Abstract
Clade A protein phosphatases type 2C (PP2Cs) are negative regulators of abscisic acid (ABA) signaling that are inhibited in an ABA-dependent manner by PYRABACTIN RESISTANCE1 (PYR1)/PYR1-LIKE (PYL)/REGULATORY COMPONENTS OF ABA RECEPTORS (RCAR) intracellular receptors. We provide genetic evidence that a previously uncharacterized member of this PP2C family in Arabidopsis (Arabidopsis thaliana), At5g59220, is a negative regulator of osmotic stress and ABA signaling and that this function was only apparent when double loss-of-function mutants with pp2ca-1/ahg3 were generated. At5g59220-green fluorescent protein and its close relative PP2CA-green fluorescent protein showed a predominant nuclear localization; however, hemagglutinin-tagged versions were also localized to cytosol and microsomal pellets. At5g59220 was selectively inhibited by some PYR/PYL ABA receptors, and close relatives of this PP2C, such as PP2CA/ABA-HYPERSENSITIVE GERMINATION3 (AHG3) and AHG1, showed a contrasting sensitivity to PYR/PYL inhibition. Interestingly, AHG1 was resistant to inhibition by the PYR/PYL receptors tested, which suggests that this seed-specific phosphatase is still able to regulate ABA signaling in the presence of ABA and PYR/PYL receptors and therefore to control the highly active ABA signaling pathway that operates during seed development. Moreover, the differential sensitivity of the phosphatases At5g59220 and PP2CA to inhibition by ABA receptors reveals a functional specialization of PYR/PYL ABA receptors to preferentially inhibit certain PP2Cs.
Abscisic acid (ABA) is a key phytohormone that regulates plant response to abiotic and biotic stress as well as plant development and growth. In seeds, ABA regulates several processes essential for seed viability and germination, including the accumulation of protein and lipid reserves, the induction of dormancy, and the acquisition of tolerance to desiccation (Cutler et al., 2010). Recently, a core signaling pathway has been established that connects ABA perception, inactivation of protein phosphatases type 2C (PP2Cs), and activation of three SUCROSE NONFERMENTING1-RELATED SUBFAMILY2 (SnRK2s) protein kinases (i.e. SnRK2.2/D, -2.3/I, and -2.6/OST1/E; Cutler et al., 2010). Under basal ABA levels, at least six clade A PP2Cs (Schweighofer et al., 2004), ABA-INSENSITIVE1 (ABI1), ABI2, ABA-HYPERSENSITIVE1 (HAB1), HAB2, ABA-HYPERSENSITIVE GERMINATION1 (AHG1), and PP2CA/AHG3, act as negative regulators of ABA signaling, either through dephosphorylation of SnRK2s or interaction with other targets (Sheen, 1998; Gosti et al., 1999; Merlot et al., 2001; Tähtiharju and Palva, 2001; Himmelbach et al., 2002; Leonhardt et al., 2004; Saez et al., 2004, 2006, 2008; Kuhn et al., 2006; Miao et al., 2006; Yoshida et al., 2006; Nishimura et al., 2007; Umezawa et al., 2009; Vlad et al., 2009). When ABA levels rise, the PYR/PYL ABA receptors inactivate PP2Cs in an ABA-dependent manner, which leads to the activation of SnRK2.2, -2.3, and -2.6/OST1 and subsequent phosphorylation of downstream targets (e.g. members of the ABF/AREB transcription factors that recognize ABRE promoter sequences or regulatory components of the stomatal aperture, such as the anion channel SLAC1; Fujii et al., 2009; Fujita et al., 2009; Geiger et al., 2009; Lee et al., 2009). In Xenopus laevis oocytes and in vitro studies, it has been also shown that ABI1 inhibits the calcium-dependent kinases CPK21 and CPK23, which have the anion channels SLAC1 and SLAH3 as substrates, and RCAR1/PYL9 restores the SLAC1 and SLAH3 phosphorylation through ABA-dependent inhibition of ABI1 (Geiger et al., 2010; 2011). Finally, genetic evidence has largely supported the negative role of PP2Cs in ABA signaling; indeed, certain triple loss-of-function pp2c mutants display a partial constitutive response to ABA (Rubio et al., 2009).
According to the sequence alignment of the catalytic phosphatase core, clade A of PP2Cs is arranged in two subgroups, one including ABI1, ABI2, HAB1, and HAB2 and a second one formed by PP2CA/AHG3, AHG1, At5g59220, At1g07430, and At2g29380 (Schweighofer et al., 2004; Supplemental Fig. S1). These three latter PP2Cs are also known as HAI1, HAI2, and HAI3, respectively, for HIGHLY ABA-INDUCED PP2C genes, and interaction of HAI1 with SnRK2.2 has been reported (Fujita et al., 2009). However, At1g07430 had been previously named AIP1 (Lee et al., 2007), for AKT1-INTERACTING PP2C, and later, At2g29380 and At5g59220 were named AIPH1 and AIPH2, for AIP1 HOMOLOGS 1 and 2, respectively (Lee et al., 2009). Therefore, the current nomenclature on At5g59220 reflects a connection with either ABA signaling or the regulation of K+ transport. Intriguingly, it has been reported that At5g59220, now named PP2CA2, plays a positive role in ABA signaling, because the corresponding T-DNA loss-of-function mutant shows an ABA-hyposensitive phenotype in ABA-mediated inhibition of germination and postgerminative growth (Guo et al., 2010). In that case, At5g59220 would represent a singular member of clade A PP2Cs, showing an opposite function to other members of the group.
According to sequence alignment, At5g59220 is closely related to PP2CA/AHG3 (Supplemental Fig. S1), which has been implicated as a key negative regulator of ABA signaling, since pp2ca mutant alleles show ABA-hypersensitive phenotypes in germination, growth, and stomatal assays. In addition to the dephosphorylation of ABA-activated SnRK2s (Lee et al., 2009; Umezawa et al., 2009), PP2CA has been reported to interact with two ion transporters localized to the plasma membrane (i.e. the K+ channel AKT2 and the anion channel SLAC1; Chérel et al., 2002; Lee et al., 2009). Finally, both PP2CA/AHG3 and AHG1 appear to play an essential role for ABA signaling during seed development and germination (Kuhn et al., 2006; Yoshida et al., 2006; Nishimura et al., 2007), but in contrast to pp2ca-1, the ahg1-1 mutant has no ABA-related phenotype in adult plants and the expression of AHG1 is restricted to seed (Nishimura et al., 2007). In this work, we have analyzed loss-of-function mutants of At5g59220, either single or double mutants with pp2ca-1, and found evidence that At5g59220 is also a negative regulator of osmotic stress and ABA signaling. Additionally, analysis of the biochemical regulation of At5g59220, PP2CA/AHG3, and AHG1 reveals a differential sensitivity to inhibition by ABA and PYR/PYL receptors. Since AHG1 appears to be immune to PYR/PYL-mediated inhibition, this PP2C might control ABA signaling during seed development, even in the presence of ABA and PYR/PYL receptors.
