Takatsuji, 1998). A CCCH-type zinc finger protein usually contains one or more tandemly arranged zinc-binding motifs characterized by three Cys residues followed by one His, with the characteristics CX5-14-CX4-5-CX3-H (where X indicates any amino acid; Blackshear, 2002; Wang et al., 2008a). The CCCH-type zinc finger genes are widely present in eukaryotes (Anderson et al., 1993; Tabara et al., 1999; Carballo et al., 2000; Li et al., 2001; Gao et al., 2002; Kong et al., 2006; Guo et al., 2009), and there are at least 68 genes in Arabidopsis (Arabidopsis thaliana) and 67 in rice (Oryza sativa; Wang et al., 2008a). This type of protein has been reported to regulate genes at the posttranscriptional or transcriptional level (Li et al., 2001; Wang et al., 2008b).
Most of the characterized CCCH-type zinc finger proteins are associated with RNA metabolism, including RNA cleavage, RNA degradation, RNA polyadenylation, or RNA export, by binding to RNA (Chen and Shyu, 1995; Bai and Tolias, 1996; Taylor et al., 1996; Lai et al., 1999, 2006; Hurt et al., 2009). In Arabidopsis, the CCCH-type protein HUA1 is involved in the processing of AGAMOUS pre-mRNA as an RNA-binding protein during flower development (Li et al., 2001; Cheng et al., 2003). Another Arabidopsis CCCH-type protein, AtTZF1, shuttling between the nucleus and cytoplasmic foci, can bind both DNA and RNA in vitro and is likely involved in GA/abscisic acid-mediated developmental and environmental responses through DNA or RNA regulation (Pomeranz et al., 2010).
Thus far, only a few CCCH-type zinc finger proteins have been reported to transcriptionally regulate gene expression. Arabidopsis PEI1, required for embryo development, can bind to DNA and functions as an embryo-specific transcription factor (Li and Thomas, 1998). Rice OsLIC (for Oryza sativa LEAF AND TILLER ANGLE INCREASED), containing only one CCCH-type zinc finger motif, can bind to both DNA and RNA and putatively controls plant architecture as a transcription activator (Wang et al., 2008b).
According to the strength of the plant response, plant resistance against pathogen invasion is divided into two major categories: the qualitative (or complete or vertical) resistance conferred by R genes, and the quantitative (or partial or horizontal) resistance mediated by multiple genes or quantitative trait loci (QTLs). Quantitative resistance is frequently a broad-spectrum and durable resistance and is the only form of resistance for plants against some types of pathogens (Hu and Wang, 2009; Kou and Wang, 2010). Although a large number of resistance QTLs have been identified, only a limited number of QTLs have been characterized recently, because of the genetic complexity of this type of resistance (Hayashi et al., 2010; Kou and Wang, 2010; Kou et al., 2010; Fu et al., 2011). A strategy of validation and functional analysis of the QTLs has been established to characterize resistance QTLs in rice; this strategy integrates the linkage map, expression profile, functional complementation analysis, and allele comparison and has provided the approach to characterize the genes underlying minor resistance QTLs (Hu et al., 2008; Kou et al., 2010; Kou and Wang, 2012).
Bacterial blight caused by Xanthomonas oryzae pv oryzae (Xoo) is one of the most devastating rice diseases worldwide. A previous study revealed that rice cDNA EI38D7 (GenBank accession no. BF108310), corresponding to C3H12 (locus identifier LOC_Os01g68860; http://rice.plantbiology.msu.edu/index.shtml) based on the reported naming system for the rice CCCH-type zinc finger family (Wang et al., 2008a), is a defense-responsive gene; its expression was induced in rice resistance against Xoo (Zhou et al., 2002). In addition, EI38D7 colocalized with a minor resistance QTL on chromosome 1, based on bioinformatic analysis (Xiong et al., 2002). These results suggest that C3H12 may be involved in quantitative resistance. To evaluate this inference, we monitored C3H12 expression, analyzed its role in the rice-Xoo interaction, and comapped it with resistance QTLs. These analyses suggest that C3H12 encodes a nucleic acid-binding protein and appears to contribute to quantitative resistance with a small effect; its mediated resistance is associated with the activation of a jasmonic acid (JA)-dependent pathway.
RESULTS
Modulating C3H12 Expression Influenced Rice Response to Xoo
Comparative analysis of the genomic and cDNA sequences reveals that C3H12 from rice variety Minghui 63 consists of seven exons and six introns and putatively encodes a protein consisting of 439 amino acids (GenBank accession no. JF799943; Supplemental Fig. S1). Sequence analysis showed that C3H12 was a CCCH-type zinc finger protein (Fig. 1). Functionally analyzed CCCH-type zinc finger proteins contain one to seven CCCH-type zinc finger motifs (Anderson et al., 1993; Taylor et al., 1996; Li et al., 2001; Tacahashi et al., 2003; Wang et al., 2008b; He et al., 2009; Hurt et al., 2009). The C3H12 protein contains five typical CX8-CX5-CX3-H zinc finger motifs (Fig. 1).
Figure 1.
Structural features of the C3H12 protein. A, Relative locations of CCCH-type zinc finger motifs in the C3H12 protein. B, Amino acid sequence alignment of CCCH-type zinc finger motifs in C3H12 (C3H12-ZF), Arabidopsis HUA1 (HUA1-ZF; National Center for Biotechnology Information [http://www.ncbi.nlm.nih.gov] protein accession no. NP_187874), rice OsDOS (OsDOS-ZF; Q9FU27), rice OsLIC (OsLIC-ZF; Q5Z807), cotton GhZFP1 (GhZFP1-ZF; AAX20386), cucumber (Cucumis sativus) CsSEF1 (CsSEF1-ZF; CAI30889), human HsTTP (HsTTP-ZF; P26651), and mouse MmTTP (MmTTP-ZF; P22893).
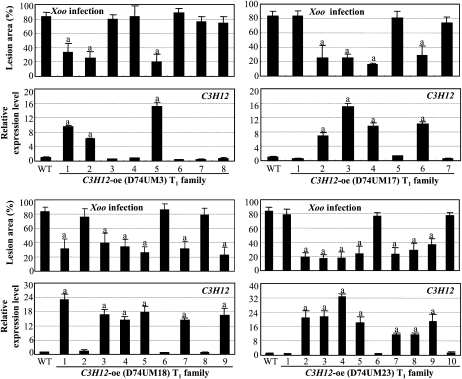
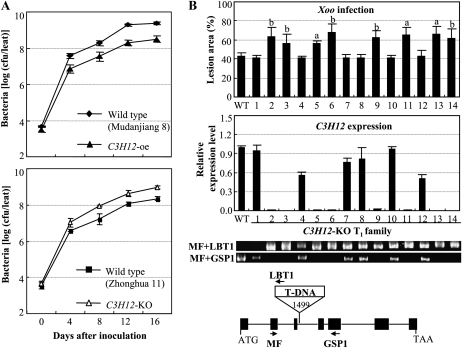
To ascertain whether C3H12 was involved in the rice-Xoo interaction, C3H12 was overexpressed in susceptible rice variety Mudanjiang 8. Twenty-seven independent positive transformants, named D74UM, were obtained. Nineteen of the 27 T0 plants showed significantly enhanced (P < 0.05) resistance to Xoo strain PXO61, with lesion areas ranging from 3% to 52% (average of 41%), compared with 65% for wild-type Mudanjiang 8 (Supplemental Table S1). To confirm that the enhanced resistance of the transgenic plants was due to overexpression of C3H12, four T1 families from D74UM3, D74UM17, D74UM18, and D74UM23 that all carried a single copy of the transgene (Supplemental Fig. S2) were further analyzed individually for their resistance to PXO61 and the C3H12 expression level. The enhanced resistance was associated with overexpression of C3H12 in all four T1 families (Fig. 2). The correlation between disease area and C3H12 expression level in the C3H12-overexpressing (oe) plants shown in Figure 2 was −0.839, significant at α = 0.01 (n = 34). The growth rate of bacteria in C3H12-oe plants was 5.4- to 10.3-fold lower than that in wild-type Mudanjiang 8 at 4 to 16 d after infection (Fig. 3A). The cosegregation of enhanced resistance and increased C3H12 expression suggest that C3H12 is involved in rice resistance against Xoo.
