Abstract
The transition to flowering in many plant species, including Arabidopsis (Arabidopsis thaliana), is marked by the elongation of internodes to make an inflorescence upon which lateral branches and flowers are arranged in a characteristic pattern. Inflorescence patterning relies in part on the activities of two three-amino-acid loop-extension homeodomain transcription factors: BREVIPEDICELLUS (BP) and PENNYWISE (PNY) whose interacting products also promote meristem function. We examine here the genetic interactions between BP-PNY whose expression is up-regulated in stems at the floral transition, and the lateral organ boundary genes BLADE-ON-PETIOLE1 (BOP1) and BOP2, whose expression is restricted to pedicel axils. Our data show that bp and pny inflorescence defects are caused by BOP1/2 gain of function in stems and pedicels. Compatible with this, inactivation of BOP1/2 rescues these defects. BOP expression domains are differentially enlarged in bp and pny mutants, corresponding to the distinctive patterns of growth restriction in these mutants leading to compacted internodes and clustered or downward-oriented fruits. Our data indicate that BOP1/2 are positive regulators of KNOTTED1-LIKE FROM ARABIDOPSIS THALIANA6 expression and that growth restriction in BOP1/2 gain-of-function plants requires KNOTTED1-LIKE FROM ARABIDOPSIS THALIANA6. Antagonism between BOP1/2 and BP is explained in part by their reciprocal regulation of gene expression, as evidenced by the identification of lignin biosynthetic genes that are repressed by BP and activated by BOP1/2 in stems. These data reveal BOP1/2 gain of function as the basis of bp and pny inflorescence defects and reveal how antagonism between BOP1/2 and BP-PNY contributes to inflorescence patterning in a model plant species.
Flowering plants display a remarkable variety of inflorescence architectures selected to optimally display flowers for pollination and seed dispersal. Formation of the aerial parts of a plant is controlled by the shoot apical meristem (SAM), a cluster of pleuripotent stem cells located at the apex of the primary shoot. The SAM produces a series of reiterative modules known as phytomers to generate the aerial parts of the plant. Each phytomer comprises an internode (stem) subtending a node, which is a leaf associated with a potential axillary meristem (Steeves and Sussex, 1989). Elaboration of the different parts of a module (leaves, internodes, and axillary meristems) varies according to the phase of development and between species to generate architectural diversity (Sussex and Kerk, 2001).
Arabidopsis (Arabidopsis thaliana) has distinct vegetative and reproductive phases. During vegetative development, the SAM generates leaf primordia on its flanks; both internode and axillary meristem formation are inhibited, resulting in a compact rosette of leaves. At the end of the vegetative phase, endogenous and environmental cues promote the transition to flowering. The SAM responds to floral inductive signals by acquiring inflorescence meristem (IM) fate. During reproductive development, internodes elongate and axillary meristems proliferate at the expense of leaves to generate lateral branches and flowers in a regular spiral pattern on the inflorescence (Bowman and Eshed, 2000; Fletcher, 2002; Barton, 2010). While the pathways that promote floral fate of axillary meristems and repress leaf development are well studied, less is known about the formation and patterning of internodes.
Internode patterning is a key determinant of inflorescence architecture, with variations in the length and pattern of internode elongation contributing to diversity in inflorescence height and organization of secondary branches and flowers on the primary stem. Formation of internodes is associated with the proliferation and elongation of cells in the region underlying the central zone of the meristem, termed the rib zone (Steeves and Sussex, 1989; Fletcher, 2002). Following their elongation, internodes are gradually fortified through the differentiation of interfascicular fibers with secondary thickened cell walls, which provides mechanical support (Nieminen et al., 2004; Ehlting et al., 2005).
Internode patterning is dependent in part on the overlapping activities of two three-amino-acid loop-extension homeodomain transcription factors: the class I KNOTTED1-like homeobox (KNOX) protein BREVIPEDICELLUS (BP; formerly KNOTTED1-LIKE FROM ARABIDOPSIS THALIANA1 [KNAT1]) and the BEL1-like (BELL) protein PENNYWISE (PNY; also called BELLRINGER, REPLUMLESS, and VAMAANA) whose interacting products also promote meristem maintenance (Douglas et al., 2002; Venglat et al., 2002; Byrne et al., 2003; Smith and Hake, 2003; Bhatt et al., 2004; Rutjens et al., 2009; for review, see Hamant and Pautot, 2010). Mutations in BP cause short internodes and downward-pointing pedicels (Douglas et al., 2002; Venglat et al., 2002) whereas mutations in PNY cause irregular internode elongation, resulting in clusters of lateral organs (branches and flowers) spaced along the inflorescence (Byrne et al., 2003; Smith and Hake, 2003). In bp pny double mutants internodes are shorter than in either single mutant, signifying that BP and PNY have only partly overlapping roles in internode elongation and patterning (Smith and Hake, 2003). In both mutants, defects in vascular differentiation also occur, resulting in changes in how lignin is deposited in stems (Douglas et al., 2002; Mele et al., 2003; Smith and Hake, 2003). Previous genetic studies have shown that two class I KNOX genes, KNAT2 and KNAT6, are misexpressed in bp and pny mutant stems and pedicels. Inactivation of these genes, primarily KNAT6, rescues bp and pny defects in inflorescence architecture (Ragni et al., 2008) however this is the extent of our current knowledge.
Here, we examine genetic interactions between BP-PNY and BLADE-ON-PETIOLE1 (BOP1) and BOP2, two BTB-ankryin transcriptional coregulators that are expressed in lateral organ boundaries (Ha et al., 2004; Hepworth et al., 2005). BOP1/2 expression is limited to the pedicel axil in inflorescence stems where their function is to promote the formation of a vestigial abscission zone (McKim et al., 2008). BOP1/2 are indirect transcriptional repressors of BP in leaves (Ha et al., 2007; Jun et al., 2010) but their genetic interactions with BP, and its partner PNY, during reproductive development have yet to be examined. We show here that BP and PNY are transcriptional repressors of BOP1/2, preventing expression in stems and pedicels. Consistent with this, inactivation of the BOP genes rescues bp and pny inflorescence defects. We further show that BOP1/2 exert their activity in part through the boundary gene KNAT6, which functions in the same genetic pathway. Finally, we show that bp and pny inflorescence defects are mimicked by BOP1/2 gain of function. To explain this, we provide evidence that the reciprocal functions of BP and BOP1/2 in the inflorescence are likely a consequence of their antagonistic regulation of downstream target genes, such as those involved in lignin biosynthesis that are repressed by BP and activated by BOP1/2 in stems. These data redefine bp and pny phenotypes as the consequence of BOP1/2 gain of function, shedding light on how interactions between BP-PNY and BOP1/2 influence inflorescence architecture in a model plant species.
RESULTS
Expression of BOP1 and BOP2 in Lateral-Organ Boundaries
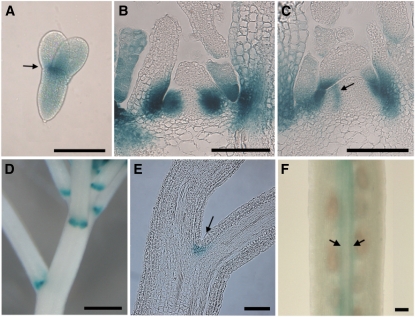
Previous analysis of BOP expression by in situ hybridization or through use of BOP1:GUS or BOP2:GUS reporter genes is consistent in showing that the BOP genes are expressed in lateral-organ boundaries formed during embryonic, vegetative, and reproductive development (Ha et al., 2004; Hepworth et al., 2005; Norberg et al., 2005; McKim et al., 2008; Xu et al., 2010). We have consolidated these data (Fig. 1). Using a BOP2:GUS reporter gene, expression was verified at the base of cotyledons in mature embryos (Fig. 1A; Ha et al., 2004). During postembryonic vegetative development, BOP2 expression was first associated with the boundary at stage 2 of leaf development, when primordia first appear as morphologically distinct from the SAM (Fig. 1, B and C, arrow indicates stage 2 leaf). As leaves expand, expression associates with the adaxial base of leaves, which gives rise to the petiole (Fig. 1, B and C; Norberg et al., 2005). Expression is also observed in the axil of pedicels (Fig. 1, D and E; McKim et al., 2008) and in the valve margins of fruit (Fig. 1F). Importantly, BOP1/2 expression is excluded from the IM and the replum of fruits, representing structures with meristematic function. While analysis of loss-of-function bop1 bop2 mutants has revealed that BOP1/2 transcriptionally repress meristematic genes in leaves (Ha et al., 2007) and floral primordia (Xu et al., 2010) relatively little is known about how BOP1/2 gain of function perturbs plant architecture.
Figure 1.
BOP2:GUS expression pattern in boundaries. A, Mature embryo; expression at base of cotyledons (arrow). B to C, Shoot apex of a short-day-grown seedling, longitudinal section; expression begins in stage 1 leaf primordia and localizes to the boundary of stage 2 leaves (arrow). As primordia expand, BOP2 expression associates with the adaxial base of leaves, which elongate to form the petiole. D, Inflorescence; horseshoe pattern of expression in the axils of floral pedicels. E, Pedicel, longitudinal section; expression in the axil (arrow). F, Silique; expression in the valve margins (arrows). Scale bars, 0.1 mm except D, 0.5 mm. [See online article for color version of this figure.]