RESULTS
An At5g59220 Loss-of-Function Mutant Reinforces the ABA-Hypersensitive Phenotype of the pp2ca-1 Mutant
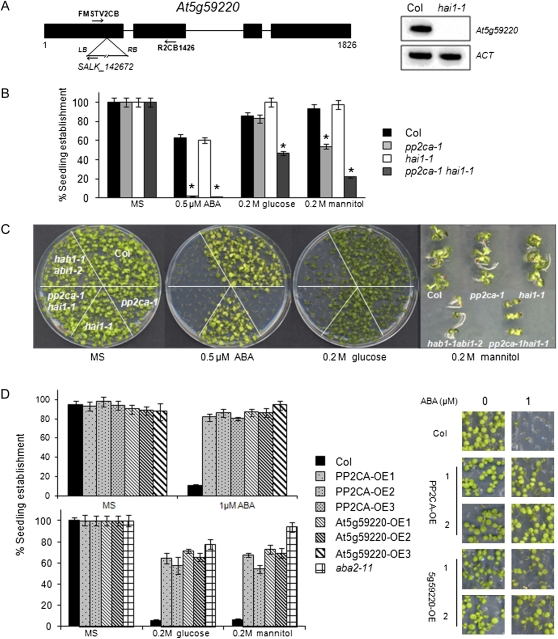
At5g59220 is not expressed in seeds (Nakabayashi et al., 2005; Yoshida et al., 2006; Nishimura et al., 2007), but it is expressed in seedlings or different tissues of adult plants according to public microarray data (Winter et al., 2007; Supplemental Fig. S2). Basal transcript levels of At5g59220 are lower than those reported for other clade A PP2Cs; however, its expression is highly induced by ABA or osmotic stress (Fujita et al., 2009; Yoshida et al., 2010; Supplemental Fig. S2), and this induction was dramatically impaired in the areb1 areb2 abf3 triple mutant (Yoshida et al., 2010). In order to investigate the relative contribution of At5g59220 to ABA signaling, we analyzed the ABA response of the At5g59220 loss-of-function mutant SALK_142672, which we have named hai1-1 (Fig. 1A). ABA-mediated inhibition of seedling establishment was similar in the single hai1-1 mutant compared with the wild type, and a double pp2ca-1 hai1-1 mutant did not show an enhanced response to ABA compared with pp2ca-1, even at low ABA concentrations (Fig. 1B; Supplemental Fig. S3). However, a double pp2ca-1 hai1-1 mutant was more sensitive to Glc- or mannitol-mediated inhibition of seedling establishment than single parental mutants (Fig. 1B; Supplemental Fig. S3).
Figure 1.
A, Schematic diagram of the At5g59220 gene showing the position of the T-DNA insertion in the hai1-1 mutant. RT-PCR analysis of mRNAs from wild-type and hai1-1 mutant seedlings is shown. Primers FMSTV2CB and R2CB1426 were used to amplify part of the At5g59220 cDNA. LB, Left border; RB, right border. B, Seedling establishment of the Columbia (Col) wild type, hai1-1, pp2ca-1, and the double mutant in medium supplemented with ABA, mannitol, or Glc. Data show the percentage of seeds that germinated and developed green cotyledons in the different media at 5 d. Values are averages ± se for three independent experiments (200 seeds each). * P < 0.01 (Student’s t test) with respect to the wild type. C, Photograph of a representative experiment taken 10 d after sowing, with a magnification of representative seedlings grown on MS plates supplemented with 0.2 m mannitol. D, Seedling establishment of wild-type, 35S:HA-PP2CA, and 35S:HA-At5g59220 lines in medium supplemented with 1 μm ABA (top panel), 0.2 m Glc, or 0.2 m mannitol (bottom panel). Approximately 200 seeds of each genotype were sown on each plate and scored 4 d later. Photographs were taken after 8 d. OE indicates overexpression lines. [See online article for color version of this figure.]
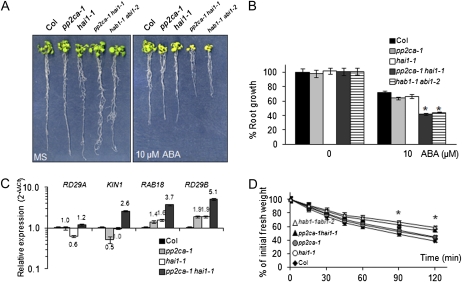
We also generated Arabidopsis (Arabidopsis thaliana) transgenic lines expressing hemagglutinin (HA)-tagged versions of the PP2Cs driven by the 35S promoter. With respect to PP2CA, we confirmed previous results obtained by Kuhn et al. (2006), which showed that overexpression of PP2CA leads to an ABA-insensitive phenotype (Fig. 1C). Likewise, 35S:At5g59220 lines showed diminished sensitivity to ABA-mediated inhibition of seedling establishment and root growth, enhanced water loss, and diminished expression of ABA-inducible genes compared with the wild type (Fig. 1C; Supplemental Fig. S4). Conversely, a characteristic feature of the pp2ca-1 hai1-1 double mutant was an enhanced sensitivity to ABA-mediated inhibition of growth compared with single mutants, which suggests that At5g59220 attenuates ABA signaling in vegetative tissue (Fig. 2, A and B). Therefore, the very moderate ABA hypersensitivity of pp2ca-1 in root assays described by Kuhn et al. (2006) is likely explained by partial redundancy with At5g59220 or other PP2Cs (Rubio et al., 2009).
Figure 2.
A, ABA-hypersensitive growth inhibition of pp2ca-1 hai1-1 and hab1-1 abi1-2 double mutants compared with the wild type and single parental mutants. Photographs show representative seedlings 10 d after the transfer of 4-d-old seedlings from MS medium to plates lacking or supplemented with 10 μm ABA. B, Quantification of ABA-mediated root growth inhibition of pp2ca-1 hai1-1 and hab1-1 abi1-2 double mutants compared with the wild type and single parental mutants. Data are averages ± se from three independent experiments (n = 15 each). * P < 0.01 (Student’s t test) with respect to the wild type. C, Relative expression of ABA-responsive genes in the pp2ca-1 hai1-1 double mutant compared with the wild type and single parental mutants. RT-qPCR analyses were performed in triplicate on RNA samples of 2-week-old seedlings that were either mock treated or 10 μm ABA treated for 3 h. Numbers indicate the expression levels of the genes in each mutant genotype with respect to the wild type in samples treated with ABA (value of 1). Expression of RD29A, KIN1, RAB18, and RD29B was up-regulated 6-, 312-, 86-, and 634-fold by ABA treatment in the wild type, respectively. D, Reduced water loss of pp2ca-1 hai1-1 compared with the Columbia (Col) wild type. Five leaves at the same developmental stage were detached from 21-d-old plants, and fresh weight was determined after submitting them to the drying atmosphere of a flow laminar hood (n = 4 plants per experiment). * P < 0.05 (Student’s t test) with respect to the wild type. [See online article for color version of this figure.]
We also analyzed the transcriptional regulation of ABA-responsive genes in the double pp2ca-1 hai1-1 mutant compared with the wild type and single parental mutants. ABA-mediated induction of the genes KIN1, RAB18, and RD29B was more than twofold higher in the double mutant compared with the other genetic backgrounds (Fig. 2C). Expression of these genes in the single parental mutants showed a less than twofold difference with respect to the wild type. Finally, by measuring the loss of fresh weight of detached leaves, we could observe a reduced water loss of the double pp2ca-1 hai1-1 mutant compared with the wild type and single parental mutants (Fig. 2D).
