Abstract
AtNAP is a NAC family transcription factor gene that plays a key role in leaf senescence but its underlying mechanisms are not known. SENESCENCE-ASSOCIATED GENE113 (SAG113), a gene encoding a Golgi-localized protein phosphatase 2C family protein phosphatase, mediates abscisic acid (ABA)-regulated stomatal movement and water loss specifically during leaf senescence. Here we report that SAG113 is a direct target gene of the AtNAP transcription factor. We found that both AtNAP and SAG113 were induced by leaf senescence and ABA. When AtNAP was chemically induced, SAG113 was also induced whereas when AtNAP was knocked out, the ABA- and senescence-induced expression of SAG113 was reduced. These data suggest that the expression of SAG113 is predominantly dependent on AtNAP. Functionally, overexpression of SAG113 restored the markedly delayed leaf senescence phenotype in atnap knockouts to wild type. Yeast (Saccharomyces cerevisiae) one-hybrid experiments and electrophoresis mobility shift assays showed that AtNAP could physically bind to the SAG113 promoter in vivo and in vitro, respectively. Site-directed mutagenesis revealed that AtNAP binds to a 9-bp core sequence of the SAG113 promoter, 5′CACGTAAGT3′. These results indicate that there is a unique regulatory chain, ABA-AtNAP-SAG113 protein phosphastase 2C, which controls stomatal movement and water loss during leaf senescence.
The terminal phase in a leaf’s life history is generally referred to as leaf senescence. It is characterized by a massive operation of programmed cell death that is accompanied by active recycling of nutrients released from the dying cells to active growing tissues or storage organs. Thus, leaf senescence represents a major developmental switch in the leaf’s lifespan from a functional photosynthetic organ to catabolic suicide. This transition, like many other developmental switches and responses to environmental cues, involves large numbers of genes that are differentially turned on or off (Guo et al., 2004; Buchanan-Wollaston et al., 2005). Transcription factors (TFs) often act as a switch to cause differential gene expression by binding to specific cis-elements of their target gene promoters, resulting in the activation and/or suppression of the target genes (Guo et al., 2004). Various TF genes have been found to be associated with leaf senescence (e.g. Guo et al., 2004; Buchanan-Wollaston et al., 2005). For example, 134 out of 1,500 TF genes in Arabidopsis (Arabidopsis thaliana) are found in senescent leaves; these senescence-associated TFs belong to more than 34 gene families based on their DNA-binding domains, including NAC, WRKY, C2H2, APE2, MYB, HB, and bZIP (Guo et al., 2004). The biological functions of these TFs in leaf senescence, however, are largely unknown except for a few genes such as AtNAP (Guo and Gan, 2006), ORS1 (Balazadeh et al., 2011), and WRKY53 (Miao et al., 2004) that have been genetically and/or physiologically proven to have a role in Arabidopsis leaf senescence. NAM-B1 (likely a ortholog of AtNAP in wheat [Triticum aestivum]; Uauy et al., 2006) and a RAV-like gene in soybean (Glycine max; Zhao et al., 2008) have also been shown to be involved in leaf senescence. AtNAP, NAM-B1, and ORS1 belong to the plant-specific NAC superfamily of TFs. A T-DNA insertion knockout of AtNAP displayed a remarkable 10-d delay in leaf senescence. In contrast, inducible overexpression of AtNAP in young leaves caused precocious senescence. Similarly, reduction of the expression of NAM-B1 homologs using RNAi delayed senescence for up to 3 weeks in wheat. These results suggest that AtNAP is a key regulator of leaf senescence (Guo and Gan, 2006). However, little is known as to how AtNAP regulates leaf senescence.
Although leaf senescence is a genetically controlled process, it can be triggered and regulated by various environmental cues (such as drought stress, nutrient deficiency, darkness, extreme temperature, and pathogen infection) and plant hormones (such as abscisic acid [ABA], ethylene, jasmonic acid, salicylic acid, cytokinins, and auxin). Comparative transcriptomic analyses of leaf senescence in Arabidopsis revealed that leaf senescence processes induced by various factors share common execution events but different hormonal, pathological, or environmental signals that lead to initiation of senescence may do so through distinct signal transduction pathways (Guo and Gan, 2012). More than 200 genes coding for components of several signal transduction pathways are associated with the processes, including the mitogen-activated protein kinase pathways, G proteins, Ca2+-dependent protein kinases, receptor-like kinases, calmodulins, and protein phosphatase (PP) 2A and 2C (Hajouj et al., 2000; Guo et al., 2004; Zhou et al., 2009; Xu et al., 2011). Protein kinases and phosphatases add or remove phosphate groups from proteins, respectively, to relay signals to modulate various biochemical and molecular processes. A MKK9-MPK6 cascade has been shown to regulate senescence in either detached or in planta Arabidopsis leaves (Zhou et al., 2009). Recently, an ABA- and senescence-up-regulated PP2C family PP named SENESCENCE-ASSOCIATED GENE113 (SAG113) has been shown to be localized in the cis-Golgi apparatus and, as a negative regulator of ABA signaling, to specifically suppress the stomata from closing such that senescing leaves lose water rapidly, leading to senescence and ultimate desiccation (Zhang et al., 2011). However, it is not known how this SAG113 PP2C gene is regulated and what relation (if any) this PP2C may have with the above-discussed AtNAP.