Figure 2.
Enhanced resistance to Xoo strain PXO61 is associated with the overexpression of C3H12 in four T1 families. Bars represent means (three to five replicates for lesion area and three replicates for gene expression) ± sd. The letter “a” above the bars indicates that a significant difference between transgenic plants and wild-type (WT) Mudanjiang 8 was detected at P < 0.01.
Figure 3.
Different transgenic plants showed different responses to Xoo infection. A, Growth of PXO61 in leaves of C3H12-oe (D74UM18; T3 generation) and C3H12-KO (04Z11KK69; T2 generation) plants. Bacterial populations were determined from three leaves at each time point by counting colony-forming units (cfu). B, The increased susceptibility of the C3H12-KO mutant to Xoo strain PXO347 was associated with insertion of T-DNA in C3H12 and marked suppression of C3H12 expression. Bars represent means (three to five replicates for lesion area and three replicates for gene expression) ± sd. The letter “a” or “b” above the bars indicates that a significant difference between mutant plants and wild-type (WT) Zhonghua 11 was detected at P < 0.01 or P < 0.05, respectively. ATG and TAA are the translation start codon and translation stop codon, respectively. The T-DNA was inserted at the 1,499 site of C3H12. Arrows indicate PCR primers used for examination of the mutant.
To further examine the role of C3H12 in the rice-Xoo interaction, a C3H12-knockout (KO) mutant (04Z11KK69), which had a T-DNA inserted into the third intron of C3H12, from the Rice Mutant Database was analyzed (Fig. 3B; Zhang et al., 2006). This C3H12-KO line had the genetic background of Zhonghua 11 (Wu et al., 2003). We obtained 14 T1 plants from the C3H12-KO mutant, including eight homozygous C3H12-KO plants, five heterozygous C3H12-KO plants, and one wild-type sibling from the C3H12-KO-segregating population, which were examined by PCR amplification using a gene-specific and T-DNA primer pair (Fig. 3B). The plants were inoculated with Xoo at booting stage. All the homozygous C3H12-KO plants (plants 2, 3, 5, 6, 9, 11, 13, and 14) showed significantly increased susceptibility (P < 0.05), with an average lesion area of 62.4% ± 4.3% compared with 42.9% ± 3.1% for the wild-type Zhonghua 11, whereas the heterozygous C3H12-KO plants (plants 4, 7, 8, 10, and 12) and wild-type siblings (plant 1) had no significant differences (P > 0.05) from wild-type plants in response to Xoo infection (Fig. 3B). The expression of C3H12 in the homozygous C3H12-KO plants was dramatically reduced, whereas C3H12 expression in heterozygous C3H12-KO plants and wild-type siblings was not influenced or only partially influenced as compared with wild-type Zhonghua 11 (Fig. 3B). The bacteria growth rate in C3H12-KO plants was 2.7- to 5.4-fold higher than that in the wild-type Zhonghua 11 at 4 to 16 d after infection (Fig. 3A). All these results confirm that C3H12 acts as a positive regulator in the rice response to Xoo infection.
C3H12 Colocalized with a Minor Disease Resistance QTL
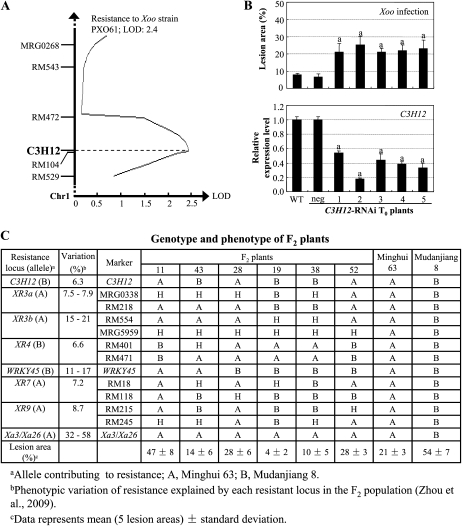
The indica rice variety Minghui 63 carries the R gene Xa3/Xa26 for resistance to Xoo strain PXO61, and the japonica variety Mudanjiang 8 is susceptible to PXO61 (Sun et al., 2004). However, Xa3/Xa26 can mediate a higher level and a more broad-spectrum resistance to Xoo in the Mudanjiang 8 background than in the Minghui 63 background (Cao et al., 2007; Zhou et al., 2009). We mapped C3H12 in an F2 population developed from a cross between Mudanjiang 8 and Minghui 63 that had been used to screen gene loci affecting genetic background-controlled disease resistance conferred by Xa3/Xa26 (Zhou et al., 2009). C3H12 colocalized with the curve peak of a minor resistance QTL against PXO61 on chromosome 1 (Fig. 4A). This QTL explained 6.3% of the phenotypic variation of resistance in the mapping population.
Figure 4.
Association of C3H12 with a bacterial resistance QTL. A, Colocalization of C3H12 and a minor resistance QTL against Xoo strain PXO61. B, Increased susceptibility to Xoo strain PXO61 was associated with suppression of C3H12 in C3H12-RNAi (38Ri) plants. neg, Negative transformant. Bars represent means (five replicates for lesion area and three replicates for gene expression) ± sd. The letter “a” above the bars indicates that a significant difference between transgenic plants and wild-type (WT) Minghui 63 was detected at P < 0.01. C, F2 plants carrying the C3H12 allele from Mudanjiang 8 were less susceptible to PXO61 than the plants carrying the C3H12 allele from Minghui 63 at relatively consistent genetic background for other resistance loci.
To further examine whether C3H12 was involved in quantitative resistance, we suppressed C3H12 in the resistant parent Minghui 63 of the mapping population using the RNA interference (RNAi) strategy. Six independent transformants, named 38Ri, were obtained. The C3H12 transcript levels in the positive C3H12-RNAi plants were 17.9% to 53.9% of that in wild-type plants. The five positive plants showed significantly increased (P < 0.01) susceptibility to Xoo strain PXO61, with lesion areas ranging from 21% to 25% versus 8% for the wild-type Minghui 63 (Fig. 4B). The increased susceptibility was associated with suppression of C3H12 (Fig. 4B). The correlation between disease area and C3H12 expression level in the C3H12-RNAi plants shown in Figure 4B was –0.975, significant at α = 0.01 (n = 6). These results suggest that C3H12 may be involved in regulating quantitative resistance against Xoo.
However, the resistance allele at the QTL was from susceptible Mudanjiang 8. Comparative sequence analysis showed that the C3H12 alleles in Minghui 63 and Mudanjiang 8 had six nucleotide substitutions in introns (Supplemental Fig. S3); thus, the C3H12 alleles of the two rice varieties encode an identical protein. The promoter region of C3H12 in Mudanjiang 8 had 12 nucleotide substitutions and an eight-nucleotide insertion as compared with that in Minghui 63 (Supplemental Fig. S3). The C3H12 allele and its promoter in rice variety Zhonghua 11, which was also used as a recipient of the transgene, had an identical sequence to that in Mudanjiang 8 (Supplemental Fig. S3). These results suggest that the C3H12 allele putatively contributing to the resistant locus may result from an expressional difference during the rice-Xoo interaction, as compared with its susceptible allele in the mapping population.