Subcellular Localization of PP2CA and At5g59220
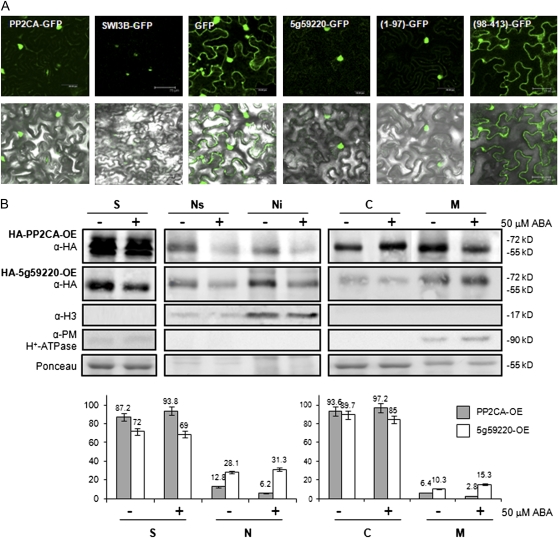
While the catalytic core of At5g59220 is closely related to PP2CA, the N-terminal sequence shows a clear divergence (Supplemental Fig. S1). Several clusters rich in Arg residues are present at the N-terminal sequence of At5g59220. Different programs for the prediction of subcellular localization reveal the presence of nuclear targeting signals in this region; indeed, two nuclear localization patterns are present, both the pattern of four basic residues (type SV40 T antigen) and the bipartite nuclear localization signal (Supplemental Fig. S1). Instead, PP2CA only displays the pattern of four basic residues, which is localized at the C terminus of the protein (Supplemental Fig. S1). In experiments where GFP fusion proteins were transiently expressed in tobacco (Nicotiana benthamiana) epidermal cells, both PP2CA and At5g59220 appeared to be predominantly localized to the nucleus, although some cytosolic expression was also observed (Fig. 3A). Deletion of the N-terminal region of At5g59220 (a construct expressing residues 98–413) led to a subcellular localization of the catalytic phosphatase core similar to GFP, whereas the fusion of residues 1 to 97 of At5g59220 to GFP rendered a nuclear GFP protein (Fig. 3A).
Figure 3.
Subcellular localization of PP2CA and At5g59220. A, Subcellular localization of PP2CA-GFP and At5g59220-GFP proteins transiently expressed in tobacco cells. Epifluorescence and bright-field images of epidermal leaf cells infiltrated with a mixture of Agrobacterium suspensions harboring the indicated constructs and the silencing suppressor p19 are shown. SWI3B is a nuclear protein that forms part of the SWI/SNF chromatin-remodeling complexes (Saez et al., 2008). The N-terminal extension (residues 1–97) and the catalytic core (residues 98–413) of At5g59220 were expressed as fusions with GFP. B, Biochemical fractionation of HA-PP2CA and HA-At5g59220 proteins. Plant material was obtained from epitope HA-tagged PP2CA or At5g59220 transgenic lines after mock treatment or 50 μm ABA treatment for 1 h. Samples were analyzed using anti-HA, anti-histone 3 (α-H3), anti-plasma membrane H+-ATPase antibodies and Ponceau staining of Rubisco. Localization of HA-PP2CA and HA-At5g59220 proteins in soluble (S), total nuclear (N), nuclear soluble (Ns), nuclear insoluble (Ni), cytosolic (C), and microsomal (M) fractions is indicated. Histograms show the relative amounts of each protein in the different fractions. OE indicates overexpression lines. [See online article for color version of this figure.]
Proper elucidation of the subcellular localization of clade A PP2Cs is an important goal to better understand their role in plant physiology; however, biochemical fractionation studies have only been reported for HAB1 (Saez et al., 2008). Since interaction of PP2CA with the plasma membrane transporters AKT2 and SLAC1 has been reported (Chérel et al., 2002; Lee et al., 2009) and interaction of PP2CA and At5g59220 with SnRK2s was localized to both nucleus and cytosol (Fujita et al., 2009), we further investigated the subcellular localization of both PP2Cs by fractionation studies. To this end, we used the Arabidopsis transgenic lines that express HA-tagged versions of the PP2Cs (Fig. 1C). Both HA-PP2CA and HA-At5g59220 proteins appeared to be functional with respect to ABA signaling, since their constitutive expression led to reduced sensitivity to ABA (Fig. 1C). Both proteins showed cytosolic and nuclear localization, and interestingly, part of the protein pool was localized to either the microsomal or nuclear insoluble (chromatin-associated) fraction (Fig. 3B). A relative quantification of the subcellular distribution of HA-PP2CA and HA-At5g59220 indicated that most of the protein was localized at the cytosol, although these data also confirmed that a significant fraction of the protein, 13% and 28% for HA-PP2CA and HA-At5g59220, respectively, was localized in the nucleus. The apparently predominant nuclear localization of transiently expressed GFP-tagged PP2Cs might be explained because the lower volume of the nucleus, compared with the cytosol, leads to a higher concentration of GFP fusion proteins, enhancing the GFP fluorescent signal (Fig. 3A).
Selective Inhibition of At5g59220, PP2CA, and AHG1 by PYR/PYL ABA receptors
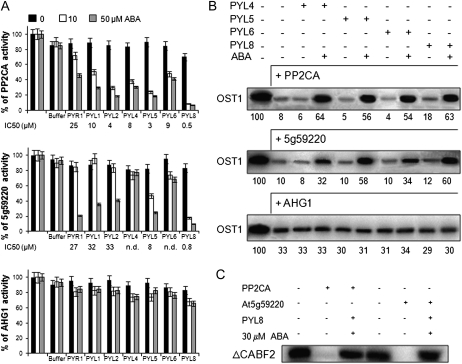
Since both At5g59220 and PP2CA regulate different aspects of ABA signaling, we analyzed its possible regulation by PYR/PYL ABA receptors. Coexpression of these PP2Cs and PYR/PYLs in seedlings, roots, or guard cells could be documented in public microarray databases (Supplemental Fig. S2; Kilian et al., 2007; Winter et al., 2007; Yang et al., 2008). Thus, we analyzed the phosphatase activity of PP2CA and At5g59220 in the presence of seven PYR/PYL receptors, which represent the dimeric class (i.e. PYR1, PYL1, and PYL2) and the monomeric class (i.e. PYL4, PYL5, PYL6, and PYL8; Dupeux et al., 2011b; Hao et al., 2011). Using a 100:1 ratio of receptor:PP2CA, between 40% and 80% of ABA-independent inhibition of PP2CA by some monomeric receptors was recently reported (Hao et al., 2011). However, in our hands, using either a 4:1 ratio in phosphopeptide-based activity assays (Fig. 4A) or a 10:1 ratio in OST1 dephosphorylation assays (Fig. 4B), we did not detect such ABA-independent inhibition of PP2CA by the PYR/PYL receptors tested. In the best case, only a 10% to 15% ABA-independent inhibition of PP2CA by PYL8 was found (Fig. 4, A and B). In the presence of ABA, PP2CA was inhibited by all the receptors, although important differences could be observed depending on the receptor considered. For instance, PP2CA inhibition by PYL8 was much more effective than by PYR1 (inhibitory concentration to obtain 50% inhibition [IC50] = 0.5 and 25 μm, respectively), whereas IC50 values for the other receptors ranged between 4 and 10 μm ABA. Such differences were not noted previously, but it is likely that they were masked by the high concentration of receptor with respect to the PP2C used by Hao et al. (2011). At5g59220 was relatively resistant to inhibition by PYL4 and PYL6 (IC50 > 50 μm), and IC50 for dimeric receptors, such as PYR1, PYL1, and PYL2, was approximately 30 μm ABA, whereas PYL5 and PYL8 where the most effective inhibitors (IC50 = 8 and 0.8 μm, respectively). Indeed, both PYL5 and PYL8 were the most effective inhibitors of PP2CA as well (IC50 = 3.7 and 0.5 μm, respectively).