A Spectrum of Inflorescence Architecture Defects Caused by BOP1/2 Gain of Function
Previous phenotypic analysis of BOP1 or BOP2 overexpression in plants has drawn attention to bp- and pny-like defects in inflorescence architecture, either short plants with floral pedicels pointing downward (Ha et al., 2007) or short bushy plants with irregular internodes (Norberg et al., 2005). Comparison of the strong activation-tagged bop1-6D line to bp pny double mutants revealed remarkably similar inflorescence architectures (Fig. 2, A–C), suggesting that BOP1/2 might antagonize both activities. This also suggested that BOP1/2 gain of function might elicit a spectrum of inflorescence defects. To examine this further, we generated transgenic plants overexpressing BOP1 or BOP2 in Columbia-0 (Col-0) and Landsberg erecta (Ler) backgrounds and scored for defects in inflorescence architecture (Fig. 2, D–I; Table I). This analysis showed that in Ler plants, downward-pointing siliques was the prevalent phenotype (up to 45% of transformants) whereas in Col-0 plants, clustered siliques was the prevalent phenotype (up to 20% of transformants). Compatible with this, the erecta mutation enhances the phenotypic severity of bp mutants (Douglas et al., 2002). Taken together, these findings suggest that BOP gain of function has variable effects on inflorescence architecture conditioned in part by ecotype. These defects may result from the antagonism of BP and/or PNY expression or activity. To examine this further, we tested the effect of bop1 bop2 loss of function on expression of bp and pny mutant phenotypes in a Col background.
Figure 2.
BOP1 gain of function causes bp- and pny-like defects in inflorescence architecture. Representative inflorescences are shown for: A, Col wild type; B, bp-2 pny double mutant. C, bop1-6D; an activation-tagged BOP1 overexpression line (with four 35S CaMV enhancers). Compact internodes similar to bp-2 pny. D, Col. E, pny mutant. F, 35S:BOP1 transformant in Col (one 35S CaMV enhancer) with clustered siliques as in pny (arrows in E and F). G, Ler wild type. H, bp-1 in Ler. I, 35S:BOP1 transformant in Ler; downward-pointing siliques as in bp-1. Scale bars, 1 cm. [See online article for color version of this figure.]
Table I.
Summary of inflorescence defects in plants overexpressing BOP1 or BOP2
| Transgene | Ecotype | Plants with Downward-Oriented Siliques | Plants with Clustered Siliques | Total No. of Transformants |
| % | ||||
| 35S:BOP1 | Col | 0.0 | 20.6 | 175 |
| 35S:BOP2 | Col | 0.0 | 10.0 | 80 |
| tCUP4:BOP1 | Col | 0.0 | 61.1 | 18 |
| 35S:BOP1 | Ler | 44.5 | 22.0 | 164 |
| 35S:BOP2 | Ler | 27.6 | 20.4 | 196 |
Inactivation of BOP1/2 Partially Rescues the bp Phenotype
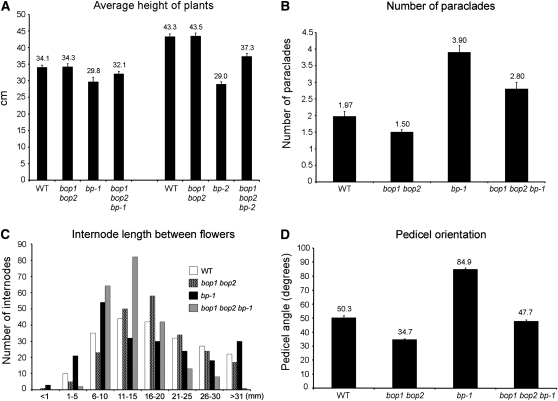
To first examine BOP1/2 interactions with BP, we generated bop1 bop2 bp-1 and bop1 bop2 bp-2 triple mutants and analyzed their phenotypes relative to wild-type and parental controls. bp mutants are characterized by short internodes, reduced apical dominance, and downward-pointing siliques (Douglas et al., 2002; Venglat et al., 2002). This analysis showed that inactivation of the BOP genes largely rescues bp inflorescence defects (Fig. 3, A–D; Supplemental Fig. S1) similar to inactivation of KNAT2 and KNAT6 (Ragni et al., 2008). Quantitative phenotypic analyses were performed on 24 plants per genotype, by measuring the average height, internode length, and number of rosette paraclades for wild type and mutants (Fig. 4, A–D). These analyses confirmed that bop1 bop2 loss of function counteracted the short stature of bp-1 and bp-2 plants (Fig. 4A) and partially restored apical dominance in bp-1 mutants (Fig. 4B). Whereas bp-1 mutants have a significant number of short internodes in the 1- to 5-mm range, the distribution in bop1 bop2 bp-1 triple mutants was similar to wild type (Fig. 4C). Whereas bp-1 pedicels point downward at an average angle of 84.9° relative to the primary stem, the average angle in bop1 bop2 bp-1 triple mutants was 47.7°, similar to wild type (Fig. 4D). Also, the average pedicel angle in bop1 bop2 double mutants was steeper than wild type (34.7° versus 50.3°), showing that BOP1/2 regulate pedicel orientation as well as abscission zone formation at the stem-pedicel junction (Fig. 4D; Supplemental Fig. S2; McKim et al., 2008). No rescue occurred in bp-2 bop1 or bp-2 bop2 double mutants (data not shown), indicating that BOP1 and BOP2 have redundant functions.
Figure 3.
Phenotypic suppression of bp and pny inflorescence defects by bop1 bop2. A, Wild-type control. B, bop1 bop2 mutant. C, bp-1 mutant; downward-pointing siliques. D, bop1 bop2 bp-1 mutant; partial rescue of bp-1 phenotype. E, pny mutant; clustered siliques (arrows). F, bop1 bop2 pny mutant; similar to wild type. Scale bars, 2 cm. [See online article for color version of this figure.]
Figure 4.
Quantitative analysis of bp phenotypic rescue by bop1 bop2. At least 24 plants for the indicated genotypes were analyzed. A, Average inflorescence height; inactivation of BOP1/2 partially rescues the short stature of bp-1 and bp-2 mutants. B, Average number of paraclades; inactivation of BOP1/2 partially restores apical dominance in bp-1 mutants. C, Distribution of internode lengths between successive siliques on the primary inflorescence. Internodes between the first and 11th siliques (counting acropetally) were measured. Distribution of internode lengths in bop1 bop2 bp-1 triple mutants is similar to wild type. D, Average orientation of pedicels; inactivation of BOP1/2 corrects pedicel orientation in bp-1 mutants. Error bars, se.
Inactivation of BOP1/2 Completely Rescues the pny Phenotype
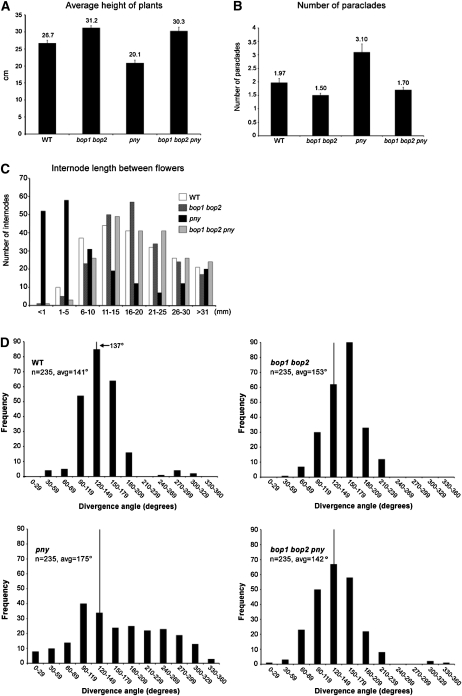
Given that BP and PNY coregulate internode patterning, we next examined the interaction of BOP1/2 with PNY by generating bop1 bop2 pny triple mutants. pny mutants are characterized by clusters of siliques due to irregular internode elongation, defects in phyllotaxy, reduced apical dominance, and replumless fruits (Byrne et al., 2003; Roeder et al., 2003; Smith and Hake, 2003; Bhatt et al., 2004). Inactivation of the BOP genes also rescued pny inflorescence defects (Fig. 3, A, E, and F). Quantitative phenotypic analyses were performed on 24 plants per genotype to further monitor this rescue, by measuring the average height, internode length, and number of rosette paraclades for wild type and mutants. These analyses confirmed that loss-of-function bop1 bop2 restored the stature of pny plants and the number of rosette paraclades to wild type (Fig. 5, A and B). Whereas pny mutants have a significant number of internodes in the 1- to 5-mm range, the distribution in bop1 bop2 pny triple mutants was similar to wild type (Fig. 5C). To quantify rescue of phyllotactic patterning in bop1 bop2 pny triple mutants, we measured divergence angles between successive floral pedicels on the primary stem (Fig. 5D; see Peaucelle et al., 2007). Whereas the distribution of divergence angles in pny was largely random (mean of 175°), the distribution in bop1 bop2 pny triple mutants was similar to wild type (mean of 142° versus 141°). Surprisingly, partial loss of BOP function was sufficient to rescue the pny phenotype since pny bop1 and pny bop2 mutant inflorescences also resembled wild type (data not shown).
Figure 5.
