Abstract
The mechanism whereby brief light pulses reset the mammalian circadian clock involves acute Per gene induction. In a previous study we investigated light-induced expression of mPer1 and mPer2 mRNA in the suprachiasmatic nuclei (SCN), with the aim of understanding the relationship between gene expression and behavioural phase shifts. In the present study, we examine the protein products of mPer1 and mPer2 genes in the core and shell region of SCN for 34 h following a phase-shifting light pulse, in order to further explore the molecular mechanism of photic entrainment. The results indicate that, during the delay zone of the phase response curve, while endogenous levels of mPER1 and mPER2 protein are falling, a light pulse produces an increase in the expression of both proteins. In contrast, during the advance zone of the phase response curve, while levels of endogenous mPER1 and mPER2 proteins are rising, a light pulse results in a further increase in mPER1 but not mPER2 protein. The regional distribution of mPER1 and mPER2 protein in the SCN follows the same pattern as their respective mRNAs, with mPER1 expression in the shell region of SCN correlated with phase advances and mPER2 in the shell region correlated with phase delays.
Keywords: circadian rhythm, light pulse, mouse, suprachiasmatic nucleus
Introduction
In mammals, the suprachiasmatic nuclei (SCN) of the hypothalamus are the locus of the master circadian clock that controls the daily variations in physiology and behaviour (Rusak & Zucker, 1979). The circadian clock has two distinct functions: to generate rhythms and to synchronize these rhythms to the environment (entrainment). The molecular mechanism underlying rhythm generation has been described as interlocked positive and negative transcription–translation-based feedback loops (Dunlap, 1999; Reppert & Weaver, 2001).
Light is the most salient cue for the entrainment of the circadian clock. It is widely accepted that resetting the mammalian circadian clock involves acute induction of Per1 and Per2 genes (Dunlap, 1999; Lowrey & Takahashi, 2000). For alterations in gene expression to be functionally meaningful, they must be translated to changes in protein. How the protein products of these genes are regulated and subsequently contribute to the core feedback loops to reset the circadian phase of the molecular clock is not well understood. While numerous studies document change in Per gene expression following a light pulse, the effect of light on PER protein has received less attention. Field et al. (2000) reported that a phase-resetting light pulse increased mPER1 expression in the SCN, but had no acute effect on other clock proteins examined, including mPER2, mCRYand mTIM. These results suggested a central role for mPER1 in photic entrainment. On the other hand, behavioural studies have demonstrated that mPer1-mutant mice exhibit phase delays and advances following appropriately timed light pulses (Cermakian et al., 2001; Bae & Weaver, 2003). Thus, it is not clear whether mPER1 is the sole mediator of photic entrainment. One of the aims of the present study is to identify the role of mPER proteins in photic entrainment by investigating mPER1 and mPER2 in the SCN following a light pulse.
SCN contains two distinct compartments, termed `core' and `shell' (Moore, 1996). We previously examined light-induced mPer1 and mPer2 gene expression in the SCN following a light pulse (Yan & Silver, 2002). We found that, in the shell SCN region, light-induced mPer1 expression was correlated with phase advances while mPer2 was correlated with phase delays. In this view, phase shifting of the molecular clock in the shell oscillator cells rather than in the core retinorecipient cells is the key step for phase-shifting circadian rhythms. This conclusion was tentative due to lack of evidence of protein expression, which is necessary for functional changes. Thus, the other aim of the present study is to further test this hypothesis at the level of protein expression, by characterizing the light response profile of mPER1 and mPER2 protein in the core and shell region of the SCN following delaying and advancing light pulses.
Materials and methods
Male C57BL/6 mice, purchased from Charles River Laboratories (Wilmington, MA, USA) at 5 weeks of age, were housed in a 12-h light–dark (LD; light 300 lux) cycle for 2–4 weeks, then were transferred to constant darkness (DD). They were used in the experiment on the 3rd day in DD. Food and water were available ad libitum. A white noise generator (91 dB sound pressure level) masked environmental noise.