Here we report our deciphering of an ABA-AtNAP-SAG113 PP2C regulatory chain that controls stomatal movement and water loss specifically in senescing leaves in Arabidopsis.
RESULTS
Both AtNAP and SAG113 Are Coexpressed during Leaf Senescence and in Response to ABA Treatment
We previously reported that AtNAP is a NAC family TF that plays a role in leaf senescence (Guo and Gan, 2006). We also found that SAG113 encodes a PP2C family PP that controls water loss in senescing leaves (Zhang et al., 2011). Both AtNAP and SAG113 are expressed in senescing Arabidopsis leaves (Fig. 1A). Both genes can also be induced by ABA treatment, and the transcript levels of AtNAP and SAG113 are significantly increased upon ABA treatment compared with controls (mock and no treatment; Fig. 1B).
Figure 1.
Both AtNAP and SAG113 are induced by leaf senescence and ABA. A, Coexpression patterns of AtNAP and SAG113 in senescing leaves. Y, Young leaves; M, mature leaves; ES, leaves at early senescence stage; LS, leaves at late senescence stage. B, Induction of AtNAP by ABA. C, Induction of SAG113 by ABA in wild-type or atnap knockout mutants. RNA gel-blot analyses were used. 0 indicates the wild-type mature leaves (35 DAG, all rosette leaves) without any treatment; M indicates the wild-type mature leaves treated with mock solution (0.005% silwett 77) for 3 h; ABA indicates the wild-type or atnap mature leaves treated with ABA (100 μm) for 3 h.
When AtNAP Is Chemically Induced, SAG113 Is Coinduced
The above coexpression patterns raised the possibility that SAG113 is a downstream target gene of the AtNAP TF. If so, SAG113 should be induced when AtNAP is chemically induced in young, nonsenescing leaves. Plasmid 7001 is a control binary vector that allows plants to express an activator (Aoyama and Chua, 1997; Guo and Gan, 2006). Plasmid 1167 is a binary vector in which the AtNAP coding region is fused to a recombinant promoter; the recombinant promoter can be bound by the activator from 7001 to direct the expression of the gene of interest (e.g. AtNAP in this case; Guo and Gan, 2006) upon dexamethasone (DEX; a synthetic hormone) treatment. As shown in Figure 2, in leaves of the control plants harboring 7001, there are no detectable AtNAP transcripts before and after DEX induction; similarly, there are also no SAG113 transcripts. In contrast, in leaves of plants that harbor both 7001 and 1167 vectors, AtNAP transcripts were readily detectable after 3 h of the DEX induction, and the transcript levels increased with prolonged DEX induction. Likewise, SAG113 is also induced by DEX in these leaves and its transcripts accumulated to higher levels with longer induction times (Fig. 2). These data suggest that SAG113 is likely regulated by the AtNAP TF.
Figure 2.
Chemical induction of AtNAP coinduces SAG113 expression. RNA gel-blot analysis of SAG113 expression after AtNAP was induced at 0, 3, 6, 12, and 24 h with DEX treatment. 7001: transgenic plants harbor the vector pTA7001; 1167/7001: transgenic plants harboring both pTA7001 and pGL1167. AtNAP can be induced in the 1167/7001 plants only; the detailed information of the inducible expression of AtNAP was shown in Guo and Gan (2006).
Senescence-Associated and ABA-Regulated Expression of SAG113 Is Predominantly Dependent on AtNAP
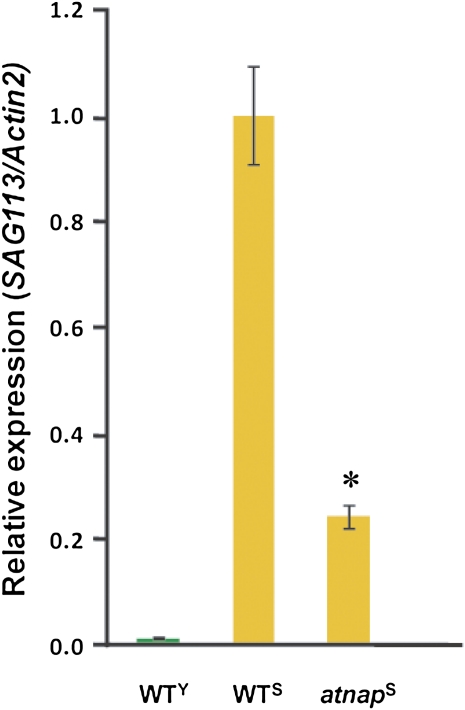
We then investigated the expression of SAG113 in the absence of AtNAP (i.e. in the atnap knockout lines). Quantitative PCR (qPCR) analysis revealed that the expression of SAG113 in senescent leaves (WTS) of wild-type Arabidopsis plants was relatively high compared with almost nondetectable expression in the young leaves (WTY; Fig. 3), which is consistent with the RNA gel-blot analysis (Fig. 1A). However, in the similarly senescent leaves (atnapS) of the atnap knockout plants, the expression levels of SAG113 was significantly reduced to 20% of that in WTS (Fig. 3), suggesting that the senescence-associated expression of SAG113 is predominantly dependent on AtNAP. Similarly, as shown in Figure 1C, the ABA-induced expression of SAG113 was also dramatically reduced in the atnap knockouts. These data strongly suggest that the ABA- and senescence-up-regulated SAG113 expression depends largely on the presence of AtNAP.