Xoo Infection Influenced C3H12 Expression
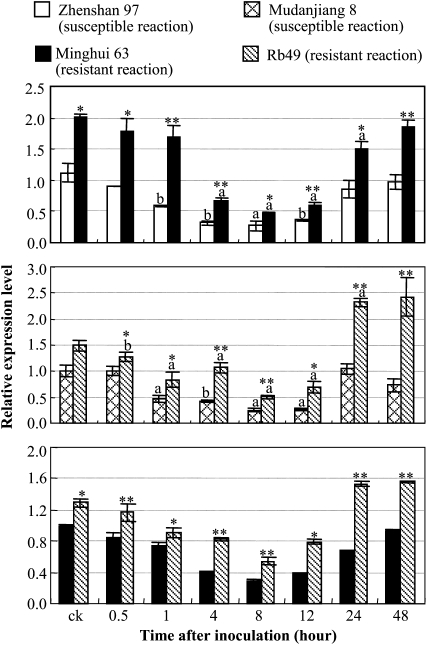
To test whether C3H12 had different expression patterns in resistant and susceptible reactions, we analyzed C3H12 expression in two pairs of rice lines after PXO61 infection. Rice variety Zhenshan 97 is susceptible to PXO61 and is an indica variety, as is the moderately resistant Minghui 63 carrying R gene Xa3/Xa26 (Cao et al., 2007). Rb49 is a transgenic line carrying a single copy of Xa3/Xa26 driven by its native promoter, with the genetic background of susceptible rice variety Mudanjiang 8; it is more resistant to Xoo than Minghui 63, the donor of Xa3/Xa26 (Sun et al., 2004; Cao et al., 2007). C3H12 showed a similar expression pattern in both resistant and susceptible rice lines after Xoo infection (Fig. 5). Its expression was rapidly suppressed at 1 to 12 h after infection and then returned to the basal level or was induced at 24 to 48 h after infection. However, the expression level of C3H12 was significantly higher (P < 0.05) in resistant lines than in susceptible lines either with or without pathogen infection. Interestingly, with the presence of Xa3/Xa26, the expression level of C3H12 in the Mudanjiang 8 background (rice line Rb49) was significantly higher (P < 0.05) than that in Minghui 63 (Fig. 5).
Figure 5.
C3H12 expression in response to pathogen infection. Plants were inoculated with Xoo strain PXO61 at booting stage. ck, Before inoculation. Bars represent means (three replicates) ± sd. The letter “a” or “b” above the bars indicates that, in the same rice line, a significant difference between Xoo-infected and noninfected plants was detected at P < 0.01 or P < 0.05, respectively. Two or one asterisks indicate that a significant difference between two rice lines with the same treatment was detected at P < 0.01 or P < 0.05, respectively.
Twenty-one randomly chosen F2 plants generated from the cross between Mudanjiang 8 and Minghui 63 and segregated for R gene Xa3/Xa26 to Xoo were further analyzed for the relationship between resistance and C3H12 expression level. Increased C3H12 expression was correlated (r = −0.5, n = 21, significant at α = 0.05) with the enhanced resistance in these F2 plants (Supplemental Fig. S4). The genotypes of some F2 plants at the resistance loci were further analyzed using gene markers or simple sequence repeat markers flanking resistance QTLs in this F2 population (Zhou et al., 2009). Several F2 plants that had relatively consistent genetic backgrounds at major resistance loci (explaining more than 10% of the phenotypic variation of resistance in the mapping population) but had the C3H12 allele from either of the parents were identified (Fig. 4C). Plant 43, which carried the C3H12 allele from Mudanjiang 8 and had significantly increased C3H12 transcripts, showed enhanced resistance to Xoo, as compared with plant 11, which carried the C3H12 allele from Minghui 63 and had significantly reduced C3H12 transcripts (Fig. 4C; Supplemental Fig. S4). There was a similar situation for plants 19 and 38, which carried the C3H12 allele from Mudanjiang 8, as compared with plants 28 and 19, which carried the C3H12 allele from Minghui 63. These results suggest that rice resistance is associated with a higher level of C3H12 transcripts and that a higher level of C3H12 expression is contributed by the allele from Mudanjiang 8. This inference is also consistent with the analysis that the resistance allele at the QTL putatively contributed by C3H12 was from the Mudanjiang 8 background in the mapping population (Fig. 4A).
C3H12 Induced the Expression of a Set of Defense-Responsive Genes
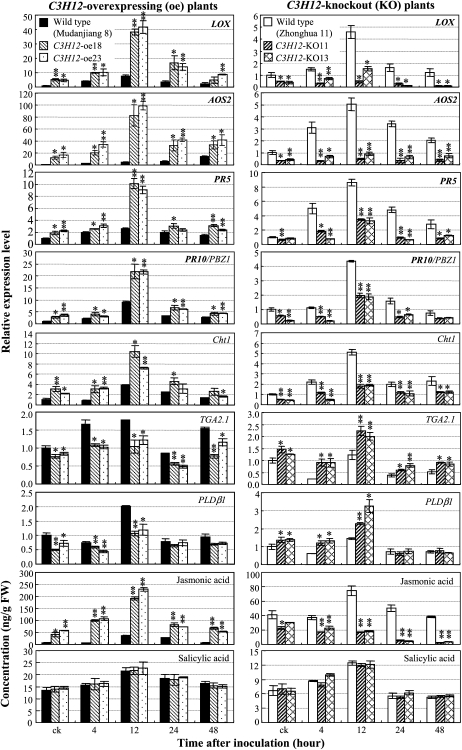
To dissect a possible defense pathway in which C3H12 was involved, we analyzed the expression of a set of pathogen-induced defense-responsive genes in different transgenic plants after infection of Xoo strain PXO61. PAL1 (for Phe ammonia lyase 1; GenBank accession no. X16099), ICS1 (for isochorismate synthase 1; AK120689), PAD4 (for phytoalexin-deficient 4; CX118864), PR1a (for acidic pathogenesis-related [PR] protein 1; AJ278436), and NH1 (for Arabidopsis NPR1 homolog 1; AY9123983) were associated with the salicylic acid (SA)-dependent pathway (Qiu et al., 2007; Yuan et al., 2007; Shen et al., 2010). LOX (for lipoxygenase; D14000) and AOS2 (for allene oxide synthase 2; AY062258) are involved in JA synthesis (Peng et al., 1994; Mei et al., 2006). PR5 (P28493), PR10/PBZ1 (D38170), and Cht1 (for chitinase 1; Q42993) appeared to function both in JA- and SA-dependent pathways (Qiu et al., 2007; Xiao et al., 2009; Shen et al., 2010). TGA2.1 (AB051295), PLDβ1 (for phospholipase D β1; AJ419630), NRR (for negative regulator of resistance; AY846391), and WRKY62 (DQ298182) were negative regulators in the rice-Xoo interaction (Chern et al., 2005; Fitzgerald et al., 2005; Peng et al., 2008; Yamaguchi et al., 2009).
PXO61 infection induced the expression of LOX, AOS2, PR5, PR10, and Cht1 both in wild-type Mudanjiang 8 and Zhonghua 11 and transgenic plants (Fig. 6). However, the expression levels of the five genes were significantly higher (P < 0.05) in C3H12-oe plants than in wild-type plants both before and after infection. In contrast, the expression levels of the five genes were significantly lower (P < 0.05) in C3H12-KO plants than in wild-type plants either before or after infection. The expression of rTGA2.1 and OsPLDβ1 was slightly influenced after PXO61 infection in both wild-type and transgenic plants (Fig. 6). Nevertheless, the expression levels of the two genes were significantly lower (P < 0.05) in C3H12-oe plants and significantly higher (P < 0.05) in C3H12-KO plants than in corresponding wild-type plants both before and after infection. The expression of PAL1, ICS1, PAD4, PR1a, NH1, NRR, and WRKY62 in transgenic plants showed no obvious difference from that in wild-type plants (data not shown). These results suggest that C3H12 may be involved in a JA-dependent signaling pathway in the rice-Xoo interaction.
Figure 6.
Transcriptionally modulating C3H12 influenced the expression of a set of defense-responsive genes and the accumulation of JA. Transgenic and wild-type plants were inoculated with Xoo strain PXO61 at booting stage. Bars represent means (three replicates) ± sd. Two or one asterisks indicate that a significant difference between transgenic and wild-type plants at the same time point was detected at P < 0.01 or P < 0.05, respectively. ck, Before inoculation; FW, fresh weight.
C3H12 Promoted the Accumulation of JA
To gain further insight into the relationship between C3H12 and a JA-dependent defense pathway, we quantified the concentrations of the endogenous JA in the leaves of the same plants used for analyzing the expression of defense-responsive genes after infection of Xoo strain PXO61 (Fig. 6). The endogenous level of JA was markedly induced by PXO61 infection in both C3H12-oe and wild-type plants, but the JA level was significantly higher (P < 0.05) in C3H12-oe plants than in wild-type Mudanjiang 8 both before and after Xoo infection. In contrast, the JA level was significantly lower (P < 0.01) in the C3H12-KO plants than in wild-type Zhonghua 11 after Xoo infection. Consistent with the expression patterns of defense-responsive genes functioning in the SA-dependent pathway, modulating C3H12 expression did not influence the endogenous level of SA, although Xoo infection slightly induced SA accumulation in both transgenic and wild-type plants (Fig. 6). These results suggest that C3H12-mediated disease resistance may be associated with the JA-dependent pathway.