To investigate the distribution of PER1 and PER2 in mouse brain, animals were killed with an overdue of pentobarbital at circadian time (CT) 14. Circadian time was determined by projection based on a mean period (23.8 h) of free running locomoter activity in this mouse strain (Schwartz & Zimmerman, 1990; Yan & Silver, 2002). To study the response to light, animals were randomly assigned to two groups, defined by the time of light pulse (LP) administration (CT16 or 22). To characterize the time course of light-induced PER1 and PER2 expression, animals (n = 4/time point) were killed at 2, 4, 6 and 8 h after the CT16 light pulse (900 lux, 30 min), or 4, 6, 8, 10, 16, 20, 30 and 34 h after the CT22 light pulse (900 lux, 30 min). Control animals (n = 2–4/time point) were treated identically, but were not exposed to the light pulse. All experimental procedures were approved by the Institutional Animal Care and Use Committee of Columbia University.
Mice were anaesthetized (pentobarbital, 200 mg/kg) and perfused intracardially with 50 mL saline followed by 100 mL 4% paraformaldehyde in 0.1 m phosphate buffer. Brains were postfixed for 12–18 h and cryoprotected in 20% sucrose overnight. Sections (30 μm) were cut through the entire SCN using a cryostat. Alternate free-floating sections were incubated with mPER1 antibody (1 : 5000, gift of Drs S. M. Reppert and D. R. Weaver, University of Massachusetts, MA, USA); or mPER2 antibody (1 : 5000, Alpha Diagnostics), and processed with the avidin–biotin–immunoperoxidase technique using DAB as the chromogen.
For quantification of mPER1 and mPER2-immunoreactive (ir) cells, cell counting of printed images of three sections from mid-SCN (Fig. 1) was conducted by two observers who were blind to the experimental conditions. The results from the two observers are highly correlated (r = 0.82, P < 0.01). SCN subregions were defined by a template using hypothalamic markers, in which the area of vasoactive intestinal peptide (VIP) mRNA containing cells was defined as the core and the rest SCN was defined as shell (for details, see Yan & Silver, 2002). To deal with potential variability in independent immunochemistry runs, animals to be compared directly were processed together in the following three batches (groups shown in Fig. 2, in Fig. 3, and those killed at 16 h and later, after exposure to a light pulse at CT22). A two-way anova (LP × time) was used to analyse the difference between light-exposed groups and control groups. Where significant LP effect and/or interaction were detected, t-tests were performed to examine the light effect at each time points.
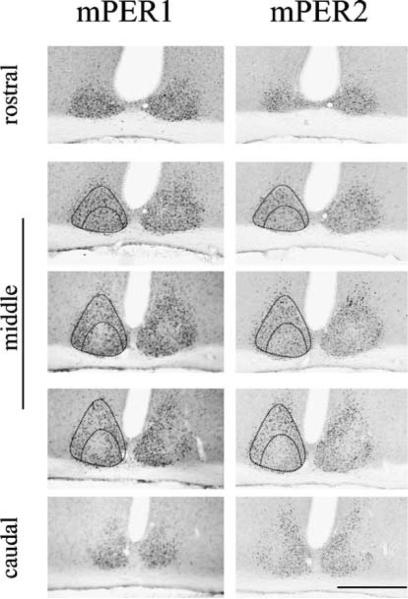
Fig. 1.
Distribution of mPER1 and mPER2 in the SCN at CT14. Micrographs of mPER1 and mPER2 staining in the SCN in serial adjacent sections arranged from rostral to caudal. mPER1 and mPER2 showed similar regional distributions in the SCN. Scale bar, 500 μm.