Figure 4.
Differential sensitivity of PP2CA, At5g59220, and AHG1 to ABA-dependent PYR/PYL-mediated inhibition. A, Phosphatase activity of the different PP2Cs was measured in vitro using a phosphopeptide substrate in the absence or presence of 0.5, 1, 5, 10, 20, 40, or 50 μm ABA and the indicated receptors. For the sake of clarity, only data for 10 and 50 μm ABA are shown as well as IC50 values (n.d., not determined at IC50 > 50 μm). Data are averages ± sd for three independent experiments. Phosphatase assays were performed in a 100-μL reaction volume containing 2.3 μg of His6-PP2CA, 2.1 μg of His6-ΔNAt5g59220, or 2.5 μg of His6-AHG1 and between 5 and 5.7 μg of the different His6-PYR/PYL proteins in order to obtain a 1:4 phosphatase:receptor stoichiometry. The activities of the PP2C recombinant proteins in the absence of ABA (100% activity) were 12.2 ± 0.3, 13.3 ± 0.2, and 11.0 ± 0.2 nmol inorganic phosphate min−1 mg−1, respectively. In order to check the effect of the HIS elution buffer on the PP2C activity, we performed an assay lacking PYR/PYL proteins but adding an equivalent volume of HIS elution buffer. B, In vitro OST1 kinase activity in the absence or presence of 30 μm ABA and the indicated receptors. A 1:10 phosphatase:receptor stoichiometry was used in this assay. The quantification of autoradiography (numbers below) shows the percentage of OST1 phosphorylation in each reaction relative to the first reaction (100%; phosphorylation of OST1 in the absence of PP2Cs). C, Dephosphorylation of ΔCABF2 by PP2CA and At5g59220 in the absence or presence of 30 μm ABA and PYL8.
Structural and genetic studies have shown the importance for the locking mechanism of the ternary receptor:ABA:PP2C complex of a conserved Trp residue of clade A PP2Cs, which establishes a water-mediated hydrogen bond with the ketone group of ABA in ternary complexes (Melcher et al., 2009; Miyazono et al., 2009; Dupeux et al., 2011a; Supplemental Fig. S1). Interestingly, AHG1 is the only clade A PP2C that lacks this conserved Trp (Dupeux et al., 2011a). Therefore, we wondered whether this seed-specific PP2C would be subjected to PYR/PYL regulation. As can be observed in Figure 4A, AHG1 phosphatase activity was not significantly affected by PYR/PYL receptors even at 50 μm ABA. This result indicates that AHG1 could negatively regulate ABA signaling even in the presence of high levels of ABA and PYR/PYL receptors.
To gain additional evidence for the biochemical regulation of the above-described PP2Cs, we also performed in vitro reconstitution of the ABA signaling cascade and tested the protection of OST1 activity by PYL4, PYL5, PYL6, and PYL8 in the presence of the different PP2Cs and ABA (Fig. 4B). Both PP2CA and At5g59220 efficiently dephosphorylated OST1, whereas AHG1 was less effective (Fig. 4). Coincubation of PP2CA in the presence of ABA either with PYL4, PYL5, PYL6 or PYL8 or of At5g59220 with PYL5 or PYL8 notably protected OST1 activity. PYL4 and PYL6 only modestly recovered OST1 activity when coincubated with At5g59220 in the presence of ABA. In agreement with the phosphatase assays described in Figure 4A, coincubation of AHG1 with PYR/PYL receptors did not prevent OST1 dephosphorylation. Finally, we used ABF2 as a substrate of OST1, and after generation of phosphorylated ABF2, we incubated it with PP2CA and At5g59220 (Fig. 4C). Both PP2Cs efficiently dephosphorylated ABF2, whereas coincubation with PYL8 in the presence of ABA abolished their activity against the transcription factor. Taking into account that a significant portion of PP2CA and At5g59220 is localized at the nucleus, these results suggest that ABFs might also be substrates of these PP2Cs.
DISCUSSION
In this work, we provide novel insights on the role of clade A PP2Cs in ABA signaling and their regulation by PYR/PYL receptors. Genetic analysis of hai1-1 indicates that At5g59220 functions as a negative regulator of ABA signaling, although this role has likely been masked by functional redundancy with other PP2Cs. Thus, compared with the wild type and single parental mutants, the pp2ca-1 hai1-1 double mutant showed enhanced ABA-mediated inhibition of growth, induction of ABA-responsive genes, and diminished water loss. Taking into account that both PP2CA and At5g59220 transcripts are themselves strongly induced by ABA or osmotic stress (Fujita et al., 2009), their up-regulation under these conditions likely exerts a negative feedback on ABA and osmotic stress signaling. Glc-mediated inhibition of seedling establishment was also notably enhanced in the pp2ca-1 hai1-1 double mutant compared with single mutants and the wild type. Although part of this effect could be attributed to the enhanced osmotic stress sensitivity of the double mutant, early seedling growth in medium supplemented with Glc was more severely inhibited than by an iso-osmotic concentration of mannitol. Indeed, 0.2 m Glc (3.6%) was relatively well tolerated by parental single mutants or wild-type seedlings, whereas a strong inhibition of early seedling growth was found in the double mutant.
Subcellular localization studies of these PP2Cs indicated that they are present both at the nucleus and cytosol, which is in agreement with their reported interaction with SnRK2s (Fujita et al., 2009). In addition to the dephosphorylation of SnRK2s (Umezawa et al., 2009; this work), these PP2Cs might efficiently dephosphorylate ABFs (Fig. 4C), although further studies are required to firmly establish this point. Interestingly, a minor portion of these PP2Cs colocalized with the nuclear insoluble fraction (chromatin associated), and interaction of PP2CA with SWI3B, a putative component of chromatin-remodeling complexes, was previously reported (Saez et al., 2008). Although a major portion of PP2CA was localized in the cytosol, the presence of PP2CA was also detected in microsomal membranes, where two PP2CA-interacting proteins are localized (i.e. AKT2 and SLAC1).