. Quantitative analysis of pny phenotypic rescue by bop1 bop2. At least 24 plants per genotype were analyzed. A, Average height of primary inflorescence; inactivation of BOP1/2 rescues short stature of pny mutants. B, Average number of rosette paraclades; inactivation of BOP1/2 restores apical dominance in pny mutants. C, Distribution of internode lengths between successive siliques on the primary inflorescence. Internodes between the first and 11th siliques (counting acropetally) were measured. The distribution of siliques in bop1 bop2 pny mutants was similar to wild type. D, Distribution of divergence angles between siliques on the primary inflorescence. At least 10 successive angles between the first and 24th siliques (counting acropetally) were measured for n ≥ 14 plants per genotype. The class containing the theoretical angle of 137° is indicated by a vertical line. Average angle, Avg. In pny plants, distribution is uniform across all classes but in bop1 bop2 pny plants, the distribution is similar to wild type.
A final defining characteristic in pny mutants is a replumless fruit (Roeder et al., 2003). Scanning electron microscopy (SEM) showed that inactivation of BOP1/2 also rescues replum formation in pny fruits (Supplemental Fig. S3, A–D), similar to inactivation of KNAT6 and consistent with coexpression of BOP1/2 and KNAT6 in valve margins (Fig. 1F; Supplemental Fig. S4; Ragni et al., 2008). We further examined the pattern of lignin deposition in fruit cross sections (Supplemental Fig. S3, E–H). In pny mutants, lignin (pink color) was detected throughout the junction between the valves, reflecting lack of the replum. In bop1 bop2 and bop1 bop2 pny triple mutants, lignin formed only at the valve margins as in wild type. Collectively, these data demonstrate complete rescue of pny defects, supporting the model that BOP1/2 antagonize BP and PNY activities in the inflorescence. These data further suggest that BOP1/2 and KNAT6 control similar developmental processes, based on their similar interactions with BP and PNY and their overlapping expression patterns in lateral organ boundaries (Ragni et al., 2008; see also Fig. 1; Supplemental Fig. S4).
BOP1/2 Expression Domains Are Expanded in bp and pny Mutants
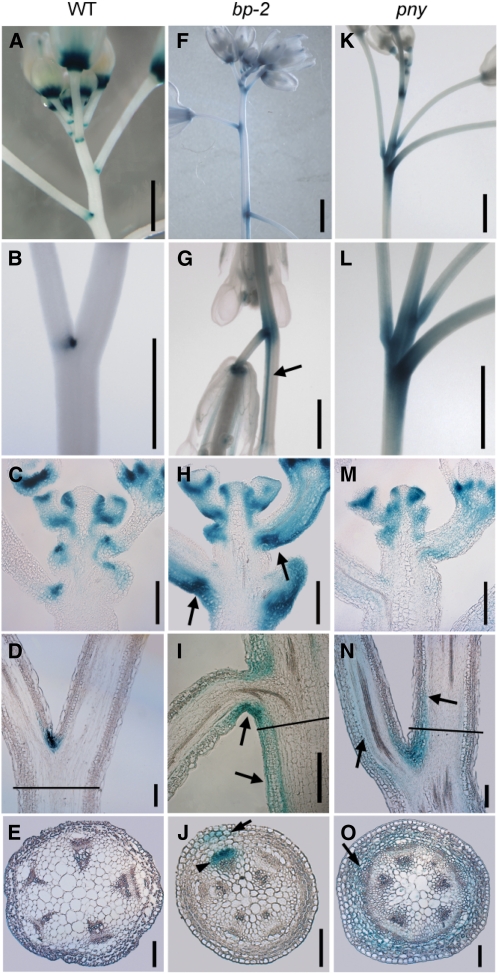
Ragni et al. (2008) showed that BP and PNY prevent KNAT2 and KNAT6 expression in stems and pedicels and that loss-of-function knat6 (and knat2 knat6) rescues bp and pny defects. This prompted us to examine if BOP1/2 expression domains are likewise expanded in bp and pny mutants, using the BOP2:GUS reporter gene (Fig. 6, A–O). In bp mutants, BOP2 expression was expanded in stems and pedicels, particularly below nodes. Expression on the abaxial side of nodes is consistent with localized growth restriction, causing pedicels to point downward. Staining was also seen in stripes of abnormal epidermal tissue that extend below the node and become ectopically lignified in mature bp stems (Fig. 6, F–I; Venglat et al., 2002; Mele et al., 2003). Stem cross sections from just below the node confirmed BOP2 misexpression in the stem cortex beneath the epidermis and in phloem regions associated with the primary vascular bundle (Fig. 6J). In pny mutants, BOP2 expression was also expanded in stems and pedicels above and below nodes, compatible with growth impairment, causing irregular internodes and silique clustering (Fig. 6, K–N). Stem cross sections near pny nodes confirmed BOP2 misexpression throughout the stem cortex (Fig. 6O). BOP1:GUS expression in bp and pny mutants showed a similar pattern (Supplemental Fig. S5). In summary, the misexpression patterns of BOP1/2 differ in bp and pny mutants, bearing resemblance to the distinct inflorescence defects that characterize these mutants.
Figure 6.
BOP2:GUS expression in wild-type, bp, and pny inflorescences. A to E, Wild type. A and B, Expression restricted to stem-pedicel axil. C, Apex; no expression in the IM, internodes, or pedicels. D, Node. E, Stem; line in D shows plane of cross section. F to J, bp-2 mutant. F to G, Expression expands beyond nodes, thin stripes of tissue extending basipetally below nodes stain strongly (arrow). H, Apex; misexpression on the abaxial side of nodes (arrows) and in pedicels. I, Node; misexpression on the abaxial side of the node (arrows). J, Stem; stripe of expression below node. Line in I shows plane of cross section. Arrow, cortical cells; arrowhead, phloem. K to O, pny mutant. K and L, Expression expands above and below nodes. M, Apex; staining strongest near the apex and in pedicels. N, Node; diffuse expression above and below the node (arrows). O, Stem; misexpression in stem cortex (arrow). Line in N shows plane of cross section. Scale bars, 1 mm except 100 μm for C to E, H to J, and M to O. [See online article for color version of this figure.]
BOP1/2 Promote KNAT6 Expression
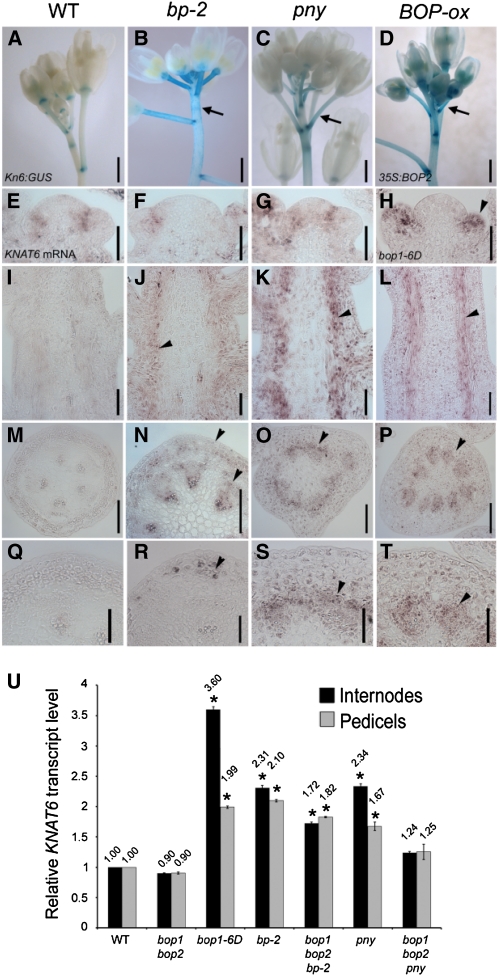
Given that BOP1/2 and KNAT6 are both required for bp and pny phenotypes and BOP1/2 gain of function produces bp- and pny-like phenotypes, we compared KNAT6:GUS expression in various BOP gain-of-function lines: bp, pny, and 35S:BOP2 or bop1-6D. Misexpression of KNAT6:GUS in stems was confirmed for all genotypes (Fig. 7, A–D). However, the reporter gene was not expressed in boundaries of the IM, indicating that some of its control sequences were missing (data not shown). We therefore used in situ hybridization to further examine KNAT6 expression in the inflorescence apex and stem (Fig. 7, E–T). In the bp mutant, KNAT6 transcript was misexpressed in the stem cortex and vascular tissue (Fig. 7, J and N) and beneath the node in a stripe pattern (Fig. 7R) similar to misexpression of BOP2 (Fig. 6, I and J). In the pny mutant, KNAT6 was misexpressed in the vascular tissue of elongated stems similar to bop1-6D mutants (Fig. 7, K, L, O, P, S, and T). Both mutants formed extra vascular bundles, resulting in a dense vascular ring with little interfascicular space (Fig. 7, O and P; Smith and Hake, 2003). KNAT6 transcript levels were also monitored in internodes and pedicels by quantitative reverse transcription (qRT)-PCR. These data confirmed 2- to 3-fold higher levels of KNAT6 transcript in bp-2, pny, and bop1-6D plants relative to wild type and bop1 bop2 controls (Fig. 7U). Higher levels of KNAT6 transcript are consistent with an expanded domain of KNAT6 expression in bop1-6D/35S:BOP2 stems. We therefore concluded that BOP1/2 promote KNAT6 expression. Consistent with this, KNAT6 transcript levels were slightly lower in bop1 bop2 bp and bop1 bop2 pny internodes and pedicels relative to bp-2 and pny single mutants (Fig. 7U). No similar up-regulation was observed for KNAT2 in bop1-6D plants (data not shown).