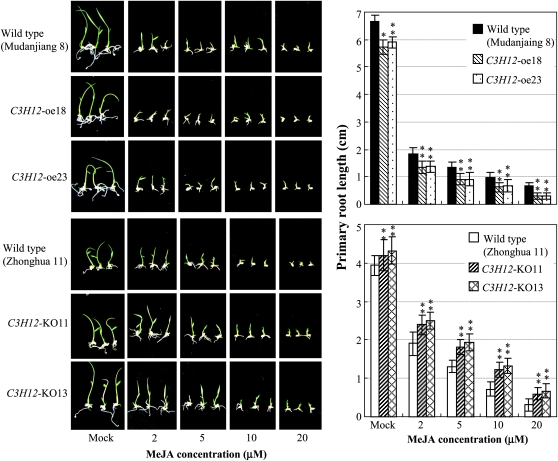
JA inhibits root elongation, and this property has been frequently used in JA synthesis- and signaling-related mutant selection (Feys et al., 1994; Lorenzo et al., 2004). The effect of methyl jasmonate (MeJA) on the root development of C3H12 transgenic plants also supports the inference that C3H12 regulates the JA-dependent pathway. A seed germination assay showed that the root elongation of the C3H12-oe seeds became more sensitive to MeJA treatment than the wild-type seeds, whereas the root elongation of C3H12-KO seeds was less influenced by MeJA treatment than the wild-type seeds (Fig. 7). Taken together, these results suggest that C3H12 appears to positively regulate the JA-dependent pathway.
Figure 7.
C3H12-oe and C3H12-KO plants showed opposite responses to MeJA treatment in primary root development. Two or one asterisks indicate that a significant difference between C3H12-oe or C3H12-KO and wild-type plants at the same time point was detected at P < 0.01 or P < 0.05, respectively. Mock, Without supplementation of MeJA.
C3H12 Had Nucleic Acid-Binding Ability
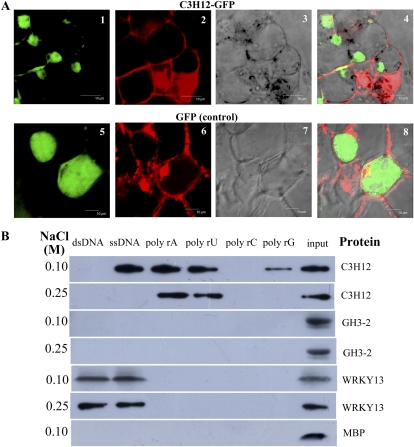
To examine the possible biochemical function of C3H12 protein, its subcellular localization was first analyzed by fusing the C3H12 coding region with the GFP gene. The C3H12-GFP fusion gene was transiently expressed in onion (Allium cepa) epidermal cells. The green fluorescent signal of the C3H12-GFP protein was localized predominantly in the nucleus of the cells, whereas the control GFP was uniformly presented throughout the cytoplasm (Supplemental Fig. S5). The C3H12-GFP fusion gene was further expressed in rice calli. A similar result as in onion epidermal cells was obtained. The C3H12-GFP protein was mainly localized in the nucleus of rice cells, whereas GFP was largely expressed in the cytoplasm of rice cells (Fig. 8A). These results suggest that C3H12 may function in the nucleus.
Figure 8.
Analyses of the biochemical function of C3H12. A, C3H12 localized in the nuclei of rice callus cells. Panel 1, C3H12-GFP expression; panel 2, staining of the cell with propidium iodide as a control; panel 3, transmission image; panel 4, overlay of panels 1, 2, and 3; panel 5, GFP expression; panel 6, staining of cell with propidium iodide as a control; panel 7, transmission image; panel 8, overlay of panels 5, 6, and 7. Bars = 10 μm. B, The MBP-tagged C3H12 protein bound to various nucleic acids at buffer containing 0.1 or 0.25 m NaCl. This experiment was repeated three times, and similar results were obtained. dsDNA, Double-stranded DNA (calf thymus DNA); ssDNA, single-stranded DNA (calf thymus DNA).
Many proteins harboring a CCCH-type zinc finger motif bind to RNA or DNA to perform their functions (Hall, 2005; Wang et al., 2008b). To understand whether C3H12 had one of these activities, we performed RNA- and DNA-binding assays using recombinant C3H12 protein purified from Escherichia coli and a transactivation activity assay in Saccharomyces cerevisiae. Under moderate salt concentrations (0.1 and 0.25 m NaCl) that resemble the in vivo situation (Li et al., 2001), the maltose-binding protein (MBP)-tagged C3H12 bound to certain types of ribohomopolymers (poly rA, rU, rC, or rG) and DNA (Fig. 8B). In the buffer containing 0.1 m NaCl, MBP-tagged C3H12 bound to single-stranded DNA, poly rA, poly rU, and poly rG but not double-stranded DNA and poly rC. In the buffer containing 0.25 m NaCl, MBP-tagged C3H12 bound only to poly rA and poly rU. The control MBP did not bind to any type of nucleic acids under the same experimental conditions. As a negative rice protein control, rice GH3-2, which is an indole-3-acetic acid-amido synthetase and catalyzes the formation of indole-3-acetic acid-amino acid (Fu et al., 2011), did not bind to any type of nucleic acid (Fig. 8B). As a positive rice protein control, rice WRKY13, which is a transcription-like regulator and binds to the promoters of some defense-responsive genes (Qiu et al., 2007, 2009), bound only to double- and single-stranded DNA but not to any type of ribohomopolymer (Fig. 8B). In the transactivation activity assay, C3H12 showed no activity of transactivation in yeast cells as compared with the positive control, rice transcription factor OsbZIP23 (Supplemental Fig. S6). This result is consistent with that showing no conserved activation domain identified in the C3H12 protein based on bioinformatic analysis. These results suggest that C3H12 may function as a nucleic acid-binding protein.
DISCUSSION
Although CCCH-type zinc finger proteins belong to a large family, their functions in plants are poorly understood. Only a few CCCH-type proteins functioning in the regulation of development, growth, or abiotic stress responses have been characterized in Arabidopsis (Li and Thomas, 1998; Li et al., 2001; Schmitz et al., 2005; Sun et al., 2007; Kim et al., 2008; Pomeranz et al., 2010) and rice (Kong et al., 2006; Wang et al., 2008b). Although the expression profiles of CCCH-type zinc finger protein genes in Arabidopsis and rice suggest that most members in one plant-specific subfamily of the CCCH-type gene family may be involved in abiotic or biotic stress tolerance (Wang et al., 2008a), the only evidence, so far, that CCCH-type protein is involved in plant-pathogen interaction involves cotton (Gossypium hirsutum) GhZFP1 (Guo et al., 2009). The GhZFP1 positively regulates resistance to the fungal pathogen Rhizoctonia solani in tobacco (Nicotiana tabacum) in addition to enhancing tobacco tolerance to salt stress. Here, we provide, to our knowledge, the first evidence that CCCH-type zinc finger protein is also involved in rice-pathogen interaction. Rice C3H12 functions as a positive regulator to mediate resistance against the bacterial pathogen Xoo.
C3H12-Mediated Disease Resistance Is Associated with Activation of the JA-Dependent Pathway
JA and SA are two well-known phytohormones involved in host-pathogen interactions. In general, plant resistance to biotrophic and hemibiotrophic pathogens is frequently controlled by the SA-dependent pathway, whereas resistance to necrotrophic pathogens is frequently regulated by the JA/ethylene-dependent pathway (Bari and Jones, 2009). Xoo is a biotrophic pathogen. The results presented here suggest that C3H12-mediated bacterial resistance may be dependent on JA but not SA. This inference is supported by the following evidence. First, the enhanced resistance of C3H12-oe plants was associated with increased transcripts of JA synthesis-related genes (LOX and AOS2) and the accumulation of JA but not with the expression of SA synthesis-related genes (PAL1 and ICS1) and SA signaling-related genes (PAD4, PR1a, and NH1) and the accumulation of SA. Second, the hypersensitivity of C3H12-oe plants and the hyposensitivity of C3H12–KO plants to MeJA treatment on root development suggest that C3H12 positively regulates a JA-dependent pathway.