Figure 3.
qPCR analysis of SAG113 in senescing leaves of wild-type and the atnap knockout plants. WTY, Wild-type young leaves (the fifth and sixth leaves, counted from bottom); WTS, the senescing fifth and sixth leaves (with 25% yellowing) of wild type; atnapS, the senescing fifth and sixth leaves (with 25% yellowing) of the atnap knockout plants. Asterisks indicate significant differences between WTY and WTS and atnapS. *, Student’s t test; P < 0.05. [See online article for color version of this figure.]
AtNAP TF Binds to the Promoter Region of SAG113 in Vivo
Based on the data presented above, we hypothesized that SAG113 is a direct target gene of AtNAP. In other words, the AtNAP TF physically binds to the promoter region of SAG113 to activate its expression at the transcriptional level. To test this hypothesis, the promoter regions and promoter deletions of SAG113 as shown in a through j of Figure 4B were cloned in front of a LacZ reporter gene construct as promoter baits to form various reporter constructs (Fig. 4A), and the AtNAP coding sequence was fused with the yeast (Saccharomyces cerevisiae) GAL4 activation domain (GAD) to form the effector AD-AtNAP construct shown in Figure 4A. Yeast cells containing both the effector AD-AtNAP (the effector Ctr AD as negative control) and the reporter constructs will express the AtNAP-GAD fusion protein, and if the AtNAP can bind to the cloned SAG113 promoter sequence of the reporter gene construct, the GAD will be able to direct LacZ expression, resulting in the blue color accumulation in the presence of substrate 5-bromo-4-chloro-3-indolyl-β-glucuronic acid. As shown in Figure 4B, these yeast one-hybrid experiments confirmed the ability of AtNAP to bind to the SAG113 promoter baits such as a through e, h, and i. These experiments also mapped the AtNAP-binding site to a 52-bp region that is located at −216 to −267 bp from the transcription start site (as shown in i of Fig. 4B).
Figure 4.
Yeast one-hybrid analysis of the binding of AtNAP to the SAG113 promoter truncations. A, Diagram of constructs used in the studies. The GAD and AtNAP fusion genes driven by the GAL1 (PGAL1) promoter serves as effectors. The GAD protein served as negative control to see if there exits the self activity of each SAG113 promoter truncations. The LacZ gene driven by SAG113 promoter truncations served as the reporter to test the binding activity of the GAD-AtNAP fusion protein to individual promoter truncations. B, The fusion protein GAD-AtNAP, but not GAD alone, can strongly bind to some of the promoter truncations of SAG113 to direct LacZ expression in yeast cells that turns 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside to blue compound. a through j contain different lengths of the SAG113 promoter sequence. The promoter sequences were amplified using primers indicated, e.g. G2923 and G2924 for the 1,001-bp fragment in a. The transcription start site was numbered as +1. Both a and b contain 119-bp 5′ untranslated region of SAG113. The shortest promoter truncation that can be bound by AtNAP is 52 bp in length as shown in i. [See online article for color version of this figure.]
AtNAP TF Binds to the Promoter Region of SAG113 in Vitro
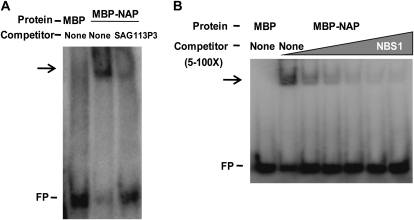
The in vivo interaction of the AtNAP TF with the 52-bp promoter sequence of SAG113 was further confirmed by electrophoresis mobility shift assay (EMSA; Fig. 5A). The recombinant protein of maltose-binding protein (MBP)-AtNAP overexpressed in, and purified from, Escherichia coli was coincubated with the 32P-labeled 52-bp promoter sequence of SAG113. MBP is from a bacterial vector that facilitates purification of the recombinant protein. If AtNAP can bind to the DNA sequence, the DNA-protein complex will migrate more slowly in gel electrophoresis than DNA alone (Fig. 5A, lane 2). When unlabeled 52-bp promoter (competitor) of SAG113 was added to the reaction, the slow migrating band was significantly reduced (Fig. 5A, lane 3), indicating that AtNAP specifically binds to the 52-bp promoter region. Subsequent experiments also showed that AtNAP was able to bind to the central 36-bp fragment (5′GAGTAGTGTTAGACTTTGATTGGTGCACGTAAGTGT3′, named as NBS1 for NAP-BINDING SEQUENCE1) of the 52-bp sequence. This binding is also specific, because the binding could be competed off with the increased amounts of the unlabeled competitor (Fig. 5B).
Figure 5.
The specific binding of AtNAP to the SAG113 promoter in vitro. A, EMSA assays show that the MBP-AtNAP fusion protein, but not MBP, can specifically bind to the 52-bp promoter sequence that was identified in Figure 4. Fifty-fold excessive unlabeled 52-bp fragment was used to compete with the 32P-labeled 52-bp probe. B, AtNAP specifically binds to a 36-bp region of the 52-bp fragment in A. The synthesized 36-bp probe was incubated with 250 ng of the MBP-AtNAP fusion protein. Competition experiments were carried out by adding 5-, 10-, 25-, 50-, and 100-fold excessive unlabeled 36-bp fragments. The arrow indicates the up-shifted bands. FP, Free probe. MBP is from the expression vector pMAL-c2.