High ABA levels and presumably active ABA signaling are temporarily correlated with the onset of maturation and the prevention of precocious germination during mid embryo development (Kanno et al., 2010). Thus, ABA levels reach a maximum in the middle of seed development (around 9–10 d after flowering [DAF]), and a second peak of ABA accumulation takes place late in development (15–16 DAF; Kanno et al., 2010). This latter peak appears to be required to regulate the synthesis of proteins involved in desiccation tolerance and the development of seed dormancy. The ABA-induced late embryogenesis-abundant proteins have been proposed to play a key role in protecting proteins and membranes from the severe water loss that occurs during seed desiccation. Different PP2Cs are expressed during seed development to regulate ABA signaling; however, both AHG3/PP2CA and AHG1 are supposed to play a major role according to the seed phenotype of ahg1 and pp2ca mutants and expression levels in seed (Nishimura et al., 2007). Even though AHG1 shares many features with AHG3/PP2CA, detailed characterization of ahg3-1/pp2ca and ahg1-1 mutants has revealed important differences, particularly enhanced ABA hypersensitivity of ahg1 in radicle emergence and deeper seed dormancy compared with ahg3 (Nishimura et al., 2007). Figure 4 shows an additional key difference, since PP2CA is regulated by PYR/PYL receptors whereas AHG1 seems to be immune to such regulation. Interestingly, whereas the expression of PP2CA remained steady during seed development, the expression of AHG1 was detected at 8 DAF and increased until 16 DAF (Nishimura et al., 2007). This expression pattern is similar to that of ABI5, which plays a key role for ABA signaling during seed development, and genetic analysis indicated that AHG1 functions upstream of ABI5 and ABI3 in the ABA pathway (Nishimura et al., 2007). The biochemical assays performed here for AHG1 indicate that this PP2C could partially dephosphorylate a SnRK2 even in the presence of high levels of ABA and PYR/PYL receptors. The presence of a PP2C resistant to inhibition might represent an adaptive response to partially control the highly active ABA signaling pathway that operates during mid and late seed development. Otherwise, since the rest of clade A PP2Cs are inhibited by PYR/PYL receptors, seed ABA signaling would operate in the absence of the negative control imposed by PP2Cs, which might impair the interplay with other hormonal pathways that also operate during seed development, such as cytokinins, auxins, and GAs (Kanno et al., 2010). Indeed, inactivation of AHG1 leads to extreme hypersensitivity to ABA-mediated inhibition of germination, and ahg1 shows a delayed germination in the absence of exogenous ABA (Nishimura et al., 2007).
The analysis of the interaction between PP2Cs and PYR/PYLs has shown that receptor complexes differ in their sensitivity to ABA-mediated inhibition (Santiago et al., 2009; Szostkiewicz et al., 2010). However, PP2Cs such as ABI1, ABI2, and HAB1 all appear to be inhibited at least more than 50% by the different PYR/PYLs tested. This situation also applies to PP2CA but not to At5g59220 (Fig. 4A), which reveals that receptors can discriminate among closely related PP2Cs and preferentially inhibit some of them. Finally, AHG1 represents an exception to the general mechanism of clade A PP2C inhibition based on ABA and PYR/PYL receptors.
ABA-independent inhibition of some PP2Cs has been recently reported for some monomeric receptors using in vitro phosphatase assays (Hao et al., 2011). However, in order to achieve high inhibition of the PP2Cs, a 100:1 ratio of receptor to PP2C was used in these assays, and only PYL10, which is not expressed in the Arabidopsis transcriptome database (http://signal.salk.edu/cgi-bin/atta?GENE=At4g27920), was effective at a 1:1 ratio (Hao et al., 2011). At the ratios used in this work, we did not find evidence for a meaningful inhibition of PP2CA or At5g59220 in the absence of ABA by PYR/PYL receptors either using phosphopeptide-based or OST1 dephosphorylation assays. These latter assays are particularly valuable in this context, because monomeric PYLs compete with SnRK2s to interact with PP2Cs (Melcher et al., 2009; Soon et al., 2011). Indeed, the recent elucidation of a SnRK2-PP2C complex reveals a striking similarity in PP2C recognition by SnRK2 and ABA-bound receptors (Soon et al., 2011). Therefore, it was important to test whether the ABA-independent interaction of monomeric PYLs with PP2Cs was strong enough to efficiently block PP2C-mediated dephosphorylation of SnRK2s (Fig. 4B). In our hands, a major restoration of OST1 activity by PYL-mediated inhibition of PP2Cs was dependent on ABA, which is in agreement with the in vivo results obtained through protoplast transfection assays (Fujii et al., 2009). In the absence of ABA, dimerization in receptors like PYR1, PYL1, and PYL2 prevents basal interactions with the PP2Cs, while monomeric receptors are able to form low-affinity complexes with PP2Cs, but these complexes lack the network of interactions that occur in the ternary complex with ABA (Dupeux et al., 2011a). For instance, they lack the hydrogen bonds established among the conserved Trp residue of clade A PP2Cs, key residues of the receptor gating loops, and the key water molecule that contacts the ketone group of ABA (Melcher et al., 2009; Miyazono et al., 2009; Dupeux et al., 2011b). Finally, biochemical analyses of a natural PP2C version lacking the conserved Trp residue, namely AHG1, or the mutants abi1W300A and hab1W385A further support the structural mechanism of ABA signaling, which indicates that ternary receptor:ABA:phosphatase complexes are required to fully inhibit PP2C activity (Melcher et al., 2009; Miyazono et al., 2009; Dupeux et al., 2011b).
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants were routinely grown under greenhouse conditions in pots containing a 1:3 vermiculite:soil mixture. For plants grown under growth chamber conditions, seeds were surface sterilized by treatment with 70% ethanol for 20 min, followed by commercial bleach (2.5% sodium hypochlorite) containing 0.05% Triton X-100 for 10 min, and finally, four washes with sterile distilled water. Stratification of the seeds was conducted in the dark at 4°C for 3 d. Then, seeds were sown on Murashige and Skoog (1962; MS) plates composed of MS basal salts, 0.1% MES, and 1% agar. The pH was adjusted to 5.7 with KOH before autoclaving. Plates were sealed and incubated in a controlled-environment growth chamber at 22°C under a 16-h-light/8-h-dark photoperiod at 80 to 100 μE m−2 s−1.
Subcellular Localization Studies
Constructs to investigate the subcellular localization of PP2CA (At3g11410) and At5g59220 were generated in Gateway-compatible vectors. To this end, the coding sequences of PP2CA, At5g59220, and the N-terminal extension (residues 1–97) and the catalytic core (residues 98–413) of At5g59220 were PCR amplified using the following primer pairs, respectively: FPP2CANcoI and RPP2CAnostopSalI; F5g59220 and Rnostop5g59; F5g59220 and RNterm2CB; FMSTV2CB and Rnostop5g59. The sequences of all primers used in this work are provided in Supplemental Table S1. The PCR products were cloned into the pCR8/GW/TOPO entry vector (Invitrogen) and recombined by Gateway LR reaction into the pMDC83 destination vector (Saez et al., 2008). The different binary vectors were introduced into Agrobacterium tumefaciens C58C1 (pGV2260) by electroporation (Deblaere et al., 1985). Transformed cells were grown in liquid Luria-Bertani medium to late exponential phase, and cells were harvested by centrifugation and resuspended in 10 mm MES-KOH, pH 5.6, containing 10 mm MgCl2 and 150 mm acetosyringone to an optical density at 600 nm of 1. These cells were mixed with an equal volume of Agrobacterium C58C1 (pCH32 35S:p19) expressing the silencing suppressor p19 of Tomato bushy stunt virus (Voinnet et al., 2003) so that the final density of Agrobacterium solution was about 1. Bacteria were incubated for 3 h at room temperature and then injected into young fully expanded leaves of 4-week-old Nicotiana benthamiana plants. Leaves were examined after 3 to 4 d with a Leica TCS-SL confocal microscope and a laser scanning confocal imaging system.