Rice resistance against Xoo appears to be regulated by multiple SA- or JA-related pathways. The enhanced rice resistance to Xoo by activating WRKY13 or suppressing OsDR10 or MPK6 that negatively regulates systemic acquired resistance and positively regulates local resistance is associated with activation of the SA-dependent pathway and suppression of the JA-dependent pathway (Qiu et al., 2007, 2008; Xiao et al., 2009; Shen et al., 2010). In addition, enhanced rice resistance to Xoo can also be achieved by activating MPK6 or suppressing WRKY45-1 or EDR1, which is associated with activation of both JA- and SA-dependent pathways (Tao et al., 2009; Shen et al., 2010, 2011). Furthermore, suppressing the auxin-dependent pathway by activating either GH3-2 or GH3-8 can enhance rice resistance against Xoo, which is accompanied by suppression of both SA- and JA-dependent pathways (Ding et al., 2008; Fu et al., 2011). Like the C3H12-oe plants, the enhanced rice resistance to Xoo by activating WRKY45-2, which is the allele of WRKY45-1 in indica rice, is associated with increased accumulation of JA but not SA (Tao et al., 2009). Interestingly, activation of WRKY45-2 did not significantly influence (P > 0.05) C3H12 expression, whereas suppression of C3H12 significantly repressed (P < 0.01) WRKY45-2 expression (Supplemental Fig. S7). These results suggest that C3H12 and WRKY45-2 may function in the same defense transduction pathway with WRKY45-2, localizing downstream of C3H12. Furthermore, multiple mechanisms may be involved in rice resistance against Xoo, although this inference needs to be confirmed by analyzing double or triple mutants.
C3H12 May Function as an RNA-Binding Protein
The zinc finger is a characterized motif for nucleic acid binging (Hall, 2005). Most of the characterized CCCH-type zinc finger proteins are associated with RNA metabolism by binding to the target mRNA (Cheng et al., 2003; Delaney et al., 2006; Lai et al., 2006; Hurt et al., 2009), and only two, Arabidopsis PEI1 and rice OsLIC, are suggested to transcriptionally regulate gene expression by binding to DNA (Li and Thomas, 1998; Wang et al., 2008b). In addition, OsLIC harbors an EELR-type activation domain for regulating gene transcription in yeast (Wang et al., 2008b). Consistent with other characterized CCCH-type proteins, C3H12 localized in the nucleus and possessed the capability of nucleic acid binding, suggesting its potential role in RNA or DNA regulation. However, C3H12 did not display transactivation activity in yeast cells and preferentially bound to poly rA and poly rU but not double- and single-stranded DNA in the buffer containing a relatively higher physiologic concentration of NaCl, suggesting that C3H12 may function as an RNA-binding protein. This assumption is also supported by the evidence that C3H12 does not harbor a known activation domain based on bioinformatic analysis. According to the nuclear localization and nucleic acid-binding specificity of the C3H12 protein, a potential role in nuclear RNA regulation is considered.
C3H12 may regulate disease resistance by promoting the cleavage or degradation of mRNAs of some defense-responsive genes whose encoded proteins function as negative regulators in the rice-Xoo interaction and thus remove the suppression of defense positive regulators. TGA2.1 is a transcriptional regulator and a negative player in rice resistance against Xoo; it functions upstream of the defense-responsive gene PR10, which positively regulates the rice defense response, by suppressing PR10 expression (Fitzgerald et al., 2005). PLDβ1, which appeared to be involved in phospholipid signaling, is also a negative regulator for the defense response; the resistance of PLDβ1-knockdown plants is associated with an increased expression of PR10 and chitinase genes, including Cht1, analyzed in this study (Yamaguchi et al., 2009). C3H12-mediated resistance was accompanied by reduced TGA2.1 and PLDβ1 transcripts and increased PR10 and Cht1 transcripts (Fig. 6). Thus, further studies may concentrate on whether C3H12 targets to the mRNA of some negative defense-responsive genes, such as TGA2.1 and PLDβ1, in the rice defense response.
C3H12 Confers Quantitative Resistance
Map-based cloning is a traditional method to find major resistance genes but is not efficient in isolating minor resistance QTLs, because of their small effect on disease resistance. According to an analysis using the strategy of validation and functional analysis of the QTL (Hu et al., 2008), we argue that C3H12 contributes to a minor resistance QTL against Xoo. This inference can be supported by the following evidence. First, C3H12 tightly colocalized with the curve peak of the resistance QTL based on the mapping analysis using a segregation population. Second, C3H12 only conferred a partial (or quantitative) resistance after activating it, suggesting its small effect on disease resistance. Third, enhanced resistance correlated with an increased expression level of C3H12 in F2 plants. Finally, suppressing C3H12 in the parent of the mapping population partially increased susceptibility to Xoo. As a positive regulator of rice resistance to Xoo, the function of C3H12 is associated with its transcriptional activation (Fig. 2). With the presence of R gene Xa3/Xa26, C3H12 showed a significantly higher expression level in the Mudanjiang 8 background than in the Minghui 63 background. This may explain why the resistance QTL underlying C3H12 was contributed by the allele from Mudanjiang 8 in the mapping population developed from the cross between Mudanjiang 8 and Minghui 63.
Genetic background influences the function of Xa3/Xa26 in resistance against Xoo; the Mudanjiang 8 background facilitates the function of Xa3/Xa26 more than does the Minghui 63 background (Sun et al., 2004; Cao et al., 2007). The function of Xa3/Xa26 is associated with its expression level: the higher its expression, the more resistant the plant; Xa3/Xa26 has a higher expression level in Mudanjiang 8 than in Minghui 63 (Cao et al., 2007). The resistance QTL underlying C3H12 has been proposed to be one of the loci that facilitate Xa3/Xa26 function in the Mudanjiang 8 background (Zhou et al., 2009). As discussed above, C3H12 may be involved in RNA regulation, and this gene is a potential candidate to study the differential regulation of Xa3/Xa26 expression in different genetic backgrounds.
Most of the characterized plant resistance QTLs, including those in rice, have small effects on disease resistance, which makes it difficult to use minor resistance QTLs that explain less than 10% of the phenotypic variation for breeding programs by marker-assisted selection (Kou and Wang, 2010). Our results here provide another example, in the limited list of the characterized resistance QTLs, that a single QTL with a minor effect may be used in breeding programs for disease resistance after manipulating its expression.
CONCLUSION
C3H12, encoding a CCCH-type zinc finger protein with nucleic acid-binding activity, confers quantitative resistance against rice bacterial blight disease, which is associated with a JA-dependent defense pathway. This research may be a pioneer for further understanding the molecular functions of CCCH-type zinc finger proteins in plant-pathogen interactions.
MATERIALS AND METHODS
Gene Isolation and Structural Analysis
To isolate the C3H12 gene, the cDNA fragment of C3H12, EI38D7 from rice (Oryza sativa indica) variety Minghui 63 (Zhang et al., 2005), was used to screen the genomic bacterial artificial chromosome library constructed with Minghui 63 tissues (Peng et al., 1998). A positive bacterial artificial chromosome clone, 16D13, was identified. A DNA fragment approximately 6 kb in length and harboring C3H12 was obtained from 16D13 by digestion with restriction enzyme HindIII and subcloned into vector pUC19. The subclone sub38a harboring C3H12 was sequenced. The structure of C3H12 was determined by comparatively sequencing the genomic DNA and cDNA. The cDNA harboring the full-length coding region was obtained using primers 38D75UF5 and 38D7stop (Supplemental Table S2), cloned into vector pGEM-T (Promega), and named 38F1c. The 5′ untranslated region was analyzed by 5′-RACE assays using the SMART RACE cDNA Amplification Kit (TaKaRa Biotechnology) using gene-specific primers (Supplemental Table S2) according to the manufacturer’s protocols. EI38D7 contained the 3′ untranslated region of C3H12.