Fine Mapping of the AtNAP-Binding Site to a 9-bp Sequence in the Promoter Region of SAG113
Further EMSA analyses were used to define the binding sequence of the AtNAP TF within the 36-bp NBS1 described above. NBS1 was subjected to site-directed mutagenesis with six transversions each time. The six mutated NBS1 segments, namely NBS1m1 through NBS1m6 (Fig. 6A), served as competitors of NBS1 in the EMSA. The competition tests revealed that the first 6-bp segment is not required for the AtNAP binding, and that the last 12-bp segment, 5′GCACGTAAGTGT3′, is particularly essential for the binding (Fig. 6A).
Figure 6.
Determination of AtNAP-binding core sequence of the SAG113 promoter using EMSA and site-directed mutagenesis. A, The effects of 6-bp transversion mutations of the SAG113 promoter on AtNAP binding. Fifty-fold excessive unlabeled 36-bp fragments (NBS1) with respective mutations were used as competitors. The mutated sequences are indicated in red. These are all transversion mutations. B, The effects of deletions on AtNAP binding. Fifty-fold unlabeled 36-bp (NBS1), 30-bp (NBS2), and 24-bp (NBS3) probes were used as competitors in EMSA. C, Diagram of a series of single base pair transversion mutations (in red) of NBS2. The 3′ end 12 bp of NBS2 in B were mutated one by one. D, The effects of single base pair transversion mutations on AtNAP binding. The mutated sequences in C were used as competitors in the EMSA. The 9-bp sequence (5′CACGTAAGT3′) immediately below the gel is the important core nucleotides for AtNAP binding. [See online article for color version of this figure.]
A series of EMSA assays involving deletions of the NBS1 were also performed to define the AtNAP-binding sequence. As shown in Figure 6B, NBS2 (30 bp in length) could effectively compete with the 32P-labeled NBS1 (36 bp) while NBS3 (24 bp) was less competitive, suggesting NBS2 has intact binding ability but NBS3 loses partial binding activity by AtNAP.
Because the last 12-bp segment was shown to be essential for the AtNAP binding (Fig. 6A) and because NBS2 (30 bp) possessed full binding capacity by AtNAP (Fig. 6B), individual transversion mutations for the respective 12 nucleotides of NBS2, namely NBS2m5-1 through NBS2m5-6 and NBS2m6-1 through NBS2m6-6, were made (Fig. 6C). These mutated fragments were used to compete with the 32P-labeled NBS2. The results showed that a 9-bp segment, 5′CACGTAAGT3′, is the core sequence involved in the interaction with AtNAP (Fig. 6D).
The Delayed Leaf Senescence Phenotype of atnap Knockout Can Be Restored to Wild Type by Overexpressing SAG113
The above data demonstrate that AtNAP can bind to the promoter region of SAG113 and regulate the expression levels of SAG113 to affect leaf senescence. To functionally test this, SAG113 was constitutively overexpressed in the atnap knockout background. atnap knockouts displayed a significantly delayed leaf senescence phenotype (Guo and Gan, 2006). The delay in leaf senescence in atnap plants was almost completely restored to the wild-type phenotype in the atnap plants overexpressing SAG113 driven by 35S promoter in planta (Fig. 7, A and B) and in detached leaves (Fig. 7, C and D). Specifically, two randomly selected transgenic plants overexpressing SAG113, lines 3119-1 and 3119-2, exhibited wild-type-like senescence phenotype in both attached and detached leaves, and the chlorophyll contents of these transgenic plants were restored to wild-type levels and were significantly lower than those of the atnap knockout mutants.
Figure 7.
Restoration of the delayed leaf senescence phenotype of atnap mutants to wild type by constitutive SAG113 overexpression. A, Phenotypes of age-matched plants (approximately 35 DAG) of wild type, atnap, 3119-1, and 3119-2. 3119-1 and 3119-2 are randomly chosen lines of atnap transformed with 35S::SAG113. The constitutive expression of SAG113 rescued the delayed leaf senescence phenotype of atnap. B, The chlorophyll contents of the fifth and sixth rosette leaves of plants in A. Mean values of four samples ± se are shown. Asterisks indicate significant differences between wild-type, atnap mutant, and transgenic plants (Student’s t test, P < 0.05). C, Phenotypes of leaves detached from the age-matched 30 DAG plants in A. The detached leaves were incubated in light for 5 d. The leaves were counted from bottom (oldest). D, The chlorophyll contents in the fifth to eighth leaves described in C. Mean values of four samples ± se are shown. Asterisks indicate significant differences between wild-type, atnap mutant, and transgenic plants (Student’s t test, P < 0.05). [See online article for color version of this figure.]
