Abstract
Histone deacetylases (HDACs) affect cell growth at the transcriptional level by regulating the acetylation status of nucleosomal histones. HDAC inhibition induces differentiation and/or apoptosis in transformed cells. We recently showed that HDAC inhibitors, such as suberoylanilide hydroxamic acid (SAHA), potently induce apoptosis of human multiple myeloma (MM) cells. In this study, we focused on MM as a model to study the transcriptional profile of HDAC inhibitor treatment on tumor cells and to address their pathophysiological implications with confirmatory mechanistic and functional assays. We found that MM cells are irreversibly committed to cell death within few hours of incubation with SAHA. The molecular profile of MM cells before their commitment to SAHA-induced cell death is hallmarked by a constellation of antiproliferative and/or proapoptotic molecular events, including down-regulation of transcripts for members of the insulin-like growth factor (IGF)/IGF-1 receptor (IGF-1R) and IL-6 receptor (IL-6R) signaling cascades, antiapoptotic molecules (e.g., caspase inhibitors), oncogenic kinases, DNA synthesis/repair enzymes, and transcription factors (e.g., XBP-1, E2F-1) implicated in MM pathophysiology. Importantly, SAHA treatment suppresses the activity of the proteasome and expression of its subunits, and enhances MM cell sensitivity to proteasome inhibition by bortezomib (PS-341). SAHA also enhances the anti-MM activity of other proapoptotic agents, including dexamethasone, cytotoxic chemotherapy, and thalidomide analogs. These findings highlight the pleiotropic antitumor effects of HDAC inhibition, and provide the framework for future clinical applications of SAHA to improve patient outcome in MM.
Keywords: oligonucleotide microarrays, cell signaling, drug resistance
Histone deacetylases (HDACs) and acetyltransferases (HATs) regulate gene expression by removal or addition, respectively, of acetyl groups to ε-amino groups in lysine residues of core nucleosomal histones. Increased histone acetylation attenuates their electrostatic interaction with the negatively charged DNA backbone, promotes unfolding of the histone–DNA complex, and modulates access of transcription factors to their sites of action and transcription of their target genes (1–6). Normal cell differentiation and function require coordinated gene transcription and appropriate balance of HAT and HDAC activities. Indeed, deletions or inactivating mutations of HATs have been implicated in development of human neoplasms (7–9), whereas inhibition of HDAC activity and increased transcriptionally active chromatin (2–4) promotes differentiation, growth arrest, and/or apoptosis of tumor cells in vitro (10, 11) and in vivo (2, 11).
Suberoylanilide hydroxamic acid (SAHA), the prototype of synthetic hydroxamic acid-based hybrid polar HDAC inhibitors, binds directly to the HDAC catalytic site, inhibits its enzymatic activity (1), and exerts antiproliferative and/or proapoptotic effects restricted to transformed cells (7). It is orally bioavailable and had objective evidence of antitumor activity with a favorable side effect profile in phase I and II clinical trials (12). Multiple myeloma (MM) cells are sensitive to SAHA (13), and therefore, in this study, we used MM as a model to delineate the molecular mechanisms of HDAC-induced antitumor activity. We show that MM cells are irreversibly committed to cell death after few hours of incubation with HDAC inhibitors. When we used oligonucleotide micro array analyses to characterize the transcriptional profile of HDAC inhibition in MM cells, we found that the commitment of MM cells to SAHA-induced apoptosis is associated with early changes in gene expression profile, including suppression of genes mediating cytokine-driven proliferation and survival, drug-resistance, cell cycle control, DNA synthesis/repair, and proteasome function. These findings shed light on the complex molecular sequelae of HDAC inhibition in tumor cells and provide a preclinical rationale for clinical evaluation of SAHA, alone and in combination with conventional or investigational antitumor therapies, to improve patient outcome in MM.
Materials and Methods
Tissue Culture and Materials. The human MM cell line MM-1S was kindly provided by Steven Rosen (Northwestern University, Chicago) and cultured as described (14). SAHA was obtained from Aton Pharma (Tarrytown, NY).
Oligonucleotide Microarray Analysis of Gene Expression. The transcriptional profile of HDAC inhibition was characterized by oligonucleotide microarray analysis of MM-1S cells treated with SAHA (1 μM for 0–48 h) vs. control cells, by using the human U133A Affymetrix GeneChip, according to previously described protocols for total RNA extraction and purification, cDNA synthesis, in vitro transcription reaction for production of biotin-labeled cRNA, hybridization of cRNA with U133A Affymetrix gene chips, scanning of image output files, and analysis of gene expression data sets by filtering of up-regulated or down-regulated transcripts based on conventional criteria for statistical significance, as well as by hierarchical and functional clustering algorithms (15, 16).
Functional Assays. The transcriptional activities of NF-κB and p53 were quantified in ELISA-based assays of nuclear extracts of SAHA-treated MM-1S, as described (15, 17–20). ELISA for IGF-1 in supernatants of SAHA-treated MM cells were performed with commercially available kit (R & D Systems). Previously described protocols were used for telomerase activity assays (21) and 20S proteasome activity assays (22).