Figure 7.
KNAT6 expression in wild type, bp-2, pny, and BOP gain-of-function mutants. A to D, KNAT6:GUS expression. Inflorescences shown for: A, wild type; B, bp-2; C, pny; D, 35S:BOP2. Expression localized to the pedicel axil in wild type (A) but misexpressed in stems and pedicels of mutants (B–D). E to T, KNAT6 mRNA detected using in situ hybridization. Inflorescence apices shown for: E, Wild type; F, bp-2; G, pny; H, bop1-6D. Transcript is correctly localized to the IM-floral meristem boundary except in bop1-6D (H) where expression is throughout the adaxial area of floral meristems. Stem longitudinal sections shown for: I, wild type; J, bp-2; K, pny; L, bop1-6D. Transcript up-regulated in the cortex of mutant stems (J–L). In K and L, the vascular cambium area shows strong expression. Stem cross sections shown for: M, wild type; N, bp-2. Expression strongest in the cortex and vascular bundles (arrowheads). O, pny. Irregular vascular bundles; vascular cambium area shows the strongest expression (arrowhead). P, bop1-6D; strong expression in vascular bundles (arrowhead). Magnified stem cross sections shown for: Q, wild type. R, bp-2; stripe of expression in cortex below node (arrowhead). S, pny; expression strongest in stripe of cells near vascular cambium (arrowhead). T, bop1-6D; expression in vascular bundles. U, qRT-PCR analysis of relative KNAT6 transcript levels in wild-type and mutant internodes and pedicels. Asterisks, Significantly different from wild type (Student’s t tests, P < 0.0001; except pny, P < 0.001). Scale bars, 50 μm except 0.5 mm for A to D and 100 μm for M to P. [See online article for color version of this figure.]
BOP1/2 Exert Their Function through KNAT6
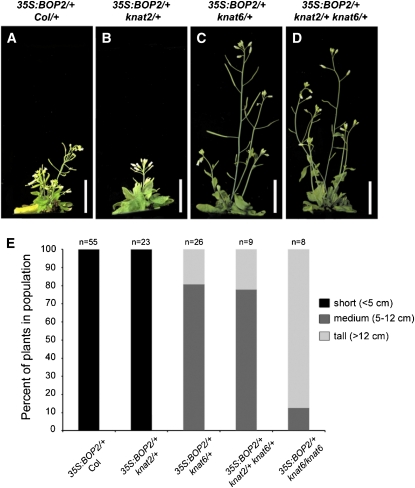
Given that BOP1/2 promote KNAT6 expression, we reasoned that BOP1/2 may exert all or part of their function through KNAT6. To examine this, we tested the effect of knat6 loss of function on the phenotype of a strong 35S:BOP2 gain-of-function line with short compact inflorescences (Norberg et al., 2005). In this experiment, plants that were homozygous for the 35S:BOP2 transgene were crossed to wild type or to lines homozygous for knat2, knat6, or knat2 knat6 mutations. The phenotypes of progeny were scored in the F1 generation. To rule out transgene silencing, we took the additional step of confirming BOP2 overexpression in F1 populations (Supplemental Fig. S6). These experiments revealed that partial knat6 loss of function (i.e. knat6/+ or knat2/+ knat6/+) was sufficient to restore internode elongation in 35S:BOP2 plants (Fig. 8, A and C–E). In contrast, no rescue occurred in control crosses to wild type or knat2 alone (Fig. 8, A, B, and E). Compatible with this, mutations in knat2 alone do not rescue bp or pny inflorescence defects (Ragni et al., 2008). These data indicate that BOP1/2 exert much of their function through KNAT6. Interestingly however, 35S:KNAT6 plants are not short and mimic 35S:BP plants with lobed leaves (Supplemental Fig. S7A; see also Lincoln et al., 1994; Dean et al., 2004), indicating that the functions of BP and KNAT6 are redundant when BOP1/2 is not comisexpressed. Thus, both BOP1/2 and KNAT6 are required to exert changes in inflorescence architecture.
Figure 8.
Inactivation of KNAT6 rescues compact internodes caused by BOP2 gain of function. Plants homozygous for a 35S:BOP2 transgene were crossed to wild-type control plants or to plants homozygous for mutations in knat2, knat6, or knat2 knat6. The inflorescences of representative F1 plants are shown. A, 35S:BOP2/+ Col. B, 35S:BOP2/+ knat2/+. C, 35S:BOP2/+ knat6/+. D, 35S:BOP2/+ knat2/+ knat6/+. E, Quantitative analysis of inflorescence height in populations of F1 plants for the genotypes indicated. Scale bars, 2 cm. [See online article for color version of this figure.]
BOP1/2 and BP/PNY Are Antagonistic Regulators of Stem Lignification
We next sought to determine how BOP1/2 gain of function antagonizes BP and PNY activities in the stem. We initially considered that BOP1/2 might function through ASYMMETRIC LEAVES2 (AS2) to inhibit BP and/or PNY expression in stems. BOP1/2 indirectly repress BP in leaves by promoting AS2 expression, whose product is a direct repressor of BP transcription (Guo et al., 2008; Jun et al., 2010). However, inactivation of AS2 failed to rescue the short stature of 35S:BOP2 plants (Ha et al., 2007) or bp and pny inflorescence defects (Supplemental Fig. S7, B and C). Moreover, no decrease in BP or PNY expression was detected in the stem of BOP1/2 loss- or gain-of-function mutants (Supplemental Fig. S8). These data indicate that BOP1/2 control of stem architecture is largely independent of AS2 and transcriptional repression of BP. We therefore examined the model that BOP1/2 function downstream of BP-PNY and have reciprocal functions in the stem based on their compartmentalized expression domains in the inflorescence.
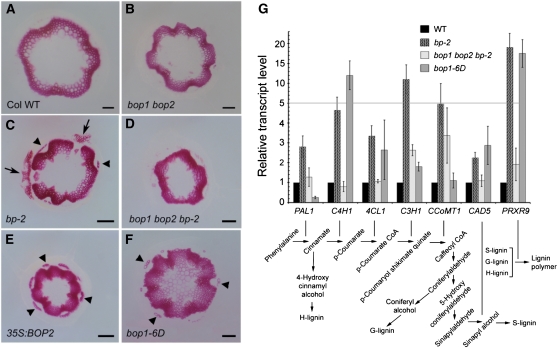
To examine this, we turned to work showing that BP is a negative regulator of lignin deposition in stems (Mele et al., 2003). In the primary inflorescence stem, formation of secondary cell walls is tightly regulated over developmental time (Ehlting et al., 2005). Cross sections were cut from the base of wild-type and mutant primary stems at the same developmental age and stained with phloroglucinol, which detects lignin deposition, a hallmark of secondary walls in vessel and fiber cells in the inflorescence stem. As expected, complex patterning changes were seen in bp mutants. Phloem fibers overlying primary vascular bundles were prematurely lignified. In addition, gaps were observed in the ring of interfascicular fiber cells with lignin abnormally deposited in the epidermis and cortex of these gaps. This pattern correlates with the position of abnormally differentiated stripes of tissue in bp stems that originate below nodes and extend basipetally (Fig. 9, A–C; Douglas et al., 2002; Venglat et al., 2002; Mele et al., 2003). Loss-of-function bop1 bop2 partially rescued bp defects, resulting in a pattern similar to wild type (Fig. 9, A, C, and D). Ectopic stem lignification also occurs in pny stems, albeit in a different pattern than for bp, which may reflect differences in where or when BOP1/2 are misexpressed. In pny stems, vascular bundles were more crowded than in wild type, resulting in a dense vascular ring (Fig. 7O; Supplemental Fig. S9; Smith and Hake, 2003). Loss-of-function bop1 bop2 also rescued pny defects, resulting in a pattern similar to wild type (Supplemental Fig. S9, A, E, and G). Importantly, stem cross sections from 35S:BOP2 and bop1-6D plants showed expanded patterns of lignification, similar to bp pny double mutants (Fig. 9E; Supplemental Fig. S9; Smith and Hake, 2003). In BOP1/2 overexpressing lines, the vascular ring was dense (similar to pny mutants) and phloem fiber cells overlying primary vascular bundles were prematurely lignified (similar to bp mutants). However, there were no gaps in the vascular ring, presumably due to uniformity in BOP1/2 misexpression. In bop1-6D mutants, parts of the pith were lignified, never observed in wild-type stem development. Thus, BOP1/2 gain of function induces lignified phloem and interfascicular fibers in a pattern reminiscent of the secondary growth that occurs in trees (Fig. 9E; Supplemental Fig. S9; Nieminen et al., 2004; Baucher et al., 2007). These data support the model BOP1/2 function downstream of BP-PNY in the stem and have a reciprocal function associated with lignin biosynthesis.
Figure 9.