DISCUSSION
Leaf senescence is a genetically well-controlled program. Although during leaf senescence the vast majority of genes such as those related to photosynthesis are down-regulated, expression of a subset of new genes termed SAGs is required for leaf cells to undergo senescence (for review, see Gan and Amasino, 1997; Gan, 2003). Much effort has been devoted to identify SAGs. Various genomic approaches have led to an estimation of approximately 10% of genes in the Arabidopsis genome or over 2,500 genes that are up-regulated during leaf senescence (He et al., 2001; Guo et al., 2004). Many of the SAGs are involved in such senescence processes as gene regulation, signal transduction, macromolecular degradation, and nutrition remobilization (He et al., 2001; Gepstein et al., 2003; Guo et al., 2004; Buchanan-Wollaston et al., 2005; Guo and Gan, 2012). With a large number of SAGs identified so far, one of the immediate questions is how the products of these genes act in a network. It has been postulated that there are multiple pathways corresponding to various environmental and internal factors that are interlinked to form a gene network of leaf senescence (figure 3 in Gan and Amasino, 1997). This hypothesis is supported by recent comparative transcriptomic analyses of gene expression profiles that are induced by developmental leaf senescence and 27 senescence-promoting hormonal, pathological, and environmental stress treatments (Guo and Gan, 2012). As an effort toward SAG networking, we investigated the relationship between one TF (AtNAP) and one signal transduction component (SAG113 PP2C family PP) and provided evidence that these two SAGs and the ABA hormone are connected together to form an ABA-AtNAP-SAG113 PP2C regulatory chain that controls stomatal closure specifically during leaf senescence in Arabidopsis (Fig. 8). The AtNAP TF acts as a master regulator of leaf senescence (Guo and Gan, 2006) and the SAG113 PP2C PP serves as a negative regulator of ABA to prevent stomatal closure, resulting in water loss from the leaf, which in turn triggers leaf senescence (Zhang et al., 2011) presumably by slowly ceasing the cell’s biochemical activities. ABA is generally synthesized in response to water deficiency, which promotes stomatal closure to prevent water loss prior to senescence (Schroeder et al., 2001; Finkelstein and Rock, 2002; Hauser et al., 2011). Numerous reports have shown that ABA levels in senescing leaves are elevated (Gepstein and Thimann, 1980; Zhang et al., 2011). However, the ABA-stimulated stomatal closure mechanism is not desirable at the onset of and during leaf senescence. It is thus reasonable to speculate that the ABA-AtNAP-SAG113 PP2C regulatory chain has evolved to overcome ABA’s promotion of stomatal closure to allow faster water loss from senescing leaves, leading to ultimate cell death and desiccation. The complete desiccation may prevent pathogen infection and proliferation. In the regulatory path, elevated ABA activates AtNAP TF, which in turn binds to the 9-bp core sequence of the promoter of SAG113, to activate this target gene at the transcriptional level; the Golgi-localized SAG113 PP2C family PP then acts as a negative regulator in the ABA signaling pathway that modulates the stomatal movement specifically in senescing leaves so that the water loss can be orchestrated as a part of the senescence processes (Fig. 8).
Figure 8.
A working model of ABA-AtNAP-SAG113 PP2C regulatory chain in leaf senescence in Arabidopsis. Elevated ABA induces AtNAP, and subsequently the AtNAP TF activates its immediate target gene SAG113 (encoding a PP2C) and other target genes. SAG113 PP2C may also be induced by ABA to a less extent. SAG113 PP2C in turn inhibits the stomatal closure in leaves, resulting in faster water loss, and consequent triggering of leaf senescence processes. [See online article for color version of this figure.]
It is generally believed that there are sophisticated genetic regulatory networks that orchestrate leaf senescence by regulating appropriate gene functions at transcriptional, posttranscriptional, translational, and posttranslational levels. TFs are responsible for gene regulation at the transcriptional levels. Approximately 130 putative TF genes are expressed during leaf senescence in Arabidopsis, 24 of them, including AtNAP, are NAC family members (Guo et al., 2004). AtNAP, AtNAP’s possible ortholog in wheat, and ORS1 have been shown to have pivotal roles in regulating leaf senescence (Guo and Gan, 2006; Uauy et al., 2006; Balazadeh et al., 2011).
To explore how the NAC family TFs, AtNAP in particular, function in the regulation of leaf senescence will be facilitated by identifying the downstream genes. In our current study, SAG113 is identified and confirmed to be a direct target gene of AtNAP by yeast one-hybrid (Fig. 4) and EMSA analyses (Fig. 5). Both AtNAP and SAG113 are expressed during leaf senescence and are induced by ABA with very similar kinetics. There exist ABA-responsive elements in the promoters of both AtNAP and SAG113 (Supplemental Information S1). The ABA- and leaf-senescence-induced SAG113 expression is predominantly dependent on AtNAP because its expression levels are significantly reduced in the atnap knockout mutants (Figs. 1C and 3). The dependence of the SAG113 expression on AtNAP is also evidenced by the fact that both AtNAP and SAG113 are not expressed in nonsenescing leaves, but when AtNAP is chemically induced in the nonsenescing leaves, SAG113 is also induced (Fig. 2). Functionally, overexpression of SAG113 in atnap could largely rescue the delayed leaf senescence phenotype of atnap (Fig. 7), suggesting that AtNAP regulates leaf senescence at least partially through the SAG113, a negative regulatory component in the ABA signaling pathway that specifically inhibits stomata from closure (Zhang et al., 2011). Compared with the phenotype of atnap, the delayed leaf senescence phenotype of SAG113 is weaker (Guo and Gan, 2006; Zhang et al., 2011), which suggests there are other direct target genes that also play important roles in leaf senescence downstream of the AtNAP TF.