Further Details. Further details of the experimental procedures and gene expression profiling analyses are provided in Supporting Text, which is published as supporting information on the PNAS web site.
Results
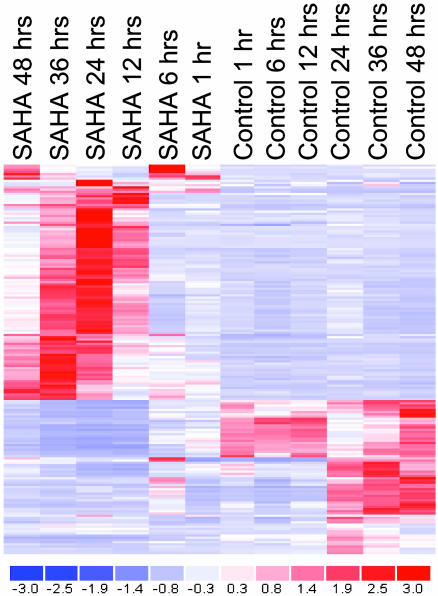
Commitment of MM Cells to SAHA-Induced Cell Death. We first investigated whether a short incubation with SAHA is sufficient to commit MM cells to cell death. In these cell death commitment assays, MM-1S cells treated with SAHA (1 μM for 0–24 h) were washed to remove SAHA from the culture medium, and were subsequently cultured with standard drug-free medium for an additional 72–96 h (total duration of culture, for all SAHA-treated and controls cells, was 96 h). 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay showed that SAHA culturefor8hor more is sufficient to irreversibly commit MM cells to apoptosis (Fig. 1 and Fig. 6, which is published as supporting information on the PNAS web site). Therefore, studies of SAHA-induced molecular sequelae in the first few hours of treatment can provide insight into the mechanisms of cell death commitment by HDAC inhibition.
Fig. 1.
Hierarchical clustering of gene expression profiling data (obtained by oligonucleotide-microarray analysis) in SAHA-treated vs. control human MM-1S cells.
Transcriptional Profile of SAHA Treatment. Increased histone acetylation increases the access of the transcriptional machinery to its target genes, and HDAC inhibition was generally considered to enhance gene transcription. The gene expression profiles of SAHA-treated MM cells were not hallmarked by global transcriptional activation. Rather, we found distinct patterns of coordinated transcriptional changes (most frequently repression) of specific functional groups of genes with well documented roles in the pathophysiology of MM and other neoplasias. Transcriptional changes (decreases or increases in gene expression of 2-fold or greater) occurred in select genes of the cytokine-induced proliferative/survival signaling cascades, oncogenes/tumor suppressor genes, regulators of apoptosis, DNA synthesis/repair and cell cycle, and proteasome/ubiquitin function (Figs. 2, 3, 4, 5). The lack of global transcriptional alterations in SAHA-treated MM cells, by using one of the most comprehensive oligonucleotide microarrays currently available, is consistent with previous reports indicating that ≈2% of expressed genes were modulated by HDAC inhibition in solid tumors (23). This selectivity of SAHA-induced transcriptional changes is useful for mechanistic studies of the consequences of such altered gene expression and may explain, at least in part, the therapeutic window of SAHA administration in clinical trials (12).
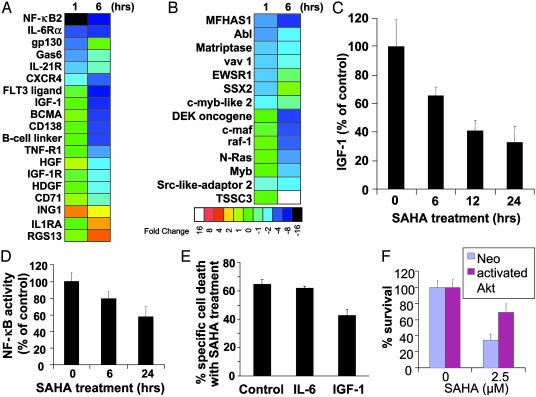
Fig. 2.
Functional clustering analysis of genes implicated in cytokine-induced proliferative/antiapoptotic signaling pathways (A) and oncogenes/tumor suppressor genes (B). SAHA down-regulates signaling pathways for MM cell proliferation and survival, including IGF/IGF-1R and IL-6R/gp130, suppresses expression of multiple oncogenes, and up-regulates several tumor suppressor genes. Color saturation is proportional to magnitude of the difference from the respective control. (C) SAHA treatment (1 μM, 0–24 h) suppresses IGF-1 autocrine production by MM-1S cells. (D) NF-κB DNA binding ELISA confirms that SAHA (1 μM, 0–24 h) suppresses NF-κB activity in MM-1S cells. (E and F) IGF/IGF-1R/Akt pathway protects against apoptosis induced by HDAC inhibition. (E) IGF-1 (200 ng/ml), but not IL-6 (200 ng/ml), reduces the percentage of specific cell death of MM-1S cells after treatment with SAHA (1 μM for 48 h) (MTT survival assay). (F) SAHA-induced cell death (quantified by MTT, mean ± SD) in MM-1S cells transfected with a vector expressing constitutively active Akt or control (neo) vector, after overnight serum starvation, and incubation with or without SAHA for additional 48 h. MM-1S cells transfected with constitutively active Akt construct have reduced sensitivity to SAHA.