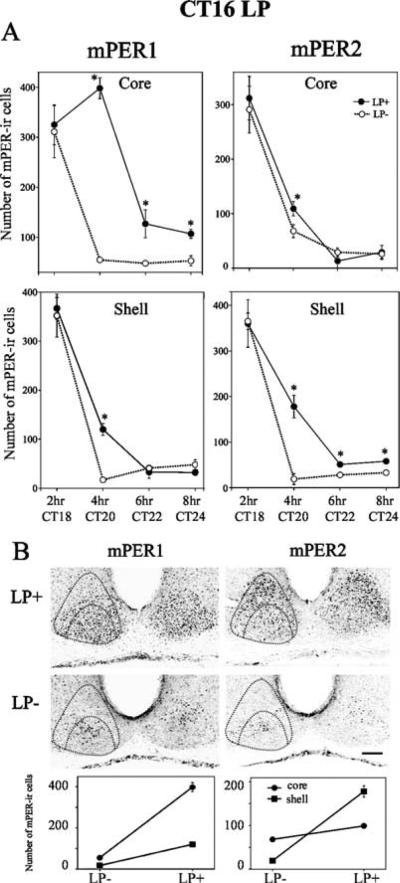
Fig. 2.
mPER1 and mPER2 expression after a phase-delaying light pulse. Animals were given a light pulse (LP) at CT16, then killed 2, 4, 6 or 8 h after the beginning of the light pulse. (A) Quantitative analysis of the number of mPER1-ir (left panels) and mPER2-ir (right panels) cells in LP+ and control (LP−) groups. The data are presented as mean ± SEM (*p<0.05, t-test). (B) Topographic (upper level) and quantitative (lower level) analysis of mPER1 and mPER2 in the SCN at 4 h in an LP+ and a control animal. In LP+ animals, the greatest increase in mPER1-ir cells occurs in the core SCN, while the increase of mPER2-ir cells is greatest in the shell SCN. Scale bar, 100 μm.
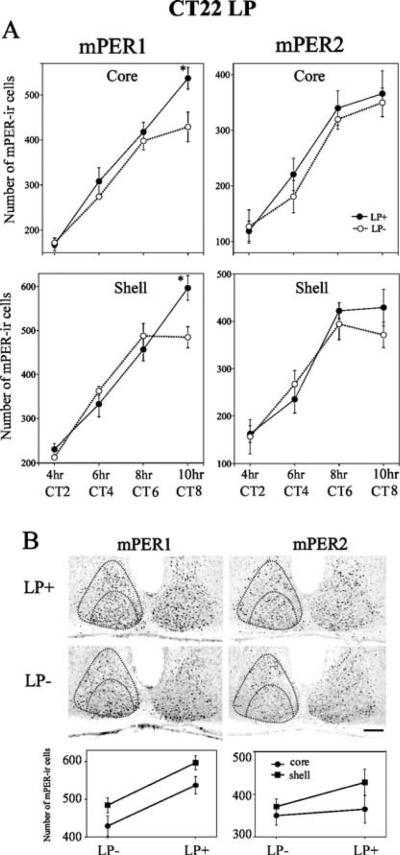
Fig. 3.
mPER1 and mPER2 expression after a phase-advancing light pulse. Animals were given a light pulse (LP) at CT22, then killed 4, 6, 8, 10, 16, 20, 30 or 34 h after the beginning of the light pulse (data from 16 to 34 h not shown). (A) Quantitative analysis of the number of mPER1-ir (left panel) and mPER2-ir (right panel) cells in LP+ and control (LP−) groups. The data are presented as mean ± SEM (*P<0.05, t-test). (B) Topographic (upper level) and quantitative (lower level) analysis of mPER1 and mPER2 in the SCN at 10 h in an LP+ and an LP− animal. Increased numbers of mPER1-ir cells were seen in each compartment of the SCN in LP+ animals. Statistically significant changes in the number of mPER2-ir cells were not observed in either core or shell of the SCN. Scale bar, 100 μm.
Results
Figure 1 shows regional differences in SCN staining for mPER1 and mPER2 at CT14. Adjacent serial sections reveal similar regional distributions for mPER1 and mPER2. Both proteins were expressed uniformly in rostral sections, but were concentrated in the shell region of mid-SCN sections.