Lignification pattern and lignin biosynthetic gene expression in wild-type and mutant stems. A to F, Cross sections from the base of fully elongated stems were stained with phloroglucinol-HCl to reveal lignin. Representative sections are shown for: A, wild type; B, bop1 bop2. C, bp-2; gaps in the vascular ring (arrows) are associated with stripes of ectopically lignified epidermal/cortical tissue. Arrowheads, Premature lignification of phloem fiber cells in primary vascular bundles. D, bop1 bop2 bp-2; similar to wild type. E, 35S:BOP2; dense vascular ring compared to wild type. Arrowheads, Premature lignification of phloem fiber cells, similar to bp-2 mutants. F, bop1-6D; similar to 35S:BOP2 but pith is also lignified. Scale bars, 100 μm. G, qRT-PCR analysis of lignin biosynthesis genes in stem tissue (same stage as above). Error bars, se of three biological replicates. Position of genes in the lignin biosynthetic pathway is depicted below (adapted from Mele et al., 2003; Zhou et al., 2009). [See online article for color version of this figure.]
BOP1/2 Activate Genes Repressed by BP
Microarray and electrophoretic mobility shift assay experiments have previously identified lignin biosynthetic genes that are directly repressed by BP in stems (Mele et al., 2003). Direct targets of PNY have not been identified to our knowledge. Therefore, qRT-PCR was used to examine whether lignin biosynthetic gene transcripts are reciprocally regulated by BP and BOP1/2 in inflorescence stems (Fig. 9G). This approach confirmed up-regulation of all four genes previously identified by Mele et al. (2003) as up-regulated in mature bp-2 stems (Phe ammonia lyase1 [PAL1]; cinnamate 4-hydroxylase1 [C4H1]; 4-coumarate CoA ligase1 [4CL1]; and PRXR9GE, a class III peroxidase) as well as several additional genes (C3H1; caffeolyl CoA 3-O-methyltransferase1 [CCoMT1]; cinnamyl alcohol dehydrogenase5 [CAD5]) in the lignin biosynthetic pathway (for review, see Boerjan et al., 2003). Mutation of bop1 bop2 in bp-2 restored all but one of these gene transcripts to near wild-type levels, supporting the observed promotive effect of BOP1/2 on lignin deposition in stems. Four of the above genes were also up-regulated in bop1-6D stems (C4H1, C3H1, CAD5, and PRXR9GE), suggesting that BOP1/2 directly or indirectly promotes the expression of genes in the lignin biosynthetic pathway. Similar results were obtained using internode tissue (data not shown). As reported by Mele et al. (2003), the class III peroxidase gene PRXR9GE showed the greatest fold-change (15- to 20-fold) over wild type in both bp-2 and bop1-6D stems, suggesting that polymerization of monolignol subunits may be a key regulatory point in the developmental control of secondary wall formation. Collectively, these data support the model that BOP1/2 and BP have reciprocal functions in the stem and show how antagonistic interactions between BOP1/2 and BP-PNY are important for patterning of cell-type differentiation in stems as well as inflorescence architecture.
DISCUSSION
Internodes are elongated at the transition to flowering as a result of expanded rib meristem activity in the IM (Steeves and Sussex, 1989; Fletcher, 2002). The meristem expression of BP diminishes with the floral transition and becomes restricted to the cortex of the inflorescence stem and pedicel, where its activity together with PNY promotes internode elongation and vascular patterning (Lincoln et al., 1994; Douglas et al., 2002; Venglat et al., 2002; Smith and Hake, 2003). In this article, we used a genetics approach to examine how interactions between BP-PNY and the lateral-organ boundary regulators BOP1/2 govern Arabidopsis inflorescence architecture. Our data show that a spectrum of bp- and pny-like defects in inflorescence architecture are caused by BOP1/2 gain of function. Our key findings are that BP and PNY restrict BOP1/2 expression to the pedicel axil together with KNAT6 to prevent their misexpression in stems and pedicels, which causes altered growth patterns in bp and pny inflorescences. Our data also indicate that BOP1/2 promote KNAT6 expression and that both activities are required to inhibit internode elongation in stems (Fig. 10). Our analysis of gain-of-function mutants demonstrates that BOP1/2 function downstream of BP-PNY in a reciprocal manner, accelerating the final steps of stem differentiation in opposition to BP.
Figure 10.
Summary of genetic interactions between BP-PNY, BOP1/2, and KNAT6 in the inflorescence. BP and PNY in the stem and pedicels are transcriptional repressors of BOP1/2 and KNAT6, limiting their expression to the pedicel axil. BOP1/2 gain-of-function mutants phenocopy bp and pny mutants because BOP1/2 function downstream of BP-PNY in an antagonistic manner. BOP1/2 are positive regulators of KNAT6 expression that depend in part on KNAT6 activity to exert changes in inflorescence architecture.
BOP1/2 Differentially Regulate KNOX Activity in Leaves and the Inflorescences
Previous work has established that BOP1/2 in leaves function together with AS1/2 to maintain repression of the class I KNOX genes BP, KNAT2, and KNAT6 during leaf development (Ori et al., 2000; Semiarti et al., 2001; Ha et al., 2003, 2007; Jun et al., 2010). In this context, BOP1/2 indirectly represses BP transcription by promoting AS2 expression in leaf petioles (Jun et al., 2010). Genetic assays show that BOP1/2 also repress BP through an AS2-independent pathway that is as-yet undefined (Ha et al., 2007; data not shown). Our data reveals an opposite regulatory pattern in inflorescences with BP and PNY functioning as transcriptional repressors of BOP1/2 and KNAT6. Comisexpression of BOP1/2 and KNAT6 permits their opposing function downstream of BP-PNY to restrict growth and promote premature differentiation of the stem. These data are compatible with BOP1/2 gain-of-function studies in moss. In this species, stabilization of BOP1/2 transcripts causes premature gametophore differentiation (Saleh et al., 2011).
Misexpression of BOP1/2 Restricts Growth to Create Variations in Inflorescence Architecture
Variations in inflorescence architecture are extensive among flowering plants, with the length and pattern of internode elongation and pedicel angle acting as key variables in the display of flowers (Steeves and Sussex, 1989; Sussex and Kerk, 2001). Short internodes and pedicels like those in bp mutants are reminiscent of species with spike-type inflorescences where internodes between successive flowers are short (Bell and Bryan, 2008). Conversely, long internodes separating whorls of flowers on the stem, like those in pny mutants, are reminiscent of species with verticillate-type inflorescences (Bell and Bryan, 2008). Our data show that a spectrum of inflorescence architectures ranging from short internodes, to downward-pointing pedicels, to clusters of flowers may be produced by differentially regulating the pattern and degree of BOP gain of function in stems. In bp mutants, ectopic BOP1/2 expression on the abaxial side of nodes leads to growth restriction and downward-pointing pedicels. BOP1/2 are also misexpressed in the stem cortical tissue where BP-PNY normally function, thereby inhibiting internode elongation and causing cells to differentiate prematurely. In pny mutants, BOP1/2 are strongly misexpressed in the pedicels and stem cortex of young internodes, leading to irregular elongation of internodes and the clustering of flowers in whorls. These defects are phenocopied to various degrees by ectopically expressing BOP1/2 under the control of single or multiple 35S Cauliflower mosaic virus (CaMV) enhancers, indicating that BOP1/2 function downstream of BP-PNY in an antagonistic manner. However, BOP1/2 gain of function does not reduce BP or PNY transcript levels in the stem (Supplemental Fig, S8), indicating that BOP1/2 likely oppose BP-PNY function posttranscriptionally.
BOP1/2 and KNAT6 Function in the Same Genetic Pathway
Our genetic assays and expression data show that misexpression of BOP1/2 is the cause of inflorescence patterning defects in bp and pny mutants. For reasons that are unclear, inactivation of BOP1/2 partially suppresses bp defects but completely suppresses pny defects. This difference may be related to the slightly different roles that bp and pny play in internode development (Hake and Smith, 2003; Peaucelle et al., 2011). These data extend the work of Ragni et al. (2008) who showed an identical pattern of rescue for bp and pny defects by inactivation of KNAT6 (and to a lesser extent KNAT2), genes that are misexpressed in an overlapping domain with BOP1/2 in bp and pny stems (Figs. 6 and 7). These studies place BOP1/2 and KNAT6 in the same genetic pathway controlling inflorescence architecture. Compatible with this, BOP1/2 gain of function promotes KNAT6 expression. However, both activities are required to restrict internode elongation since inactivation of KNAT6 restores internode elongation in 35S:BOP2 plants and 35S:KNAT6 internodes are not short (Fig. 8; Supplemental Fig. S7; Dean et al., 2004). Despite several attempts with 35S:BOP1-GR transgenic plants and chromatin immunoprecipitation assays, we have yet to determine if BOP1/2 directly regulate KNAT6. Given that BP and KNAT6 are highly related proteins with the same gain-of-function phenotype (Lincoln et al., 1994; Chuck et al., 1996; Dean et al., 2004) they may regulate some of the same genes. However, KNAT6 with BOP1/2 function in opposition to BP. A physical complex between BOP1/2 and KNAT6 was not detected in yeast (Saccharomyces cerevisiae; data not shown). We therefore favor a model in which BOP1/2 bind independently to the same promoters as KNAT6 or induce the expression of a KNAT6 cofactor to exert their effect.
In fruits, BOP1/2 and KNAT6 likewise function in the same genetic pathway as evidenced by rescue of replum formation in pny mutants by either bop1 bop2 or knat6 loss of function (Ragni et al., 2008; this study). BOP1/2 and KNAT6 may also share a role in floral-organ abscission based on recent evidence that IDA-dependent signaling inhibits BP activity, allowing KNAT2 and KNAT6 to promote abscission (McKim et al., 2008; Shi et al., 2011). Thus, antagonism between BP-PNY and a genetic pathway involving BOP1/2 and KNAT6 is likely to be a conserved module in plant development.