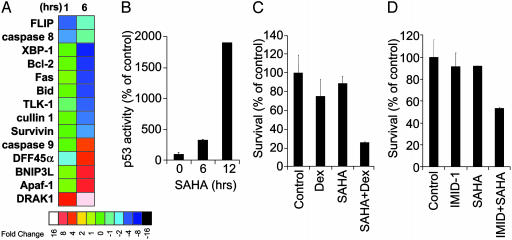
Fig. 3.
Effects of SAHA on regulators of apoptosis and sensitivity to caspase-dependent drug-induced apoptosis. (A) Functional clustering analysis for changes in gene expression of regulators of apoptosis. (B) p53 DNA-binding ELISA confirms that SAHA treatment (1 μM0–24 h) induces activation of p53. (C and D) MTT assays confirm that SAHA pretreatment (50 nM for 24 h) enhances sensitivity of MM-1S cells to dexamethasone (0.1 μM for an additional 48 h) (C) or the immunomodulatory thalidomide derivative IMID-1 (CC-4047) (0.01 μM for 48 h) (D).
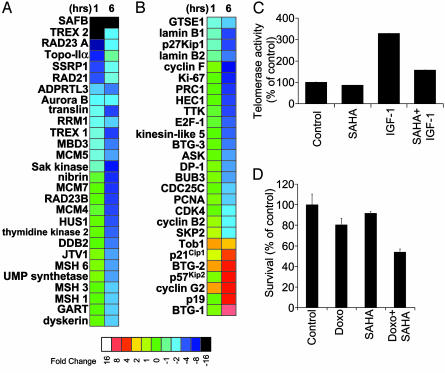
Fig. 4.
Effects of SAHA on DNA synthesis and repair, cell cycle regulation telomerase activity, and chemosensitivity. (A and B) Functional clustering analysis for genes involved in cell cycle regulation, DNA synthesis and repair (A), and cell cycle regulation (B). (C) Telomeric repeat amplification protocol assay for quantification of telomerase (hTERT) activity indicates that SAHA suppresses both constitutive and IGF-1 (200 ng/ml)-induced hTERT activity. (D) MTT assays confirm that SAHA pretreatment (50 nM for 24 h) enhances the sensitivity of MM-1S cells to doxorubicin (25 ng/ml for additional 48 h).
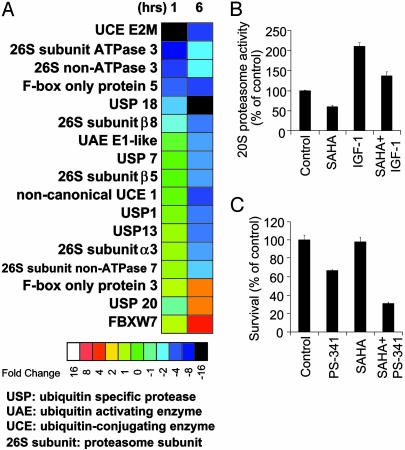
Fig. 5.
Functional impact of SAHA on the ubiquitin/proteasome pathway. (A) Functional clustering analysis for genes of the ubiquitin/proteasome pathway. (B) 20S proteasome activity assays confirm that HDAC inhibition by SAHA (1 μM, 24 h incubation) suppresses both constitutive and IGF (200 ng/ml)-induced activity of the proteasome. (C) MTT assays confirm that SAHA pretreatment (50 nM for 24 h) enhances sensitivity of MM-1S cells to proteasome inhibitor PS-341 (bortezomib) (5 nM for additional 24 h).
Effects of SAHA on Cytokine-Induced Proliferative/Antiapoptotic Signaling Pathways. SAHA treatment suppresses transcription of genes implicated in cytokine-induced proliferative/antiapoptotic signaling pathways (Fig. 2A) and oncogenic transformation (Fig. 2B). SAHA down-regulates insulin-like growth factor-1 (IGF-1) and its receptor IGF-1R (CD221); down-regulates IL-6R receptor and its key signal transducer gp130 (24); suppresses expression of members of the signaling axis of the tumor necrosis factor (TNF) receptor (TNF-R) superfamily and NF-κB pathway, including TNF-receptor-1 (TNF-R1), the proliferative TNF-R family member BCMA (which is up-regulated in MM cells vs. normal B cells or plasma cells, ref. 25), and NF-κB2 itself; suppresses expression of other cytokines with known or proposed roles in stimulating the proliferation and/or survival of malignant cells from MM or other neoplasias, including hepatocyte growth factor (HGF) [which is expressed at higher levels in sera of MM patients than healthy age-matched controls (26) and functions as an autocrine/paracrine proliferation factor for MM cells], hepatoma-derived growth factor (HDGF) (a known mitogen for various tumors, including hepatocellular carcinoma; ref. 27) and growth arrest-specific 6 (Gas6, the ligand for the proliferative cell surface tyrosine kinase receptor Axl; ref. 28). SAHA also suppresses expression of receptors that have been shown to trigger MM cell proliferation, survival, and/or migration, such as CD138 (syndecan-1), CD71 (transferrin receptor), IL-21R, and CXCR-4 (29–32). In addition, SAHA upregulates negative regulators of cell proliferation, such as ING-1 (inhibitor of growth family member 1) (33) and regulator of G protein signaling 13 (34) and also up-regulates IL-1 receptor (IL-1R) antagonist, which inhibits IL-1-induced signaling in MM (35) (Fig. 2A).