To examine the spatial and temporal patterns of mPER1 and mPER2 expression in core and shell SCN following a phase-delaying light pulse at CT16, experimental and control animals were sampled at CT18, 20, 22 and 24 (Fig. 2). Light exposure increased the number of mPER1-ir cells in both core and shell SCN compared to controls killed at the same circadian time (Fig. 2A left panels; core: LP effect, P < 0.0001, shell: LP effect, P < 0.05). The effect of light was significant at 4, 6 and 8 h in the core and at 4 h in the shell (P < 0.05). In both groups the number of mPER1-ir cells decreased from CT18 to CT24 in both core and shell regions (time effect, P < 0.0001). For mPER2, light exposure increased the number of mPER2-ir cells at 4 h in the core SCN (Fig. 2A upper right panel: LP effect, P > 0.05; LP × time interaction, P < 0.05; t-test, P < 0.05 at 4 h). In the shell, light exposure increased the number of mPER2-ir cells at 4, 6 and 8 h (Fig. 2A lower right panel: LP effect, P < 0.0001; t-test, P < 0.05 at 4, 6 and 8 h). The number of mPER2-ir cells also decreased from CT18 to CT24 in both core and shell regions (time effect, P < 0.0001).
To compare expression in the core and shell SCN, mPER1 and mPER2 were re-examined at 4 h after the light pulse, when the experimental and control animals were maximally different. Figure 2B shows mPER1 and mPER2 expression at CT20 in representative photomicrographs (upper panels) and their quantification (lower panels). In LP+ animals, mPER1-positive cells were seen in the core and in a thin region of the shell surrounding the core SCN, whereas mPER2-positive cells were observed throughout the core and shell region of the SCN. Light exposure significantly increased the number of mPER1 cells in both core and shell SCN regions (LP effect, P < 0.0001), the increase in the core being much greater than that in the shell (LP × region interaction, P < 0.0001). The numbers of mPER2 cells in light-exposed animals also increased in both core and shell regions compared to unexposed controls (LP effect, P < 0.0001). In this case, the increase was much greater in the shell than that in the core SCN (LP × region interaction, P < 0.0001).
To examine the spatial and temporal patterns of mPER1 and mPER2 expression in core and shell SCN following a phase-advancing light pulse at CT22, experimental and control animals were sampled at CT2, 4, 6 and 8 (Fig. 3). Light exposure increased the number of mPER1 cells in the core region (Fig. 3A, upper left panel; LP effect, P < 0.01). In the shell region, the effect of light was not statistically significant overall, and varied among time points (LP effect, P > 0.05; LP × time interaction, P < 0.01). The effect of light was significant at 10h after the LP in both core and shell region (P < 0.05). The number of mPER1-ir cells increased from CT2 to CT8 in both groups (time effect, P < 0.0001). For mPER2 (Fig. 3A, right panels), the difference between the light-exposed and control groups was not significant in either core or shell SCN (core: LP effect, P > 0.05; LP × time interaction, P > 0.05; shell: LP effect, P > 0.05; LP × time interaction, P > 0.05). The number of mPER2-ir cells increased from CT2 to CT8 in both groups in core and shell SCN (time effect, P < 0.0001). In a separate experimental run, mPER1 and mPER2 expression was examined at 16, 20, 30 and 34 h after the CT22 light pulse (data not shown). There was a significant effect of time but no effect for light observed (time effect, P < 0.0001; LP effect, P > 0.05; LP × time interaction, P > 0.05).
To compare expression in the core and shell SCN, mPER1 and mPER2 were re-analysed at CT8, corresponding to the time point at 10 h following the light pulse. Figure 3B shows mPER1 and mPER2 expression at CT8 in representative photomicrographs (upper panels) and their quantification (lower panels). The increase in the number of mPER1-ir cells in the light-exposed animals was observed in both core and shell regions of the SCN (LP effect, P < 0.0001; LP × region interaction, P > 0.05). In contrast, there was no significant difference in the number of mPER2-ir cells between the light-exposed and nonexposed animals (LP effect, P > 0.05; LP × region interaction, P > 0.05).