Plant Transformation
The overexpressing construct of C3H12 was made by inserting a 6-kb DNA fragment (Supplemental Fig. S1) digested with KpnI and BamHI from subclone sub38a into vector pU1301, which contained a maize (Zea mays) ubiquitin gene promoter (Cao et al., 2007). To construct an RNAi vector of C3H12, a 538-nucleotide fragment amplified from Minghui 63 cDNA using primers 38D7RIF and 38D7RIR (Supplemental Table S2) was inserted into the pDS1301 vector (Yuan et al., 2007). The constructs were introduced into Agrobacterium tumefaciens strain EHA105 by electroporation. Agrobacterium-mediated transformation was performed according to a published protocol (Lin and Zhang, 2005). The C3H12-oe construct was transferred into rice variety Mudanjiang 8 (Oryza sativa japonica). The C3H12-RNAi construct was transferred into rice variety Minghui 63.
The copy number of transgenes in plants was determined by DNA gel-blot analysis using probe amplified from transformation marker gene Hygromycin Phosphotransferase (Supplemental Table S2). Total DNA isolated from transgenic plants was digested with restriction enzyme BamHI before electrophoresis.
Pathogen Inoculation
To evaluate bacterial blight disease, plants were inoculated with Xanthomonas oryzae pv oryzae strain PXO61 at the seedling or booting (panicle development) stage by the leaf-clipping method (Chen et al., 2002). Disease was scored by measuring the percentage lesion area (lesion length/leaf length) at 2 weeks after inoculation. The bacterial growth rate in rice leaves was determined by counting colony-forming units (Sun et al., 2004).
Analysis of Gene Expression
RNA gel-blot analysis was performed as described previously (Zhou et al., 2002). In brief, 20 μg of total RNA was used for this analysis. The cDNA fragment (EI38D7) of C3H12 was used as a hybridization probe. For analyzing gene expression after Xoo infection, 2-cm leaf fragments next to bacterial infection sites were used for RNA isolation. Quantitative reverse transcription (qRT)-PCR analysis was conducted as described previously (Qiu et al., 2007) using gene-specific primers (Supplemental Table S3). The expression level of the rice actin gene was used to standardize the RNA sample for each qRT-PCR. The expression level relative to the control is presented. For each gene, qRT-PCR assays were repeated at least twice, with each repeat having three replicates. When similar results were obtained in repeated experiments, only the result in one repetition is presented.
JA Treatment
Rice seeds used for germination assays were sterilized with 75% ethanol and 0.15% HgCl2 and pregerminated on half-strength Murashige and Skoog medium for 2 d. The identically sprouted seeds were transplanted on half-strength Murashige and Skoog plates supplemented with MeJA or without supplementation of MeJA (mock control) for 7 d. The seedlings were photographed, and the lengths of the primary roots were measured.
Subcellular Localization of C3H12
To produce the C3H12-GFP construct, the coding region of C3H12, obtained by PCR using gene-specific primers (Supplemental Table S2) and cDNA clone 38F1c as template, was cloned into vector pU1391, which carries a PUbi:GFP cassette (Shen et al., 2010). Transient expression of the fusion genes in white onion (Allium cepa) epidermal cells was performed by Agrobacterium-mediated transformation as described previously (Shen et al., 2010). The transformed epidermal cells were stained with 4,6′-diamidino-2-phenylindole, and the image was observed using a confocal microscope. The C3H12-GFP construct was also expressed in rice callus cells by Agrobacterium-mediated transformation (Yuan et al., 2011). The sliced calli were stained with propidium iodide before observation with a confocal microscope.
Comapping of C3H12 and Resistance QTLs
An F2 population consisting of 146 individuals developed from a cross between susceptible Mudanjiang 8 and Minghui 63 was used for analyzing the colocalization of C3H12 and resistance QTLs. This population had been used to study the quantitative disease resistance to Xoo strain PXO61, and a molecular linkage map containing 136 markers spanning a total of 1,631 centimorgan was developed using this population (Zhou et al., 2009). C3H12 was mapped on the molecular linkage map using a PCR-based derived cleaved amplification polymorphism sequence marker. The polymorphic PCR fragments were determined by electrophoresis of XhoI-digested PCR products amplified using primers 38D7dCAPSF and 38D7dCAPSR (Supplemental Table S2). QTL analysis was conducted using the computer program Windows QTL Cartographer version 2.0 for composite interval mapping at a threshold of logarithm of odds 2.0 (Wang and Zeng, 2003). The genotypes at resistance loci in some F2 plants of this population were analyzed by PCR amplification of polymorphic fragments using gene-specific primers (Supplemental Table S2) or primers for simple sequence repeat markers flanking resistance QTLs (Zhou et al., 2009).
In Vitro Nucleic Acid-Binding Assay
The coding region of C3H12 was obtained by PCR using primers 38D7CF2 and 38D7CR2 (Supplemental Table S2) and cDNA clone 38F1c as template and was cloned into the pMAL-c2X protein expression vector that harbors a MBP gene at the 5′ end of the multiple cloning site (New England Biolabs). The GH3-2 and WRKY13 vectors were from previous studies (Qiu et al., 2007; Fu et al., 2011). The MBP-tagged C3H12, MBP, WRKY13, and GH3-2 proteins were purified from Escherichia coli using QIAexpressionist (Qiagen). An in vitro nucleic acid-binding assay was performed as described previously (Yang et al., 1998). In brief, 500 ng of proteins was incubated with 20 μL of agarose bead-labeled poly rA, poly rU, poly rG, or poly rC or cellulose bead-labeled double-stranded and single-stranded calf thymus DNA (Sigma-Aldrich) in 500 μL of ribohomopolymer assay binding buffer (10 mm Tris, pH 7.4, 2.5 mm MgCl2, 0.5% Triton X-100, and 0.1 or 0.25 m NaCl) with 1 mg mL−1 heparin. After incubation at 4°C for 10 min, the beads were washed five times in RHPA buffer and boiled in SDS protein-loading buffer. The proteins were detected with the anti-MBP antibody (ProteinTech Group) after separation by SDS-PAGE.
Transactivation Activity Assay
The transactivation activity of C3H12 was analyzed in yeast cells using the known rice transcription factor OsbZIP23 (Xiang et al., 2008) as a control. The coding regions of C3H12 and rice OsbZIP23 were amplified using primer pairs 38D7CF1/38D7CR and OsbZIP23F/OsbZIP23R, respectively (Supplemental Table S2). PCR product was sequenced, digested using NcoI and BamHI, and ligated into the pGBKT7 vector. The recombinant vector and the pGBKT7 empty vector (control) were transferred into Saccharomyces cerevisiae strain AH109 by yeast LiAc-mediated transformation according to the manufacturer’s protocol (BD Biosciences Clontech). Yeast transformants were screened by culture in synthetic dextrose-Trp-His-Ade medium.
Sequence Analysis
The CCCH-type zinc finger motif of C3H12 and other proteins was predicted by searching the Conserved Domains database (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). The sequence alignment was performed using Genedoc version 3.2 (http://www.psc.edu/biomed/genedoc).
Statistical Analysis
The significant differences between control and treatment of the samples were analyzed by the pairwise t test installed in the Microsoft Office Excel program. The correlation analysis between disease area and C3H12 expression level was performed using the CORREL analysis installed in the Microsoft Office Excel program.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number JF799943 (Minghui 63) for C3H12.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. The structures of C3H12 gene and rice DNA fragments for transformation.
Supplemental Figure S2. Southern-blot analysis of the copy numbers of transgene C3H12.
Supplemental Figure S3. Sequence comparison of the C3H12 gene and its promoter region from rice varieties Minghui 63, Mudanjiang 8, and Zhonghua 11.
Supplemental Figure S4. Relationship of C3H12 expression level and the resistance level in F2 plants.
Supplemental Figure S5. C3H12 localized in the nuclei of onion epidermal cells.
Supplemental Figure S6. C3H12 displayed no transactivation activity as compared with the positive control.