Functional Studies of the Effect of SAHA on Cytokine-Induced Proliferative/Antiapoptotic Signaling Pathways. Recent studies indicate that the IGF/IGF-1R cascade plays critical role in MM cell proliferation and survival (17, 36). Furthermore, the TNF receptor superfamily and the antiapoptotic activities of NF-κB play a key role in MM pathophysiology, and NF-κB activity itself is major target for multiple emerging therapeutic strategies for MM (17, 19, 36, 37). Because SAHA had significant early impact on members of these cascades, we investigated their functional role in modulating HDAC inhibitor activity. We first observed, by ELISA-based assays, that SAHA suppresses autocrine IGF-1 production by MM cells (Fig. 2C) and suppresses the binding of NF-κB to its consensus DNA binding sequences (Fig. 2D). The SAHA-induced suppression of IGF-1 expression was also confirmed in two primary tumor samples from MM patients with resistance to both conventional and novel (thalidomide and proteasome inhibitor) treatments, and the SAHA-induced suppression of NF-κB was confirmed in the OPM-1 cell line model (data not shown). To specifically address whether SAHA-induced suppression of the IGF/IGF-1R pathway contributes to the anti-MM effect of the drug, we determined the effect of IGF-1 (200 ng/ml) on SAHA-induced cell death of MM cells in vitro. Exogenous IGF-1 attenuated the sensitivity of MM cells to SAHA (Fig. 2E), consistent with the binding that SAHA down-regulates autocrine IGF-1/IGF-1R activity in MM cells. We performed stable transfection of MM-1S with construct for constitutively activated (myristoylated) Akt kinase, to allow for Akt activation despite inhibition of IGF-1/IGF-1R upstream signaling. The sensitivity of MM-1S-myr-Akt cells to SAHA was decreased relative to empty vector transfected cells (Fig. 2F), indicating that downstream constitutive activation of IGF-1R pathway can overcome the SAHA-induced inhibition of upstream components of this cascade. These data suggest that the antitumor effect of SAHA may be enhanced when it is combined with other agents targeting the IGF-1/IGF-1R pathway. The fact that IGF-1, but not IL-6, can attenuate SAHA-induced apoptosis, even though both cytokines activate Akt, is probably due to the fact that IGF-1 induces more pronounced and sustained activation of Akt and its downstream targets than IL-6 (17). This explanation would also be consistent with the ability of IGF-1 to stimulate, in a PI-3K/Akt-dependent manner, the up-regulation of a broader spectrum of antiapoptotic events, e.g., up-regulation of more inhibitors of apoptosis (IAPs) than IL-6, and thus protect MM cells against proapoptotic drugs, e.g., Apo2L/TRAIL (TNF-related apoptosis-inducing ligand) or the proteasome inhibition PS-341 (15, 17), which overcome the protective effects of IL-6.
Effect of SAHA on Oncogenes and Tumor Suppressor Genes. SAHA treatment modulates genes with documented role in oncogenic transformation in MM and/or other neoplasias (Fig. 2B). Myb (25) and myb-like 2 (c-MybL2), which have higher levels of expression in MM cells and N-Ras, which is frequently mutated in MM cells, were down-regulated (38). HDAC inhibition also suppressed oncogenes implicated in other disease models, including raf-1, abl (which is involved in the bcr/abl chimeric kinase in chronic myelogenous leukemia and a subset of acute lymphoblastic leukemias, vav1 (a proto-oncogene activated by IGF/IGF-1R signaling), DEK (a proto-oncogene overexpressed in various forms of leukemia), SSX2 (synovial sarcoma X2, an oncogene originally identified in sarcoma cells, but recently described to be over-expressed in a subset of MM cases; refs. 39–41),§§ and EWSR1 (43) or MFHAS1 (malignant fibrous histiocytoma amplified sequence 1) (44). SAHA induced up-regulation of tumor suppressor genes, including TSSC3 (tumor suppressing subtransferable candidate 3), an imprinted apoptosis-related gene located within the tumor suppressor region of 11p15, which is implicated in various forms of cancer (45).