The AtNAP-SAG113 regulatory node is further supported by the fact that the AtNAP TF is indeed binding to the promoter region of SAG113 as revealed using the yeast one-hybrid system (Fig. 4) and EMSA analyses (Figs. 5 and 6). As shown in Figure 6A, AtNAP binds to a 30-bp segment (GTTAGACTTTGATTGGTGCACGTAAGTGT) of the SAG113 promoter region. Further analyses revealed that a 9-bp segment (CACGTAAGT) of the 30-bp fragment constitutes the core-binding region (Fig. 6D). In addition to the core-binding motif, this study also revealed that the first six nucleotides TGTTAG flanking the core sequence (NBS2 in Fig. 6A) are also important for tighter and perhaps accurate binding of the TF to the SAG113 promoter. The identification of the binding sequence may facilitate the identification of other potential target genes of AtNAP.
To our knowledge, this 9-bp binding sequence (CACGTAAGT) represents a new senescence-specific motif in gene regulation during leaf senescence. This sequence is distinctive from the potential binding sequences ([AG]CGT[AG](4-5n)[AG][CT]ACGCAA) of the hydrogen-peroxide-responsive NAC gene named ORS1 that have been recently reported (Balazadeh et al., 2011). The 9-bp sequence is also quite different from several other sequences that are bound by non-senescence-related NAC family TFs. For example, ANAC055, a NAC family protein that functions in drought stress, was found to interact with a 4-bp core sequence (CACG; Tran et al., 2004). The Arabidopsis NAC1 protein was shown to bind to a 21-bp fragment (CTGACGTAAGGGATGACGCAC) of the 35S-90 promoter (Xie et al., 2000). Duval et al. (2002) also independently demonstrated that purified AtNAM recombinant protein protected the region of the cauliflower mosaic virus 35S promoter between −70 and −76, which is the underlined sequence within the 21-bp fragment (Duval et al., 2002). The 35S promoter region does not contain the AtNAP core-binding motif.
CONCLUSION
The leaf-senescence-specific TF AtNAP binds to a 9-bp core sequence of the promoter region of its direct target gene named SAG113, which encodes a PP2C family PP, to form an AtNAP-SAG113 PP2C regulatory node. This TF node mediates ABA-regulated inhibition of stomatal closure specifically in senescing leaves in Arabidopsis.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia was used in the study. The atnap knockout mutants and the related AtNAP-inducible expression lines are in Columbia background (Guo and Gan, 2006). Seeds were sterilized with three rinses in 70% ethanol containing 0.01% Triton X-100, and then sown on petri dishes containing Murashige and Skoog salts with 0.7% w/v phytoagar (Sigma) and appropriate antibiotics. The dishes were kept at 4°C for 2 d and then moved to a growth chamber at 22°C with 60% relative humidity under continuous light (110 μmol m−1 s−1) from a mixture of fluorescent and incandescent bulbs. Approximately 8 d after germination (DAG), seedlings were transplanted to Cornell mix soils (3:2:1 peat moss:vermiculite:perlite, v/v/v) and grew in a growth chamber. The mutants, transgenic plants, and wild type were grown side by side.
Plasmid Construction
For constructing pGL3182 (for producing the AtNAP protein in Escherichia coli for EMSA assays), the coding region of AtNAP was amplified using primers G2909 (5′-GATGAATTCATGGAAGTAACTTCCCAATC-3′, the underlined section is an engineered EcoRI site) and G2910 (5′-GATCTCGAGCTA AAACTTAAACATCGC TT-3′, the underlined section is an engineered XhoI site) on the template of cDNA from senescence leaves. The PCR products were cloned into pGEM-T (Promega) to form pGL3170. The coding sequence was then released from pGL3170 with EcoRI and XhoI, and was subcloned into EcoRI and SalI sites of pMAL-c2 (New England Biolabs) to form pGL3182 for producing recombinant MBP-AtNAP fusion protein. MBP is from pMAL-c2 for facilitating purification of the fusion protein.
Yeast (Saccharomyces cerevisiae) one-hybrid assay related constructs: To construct plasmid pGL3175 (for producing GAD-AtNAP fusion protein in yeast), the AtNAP coding sequence was released from pGL3170 with EcoRI and XhoI, and was subcloned into pAD42 to form pGL3175. To construct SAG113P::LacZ reporter gene, the 1,001-bp SAG113 promoter region (including 119 bp of 5′ untranslated region sequence of SAG113) was amplified from genomic DNA using primers G2923(5′-GATGAATTCCTAAGAAGTATTCACGCACC-3′, the underlined section is an engineered EcoRI site) and G2924 (5′-GATCTCGAGATCGTCAAAACTATTCAACA-3′, the underlined section is an engineered XhoI site). The amplified fragment was ligated to pGEM-T easy vector (Promega) and sequenced. The correct promoter sequence was released from the plasmid with EcoRI and XhoI, and cloned into pLacZi2μ vector (Lin et al., 2007) to form pGL3183. Other LacZ reporter gene plasmids containing various truncated SAG113 promoter were similarly constructed using the primers listed in Supplemental Table S1.