Supplemental Figure S7. C3H12 influenced the expression of defense-responsive gene WRKY45-2.
Supplemental Table S1. Resistance of T0 C3H12-overexpressing plants (D74UM) to Xoo strain PXO61 at booting stage.
Supplemental Table S2. PCR primers used for the construction of vectors, gene structure analysis, gene mapping, and transgene copy number analysis.
Supplemental Table S3. Primers used for quantitative PCR in gene expression analysis.
Supplementary Material
References
- Anderson JT, Wilson SM, Datar KV, Swanson MS. (1993) NAB2: a yeast nuclear polyadenylated RNA-binding protein essential for cell viability. Mol Cell Biol 13: 2730–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai C, Tolias PP. (1996) Cleavage of RNA hairpins mediated by a developmentally regulated CCCH zinc finger protein. Mol Cell Biol 16: 6661–6667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R, Jones JD. (2009) Role of plant hormones in plant defence responses. Plant Mol Biol 69: 473–488 [DOI] [PubMed] [Google Scholar]
- Berg JM, Shi Y. (1996) The galvanization of biology: a growing appreciation for the roles of zinc. Science 271: 1081–1085 [DOI] [PubMed] [Google Scholar]
- Blackshear PJ. (2002) Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochem Soc Trans 30: 945–952 [DOI] [PubMed] [Google Scholar]
- Cao Y, Ding X, Cai M, Zhao J, Lin Y, Li X, Xu C, Wang S. (2007) The expression pattern of a rice disease resistance gene xa3/xa26 is differentially regulated by the genetic backgrounds and developmental stages that influence its function. Genetics 177: 523–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballo E, Lai WS, Blackshear PJ. (2000) Evidence that tristetraprolin is a physiological regulator of granulocyte-macrophage colony-stimulating factor messenger RNA deadenylation and stability. Blood 95: 1891–1899 [PubMed] [Google Scholar]
- Chen CY, Shyu AB. (1995) AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci 20: 465–470 [DOI] [PubMed] [Google Scholar]
- Chen H, Wang S, Zhang Q. (2002) New gene for bacterial blight resistance in rice located on chromosome 12 identified from Minghui 63, an elite restorer line. Phytopathology 92: 750–754 [DOI] [PubMed] [Google Scholar]
- Cheng Y, Kato N, Wang W, Li J, Chen X. (2003) Two RNA binding proteins, HEN4 and HUA1, act in the processing of AGAMOUS pre-mRNA in Arabidopsis thaliana. Dev Cell 4: 53–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chern M, Canlas PE, Fitzgerald HA, Ronald PC. (2005) Rice NRR, a negative regulator of disease resistance, interacts with Arabidopsis NPR1 and rice NH1. Plant J 43: 623–635 [DOI] [PubMed] [Google Scholar]
- Delaney KJ, Xu R, Zhang J, Li QQ, Yun KY, Falcone DL, Hunt AG. (2006) Calmodulin interacts with and regulates the RNA-binding activity of an Arabidopsis polyadenylation factor subunit. Plant Physiol 140: 1507–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Cao Y, Huang L, Zhao J, Xu C, Li X, Wang S. (2008) Activation of the indole-3-acetic acid-amido synthetase GH3-8 suppresses expansin expression and promotes salicylate- and jasmonate-independent basal immunity in rice. Plant Cell 20: 228–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys B, Benedetti CE, Penfold CN, Turner JG. (1994) Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6: 751–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald HA, Canlas PE, Chern MS, Ronald PC. (2005) Alteration of TGA factor activity in rice results in enhanced tolerance to Xanthomonas oryzae pv. oryzae. Plant J 43: 335–347 [DOI] [PubMed] [Google Scholar]
- Fu J, Liu H, Li Y, Yu H, Li X, Xiao J, Wang S. (2011) Manipulating broad-spectrum disease resistance by suppressing pathogen-induced auxin accumulation in rice. Plant Physiol 155: 589–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G, Guo X, Goff SP. (2002) Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein. Science 297: 1703–1706 [DOI] [PubMed] [Google Scholar]
- Guo YH, Yu YP, Wang D, Wu CA, Yang GD, Huang JG, Zheng CC. (2009) GhZFP1, a novel CCCH-type zinc finger protein from cotton, enhances salt stress tolerance and fungal disease resistance in transgenic tobacco by interacting with GZIRD21A and GZIPR5. New Phytol 183: 62–75 [DOI] [PubMed] [Google Scholar]
- Hall TM. (2005) Multiple modes of RNA recognition by zinc finger proteins. Curr Opin Struct Biol 15: 367–373 [DOI] [PubMed] [Google Scholar]
- Hayashi N, Inoue H, Kato T, Funao T, Shirota M, Shimizu T, Kanamori H, Yamane H, Hayano-Saito Y, Matsumoto T, et al. (2010) Durable panicle blast-resistance gene Pb1 encodes an atypical CC-NBS-LRR protein and was generated by acquiring a promoter through local genome duplication. Plant J 64: 498–510 [DOI] [PubMed] [Google Scholar]
- He F, Dang W, Abe C, Tsuda K, Inoue M, Watanabe S, Kobayashi N, Kigawa T, Matsuda T, Yabuki T, et al. (2009) Solution structure of the RNA binding domain in the human muscleblind-like protein 2. Protein Sci 18: 80–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu K, Wang S. (2009) Rice disease resistance resources and genetic improvement. Zhang Q, , Strategies and Practice for Developing Green Super Rice. Science Press, Beijing, pp 35–57 [Google Scholar]
- Hu KM, Qiu DY, Shen XL, Li XH, Wang SP. (2008) Isolation and manipulation of quantitative trait loci for disease resistance in rice using a candidate gene approach. Mol Plant 1: 786–793 [DOI] [PubMed] [Google Scholar]
- Hurt JA, Obar RA, Zhai B, Farny NG, Gygi SP, Silver PA. (2009) A conserved CCCH-type zinc finger protein regulates mRNA nuclear adenylation and export. J Cell Biol 185: 265–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kim DH, Yamaguchi S, Lim S, Oh E, Park J, Hanada A, Kamiya Y, Choi G. (2008) SOMNUS, a CCCH-type zinc finger protein in Arabidopsis, negatively regulates light-dependent seed germination downstream of PIL5. Plant Cell 20: 1260–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Z, Li M, Yang W, Xu W, Xue Y. (2006) A novel nuclear-localized CCCH-type zinc finger protein, OsDOS, is involved in delaying leaf senescence in rice. Plant Physiol 141: 1376–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kou Y, Li X, Xiao J, Wang S. (2010) Identification of genes contributing to quantitative disease resistance in rice. Sci China Life Sci 53: 1263–1273 [DOI] [PubMed] [Google Scholar]
- Kou Y, Wang S. (2010) Broad-spectrum and durability: understanding of quantitative disease resistance. Curr Opin Plant Biol 13: 181–185 [DOI] [PubMed] [Google Scholar]
- Kou Y, Wang S. (2012) Toward an understanding of the molecular basis of quantitative disease resistance in rice. J Biotechnol (in press) [DOI] [PubMed] [Google Scholar]
- Lai WS, Carballo E, Strum JR, Kennington EA, Phillips RS, Blackshear PJ. (1999) Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol Cell Biol 19: 4311–4323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai WS, Parker JS, Grissom SF, Stumpo DJ, Blackshear PJ. (2006) Novel mRNA targets for tristetraprolin (TTP) identified by global analysis of stabilized transcripts in TTP-deficient fibroblasts. Mol Cell Biol 26: 9196–9208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Jia D, Chen X. (2001) HUA1, a regulator of stamen and carpel identities in Arabidopsis, codes for a nuclear RNA binding protein. Plant Cell 13: 2269–2281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZS, Thomas TL. (1998) PEI1, an embryo-specific zinc finger protein gene required for heart-stage embryo formation in Arabidopsis. Plant Cell 10: 383–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YJ, Zhang Q. (2005) Optimising the tissue culture conditions for high efficiency transformation of indica rice. Plant Cell Rep 23: 540–547 [DOI] [PubMed] [Google Scholar]