Generation of Overexpressing Lines for PP2CA and At5g59220
The coding sequence of PP2CA was amplified by PCR using the primers FPP2CANcoI and RPP2CASalI. The coding sequence of At5g59220 was amplified by PCR using the primers F5g59220 and Rnosto5g59. Next, both were cloned into pCR8/GW/TOPO and recombined by LR reaction into the ALLIGATOR2 destination vector (Bensmihen et al., 2004). The ALLIGATOR2-35S:3HA-PP2CA and 35S:3HA-At5g59220 constructs were transferred to Agrobacterium C58C1 (pGV2260; Deblaere et al., 1985) by electroporation and used to transform Columbia wild-type plants by the floral dip method. T1 transgenic seeds were selected based on GFP visualization, and T3 progeny homozygous for the selection marker were used for further studies.
Seedling Establishment and Root Growth Assays
To determine the sensitivity to inhibition of seedling establishment by ABA, Glc, or mannitol, the MS medium was supplemented with the indicated concentrations of these compounds. The percentage of seeds that had germinated and developed fully green expanded cotyledons was determined. Approximately 200 seeds of each genotype were sown in each medium and scored for germination and early growth at 3, 5, 7, and 10 d later. For root growth assays, seedlings were grown on vertically oriented MS medium plates for 4 to 5 d. Afterward, 20 plants were transferred to new plates containing MS medium lacking or supplemented with the indicated concentrations of ABA. After the indicated periods of time, the plates were scanned on a flatbed scanner to produce image files suitable for quantitative analysis using the NIH Image software ImageJ version 1.37.
Biochemical Fractionation
Nuclear fractionation was performed according to techniques described by Bowler et al. (2004) and Cho et al. (2006). Arabidopsis leaves of epitope HA-tagged PP2CA or At5g59220 transgenic lines were ground in lysis buffer (20 mm Tris-HCl, pH 7.4, 25% glycerol, 20 mm KCl, 2 mm EDTA, 2.5 mm MgCl2, and 250 mm Suc) containing protease inhibitor cocktail (Roche) and 1 mm phenylmethylsulfonyl fluoride (PMSF). The lysate was filtered through four layers of Miracloth (Calbiochem) and centrifuged at 1,000g for 10 min to pellet the nuclei. The cytosolic fraction was removed, and the pellet was washed in nuclei resuspension buffer (20 mm Tris-HCl, 25% glycerol, 2.5 mm MgCl2, and 0.5% Triton X-100) to solubilize most proteins from the organelles. The nuclear pellet was resuspended in 5 volumes of medium salt buffer (Bowler et al., 2004; 20 mm Tris-HCl, 0.4 m NaCl, 1 mm EDTA, 5% glycerol, 1 mm 2-mercaptoethanol, 0.1% Triton X-100, 0.5 mm PMSF, and protease inhibitor cocktail [Roche]) and then frozen and thawed. After incubation with gentle mixing for 15 min at 4°C, the nuclear insoluble fraction containing the major protein histones was precipitated by centrifugation at 10,000g for 10 min, whereas the supernatant contained the nuclear soluble fraction. Detection of PP2CA or At5g59220 was performed using anti-HA-peroxidase conjugate (Roche). The purity of the different fractions was demonstrated using antibodies against histone H3 (Abcam), plasma membrane H+-ATPase (Dr. Ramón Serrano, Universidad Politecnica de Valencia), and Ponceau staining of the ribulose-1,6-bisphosphate carboxylase.
A second fractionation procedure was used to analyze the presence of PP2CA and At5g59220 in cytosol or microsomal pellets (Hua et al., 2001). Arabidopsis leaves of epitope HA-tagged PP2CA or At5g59220 transgenic lines were ground in lysis buffer (50 mm Tris, pH 8, 2 mm EDTA, 20% glycerol, 5 mm MgCl2, 1 mm dithiothreitol [DTT], and 25 mm CaCl2) containing protease inhibitor cocktail (Roche) and 1 mm PMSF. The lysate was filtered through Miracloth and centrifuged at 5,000g for 5 min to remove organelles and debris. Supernatants were centrifuged at 100,000g for 45 min to pellet microsomal membranes and to obtain the cytosolic soluble fraction. The resulting microsomal pellet was solubilized in resuspension buffer (25 mm Tris, pH 7.2, 10% Suc, 2 mm EDTA, 5 mm MgCl2, 1 mm DTT, protease inhibitor cocktail, 0.1 mm PMSF, and 25 mm CaCl2) using a 2-mL glass homogenizer.
RNA Analysis
After mock or ABA treatment, plant material was collected and immediately frozen in liquid nitrogen. Total RNA was extracted using a Qiagen RNeasy Plant Mini Kit, and 1 μg of the RNA obtained was reverse transcribed using 0.1 μg of oligo(dT)15 primer and Moloney murine leukemia virus reverse transcriptase (Roche) to finally obtain a 40-μL cDNA solution. Reverse transcription-quantitative (RT-q)PCR amplifications and measurements were performed using an ABI PRISM 7000 Sequence Detection System (Perkin-Elmer Applied Biosystems), and they were monitored using the Eva-Green fluorescent stain (Biotium). Relative quantification of gene expression data was carried out using the 2−ΔΔCT or comparative cycle threshold (CT) method (Livak and Schmittgen, 2001). Expression levels were normalized using the CT values obtained for the β-Actin8 gene. Gene induction ratios were calculated as the expression ratio between ABA-treated plantlets and mock-treated plantlets. The presence of a single PCR product was further verified by dissociation analysis in all amplifications. All quantifications were made in triplicate on RNA samples obtained from three independent experiments. The sequences of the primers used for RT-qPCR amplification have been described previously (Rubio et al., 2009).
Purification of Recombinant Proteins
The coding sequence of PP2CA was amplified by PCR using the primers F2CASalINdeI and R2CASmaISalI. The full-length At5g59220 protein was not soluble; therefore, we generated a ΔN-At5g59220 version, since according to the structure of ternary complexes reported so far, N-terminal deletions of PP2Cs are able to interact with PYR/PYL proteins (Melcher et al., 2009; Miyazono et al., 2009; Dupeux et al., 2011a). To this end, the coding sequence encompassing the catalytic core of At5g59220, residues 98 to 413, was amplified by PCR using the primers FMSTV2CB and RHAstopP2B. PCR products were cloned into pCR8/GW/TOPO, then the coding sequence of PP2CA was excised from this plasmid using NdeI/SalI double digestion and subcloned into pET28a, whereas the coding sequence of At5g5922098–413 was excised using EcoRI digestion and subcloned into pET28a. The coding sequence of AHG1 was excised from a pACT2 construct (kindly provided by Dr. J.F. Quintero, Consejo Superior de Investigaciones Cientificas) using NcoI/BamHI double digestion and subcloned into pETM11. Escherichia coli BL21 (DE3) cells transformed with the corresponding pET28a/pETM11 construct were grown in 50 mL of Luria-Bertani medium supplemented with 50 μg mL−1 kanamycin to an optical density at 600 nm of 0.6 to 0.8. Then, 1 mm isopropylthio-β-galactoside was added, and the cells were harvested 3 h after induction and stored at −80°C before purification. The pellet was resuspended in 2 mL of buffer HIS (50 mm Tris-HCl, pH 7.6, 250 mm KCl, 10% glycerol, 0.1% Tween 20, and 10 mm mercaptoethanol), and the cells were sonicated in a Branson Sonifier. A cleared lysate was obtained after centrifugation at 14,000g for 15 min, and it was diluted with 2 volumes of buffer HIS. The protein extract was applied to a 0.5 mL nickel-nitrilotriacetic acid agarose column, and the column was washed with 10 mL of buffer HIS supplemented with 20% glycerol and 30 mm imidazol. Bound protein was eluted with buffer HIS supplemented with 20% glycerol and 250 mm imidazol.