Discussion
Endogenously rhythmic mPER1 and mPER2 have overlapping distributions within the SCN (Fig. 1). This is consistent with previous findings showing that the two proteins have the same circadian profile (Field et al., 2000; Nuesslein-Hildesheim et al., 2000), and with the notion that they form a heterodimeric complex which is then translocated into the nucleus (Reppert & Weaver, 2001). Furthermore, the endogenously rhythmic mPER1 and mPER2 expressions change significantly over time in both core and shell SCN regions (Figs 2A and 3A). In contrast, in response to a light pulse, the spatial expression pattern of mPER1 and mPER2 changes depending on the time of light pulse administration. After a phase-delaying light pulse at CT16, compared to controls, the number of mPER1 cells was increased mostly in the core region, while for mPER2 cells the greatest increase was in the shell (Fig. 2). After a phase-advancing light pulse, mPER1 expression was increased in both the core and shell region, whereas there was no significant increase in mPER2 in either core or shell region (Fig. 3). In the shell SCN region, increased mPER2 expression was correlated with phase delays and mPER1 was correlated with phase advances. This regionally specific light response profile of mPER1 and mPER2 is consistent with our previous data on mRNA (Yan & Silver, 2002). A phase-delaying light pulse produces increased mPER1 and mPER2 within 4–6 h (present study; von Gall et al., 2003). Surprisingly, we found that the effect of a phase-advancing light pulse on mPER1 expression was significant at 10 h, but not earlier (4–8 h) or later (16–34 h). It may be that PER1 expression had not shifted, but only increased transiently; alternatively, the sampling frequency and the semiquantitative method used to assess protein changes may not have had sufficient temporal resolution and sensitivity to detect the changes in mPER expression that may have occurred during phase advances.
Rhythmic Per gene and protein expression in the core SCN have been reported (present study; Yan & Okamura, 2002; Nagano et al., 2003), while other studies demonstrate an SCN subregion or specific SCN cells that lack rhythmic expression of Per genes and proteins (Hamada et al., 2001; Karatsoreos et al., 2004). This apparent discrepancy can be explained by the different methods used to delineate the `core' SCN region. When the hamster `core' SCN is delineated by the calbindin-containing region (Hamada et al., 2001), or when gastrin-release peptide (GRP) cells are studied in mouse (Karatsoreos et al., 2004), one detects no rhythm in Per gene expression. However, if the `core' region is defined as entire ventrolateral region containing VIP mRNA-expressing cells, one sees a rhythm in Per gene expression as has been reported for the mouse and rat SCN (present study; Yan & Okamura, 2002; Nagano et al., 2003). The relationship between the core SCN and retinorecipient region in mouse SCN has been documented in Abrahamson & Moore (2001) and discussed in Yan & Silver (2002).
While our results support the idea that mPER2 is important for phase delays and mPER1 for phase advances, the work of Field et al. (2000) highlights a central role of mPER1 in phase shifts. Furthermore, we found that expression of PER1 peaked at 4 h after the light pulse in the delay zone and at 10 h in the advance zone, while Field et al. (2000) reported values of 9 h and 4 h, respectively, for these measures. These results are probably due to several methodological differences between our work and that of Field et al. (2000) including experimental conditions (free-running vs. entrained), analysis (core and shell SCN regions vs. c-FOS-ir region), timing of the light pulse (CT16 vs. ZT14), duration of the light pulse (30 vs. 15 min) and the mouse strains (C57BL/6 vs. CD1). The distinct significance of each of these differences in experimental methods remains to be established.