For constructing pGL3119 (for overexpression of the SAG113 protein in atnap mutant background), the coding region of SAG113 was amplified using primers G2786 (5′-CTGCAGATGGCTGAGATTTGTTACGA-3′, the underlined section is an engineered PstI site) and G2787 (5′-TCTAGACTACGTGTCTCGTCGTAGAT-3′, the underlined section is an engineered XbaI site). The PCR products were cloned into pGEM-T (Promega) to form pGL3116. The coding sequence was then released from pGL3116 with PstI and XbaI, and was subcloned into PstI and XbaI sites of pGL800 (a modified 35S overexpression vector in pZP211 backbone) to form pGL3119. All the constructs were confirmed by sequencing.
Yeast One-Hybrid Assay
Yeast one-hybrid assays were performed as described by Lin et al. (2007). Plasmid for GAD::AtNAP fusion (pGL3175) was cotransformed with different LacZ reporter gene constructs containing different lengths of the SAG113 promoter fragments into the yeast strain EGY48 using standard transformation techniques. Transformants were grown on proper drop-out plates containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside for the blue color development.
EMSA Assay
The EMSA assay was performed according to Zhu et al. (2003) with minor modifications. The 52-bp nucleotide sequence (5′TGAGTAGTGTTAGACTTTGATTGGTGCACGTAAGTGTTTCGTATCGCGATTT3′) was PCR amplified using a pair of primers G3202 and G3205 on the template of pGL3183 described above (Supplemental Table S2). The amplified fragments were digested with EcoRI/XhoI, recovered, and then labeled with Klenow Fragment (3′→5′ exo−; New England Biolabs) with 32P. The 36-, 30-, and 24-bp nucleotide sequences and their mutation variation with EcoRI and XhoI ends (Supplemental Table S2) were synthesized (Integrated DNA Technologies). The synthesized fragments were annealed and labeled as described above. DNA-protein-binding reactions (30 μL each reaction) were performed as follows: 10 fmol of labeled probe was mixed with 0.2 μg recombinant protein, 6 μg of herring DNA, with or without cold competitor DNA. The DNA-binding reaction buffer contained 20 mm Tris-HCl (pH 8.5), 50 mm KCl, 5 mm MgCl2, 1 mm EDTA, 1 mm dithiothreitol, 200 ng/μL bovine serum albumin, and 10% glycerol. The DNA-binding mixtures were incubated at 25°C for 20 min and then subjected to electrophoresis on 6% acrylamide gels containing 22.5 mm Tris borate, pH 8.0, 0.25 mm EDTA. The gels were subsequently dried and exposed to x-ray film.
Chlorophyll Assay and Transcript Analysis
Chlorophyll was extracted and quantified as described previously (He and Gan, 2002). Total RNA extractions from Arabidopsis leaves and RNA gel-blot analyses were performed according to He and Gan (2002). The probes were labeled using a prime-a-gene kit (Promega) and subsequent hybridizations were performed at 65°C. The DNA templates for probe labeling were amplified using various pairs of primers (Supplemental Table S2). The ethidium-bromide-stained RNA gels were used for loading controls. The DNA template for the SAG113 probe labeling was amplified using primers G2575 (5′-ACTAGTATGGCTGAGATTTGTTACGA-3′) and G2576 (5′-CTGCAGCTACGTGTCTCGTCGTAGAT-3′). The DNA template for the AtNAP probe labeling were amplified using primers G10100 (5′-CACTAGTTCCTGTTCTATTAGATTG-3′) and G10101 (5′-GCTGCAGTAACT TTTCAAGCACATC-3′).
First-strand cDNA was synthesized from 3 μg of total RNA (treated with DNase) at 42°C with Promega MV-reverse transcriptase. For each reverse transcription-PCR, 1 μL of each diluted sample was used as a template in a 25-μL reaction following the standard methods. For real-time PCR, all PCR reactions were performed on a Bio-Rad IQ-5 thermocycler with 40 cycles and an annealing temperature of 55°C. Cycle threshold values were determined by the IQ-5 Bio-Rad software assuming 100% primer efficiency. The primers G3221 (5′-CGGGTGGTCGTGTTATCTACTG-3′) and G3222 (5′-CCTCCGGTCTGCTGATTACATAC-3′) were used for SAG113 gene qPCR assay. The primers G3053 (5′-AGTGGTCGTACAACCGGTATTGT-3′) and G3054 (5′-GATGGCATGAGGAAGAGAGAAAC-3′) were used for Action 2 gene qPCR assay. Three repetitions were performed for each combination of cDNA samples and primer pairs.
Plant Transformation
Various binary vectors were transferred into Agrobacterium tumefaciens strain ABI1 and the transformed Agrobacterium cells were used to transform atnap mutant (Guo and Gan, 2006) via the floral-dip method (Clough and Bent, 1998). Approximately 30 antibiotics-resistant T1 transgenic lines for each transgene were selected; phenotypic analyses were performed in the T2 generation. Homozygous plants were used in all experiments.
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: AtNAP (AT1G69490), SAG113 (AT5G59220), and Actin 2 (AT3G18780).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Primers used in SAG113 promoter truncation.
Supplemental Table S2. Primers used in EMSA assays.
Supplemental Information S1. ABRE motif (ACGTG) in the promoter region of AtNAP and SAG113.