Effect of SAHA on Genes Involved in Regulation of Apoptosis/Survival and Drug Resistance. SAHA induces up-regulation of several proapoptotic and down-regulation of antiapoptotic genes (Fig. 3A). SAHA induces up-regulation of the proapoptotic molecules Apaf-1 and caspase-9 (15), the apoptosis-inducing serine/threonine kinase DRAK1, and DFF45 (DNA fragmentation factor 45 kDa-α, a mediator of apoptotic signaling in myeloma; ref. 18). SAHA increased the transcriptional activity of p53 protein, as shown by up-regulation of p53-responsive genes, such as TP53TG1 (p53 target gene 1) (data not shown) and an increase in p53 DNA-binding activity (Fig. 3B). SAHA down-regulates expression of antiapoptotic molecules, including the intracellular inhibitors of apoptosis FLIP and survivin (which block the proapoptotic activity of caspases-8 and -3, respectively, and contribute to reduced MM cell sensitivity to proapoptotic therapies, such as Apo2L/TRAIL or dexamethasone; ref. 17); as well as tousled-like kinase 1 (TLK-1, which is associated with resistance to radiation therapy) (46) and XBP-1 (X-box-binding protein-1), a key transcription factor for plasma cell differentiation, which was recently proposed to function as regulator of MM cell proliferation, survival, and drug-resistance (47). These results are consistent with our previous finding that SAHA sensitizes MM cells to caspase-dependent apoptosis, e.g., by Fas ligation or Apo2L/TRAIL (13). Furthermore, culture of MM-1S cells with SAHA for 24 h increased their sensitivity to dexamethasone (Fig. 3C) and the immunomodulatory thalidomide derivative 1 (IMID-1) (CC-4047) (Fig. 3D), anti-MM agents that induce caspase-9- and caspase-8-dependent apoptosis, respectively (20, 48). The SAHA-induced sensitization to dexamethasone- and IMID-induced apoptosis was also confirmed in two primary tumor samples of MM patients resistant to conventional therapies (including dexamethasone- or thalidomide-based regimens) (data not shown).
Effects of SAHA on Cell Cycle Regulation, DNA Synthesis, and Repair. HDAC inhibition of MM cells modulates the expression of genes involved in DNA synthesis/repair (Fig. 4A) and cell cycle regulation (Fig. 4B). SAHA up-regulated genes with growth arrest/antiproliferative function (e.g., p21Cip1, p57Kip2 and p19, BTG-1, and Tob-1); down-regulated several genes required for cell proliferation and frequently overexpressed in tumor cells, including CDK4, ASK, CDC25C; various cyclins, proliferating cell nuclear antigen (PCNA), Ki-67, and SKP2 (49), E2F-1 and its dimerization partner DP1 (which activate transcription of genes necessary for DNA replication; ref. 50). SAHA down-regulated genes involved in maintaining chromosomal integrity and stability, such as HEC1 (highly expressed in cancer-1), PRC1 (protein regulator of cytokinesis 1), BUB3 (budding uninhibited by benzimidazoles 3, a gene essential for mitosis; ref. 51), MCM-4, -5, and -7 or several RAD homologs. SAHA repressed transcription of genes implicated in DNA metabolism, including uridine monophosphate synthetase, and genes implicated in DNA damage repair, including damage-specific DNA binding protein 2 (DDB2); TREX-1 and -2, and MSH-2, -3, and -6.
Functional Studies of the Effects of SAHA on Genes Regulating Cell Growth. The finding that SAHA causes a down-regulation of expression of IGF/IGF-1R and IL-6R genes, which modulate telomerase function (21), led us to examine the effect of HDAC inhibition on telomerase activity. By using telomeric repeat amplification protocol (TRAP) assays, we determined the activity of the catalytic subunit of telomerase (hTERT) and found that SAHA induces profound suppression of both constitutive and cytokine (e.g., IGF-1)-induced activity of hTERT (Fig. 4C). The impact of SAHA on transcription of genes related to DNA damage repair and chromosomal integrity suggested that SAHA may alter the chemosensitivity of tumor cells. Treatment with 50 nM SAHA for 24 h increased the doxorubicin-sensitivity of MM-1S cells (Fig. 4D) and RPMI-8226/S cells (data not shown), providing evidence of functional impact of HDAC inhibition on MM cell chemosensitivity.
Effect of SAHA Inhibition on Gene Transcription of the Ubiquitin/Proteasome Pathway. In contrast to proteasome inhibition, which triggers a stress response of increased gene transcription of ubiquitin/proteasome pathway members (15), SAHA represses expression of genes of this pathway (Fig. 5A), including 26S proteasome subunits (α3, β5, β8, ATPase 3, and non-ATPases 3 and 7), and several ubiquitin conjugating enzymes. To investigate the functional impact of these molecular events, we measured the proteasome activity in SAHA-treated MM-1S cells by using a 20S proteasome chymotryptic activity assay. SAHA suppressed both constitutive and IGF-1-induced 20S proteasome activity (Fig. 5B). This finding suggests that the proteasome can be inhibited not only by small molecules (e.g., PS-341), which directly target the active site(s) of its proteolytic function, but also by depletion of its structural components or targeting of its upstream regulators. Because HDAC and PS-341 inhibit different targets in the proteasome pathway, we examined whether their combination could enhance suppression of proteasome activity and anti-MM effects. Pretreatment of MM-1S cells with 50 nM SAHA for 24 h before treatment with 5 nM PS-341 for additional 24 h led to enhanced anti-MM effect of this combination, compared to either drug alone (Fig. 5C). The SAHA-induced sensitization to PS-341 was also confirmed in three primary tumor specimens from MM patients.