ARABIDOPSIS THALIANA HOMEOBOX GENE1 Is a Potential KNAT6 Cofactor
The BELL homeodomain protein encoded by ARABIDOPSIS THALIANA HOMEOBOX GENE1 (ATH1) is another potential member of the BOP1/2 and KNAT6 genetic pathway that will be important to investigate. KNOX homeodomain proteins like KNAT6 perform many of their functions as heterodimers with BELL proteins (e.g. Byrne et al., 2003; Bhatt et al., 2004; Kanrar et al., 2006; Rutjens et al., 2009). These partnerships can influence protein-protein interactions, nuclear localization of the KNOX partner, and/or binding-site selection (Smith et al., 2002; Hackbusch et al., 2005; Cole et al., 2006; Rutjens et al., 2009). Interestingly, loss-of-function ath1-1 rescues pny inflorescence defects (like bop1 bop2 and knat6) whereas ATH1 gain of function causes short internodes (Gómez-Mena and Sablowski, 2008; Rutjens et al., 2009). Given that ATH1 transcripts are highly up-regulated in bop1-6D internodes (data not shown), an ATH1-KNAT6 complex may restrict stem growth. Short internodes are typical of defects in gibberellic acid (GA) biosynthesis (Achard and Genschik, 2009; Schwechheimer and Willige, 2009) of which BP is a repressor (Hay et al., 2002). However, GA 20-oxidase transcript levels in 35S:BOP2 and bop1-6D stems are not significantly different from wild type (data not shown) and spray treatment of 35S:BOP2 plants with GA did not restore internode elongation (data not shown), making it uncertain if defects in GA biosynthesis or catabolism are at play.
BP and BOP1/2 Antagonistically Regulate Secondary Cell Wall Biosynthesis
Lignin biosynthesis is one of the major components of secondary cell wall formation, essential for water transport and the structural support of plants. In Arabidopsis, abundant interfascicular fibers with secondarily thickened cell walls develop in the primary stem as the inflorescence matures (Nieminen et al., 2004; Ehlting et al., 2005). In bp mutants, lignin deposition in interfascicular fibers and phloem occurs prematurely, showing that part of the function of BP is to delay terminal cell differentiation, potentially until internode elongation is complete (Mele et al., 2003). However, these defects are alleviated by bop1 bop2 mutation, showing that BOP1/2 promotes terminal cell fate and is a developmental regulator of lignin formation. Although BOP1/2 are not normally expressed in Arabidopsis stems, boundaries such as the valve margin of fruits and the base of floral organs or leaves following abscission are lignified in aid of cell separation and scar fortification, respectively (Sexton, 1976; Lewis et al., 2006; Lee et al., 2008). Interestingly, publically available poplar (Populus spp.) microarray data indicates that two potential BOP orthologs are highly expressed in xylem (http://www.bar.utoronto.ca), which suggests a conserved role for BOP1/2 in promoting secondary growth in trees.
Mele et al. (2003) identified four lignin biosynthetic genes whose expression was up-regulated in bp-9 stems. Our study confirmed up-regulation of these genes (PAL1, C4H1, 4CL1, PRXR9GE) as well as several others (C3H1, CAD5, HCT) in bp-2 and bop1-6D stems and internodes. Given that BP binds directly to the promoters of lignin genes (Mele et al., 2003) it will be interesting to confirm biochemically whether BOP1/2 and BP directly regulate a common set of genes to exert their antagonistic functions. Of the lignin genes surveyed, the class III cell wall peroxidize transcript PRXR9GE shows the most dramatic up-regulation in bp-2 and bop1-6D lines (15-fold or more) relative to wild-type control plants. Class III peroxidases are one of several classes of cell wall enzymes that use hydrogen peroxide as an oxidant to generate monolignol phenoxy radicals, thus allowing the spontaneous coupling of monolignols into their polymer form (Boerjan et al., 2003; Passardi et al., 2004). Peroxidase activity is low in seedlings and increases with age in the aerial parts of the plant (Mele et al., 2003; Cosio and Dunand, 2010). Thus, the final step of lignin formation may be a key point of developmental control, making the transcriptional regulation of PRXR9GE an interesting case study.
In conclusion, our data establish BOP1/2 gain of function as the basis of bp and pny inflorescence defects. Our study shows that ectopic BOP1/2 activity in stems both restricts growth and promotes terminal cell differentiation, dramatically altering inflorescence architecture. Future studies will establish the molecular basis of antagonism between BP-PNY and BOP1/2. Ultimately, this work will provide important insight into how changes in the interplay between KNOX-BELL factors in the meristem and BOP1/2 in lateral organ boundaries drives species variation in inflorescence architecture.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Plants were grown in growth chambers on agar plates and/or in soil at 21°C in 24-h light (100 μmol m−2s−1). Wild type was the Col-0 ecotype of Arabidopsis (Arabidopsis thaliana) unless stated otherwise. The double mutant bop1-3 bop2-1 was described previously (Hepworth et al., 2005). Mutant alleles of as2-1 (CS3117), pny-40126 (SALK_40126), knat2-5 (SALK_099837), and knat6-2 (SALK_054482) were obtained from the Arabidopsis Biological Resource Center and described previously (Byrne et al., 2000; Iwakawa et al., 2002; Smith and Hake, 2003; Belles-Boix et al., 2006). Mutant alleles of bp-1 and bp-2 (introgressed from RLD into Col-0) were provided by Raju Datla (Venglat et al., 2002). The strong 35S:BOP2 line and activation-tagged overexpression line bop1-6D were kindly provided by O. Nilsson (Norberg et al., 2005). The reporter lines KNAT2:GUS (C24 ecotype) and KNAT6:GUS (Wassilewskija ecotype) were gifts from Veronique Pautot (Dockx et al., 1995; Belles-Boix et al., 2006). The reporter line BLR:GUS (Ler ecotype, here called PNY:GUS) was provided by Mary Byrne (Byrne et al., 2003). Control crosses to Col determined that ecotype does not affect the expression pattern of GUS reporter genes. The reporter line BOP2:GUS is described elsewhere (Xu et al., 2010). All mutant combinations were constructed by crossing and confirmed by PCR genotyping where possible.
Primers and Genotyping
Primers used for genotyping, plasmid construction, and transcript analysis are listed in Supplemental Table S1. The strategy for genotyping bop1-3, bop2-1, pny-40126, knat2-5, and knat6-2 Salk T-DNA insertion mutants was as described (www.signal.salk.edu). For genotyping bp-2, primers bp-2dCAPS-F1 and bp-2dCAPs-R1 were used to amplify products from wild-type and bp-2 genomic DNA. The product from Col wild type is slightly larger than the corresponding product from bp-2, allowing their resolution on a 3.5% agarose gel.
Construction of 35S:BOP1, 35S:BOP2, and tCUP4:BOP1 Transgenic Lines
To create pBAR/35S:BOP1/2 constructs, a fragment containing one copy of the viral 35S promoter was excised from p35S:BOP2 (Norberg et al., 2005) by digestion with EcoR1 and BamHI and cloned into the corresponding site of pBAR1 (a gift from the Dangl Lab, University of North Carolina) to create the intermediate plasmid pBAR/35S. Primer pairs B1-1/B1-2 and B2-1/B2-2 1 incorporating BamHI restriction sites were used to amplify BOP1 and BOP2 coding sequences, respectively, from cloned cDNA templates. The resulting PCR products were digested with BamHI and ligated into the corresponding site in pBAR to generate pBAR/35S-BOP1 and pBAR/35S-BOP2. The EntCUP4 promoter is an alternative constitutive promoter (Malik et al., 2002). To create ptCUP4:BOP1, a DNA fragment containing the BOP1 coding sequence was amplified by PCR from cloned cDNA template using EcoR1-BOP1-F1 and BOP1-RR as the primers. The resulting fragment was digested with EcoRI and BamHI and ligated into the corresponding sites of pBAR1 to generate the intermediate plasmid pBAR1/BOP1. A 0.5-kb DNA fragment containing the EntCUP4 promoter was then amplified by PCR using pEntCUP4-nos-GUS as a plasmid template and EcoR1-tCUP-F1 and EcoR1-tCUP-R1 as the primers. The resulting fragment was digested with EcoR1 and ligated into the corresponding sites of pBAR/BOP1 to create ptCUP4:BOP1. Wild-type plants were transformed by floral dipping (Clough and Bent, 1998) using the Agrobacterium strain C58C1 pGV101 pMP90 (Koncz and Schell, 1986). Basta-resistant transformants were selected on soil using the herbicide Finale (AgrEvo). Phenotypes were scored in the T1 generation.
Phenotypic Analysis of Inflorescence Structure
Quantitative phenotypic analyses of 6-week-old plants were performed as described (Ragni et al., 2008). Phyllotaxy measurements were obtained as previously described (Peaucelle et al., 2007). The divergence angle between the insertion points of two successive floral pedicels along the main inflorescence was measured. Divergence angles were measured for the first 15 siliques of each inflorescence (counting acropetally) according to the orientation that resulted in the smallest average divergence angle. Angle of pedicel orientation was determined using a protractor to measure the angle of pedicel attachment relative to the stem. Orientation was measured for the first 11 siliques of each inflorescence (counting acropetally).