PP2C and OST1 in Vitro Activity Assays
Phosphatase activity was measured using the RRA(phosphoT)VA peptide as substrate, which has a Km of 0.5 to 1 μm for eukaryotic PP2Cs (Donella Deana et al., 1990). Assays were performed in a 100-μL reaction volume containing 25 mm Tris-HCl, pH 7.5, 10 mm MgCl2, 1 mm DTT, 25 μm peptide substrate, and 0.5 μm PP2C. When indicated, PYR-PYL recombinant proteins and ABA were included in the PP2C activity assay. ABA concentrations were 0.5, 1, 5, 10, 20, 40, and 50 μm. After incubation for 60 min at 30°C, the reaction was stopped by the addition of 30 μL of molybdate dye (Baykov et al., 1988), and the absorbance was read at 630 nm with a 96-well plate reader.
Phosphatase activity was also measured using phosphorylated OST1 and ΔCABF2 (amino acids 1–173, containing the C1, C2, and C3 protein kinase targets) as substrates (Vlad et al., 2009; Dupeux et al., 2011a). Autophosphorylated OST1 or transphosphorylated ΔCABF2 were prepared in a 60-min reaction. Dephosphorylation of OST1 or ΔCABF2 was achieved by incubation with the different PP2Cs. Assays to test the recovery of OST1 activity were done by previous incubation of the PP2C for 10 min in the absence or presence of 30 μm ABA and the indicated PYR/PYL. Next, the reaction mixture was incubated for 50 min at room temperature in 30 μL of kinase buffer: 20 mm Tris-HCl, pH 7.8, 20 mm MgCl2, 2 mm MnCl2, and 3.5 μCi of [γ-32P]ATP (3,000 Ci mmol−1). The reaction was stopped by adding Laemmli buffer. After the reaction, proteins were separated by SDS-PAGE using an 8% acrylamide gel and transferred to an Immobilon-P membrane (Millipore). Radioactivity was detected using a phosphorimage system (FLA5100; Fujifilm). After scanning, the same membrane was used for Ponceau staining. The data presented are averages of at least three independent experiments.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Cladogram and nomenclature of clade A PP2Cs.
Supplemental Figure S2. Up-regulation of At5g59220 gene expression by osmotic stress and ABA.
Supplemental Figure S3. Glc-hypersensitive growth inhibition of pp2ca-1 hai1-1 and hab1-1 abi1-2 double mutants.
Supplemental Figure S4. Analysis of water loss, ABA-mediated growth inhibition, and expression of two ABA-responsive genes in 35S:HAB1 and 35S:At5g59220 lines.
Supplemental Table S1. List of oligonucleotides used in this work.
Supplementary Material
Acknowledgments
We thank Joseph Ecker and the Salk Institute Genomic Analysis Laboratory for providing the sequence-indexed Arabidopsis T-DNA insertion mutants and the Arabidopsis Biological Resource Center/Nottingham Arabidopsis Stock Centre for distributing the seeds.
References
- Baykov AA, Evtushenko OA, Avaeva SM. (1988) A malachite green procedure for orthophosphate determination and its use in alkaline phosphatase-based enzyme immunoassay. Anal Biochem 171: 266–270 [DOI] [PubMed] [Google Scholar]
- Bensmihen S, To A, Lambert G, Kroj T, Giraudat J, Parcy F. (2004) Analysis of an activated ABI5 allele using a new selection method for transgenic Arabidopsis seeds. FEBS Lett 561: 127–131 [DOI] [PubMed] [Google Scholar]
- Bowler C, Benvenuto G, Laflamme P, Molino D, Probst AV, Tariq M, Paszkowski J. (2004) Chromatin techniques for plant cells. Plant J 39: 776–789 [DOI] [PubMed] [Google Scholar]
- Chérel I, Michard E, Platet N, Mouline K, Alcon C, Sentenac H, Thibaud JB. (2002) Physical and functional interaction of the Arabidopsis K+ channel AKT2 and phosphatase AtPP2CA. Plant Cell 14: 1133–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YH, Yoo SD, Sheen J. (2006) Regulatory functions of nuclear hexokinase1 complex in glucose signaling. Cell 127: 579–589 [DOI] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. (2010) Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol 61: 651–679 [DOI] [PubMed] [Google Scholar]
- Deblaere R, Bytebier B, De Greve H, Deboeck F, Schell J, Van Montagu M, Leemans J. (1985) Efficient octopine Ti plasmid-derived vectors for Agrobacterium-mediated gene transfer to plants. Nucleic Acids Res 13: 4777–4788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donella Deana A, Mac Gowan CH, Cohen P, Marchiori F, Meyer HE, Pinna LA. (1990) An investigation of the substrate specificity of protein phosphatase 2C using synthetic peptide substrates: comparison with protein phosphatase 2A. Biochim Biophys Acta 1051: 199–202 [DOI] [PubMed] [Google Scholar]
- Dupeux F, Antoni R, Betz K, Santiago J, Gonzalez-Guzman M, Rodriguez L, Rubio S, Park SY, Cutler SR, Rodriguez PL, et al. (2011a) Modulation of abscisic acid signaling in vivo by an engineered receptor-insensitive protein phosphatase type 2C allele. Plant Physiol 156: 106–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupeux F, Santiago J, Betz K, Twycross J, Park SY, Rodriguez L, Gonzalez-Guzman M, Jensen MR, Krasnogor N, Blackledge M, et al. (2011b) A thermodynamic switch modulates abscisic acid receptor sensitivity. EMBO J 30: 4171–4184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK. (2009) In vitro reconstitution of an abscisic acid signalling pathway. Nature 462: 660–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Nakashima K, Yoshida T, Katagiri T, Kidokoro S, Kanamori N, Umezawa T, Fujita M, Maruyama K, Ishiyama K, et al. (2009) Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol 50: 2123–2132 [DOI] [PubMed] [Google Scholar]
- Geiger D, Maierhofer T, Al-Rasheid KA, Scherzer S, Mumm P, Liese A, Ache P, Wellmann C, Marten I, Grill E, et al. (2011) Stomatal closure by fast abscisic acid signaling is mediated by the guard cell anion channel SLAH3 and the receptor RCAR1. Sci Signal 4: ra32. [DOI] [PubMed] [Google Scholar]
- Geiger D, Scherzer S, Mumm P, Marten I, Ache P, Matschi S, Liese A, Wellmann C, Al-Rasheid KA, Grill E, et al. (2010) Guard cell anion channel SLAC1 is regulated by CDPK protein kinases with distinct Ca2+ affinities. Proc Natl Acad Sci USA 107: 8023–8028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger D, Scherzer S, Mumm P, Stange A, Marten I, Bauer H, Ache P, Matschi S, Liese A, Al-Rasheid KA, et al. (2009) Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc Natl Acad Sci USA 106: 21425–21430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosti F, Beaudoin N, Serizet C, Webb AA, Vartanian N, Giraudat J. (1999) ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell 11: 1897–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo XH, Deng KQ, Wang J, Yu DS, Zhao Q, Liu XM. (2010) Mutational analysis of Arabidopsis PP2CA2 involved in abscisic acid signal transduction. Mol Biol Rep 37: 763–769 [DOI] [PubMed] [Google Scholar]
- Hao Q, Yin P, Li W, Wang L, Yan C, Lin Z, Wu JZ, Wang J, Yan SF, Yan N. (2011) The molecular basis of ABA-independent inhibition of PP2Cs by a subclass of PYL proteins. Mol Cell 42: 662–672 [DOI] [PubMed] [Google Scholar]