Supplementary Material
Acknowledgments
We thank Dr. Haiyang Wang (Yale University) for providing pAD42 and pLacZi2μ plasmids and yeast strain EGY48. We also thank William Gan (Cornell University) and the members in the Gan lab for critical reading of the manuscript and useful discussions.
References
- Aoyama T, Chua NH. (1997) A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J 11: 605–612 [DOI] [PubMed] [Google Scholar]
- Balazadeh S, Kwasniewski M, Caldana C, Mehrnia M, Zanor MI, Xue G-P, Mueller-Roeber B. (2011) ORS1, an H2O2-responsive NAC transcription factor, controls senescence in Arabidopsis thaliana. Mol Plant 4: 346–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan-Wollaston V, Page T, Harrison E, Breeze E, Lim PO, Nam HG, Lin JF, Wu SH, Swidzinski J, Ishizaki K, et al. (2005) Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J 42: 567–585 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Duval M, Hsieh TF, Kim SY, Thomas TL. (2002) Molecular characterization of AtNAM: a member of the Arabidopsis NAC domain superfamily. Plant Mol Biol 50: 237–248 [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Rock CD. (2002) Abscisic acid biosynthesis and response. The Arabidopsis Book 1: e0058. doi:0010.1199/tab.0058
- Gan S-S. (2003) Mitotic and postmitotic senescence in plants. Sci SAGE KE 2003: re7 [DOI] [PubMed] [Google Scholar]
- Gan S-S, Amasino RM. (1997) Making sense of senescence: molecular genetic regulation and manipulation of leaf senescence. Plant Physiol 113: 313–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gepstein S, Sabehi G, Carp M-J, Hajouj T, Nesher MFO, Yariv I, Dor C, Bassani M. (2003) Large-scale identification of leaf senescence-associated genes. Plant J 36: 629–642 [DOI] [PubMed] [Google Scholar]
- Gepstein S, Thimann KV. (1980) Changes in the abscisic acid content of oat leaves during senescence. Proc Natl Acad Sci USA 77: 2050–2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Cai Z, Gan S-S. (2004) Transcriptome of Arabidopsis leaf senescence. Plant Cell Environ 27: 521–549 [Google Scholar]
- Guo Y, Gan S-S. (2006) AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J 46: 601–612 [DOI] [PubMed] [Google Scholar]
- Guo Y, Gan S-S. (2012) Convergence and divergence in gene expression profiles induced by leaf senescence and 27 senescence-promoting hormonal, pathological and environmental stress treatments. Plant Cell Environ 35: 644–655 [DOI] [PubMed] [Google Scholar]
- Hajouj T, Michelis R, Gepstein S. (2000) Cloning and characterization of a receptor-like protein kinase gene associated with senescence. Plant Physiol 124: 1305–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser F, Waadt R, Schroeder JI. (2011) Evolution of abscisic acid synthesis and signaling mechanisms. Curr Biol 21: R346–R355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Gan S-S. (2002) A gene encoding an acyl hydrolase is involved in leaf senescence in Arabidopsis. Plant Cell 14: 805–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Tang W, Swain JD, Green AL, Jack TP, Gan S-S. (2001) Networking senescence-regulating pathways by using Arabidopsis enhancer trap lines. Plant Physiol 126: 707–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Ding L, Casola C, Ripoll DR, Feschotte C, Wang H. (2007) Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science 318: 1302–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y, Laun T, Zimmermann P, Zentgraf U. (2004) Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol Biol 55: 853–867 [DOI] [PubMed] [Google Scholar]
- Schroeder JI, Allen GJ, Hugouvieux V, Kwak JM, Waner D. (2001) Guard cell signal transduction. Annu Rev Plant Physiol Plant Mol Biol 52: 627–658 [DOI] [PubMed] [Google Scholar]
- Tran LS, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. (2004) Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16: 2481–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uauy C, Distelfeld A, Fahima T, Blechl A, Dubcovsky J. (2006) A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 314: 1298–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q, Frugis G, Colgan D, Chua NH. (2000) Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev 14: 3024–3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Meng T, Li P, Yu Y, Cui Y, Wang Y, Gong Q, Wang NN. (2011) A soybean dual-specificity kinase, GmSARK, and its Arabidopsis homolog, AtSARK, regulate leaf senescence through synergistic actions of auxin and ethylene. Plant Physiol 157: 2131–2153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Xia X, Zhang Y, Gan S-S. (December 1, 2011) An ABA-regulated and Golgi-localized protein phosphatase controls water loss during leaf senescence in Arabidopsis. Plant J http://dx.doi.org/10.1111/j.1365-313X.2011.04821.x [DOI] [PubMed] [Google Scholar]
- Zhao L, Luo Q, Yang C, Han Y, Li W. (2008) A RAV-like transcription factor controls photosynthesis and senescence in soybean. Planta 227: 1389–1399 [DOI] [PubMed] [Google Scholar]
- Zhou C, Cai Z, Guo Y, Gan S-S. (2009) An Arabidopsis mitogen-activated protein kinase cascade, MKK9-MPK6, plays a role in leaf senescence. Plant Physiol 150: 167–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Cai XL, Wang ZY, Hong MM. (2003) An interaction between a MYC protein and an EREBP protein is involved in transcriptional regulation of the rice Wx gene. J Biol Chem 278: 47803–47811 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.