In Situ Hybridization and Localization of GUS Activity
Tissues were fixed and analyzed for GUS activity essentially as described by Sieburth and Meyerowitz (1997). Tissues were stained for 2 to 18 h at 37°C and cleared overnight with 70% ethanol prior to imaging. Alternatively, stained tissues were embedded in Paraplast Plus (Sigma). Sections (10 μm) cut with a microtome were affixed to glass slides and dewaxed with tert-butanol and xylene prior to imaging. In situ hybridizations were performed as described (Xu et al., 2010). Primers used to make BP and KNAT6 antisense probes are listed in Supplemental Table S1.
SEM
Samples were prepared for SEM as described in Hepworth et al. (2005). Images were collected using a Vega-II XMU variable pressure SEM (Tescan).
Lignin Staining
Tissue sections (25 μm) were cut from paraffin-embedded mature green siliques to analyze replum patterning or from elongated internodes between the third and fourth siliques on the primary stem to analyze stem patterning. Tissue sections affixed to glass slides were dewaxed and dehydrated prior to addition of 2% phloroglucinol (in 95% ethanol) followed by 6N HCl for color development. For the analysis of lignin at stem bases, cross sections were cut from the base of 32-d-old flowering plants with a razor blade and placed in 3 mL of 2% phloroglucinol solution. After 5 min, five drops of concentrated HCl were added. Two minutes were allowed for color development and images were immediately collected.
qRT-PCR
Total RNA was isolated from leaves, pedicels, internodes, or the base of bolting stems (bottom 2.5 cm of 32-d-old flowering plants) using Trizol reagent (Invitrogen). cDNA was generated using 1 μg of total RNA as the template and Superscript III RT (Invitrogen) as the polymerase. qPCR was performed in triplicate using 2 μL of 10-fold diluted cDNA as the template in reactions containing SYBR Green and IQ Supermix (BioRad) using a Rotor-Gene 6000 thermocycler (Qiagen). Annealing conditions were optimized for each primer pair and data quality was verified by melting curve analysis. Relative transcript levels were calculated as described (Murmu et al., 2010). Values were normalized to GAPC and then to the wild-type control. For Figure 9G only, cDNA was generated using 2 μg of total RNA as the template and diluted 20-fold. ACTIN2 was used as a normalization control. Reactions were performed in triplicate using an annealing temperature of 55°C. All experiments were repeated at least twice with independently isolated RNA with similar results obtained. Primers for the analysis of lignin genes are given in Supplemental Table S2.
Sequence data for genes described in this article can be found in the GenBank/EMBL data libraries under the accession numbers: At2g41370 (BOP1), At3g57130 (BOP2), At1g70510 (KNAT2), At1g23380 (KNAT6), At4g08150 (BP), At5g02030 (PNY), At3g04120 (GAPC), At2g37040 (PAL1), At2g30490 (C4H1), At1g51680 (4CL1), At2g40890 (C3H1), At4g34050 (CCoMT1), At4g34230 (CAD5), At3g21770 (AtPRXR9GE), and At3g18780 (ACT2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phenotypic suppression of bp-2 inflorescence defects by bop1 bop2.
Supplemental Figure S2. Comparison of stem-pedicel junctions in wild type and mutants.
Supplemental Figure S3. Loss-of-function bop1 bop2 restores replum formation in pny mutants.
Supplemental Figure S4. KNAT6:GUS and KNAT2:GUS expression patterns in wild-type plants.
Supplemental Figure S5. BOP1:GUS expression pattern in wild type and mutants.
Supplemental Figure S6. Quantitative analysis of BOP2 transcript in 35S:BOP2 lines crossed to wild type and mutants.
Supplemental Figure S7. Inflorescence phenotype of 35S:KNAT6 transgenic plants and the double mutants bp-2 as2-1 and pny as2-1.
Supplemental Figure S8. Comparison of BP and PNY expression levels in wild type, bop1 bop2, and bop1-6D inflorescence stems.
Supplemental Figure S9. Analysis of stem lignification pattern in wild type and mutants with pny.
Supplemental Table S1. List of general primers.
Supplemental Table S2. List of primers for qPCR analysis of lignin genes.
Supplementary Material
Acknowledgments
We are grateful to the colleagues mentioned in the text for providing mutant alleles and reporter lines and to the The Arabidopsis Information Resource database for genome information. We are also grateful to Eryang Li for critical comments on the manuscript.
References
- Achard P, Genschik P. (2009) Releasing the brakes of plant growth: how GAs shutdown DELLA proteins. J Exp Bot 60: 1085–1092 [DOI] [PubMed] [Google Scholar]
- Barton MK. (2010) Twenty years on: the inner workings of the shoot apical meristem, a developmental dynamo. Dev Biol 341: 95–113 [DOI] [PubMed] [Google Scholar]
- Baucher M, El Jaziri M, Vandeputte O. (2007) From primary to secondary growth: origin and development of the vascular system. J Exp Bot 58: 3485–3501 [DOI] [PubMed] [Google Scholar]
- Bell AD, Bryan A. (2008) Plant Form: An Illustrated Guide to Flowering Plant Morphology. Timber Press, London, pp 170–173 [Google Scholar]
- Belles-Boix E, Hamant O, Witiak SM, Morin H, Traas J, Pautot V. (2006) KNAT6: an Arabidopsis homeobox gene involved in meristem activity and organ separation. Plant Cell 18: 1900–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt AM, Etchells JP, Canales C, Lagodienko A, Dickinson H. (2004) VAAMANA—a BEL1-like homeodomain protein, interacts with KNOX proteins BP and STM and regulates inflorescence stem growth in Arabidopsis. Gene 328: 103–111 [DOI] [PubMed] [Google Scholar]
- Boerjan W, Ralph J, Baucher M. (2003) Lignin biosynthesis. Annu Rev Plant Biol 54: 519–546 [DOI] [PubMed] [Google Scholar]
- Bowman JL, Eshed Y. (2000) Formation and maintenance of the shoot apical meristem. Trends Plant Sci 5: 110–115 [DOI] [PubMed] [Google Scholar]
- Byrne ME, Barley R, Curtis M, Arroyo JM, Dunham M, Hudson A, Martienssen RA. (2000) Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408: 967–971 [DOI] [PubMed] [Google Scholar]
- Byrne ME, Groover AT, Fontana JR, Martienssen RA. (2003) Phyllotactic pattern and stem cell fate are determined by the Arabidopsis homeobox gene BELLRINGER. Development 130: 3941–3950 [DOI] [PubMed] [Google Scholar]
- Chuck G, Lincoln C, Hake S. (1996) KNAT1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis. Plant Cell 8: 1277–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cole M, Nolte C, Werr W. (2006) Nuclear import of the transcription factor SHOOT MERISTEMLESS depends on heterodimerization with BLH proteins expressed in discrete sub-domains of the shoot apical meristem of Arabidopsis thaliana. Nucleic Acids Res 34: 1281–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosio C, Dunand C. (2010) Transcriptome analysis of various flower and silique development stages indicates a set of class III peroxidase genes potentially involved in pod shattering in Arabidopsis thaliana. BMC Genomics 11: 528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean G, Casson S, Lindsey K. (2004) KNAT6 gene of Arabidopsis is expressed in roots and is required for correct lateral root formation. Plant Mol Biol 54: 71–84 [DOI] [PubMed] [Google Scholar]
- Dockx J, Quaedvlieg N, Keultjes G, Kock P, Weisbeek P, Smeekens S. (1995) The homeobox gene ATK1 of Arabidopsis thaliana is expressed in the shoot apex of the seedling and in flowers and inflorescence stems of mature plants. Plant Mol Biol 28: 723–737 [DOI] [PubMed] [Google Scholar]
- Douglas SJ, Chuck G, Dengler RE, Pelecanda L, Riggs CD. (2002) KNAT1 and ERECTA regulate inflorescence architecture in Arabidopsis. Plant Cell 14: 547–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlting J, Mattheus N, Aeschliman DS, Li E, Hamberger B, Cullis IF, Zhuang J, Kaneda M, Mansfield SD, Samuels L, et al. (2005) Global transcript profiling of primary stems from Arabidopsis thaliana identifies candidate genes for missing links in lignin biosynthesis and transcriptional regulators of fiber differentiation. Plant J 42: 618–640 [DOI] [PubMed] [Google Scholar]
- Fletcher JC. (2002) Shoot and floral meristem maintenance in Arabidopsis. Annu Rev Plant Biol 53: 45–66 [DOI] [PubMed] [Google Scholar]
- Gómez-Mena C, Sablowski R. (2008) ARABIDOPSIS THALIANA HOMEOBOX GENE1 establishes the basal boundaries of shoot organs and controls stem growth. Plant Cell 20: 2059–2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Thomas J, Collins G, Timmermans MC. (2008) Direct repression of KNOX loci by the ASYMMETRIC LEAVES1 complex of Arabidopsis. Plant Cell 20: 48–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha CM, Jun JH, Nam HG, Fletcher JC. (2004) BLADE-ON-PETIOLE1 encodes a BTB/POZ domain protein required for leaf morphogenesis in Arabidopsis thaliana. Plant Cell Physiol 45: 1361–1370 [DOI] [PubMed] [Google Scholar]