Questions remain on how the altered expression level of mPER1 and mPER2 can affect the interlocked positive and negative feedback loops which form the molecular basis of circadian rhythm generation (Reppert & Weaver, 2001). In the negative-feedback loop, the rhythmic transcription of the mPer and mCry genes is driven by the CLOCK/BMAL1 heterodimer. The protein products of these genes, mPER and mCRY, form multimeric complexes and then translocate to the nucleus to act as negative regulators that inhibit their own transcription. At the same time, mPER2 can activate the transcription of Bmal1, forming a positive-feedback loop. In this molecular feedback loop, the involvement of the translational and post-translational mechanisms has also been well studied both in vivo and in vitro. Peak expression of mPer1 and mPer2 is found in the subjective day; however, nuclear accumulation of their protein products occurs after 6–8 h (Reppert & Weaver, 2001). Phosphorylation, nuclear transportation and ubiquitination of PER proteins are likely to be important causes of this time lag (Okamura et al., 2002). The monomers of mPER1 and mPER2 are phosphorylated by casein kinase Iε and then degraded rapidly until they reach a sufficient concentration to form the complex with each other and/or other clock proteins (Lowrey et al., 2000; Vielhaber et al., 2000; Lee et al., 2001). There is a codependency between the mPER and mCRY proteins for nuclear translocation (Vitaterna et al., 1999; Lee et al., 2001; Yagita et al., 2002). In contrast to these detailed studies on the mechanisms underlying rhythm generation, less is known on translational and post-translational regulation during entrainment. Based on the delayed nuclear accumulation of mPER proteins and the delayed increase of mCRY1 in the SCN after light exposure (Field et al., 2000; present study) it is reasonable to speculate that similar events of phosphorylation and protein interaction occur during entrainment as during endogenous clock function. However, it is possible that additional and/or modified mechanisms exist for entrainment. Finally, the different time course of mPER1 and mPER2 between phase delays and advances suggests different post-translational regulation for each protein underlying such processes. In conclusion, the present results support the hypothesis that light acts on cells in the SCN core, and that the outputs of these cells reset pacemaker cells in the SCN shell. This is consistent with previous work (Albrecht et al., 2001; Yan & Silver, 2002) suggesting distinct roles of mPER1 on phase advances and mPER2 on phase delays.
Acknowledgements
We thank Drs Michael Antle and Joseph LeSauter for critical reading of the manuscript. This work was supported by NIH grants NS37919 and IU54NS41069 to R.S.
Abbreviations
- CT
circadian time
- DD
constant darkness
- ir
immunoreactive
- LD
light–dark
- LP
light pulse
- SCN
suprachiasmatic nucleus
- VIP
vasoactive intestinal peptide
References
- Abrahamson EE, Moore RY. Suprachiasmatic nucleus in the mouse: retinal innervation, intrinsic organization and efferent projections. Brain Res. 2001;916:172–191. doi: 10.1016/s0006-8993(01)02890-6. [DOI] [PubMed] [Google Scholar]
- Albrecht U, Zheng B, Larkin D, Sun ZS, Lee CC. MPer1 and mPer2 are essential for normal resetting of the circadian clock. J. Biol. Rhythms. 2001;16:100–104. doi: 10.1177/074873001129001791. [DOI] [PubMed] [Google Scholar]
- Bae K, Weaver DR. Light-induced phase shifts in mice lacking mPER1 or mPER2. J. Biol. Rhythms. 2003;18:123–133. doi: 10.1177/0748730403252248. [DOI] [PubMed] [Google Scholar]
- Cermakian N, Monaco L, Pando MP, Dierich A, Sassone-Corsi P. Altered behavioral rhythms and clock gene expression in mice with a targeted mutation in the Period1 gene. EMBO J. 2001;20:3967–3974. doi: 10.1093/emboj/20.15.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- Field MD, Maywood ES, O'Brien J, Weaver DR, Reppert S, Hastings MH. Analysis of clock proteins in mouse SCN demonstrates phylogenic divergence of the circadian clockwork and resetting mechanisms. Neuron. 2000;25:437–447. doi: 10.1016/s0896-6273(00)80906-x. [DOI] [PubMed] [Google Scholar]