- Himmelbach A, Hoffmann T, Leube M, Höhener B, Grill E. (2002) Homeodomain protein ATHB6 is a target of the protein phosphatase ABI1 and regulates hormone responses in Arabidopsis. EMBO J 21: 3029–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua J, Grisafi P, Cheng SH, Fink GR. (2001) Plant growth homeostasis is controlled by the Arabidopsis BON1 and BAP1 genes. Genes Dev 15: 2263–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno Y, Jikumaru Y, Hanada A, Nambara E, Abrams SR, Kamiya Y, Seo M. (2010) Comprehensive hormone profiling in developing Arabidopsis seeds: examination of the site of ABA biosynthesis, ABA transport and hormone interactions. Plant Cell Physiol 51: 1988–2001 [DOI] [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D’Angelo C, Bornberg-Bauer E, Kudla J, Harter K. (2007) The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J 50: 347–363 [DOI] [PubMed] [Google Scholar]
- Kuhn JM, Boisson-Dernier A, Dizon MB, Maktabi MH, Schroeder JI. (2006) The protein phosphatase AtPP2CA negatively regulates abscisic acid signal transduction in Arabidopsis, and effects of abh1 on AtPP2CA mRNA. Plant Physiol 140: 127–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, Lan W, Buchanan BB, Luan S. (2009) A protein kinase-phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proc Natl Acad Sci USA 106: 21419–21424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, Lan WZ, Kim BG, Li L, Cheong YH, Pandey GK, Lu G, Buchanan BB, Luan S. (2007) A protein phosphorylation/dephosphorylation network regulates a plant potassium channel. Proc Natl Acad Sci USA 104: 15959–15964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt N, Kwak JM, Robert N, Waner D, Leonhardt G, Schroeder JI. (2004) Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant. Plant Cell 16: 596–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Melcher K, Ng LM, Zhou XE, Soon FF, Xu Y, Suino-Powell KM, Park SY, Weiner JJ, Fujii H, Chinnusamy V, et al. (2009) A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature 462: 602–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlot S, Gosti F, Guerrier D, Vavasseur A, Giraudat J. (2001) The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J 25: 295–303 [DOI] [PubMed] [Google Scholar]
- Miao Y, Lv D, Wang P, Wang XC, Chen J, Miao C, Song CP. (2006) An Arabidopsis glutathione peroxidase functions as both a redox transducer and a scavenger in abscisic acid and drought stress responses. Plant Cell 18: 2749–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazono K, Miyakawa T, Sawano Y, Kubota K, Kang HJ, Asano A, Miyauchi Y, Takahashi M, Zhi Y, Fujita Y, et al. (2009) Structural basis of abscisic acid signalling. Nature 462: 609–614 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant 15: 473–497 [Google Scholar]
- Nakabayashi K, Okamoto M, Koshiba T, Kamiya Y, Nambara E. (2005) Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: epigenetic and genetic regulation of transcription in seed. Plant J 41: 697–709 [DOI] [PubMed] [Google Scholar]
- Nishimura N, Yoshida T, Kitahata N, Asami T, Shinozaki K, Hirayama T. (2007) ABA-Hypersensitive Germination1 encodes a protein phosphatase 2C, an essential component of abscisic acid signaling in Arabidopsis seed. Plant J 50: 935–949 [DOI] [PubMed] [Google Scholar]
- Rubio S, Rodrigues A, Saez A, Dizon MB, Galle A, Kim TH, Santiago J, Flexas J, Schroeder JI, Rodriguez PL. (2009) Triple loss of function of protein phosphatases type 2C leads to partial constitutive response to endogenous abscisic acid. Plant Physiol 150: 1345–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez A, Apostolova N, Gonzalez-Guzman M, Gonzalez-Garcia MP, Nicolas C, Lorenzo O, Rodriguez PL. (2004) Gain-of-function and loss-of-function phenotypes of the protein phosphatase 2C HAB1 reveal its role as a negative regulator of abscisic acid signalling. Plant J 37: 354–369 [DOI] [PubMed] [Google Scholar]
- Saez A, Robert N, Maktabi MH, Schroeder JI, Serrano R, Rodriguez PL. (2006) Enhancement of abscisic acid sensitivity and reduction of water consumption in Arabidopsis by combined inactivation of the protein phosphatases type 2C ABI1 and HAB1. Plant Physiol 141: 1389–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez A, Rodrigues A, Santiago J, Rubio S, Rodriguez PL. (2008) HAB1-SWI3B interaction reveals a link between abscisic acid signaling and putative SWI/SNF chromatin-remodeling complexes in Arabidopsis. Plant Cell 20: 2972–2988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago J, Rodrigues A, Saez A, Rubio S, Antoni R, Dupeux F, Park SY, Márquez JA, Cutler SR, Rodriguez PL. (2009) Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J 60: 575–588 [DOI] [PubMed] [Google Scholar]
- Schweighofer A, Hirt H, Meskiene I. (2004) Plant PP2C phosphatases: emerging functions in stress signaling. Trends Plant Sci 9: 236–243 [DOI] [PubMed] [Google Scholar]
- Sheen J. (1998) Mutational analysis of protein phosphatase 2C involved in abscisic acid signal transduction in higher plants. Proc Natl Acad Sci USA 95: 975–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soon FF, Ng LM, Zhou XE, West GM, Kovach A, Tan MH, Suino-Powell KM, He Y, Xu Y, Chalmers MJ, et al. (2011) Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases. Science 335: 85–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostkiewicz I, Richter K, Kepka M, Demmel S, Ma Y, Korte A, Assaad FF, Christmann A, Grill E. (2010) Closely related receptor complexes differ in their ABA selectivity and sensitivity. Plant J 61: 25–35 [DOI] [PubMed] [Google Scholar]
- Tähtiharju S, Palva T. (2001) Antisense inhibition of protein phosphatase 2C accelerates cold acclimation in Arabidopsis thaliana. Plant J 26: 461–470 [DOI] [PubMed] [Google Scholar]
- Umezawa T, Sugiyama N, Mizoguchi M, Hayashi S, Myouga F, Yamaguchi-Shinozaki K, Ishihama Y, Hirayama T, Shinozaki K. (2009) Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci USA 106: 17588–17593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlad F, Rubio S, Rodrigues A, Sirichandra C, Belin C, Robert N, Leung J, Rodriguez PL, Laurière C, Merlot S. (2009) Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell 21: 3170–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D. (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Costa A, Leonhardt N, Siegel RS, Schroeder JI. (2008) Isolation of a strong Arabidopsis guard cell promoter and its potential as a research tool. Plant Methods 4: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Fujita Y, Sayama H, Kidokoro S, Maruyama K, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. (2010) AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J 61: 672–685 [DOI] [PubMed] [Google Scholar]
- Yoshida T, Nishimura N, Kitahata N, Kuromori T, Ito T, Asami T, Shinozaki K, Hirayama T. (2006) ABA-hypersensitive germination3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiol 140: 115–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.