- Ha CM, Jun JH, Nam HG, Fletcher JC. (2007) BLADE-ON-PETIOLE 1 and 2 control Arabidopsis lateral organ fate through regulation of LOB domain and adaxial-abaxial polarity genes. Plant Cell 19: 1809–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha CM, Kim G-T, Kim BC, Jun JH, Soh MS, Ueno Y, Machida Y, Tsukaya H, Nam HG. (2003) The BLADE-ON-PETIOLE 1 gene controls leaf pattern formation through the modulation of meristematic activity in Arabidopsis. Development 130: 161–172 [DOI] [PubMed] [Google Scholar]
- Hackbusch J, Richter K, Müller J, Salamini F, Uhrig JF. (2005) A central role of Arabidopsis thaliana ovate family proteins in networking and subcellular localization of 3-aa loop extension homeodomain proteins. Proc Natl Acad Sci USA 102: 4908–4912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamant O, Pautot V. (2010) Plant development: a TALE story. C R Biol 333: 371–381 [DOI] [PubMed] [Google Scholar]
- Hay A, Kaur H, Phillips A, Hedden P, Hake S, Tsiantis M. (2002) The gibberellin pathway mediates KNOTTED1-type homeobox function in plants with different body plans. Curr Biol 12: 1557–1565 [DOI] [PubMed] [Google Scholar]
- Hepworth SR, Zhang Y, McKim S, Li X, Haughn GW. (2005) BLADE-ON-PETIOLE-dependent signaling controls leaf and floral patterning in Arabidopsis. Plant Cell 17: 1434–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakawa H, Ueno Y, Semiarti E, Onouchi H, Kojima S, Tsukaya H, Hasebe M, Soma T, Ikezaki M, Machida C, et al. (2002) The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol 43: 467–478 [DOI] [PubMed] [Google Scholar]
- Jun JH, Ha CM, Fletcher JC. (2010) BLADE-ON-PETIOLE1 coordinates organ determinacy and axial polarity in Arabidopsis by directly activating ASYMMETRIC LEAVES2. Plant Cell 22: 62–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanrar S, Onguka O, Smith HMS. (2006) Arabidopsis inflorescence architecture requires the activities of KNOX-BELL homeodomain heterodimers. Planta 224: 1163–1173 [DOI] [PubMed] [Google Scholar]
- Koncz C, Schell J. (1986) The promoter of TL-DNA genes controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet 204: 383–396 [Google Scholar]
- Lee Y, Derbyshire P, Knox JP, Hvoslef-Eide AK. (2008) Sequential cell wall transformations in response to the induction of a pedicel abscission event in Euphorbia pulcherrima (poinsettia). Plant J 54: 993–1003 [DOI] [PubMed] [Google Scholar]
- Lewis MW, Leslie ME, Liljegren SJ. (2006) Plant separation: 50 ways to leave your mother. Curr Opin Plant Biol 9: 59–65 [DOI] [PubMed] [Google Scholar]
- Lincoln C, Long J, Yamaguchi J, Serikawa K, Hake S. (1994) A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell 6: 1859–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik K, Wu K, Li XQ, Martin-Heller T, Hu M, Foster E, Tian L, Wang C, Ward K, Jordan M, et al. (2002) A constitutive gene expression system derived from the tCUP cryptic promoter elements. Theor Appl Genet 105: 505–514 [DOI] [PubMed] [Google Scholar]
- McKim SM, Stenvik G-E, Butenko MA, Kristiansen W, Cho SK, Hepworth SR, Aalen RB, Haughn GW. (2008) The BLADE-ON-PETIOLE genes are essential for abscission zone formation in Arabidopsis. Development 135: 1537–1546 [DOI] [PubMed] [Google Scholar]
- Mele G, Ori N, Sato Y, Hake S. (2003) The knotted1-like homeobox gene BREVIPEDICELLUS regulates cell differentiation by modulating metabolic pathways. Genes Dev 17: 2088–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murmu J, Bush MJ, DeLong C, Li S, Xu M, Khan M, Malcolmson C, Fobert PR, Zachgo S, Hepworth SR. (2010) Arabidopsis basic leucine-zipper transcription factors TGA9 and TGA10 interact with floral glutaredoxins ROXY1 and ROXY2 and are redundantly required for anther development. Plant Physiol 154: 1492–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieminen KM, Kauppinen L, Helariutta Y. (2004) A weed for wood? Arabidopsis as a genetic model for xylem development. Plant Physiol 135: 653–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norberg M, Holmlund M, Nilsson O. (2005) The BLADE ON PETIOLE genes act redundantly to control the growth and development of lateral organs. Development 132: 2203–2213 [DOI] [PubMed] [Google Scholar]
- Ori N, Eshed Y, Chuck G, Bowman JL, Hake S. (2000) Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 127: 5523–5532 [DOI] [PubMed] [Google Scholar]
- Passardi F, Penel C, Dunand C. (2004) Performing the paradoxical: how plant peroxidases modify the cell wall. Trends Plant Sci 9: 534–540 [DOI] [PubMed] [Google Scholar]
- Peaucelle A, Louvet R, Johansen JN, Salsac F, Morin H, Fournet F, Belcram K, Gillet F, Höfte H, Laufs P, et al. (2011) The transcription factor BELLRINGER modulates phyllotaxis by regulating the expression of a pectin methylesterase in Arabidopsis. Development 138: 4733–4741 [DOI] [PubMed] [Google Scholar]
- Peaucelle A, Morin H, Traas J, Laufs P. (2007) Plants expressing a miR164-resistant CUC2 gene reveal the importance of post-meristematic maintenance of phyllotaxy in Arabidopsis. Development 134: 1045–1050 [DOI] [PubMed] [Google Scholar]
- Ragni L, Belles-Boix E, Günl M, Pautot V. (2008) Interaction of KNAT6 and KNAT2 with BREVIPEDICELLUS and PENNYWISE in Arabidopsis inflorescences. Plant Cell 20: 888–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder AHK, Ferrándiz C, Yanofsky MF. (2003) The role of the REPLUMLESS homeodomain protein in patterning the Arabidopsis fruit. Curr Biol 13: 1630–1635 [DOI] [PubMed] [Google Scholar]
- Rutjens B, Bao D, van Eck-Stouten E, Brand M, Smeekens S, Proveniers M. (2009) Shoot apical meristem function in Arabidopsis requires the combined activities of three BEL1-like homeodomain proteins. Plant J 58: 641–654 [DOI] [PubMed] [Google Scholar]
- Saleh O, Issman N, Seumel GI, Stav R, Samach A, Reski R, Frank W, Arazi T. (2011) MicroRNA534a control of BLADE-ON-PETIOLE 1 and 2 mediates juvenile-to-adult gametophyte transition in Physcomitrella patens. Plant J 65: 661–674 [DOI] [PubMed] [Google Scholar]
- Schwechheimer C, Willige BC. (2009) Shedding light on gibberellic acid signalling. Curr Opin Plant Biol 12: 57–62 [DOI] [PubMed] [Google Scholar]
- Semiarti E, Ueno Y, Tsukaya H, Iwakawa H, Machida C, Machida Y. (2001) The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates lamina formation, establishment of venation and repression of meristem-related homeobox genes in the leaf. Development 128: 1771–1783 [DOI] [PubMed] [Google Scholar]
- Sexton R. (1976) Some ultrastructural observations on the nature of foliar abscission in Impatiens sultani. Planta 128: 49–58 [DOI] [PubMed] [Google Scholar]
- Shi C-L, Stenvik G-E, Vie AK, Bones AM, Pautot V, Proveniers M, Aalen RB, Butenko MA. (2011) Arabidopsis class I KNOTTED-like homeobox proteins act downstream in the IDA-HAE/HSL2 floral abscission signaling pathway. Plant Cell 23: 2553–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth LE, Meyerowitz EM. (1997) Molecular dissection of the AGAMOUS control region shows that cis elements for spatial regulation are located intragenically. Plant Cell 9: 355–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith HM, Boschke I, Hake S. (2002) Selective interaction of plant homeodomain proteins mediates high DNA-binding affinity. Proc Natl Acad Sci USA 99: 9579–9584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith HMS, Hake S. (2003) The interaction of two homeobox genes, BREVIPEDICELLUS and PENNYWISE, regulates internode patterning in the Arabidopsis inflorescence. Plant Cell 15: 1717–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeves TA, Sussex IM. (1989) Patterns in Plant Development. Academic, New York [Google Scholar]
- Sussex IM, Kerk NM. (2001) The evolution of plant architecture. Curr Opin Plant Biol 4: 33–37 [DOI] [PubMed] [Google Scholar]
- Venglat SP, Dumonceaux T, Rozwadowski K, Parnell L, Babic V, Keller W, Martienssen R, Selvaraj G, Datla R. (2002) The homeobox gene BREVIPEDICELLUS is a key regulator of inflorescence architecture in Arabidopsis. Proc Natl Acad Sci USA 99: 4730–4735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Hu T, McKim SM, Murmu J, Haughn GW, Hepworth SR. (2010) Arabidopsis BLADE-ON-PETIOLE1 and 2 promote floral meristem fate and determinacy in a previously undefined pathway targeting APETALA1 and AGAMOUS-LIKE24. Plant J 63: 974–989 [DOI] [PubMed] [Google Scholar]
- Zhou J, Lee C, Zhong R, Ye Z-H. (2009) MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell 21: 248–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.