- von Gall C, Noton E, lee C, Weaver DR. Light does not degrade the constitutively expressed BMAL1 protein in the mouse suprachiasmatic nucleus. Eur. J. Neurosci. 2003;18:125–133. doi: 10.1046/j.1460-9568.2003.02735.x. [DOI] [PubMed] [Google Scholar]
- Hamada T, LeSauter J, Venuti JM, Silver R. Expression of Period genes: rhythmic and nonrhythmic compartments of the suprachiasmatic nucleus pacemaker. J. Neurosci. 2001;21:7742–7750. doi: 10.1523/JNEUROSCI.21-19-07742.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karatsoreos IN, Yan L, LeSauter J, Silver R. Phenotype matters: Identification of light-response cells in mouse SCN. J. Neurosci. 2004;24:68–75. doi: 10.1523/JNEUROSCI.1666-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107:855–867. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- Lowrey PL, Shimomura K, Antoch MP, Yamazaki S, Zemenides PD, Ralph MR, Menaker M, Takahashi JS. Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science. 2000;288:483–492. doi: 10.1126/science.288.5465.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowrey PL, Takahashi JS. Genetics of the mammalian circadian system: Photic entrainment, circadian pacemaker mechanisms, and post-translational regulation. Annu. Rev. Genet. 2000;34:533–562. doi: 10.1146/annurev.genet.34.1.533. [DOI] [PubMed] [Google Scholar]
- Moore RY. Entrainment pathways and the functional organization of the circadian system. Prog. Brain Res. 1996;111:103–119. doi: 10.1016/s0079-6123(08)60403-3. [DOI] [PubMed] [Google Scholar]
- Nagano M, Adachi A, Nakahama K, Nakamura T, Tamada M, Meyer-Bernstein E, Sehgal A, Shigeyoshi Y. An abrupt shift in the day/night cycle causes desynchrony in the mammalian circadian center. J. Neurosci. 2003;23:6141–6151. doi: 10.1523/JNEUROSCI.23-14-06141.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuesslein-Hildesheim B, O'Brien JA, Ebling FJ, Maywood ES, Hastings MH. The circadian cycle of mPER clock gene products in the suprachiasmatic nucleus of the siberian hamster encodes both daily and seasonal time. Eur. J. Neurosci. 2000;12:2856–2864. doi: 10.1046/j.1460-9568.2000.00173.x. [DOI] [PubMed] [Google Scholar]
- Okamura H, Yamaguchi S, Yagita K. Molecular machinery of the circadian clock in mammals. Cell Tissue Res. 2002;309:47–56. doi: 10.1007/s00441-002-0572-5. [DOI] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR. Molecular analysis of mammalian circadian rhythms. Annu. Rev. Physiol. 2001;63:647–676. doi: 10.1146/annurev.physiol.63.1.647. [DOI] [PubMed] [Google Scholar]
- Rusak B, Zucker I. Neural regulation of circadian rhythms. Physiol. Rev. 1979;59:449–526. doi: 10.1152/physrev.1979.59.3.449. [DOI] [PubMed] [Google Scholar]
- Schwartz WJ, Zimmerman P. Circadian timekeeping in BALB/c and C57BL/6 inbred mouse strains. J. Neurosci. 1990;10:3685–3694. doi: 10.1523/JNEUROSCI.10-11-03685.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vielhaber E, Eide E, Rivers A, Gao Z-H, Virshup DM. Nuclear entry of the circadian regulator mPER1 is controlled by mammalian casein kinase 1ε. Mol. Cell. Biol. 2000;20:4888–4899. doi: 10.1128/mcb.20.13.4888-4899.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitaterna MH, Selby CP, Todo T, Niwa H, Thompson C, Fruechte EM, Hitomi K, Thresher RJ, Ishikawa T, Miyazaki J, Takahashi JS, Sancar A. Differential regulation of mammalian Period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc. Natl Acad. Sci. USA. 1999;96:12114–12119. doi: 10.1073/pnas.96.21.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagita K, Tamanini F, Yasuda M, Hoeijmakers JH, van Der Horst GT, Okamura H. Nucleocytoplasmic shuttling and mCRY-dependent inhibition of ubiquitylation of the mPER2 clock protein. EMBO J. 2002;21:1301–1314. doi: 10.1093/emboj/21.6.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Okamura H. Gradients in the circadian expression of Per1 and Per2 genes in the rat suprachiasmatic nucleus. Eur. J. Neurosci. 2002;15:1153–1162. doi: 10.1046/j.1460-9568.2002.01955.x. [DOI] [PubMed] [Google Scholar]
- Yan L, Silver R. Differential induction and localization of mPer1 and mPer2 during advancing and delaying phase shifts. Eur. J. Neurosci. 2002;16:1531–1540. doi: 10.1046/j.1460-9568.2002.02224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]