Discussion
Microarray-based gene expression profiling of drug-treated tumor cells is a powerful tool to delineate the mechanisms of action of antitumor agents (15) and is particularly warranted for the study of antitumor agents, e.g., HDAC inhibitors, which directly target the transcriptional machinery of tumor cells. The encouraging results of phase I and phase II clinical trials of SAHA (12) and our recent studies showing that clinically relevant doses of SAHA potently induce apoptosis of MM cells (13), provided the impetus to study the gene expression profile of SAHA-treated MM cells, as a model to further define its mechanism of antitumor activity.
Culture of MM cells with SAHA for at least 8 h irreversibly commits them to cell death. In the present study, we examined the effect of SAHA on gene expression at 1 and 6 h of culture. SAHA causes selective alterations of gene expression that involves activation or suppression of functional clusters of genes with known specific roles in the pathophysiology of tumor cells, in general, or MM cells, in particular. The constellation of cellular pathways targeted by SAHA include IGF/IGF-1R/Akt (which is critical for proliferation and survival of MM and other malignancies; ref. 36), IL-6/gp130, oncogenes, proliferative/antiapoptotic transcription factors (e.g., NF-κB, XBP-1, E2F-1), and cell cycle regulators.
Contrary to previous hypotheses, SAHA did not up-regulate genes implicated in cell differentiation, but mainly suppressed genes responsible for uncontrolled proliferation and inappropriate resistance to proapoptotic stimuli. The differential impact of SAHA on gene expression in different diseases and in different gene groups within the same disease, e.g., suppression of specific functional clusters of genes vs. up-regulation of others, may be due to the fact that SAHA, by interacting directly with the catalytic site of HDACs can potentially affect the transcription rate of specific genes in a differential manner, depending on the context in which HDACs regulate the accessibility of the promoter region of these genes and/or alter the postulated “histone code” (52, 53) in areas with positive or negative regulatory role of the expression of the respective transcripts.
One of the objectives of this study of the molecular profile of SAHA-treated tumor cells was to establish a framework for design of combination therapies with conventional and/or investigational antitumor agents. This study identified the enhanced antitumor activity of combinations of SAHA with conventional anti-MM agents, such as dexamethasone or doxorubicin, as well as recently introduced anti-MM therapies, such as the proteasome inhibitor bortezomib (PS-341) and the IMIDs. Indeed, the SAHA-induced suppression of caspase inhibitors (e.g., FLIP and survivin) provides, at least in part, an explanation for the increased sensitivity of SAHA-treated MM cells to caspase-dependent apoptosis by soluble Apo2L/TRAIL or Fas-ligation (14), as well as by dexamethasone and thalidomide analogs, which trigger caspase-9- and caspase-8-dependent apoptosis, respectively. The ability of SAHA to suppress genes involved in DNA synthesis and maintenance of its structural integrity constitute a molecular basis for the chemosensitizing effect of HDAC inhibition.
The pleiotropic molecular sequelae of HDAC inhibition do not allow us to conclude which pathway(s) is/are most important for its antitumor effect, but potentially offer a major therapeutic advantage: namely, the simultaneous targeting of different proliferative/antiapoptotic pathways in tumor cells. Therapeutic strategies targeting single molecular lesions pathognomonic for specific tumor types have had significant recent success (54), but their effect is often circumvented by mutations/amplifications of the respective target and/or other compensatory collateral or downstream mechanisms for sustained tumor cell survival (42). The emergence of such resistance may be less probable or delayed in the setting of SAHA treatment, because its effect may neutralize the pathways potentially responsible for the ability of tumor cells to evade cell death. This hypothesis is supported by the potent activity of SAHA even against MM cells resistant to several conventional or investigational agents (13). Furthermore, because SAHA targets molecular pathways that function abnormally in a wide range of tumors, this may explain why it has had a broad spectrum of activity across different neoplasias (7, 12).
Supplementary Material
Acknowledgments
This work was supported by the Multiple Myeloma Research Foundation (C.S.M. and N.M.), the Lauri Strauss Leukemia Foundation (C.S.M. and N.M.), the International Waldenstrom's Macroglobulinemia Foundation (C.S.M.), The Cure for Myeloma Fund (K.C.A.), The Fulbright Commission (C.J.M.), National Institutes of Health Grants RO-1 50947 and PO-1 78378, and Doris Duke Distinguished Clinical Research Scientist Award (to K.C.A.), as well as the Susan and Jack Rudin Foundation, the Kleberg Foundation, the David Koch Foundation, the DeWitt Wallace Fund for Memorial Sloan–Kettering Cancer Center, and the National Cancer Institute (P.A.M.). C.S.M. is a Special Fellow of the Leukemia and Lymphoma Society.