- Lorenzo O, Chico JM, Sánchez-Serrano JJ, Solano R. (2004) JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16: 1938–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei CS, Qi M, Sheng GY, Yang YL. (2006) Inducible overexpression of a rice allene oxide synthase gene increases the endogenous jasmonic acid level, PR gene expression, and host resistance to fungal infection. Mol Plant Microbe Interact 19: 1127–1137 [DOI] [PubMed] [Google Scholar]
- Peng KM, Zhang HB, Zhang Q. (1998) A BAC library constructed to the rice cultivar “Minghui63” for cloning genes of agronomic importance. Acta Bot Sin 40: 1108–1114 [Google Scholar]
- Peng Y, Bartley LE, Chen X, Dardick C, Chern M, Ruan R, Canlas PE, Ronald PC. (2008) OsWRKY62 is a negative regulator of basal and Xa21-mediated defense against Xanthomonas oryzae pv. oryzae in rice. Mol Plant 1: 446–458 [DOI] [PubMed] [Google Scholar]
- Peng YL, Shirano Y, Ohta H, Hibino T, Tanaka K, Shibata D. (1994) A novel lipoxygenase from rice: primary structure and specific expression upon incompatible infection with rice blast fungus. J Biol Chem 269: 3755–3761 [PubMed] [Google Scholar]
- Pomeranz MC, Hah C, Lin PC, Kang SG, Finer JJ, Blackshear PJ, Jang JC. (2010) The Arabidopsis tandem zinc finger protein AtTZF1 traffics between the nucleus and cytoplasmic foci and binds both DNA and RNA. Plant Physiol 152: 151–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu D, Xiao J, Ding X, Xiong M, Cai M, Cao Y, Li X, Xu C, Wang S. (2007) OsWRKY13 mediates rice disease resistance by regulating defense-related genes in salicylate- and jasmonate-dependent signaling. Mol Plant Microbe Interact 20: 492–499 [DOI] [PubMed] [Google Scholar]
- Qiu D, Xiao J, Xie W, Liu H, Li X, Xiong L, Wang S. (2008) Rice gene network inferred from expression profiling of plants overexpressing OsWRKY13, a positive regulator of disease resistance. Mol Plant 1: 538–551 [DOI] [PubMed] [Google Scholar]
- Schmitz RJ, Hong L, Michaels S, Amasino RM. (2005) FRIGIDA-ESSENTIAL 1 interacts genetically with FRIGIDA and FRIGIDA-LIKE 1 to promote the winter-annual habit of Arabidopsis thaliana. Development 132: 5471–5478 [DOI] [PubMed] [Google Scholar]
- Shen X, Yuan B, Liu H, Li X, Xu C, Wang S. (2010) Opposite functions of a rice mitogen-activated protein kinase during the process of resistance against Xanthomonas oryzae. Plant J 64: 86–99 [DOI] [PubMed] [Google Scholar]
- Sun J, Jiang H, Xu Y, Li H, Wu X, Xie Q, Li C. (2007) The CCCH-type zinc finger proteins AtSZF1 and AtSZF2 regulate salt stress responses in Arabidopsis. Plant Cell Physiol 48: 1148–1158 [DOI] [PubMed] [Google Scholar]
- Sun X, Cao Y, Yang Z, Xu C, Li X, Wang S, Zhang Q. (2004) Xa26, a gene conferring resistance to Xanthomonas oryzae pv. oryzae in rice, encodes an LRR receptor kinase-like protein. Plant J 37: 517–527 [DOI] [PubMed] [Google Scholar]
- Tabara H, Hill RJ, Mello CC, Priess JR, Kohara Y. (1999) pos-1 encodes a cytoplasmic zinc-finger protein essential for germline specification in C. elegans. Development 126: 1–11 [DOI] [PubMed] [Google Scholar]
- Tacahashi Y, Helmling S, Moore CL. (2003) Functional dissection of the zinc finger and flanking domains of the Yth1 cleavage/polyadenylation factor. Nucleic Acids Res 31: 1744–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsuji H. (1998) Zinc-finger transcription factors in plants. Cell Mol Life Sci 54: 582–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Z, Liu H, Qiu D, Zhou Y, Li X, Xu C, Wang S. (2009) A pair of allelic WRKY genes play opposite roles in rice-bacteria interactions. Plant Physiol 151: 936–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor GA, Carballo E, Lee DM, Lai WS, Thompson MJ, Patel DD, Schenkman DI, Gilkeson GS, Broxmeyer HE, Haynes BF, et al. (1996) A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity 4: 445–454 [DOI] [PubMed] [Google Scholar]
- Thomma BP, Nürnberger T, Joosten MH. (2011) Of PAMPs and effectors: the blurred PTI-ETI dichotomy. Plant Cell 23: 4–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Guo Y, Wu C, Yang G, Li Y, Zheng C. (2008a) Genome-wide analysis of CCCH zinc finger family in Arabidopsis and rice. BMC Genomics 9: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Xu YY, Zhang C, Ma QB, Joo SH, Kim SK, Xu ZH, Chong K. (2008b) OsLIC, a novel CCCH-type zinc finger protein with transcription activation, mediates rice architecture via brassinosteroids signaling. PLoS ONE 3: e3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Zeng ZB. (2003) Windows QTL Cartographer V2.0. North Carolina State University, Raleigh, NC: http://statgen.ncsu.edu/qtlcart/WQTLCart.htm (July 2009) [Google Scholar]
- Wu C, Li X, Yuan W, Chen G, Kilian A, Li J, Xu C, Li X, Zhou D-X, Wang S, et al. (2003) Development of enhancer trap lines for functional analysis of the rice genome. Plant J 35: 418–427 [DOI] [PubMed] [Google Scholar]
- Xiang Y, Tang N, Du H, Ye HY, Xiong LZ. (2008) Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiol 148: 1938–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W, Liu H, Li Y, Li X, Xu C, Long M, Wang S. (2009) A rice gene of de novo origin negatively regulates pathogen-induced defense response. PLoS ONE 4: e4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong M, Wang S, Zhang Q. (2002) Coincidence in map positions between pathogen-induced defensive-responsive genes and quantitative resistance loci in rice. Sci China C Life Sci 45: 518–526 [DOI] [PubMed] [Google Scholar]
- Yamaguchi T, Kuroda M, Yamakawa H, Ashizawa T, Hirayae K, Kurimoto L, Shinya T, Shibuya N. (2009) Suppression of a phospholipase D gene, OsPLDbeta1, activates defense responses and increases disease resistance in rice. Plant Physiol 150: 308–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YY, Yin GL, Darnell RB. (1998) The neuronal RNA-binding protein Nova-2 is implicated as the autoantigen targeted in POMA patients with dementia. Proc Natl Acad Sci USA 95: 13254–13259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan B, Shen X, Li X, Xu C, Wang S. (2007) Mitogen-activated protein kinase OsMPK6 negatively regulates rice disease resistance to bacterial pathogens. Planta 226: 953–960 [DOI] [PubMed] [Google Scholar]
- Yuan T, Li X, Xiao J, Wang S. (2011) Characterization of Xanthomonas oryzae-responsive cis-acting element in the promoter of rice race-specific susceptibility gene Xa13. Mol Plant 4: 300–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Feng Q, Jin C, Qiu D, Zhang L, Xie K, Yuan D, Han B, Zhang Q, Wang S. (2005) Features of the expressed sequences revealed by a large-scale analysis of ESTs from a normalized cDNA library of the elite indica rice cultivar Minghui 63. Plant J 42: 772–780 [DOI] [PubMed] [Google Scholar]
- Zhang J, Li C, Wu C, Xiong L, Chen G, Zhang Q, Wang S. (2006) RMD: a rice mutant database for functional analysis of the rice genome. Nucleic Acids Res 34: D745–D748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Peng K, Zhaohui C, Wang S, Zhang Q. (2002) The defense-responsive genes showing enhanced and repressed expression after pathogen infection in rice (Oryza sativa L.). Sci China C Life Sci 45: 449–467 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Cao Y, Huang Y, Xie W, Xu C, Li X, Wang S. (2009) Multiple gene loci affecting genetic background-controlled disease resistance conferred by R gene Xa3/Xa26 in rice. Theor Appl Genet 120: 127–138 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.