Abbreviations: MM, multiple myeloma; HDAC, histone deacetylase; SAHA, suberoylanilide hydroxamic acid; IGF-1, insulin-like growth factor 1; IGF-1R, IGF-1 receptor; TNF, tumor necrosis factor; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide; TRAIL, TNF-related apoptosis-inducing ligand; IMID-1, immunomodulatory thalidomide derivative 1.
Footnotes
Taylor, B. J., Pittman, J., Reiman, T., Szydlowski, J., Keats, J., Belch, A. R. & Pilarski, L. M. (2003) in Proceedings of IX International Myeloma Workshop, Salamanca Spain (Nature Publishing Group, London), p. P52 (abstr.).
References
- 1.Finnin, M. S., Donigian, J. R., Cohen, A., Richon, V. M., Rifkind, R. A., Marks, P. A., Breslow, R. & Pavletich, N. P. (1999) Nature 401, 188–193. [DOI] [PubMed] [Google Scholar]
- 2.Richon, V. M., Emiliani, S., Verdin, E., Webb, Y., Breslow, R., Rifkind, R. A. & Marks, P. A. (1998) Proc. Natl. Acad. Sci. USA 95, 3003–3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marks, P. A., Richon, V. M. & Rifkind, R. A. (2000) J. Natl. Cancer Inst. 92, 1210–1216. [DOI] [PubMed] [Google Scholar]
- 4.Marks, P. A., Richon, V. M., Breslow, R. & Rifkind, R. A. (2001) Curr. Opin. Oncol. 13, 477–483. [DOI] [PubMed] [Google Scholar]
- 5.Richon, V. M., Zhou, X., Rifkind, R. A. & Marks, P. A. (2001) Blood Cells Mol. Dis. 27, 260–264. [DOI] [PubMed] [Google Scholar]
- 6.Richon, V. M., Sandhoff, T. W., Rifkind, R. A. & Marks, P. A. (2000) Proc. Natl. Acad. Sci. USA 97, 10014–10019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marks, P., Rifkind, R. A., Richon, V. M., Breslow, R., Miller, T. & Kelly, W. K. (2001) Nat. Rev. Cancer 1, 194–202. [DOI] [PubMed] [Google Scholar]
- 8.Murata, T., Kurokawa, R., Krones, A., Tatsumi, K., Ishii, M., Taki, T., Masuno, M., Ohashi, H., Yanagisawa, M., Rosenfeld, M. G., et al. (2001) Hum. Mol. Genet. 10, 1071–1076. [DOI] [PubMed] [Google Scholar]
- 9.Lin, R. J., Nagy, L., Inoue, S., Shao, W., Miller, W. H., Jr., & Evans, R. M. (1998) Nature 391, 811–814. [DOI] [PubMed] [Google Scholar]
- 10.Glick, R. D., Swendeman, S. L., Coffey, D. C., Rifkind, R. A., Marks, P. A., Richon, V. M. & La Quaglia, M. P. (1999) Cancer Res. 59, 4392–4399. [PubMed] [Google Scholar]
- 11.Butler, L. M., Agus, D. B., Scher, H. I., Higgins, B., Rose, A., Cordon-Cardo, C., Thaler, H. T., Rifkind, R. A., Marks, P. A. & Richon, V. M. (2000) Cancer Res. 60, 5165–5170. [PubMed] [Google Scholar]
- 12.Kelly, W. K., Richon, V. M., O'Connor, O., Curley, T., MacGregor-Curtelli, B., Tong, W., Klang, M., Schwartz, L., Richardson, S., Rosa, E., et al. (2003) Clin. Cancer Res. 9, 3578–3588. [PubMed] [Google Scholar]
- 13.Mitsiades, N., Mitsiades, C. S., Richardson, P. G., McMullan, C., Poulaki, V., Fanourakis, G., Schlossman, R., Chauhan, D., Munshi, N. C., Hideshima, T., et al. (2003) Blood 101, 4055–4062. [DOI] [PubMed] [Google Scholar]
- 14.Moalli, P. A., Pillay, S., Weiner, D., Leikin, R. & Rosen, S. T. (1992) Blood 79, 213–222. [PubMed] [Google Scholar]
- 15.Mitsiades, N., Mitsiades, C. S., Poulaki, V., Chauhan, D., Fanourakis, G., Gu, X., Bailey, C., Joseph, M., Libermann, T. A., Treon, S. P., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 14374–14379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitsiades, N., Mitsiades, C. S., Richardson, P. G., Poulaki, V., Tai, Y. T., Chauhan, D., Fanourakis, G., Gu, X., Bailey, C., Joseph, M., et al. (2003) Blood 101, 2377–2380. [DOI] [PubMed] [Google Scholar]
- 17.Mitsiades, C. S., Mitsiades, N., Poulaki, V., Richardson, P. G., Schlossman, R., Akiyama, M., Chauhan, D., Hideshima, T., Munshi, N. C., Treon, S. P. & Anderson, K. C. (2002) Oncogene 21, 5673–5683. [DOI] [PubMed] [Google Scholar]
- 18.Mitsiades, N., Mitsiades, C. S., Poulaki, V., Anderson, K. C. & Treon, S. P. (2002) Blood 99, 2162–2171. [DOI] [PubMed] [Google Scholar]
- 19.Mitsiades, N., Mitsiades, C. S., Poulaki, V., Chauhan, D., Richardson, P. G., Hideshima, T., Munshi, N., Treon, S. P. & Anderson, K. C. (2002) Blood 99, 4079–4086. [DOI] [PubMed] [Google Scholar]
- 20.Mitsiades, N., Mitsiades, C. S., Poulaki, V., Chauhan, D., Richardson, P. G., Hideshima, T., Munshi, N. C., Treon, S. P. & Anderson, K. C. (2002) Blood 99, 4525–4530. [DOI] [PubMed] [Google Scholar]
- 21.Akiyama, M., Hideshima, T., Hayashi, T., Tai, Y. T., Mitsiades, C. S., Mitsiades, N., Chauhan, D., Richardson, P., Munshi, N. C. & Anderson, K. C. (2002) Cancer Res. 62, 3876–3882. [PubMed] [Google Scholar]
- 22.Shringarpure, R., Grune, T., Mehlhase, J. & Davies, K. J. (2003) J. Biol. Chem. 278, 311–318. [DOI] [PubMed] [Google Scholar]
- 23.Mariadason, J. M., Corner, G. A. & Augenlicht, L. H. (2000) Cancer Res. 60, 4561–4572. [PubMed] [Google Scholar]
- 24.Tani, Y., Nishimoto, N., Ogata, A., Shima, Y., Yoshizaki, K. & Kishimoto, T. (1995) Curr. Top. Microbiol. Immunol. 194, 229–233. [DOI] [PubMed] [Google Scholar]
- 25.Davies, F. E., Dring, A. M., Li, C., Rawstron, A. C., Shammas, M. A., O'Connor, S. M., Fenton, J. A., Hideshima, T., Chauhan, D., Tai, I. T., et al. (2003) Blood, in press.
- 26.Seidel, C., Borset, M., Turesson, I., Abildgaard, N., Sundan, A. & Waage, A. (1998) Blood 91, 806–812. [PubMed] [Google Scholar]
- 27.Kishima, Y., Yoshida, K., Enomoto, H., Yamamoto, M., Kuroda, T., Okuda, Y., Uyama, H. & Nakamura, H. (2002) Hepatogastroenterology 49, 1639–1644. [PubMed] [Google Scholar]
- 28.Goruppi, S., Chiaruttini, C., Ruaro, M. E., Varnum, B. & Schneider, C. (2001) Mol. Cell. Biol. 21, 902–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ridley, R. C., Xiao, H., Hata, H., Woodliff, J., Epstein, J. & Sanderson, R. D. (1993) Blood 81, 767–774. [PubMed] [Google Scholar]
- 30.Ng, P. P., Dela Cruz, J. S., Sorour, D. N., Stinebaugh, J. M., Shin, S. U., Shin, D. S., Morrison, S. L. & Penichet, M. L. (2002) Proc. Natl. Acad. Sci. USA 99, 10706–10711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brenne, A. T., Baade Ro, T., Waage, A., Sundan, A., Borset, M. & Hjorth-Hansen, H. (2002) Blood 99, 3756–3762. [DOI] [PubMed] [Google Scholar]
- 32.Hideshima, T., Chauhan, D., Hayashi, T., Podar, K., Akiyama, M., Gupta, D., Richardson, P., Munshi, N. & Anderson, K. C. (2002) Mol. Cancer Ther. 1, 539–544. [PubMed] [Google Scholar]
- 33.Garkavtsev, I., Kazarov, A., Gudkov, A. & Riabowol, K. (1996) Nat. Genet. 14, 415–420. [DOI] [PubMed] [Google Scholar]
- 34.Johnson, E. N. & Druey, K. M. (2002) J. Biol. Chem. 277, 16768–16774. [DOI] [PubMed] [Google Scholar]
- 35.Carter, D. B., Deibel, M. R., Jr., Dunn, C. J., Tomich, C. S., Laborde, A. L., Slightom, J. L., Berger, A. E., Bienkowski, M. J., Sun, F. F., McEwan, R. N., et al. (1990) Nature 344, 633–638. [DOI] [PubMed] [Google Scholar]
- 36.Mitsiades, C. S., Mitsiades, N., Kung, A. L., Shringapurne, R., Poulaki, V., Richardson, P. G., Liberman, T. A., Munshi, N. C., Loukopoulos, D. & Anderson, K. C. (2002) Blood 100, 637. [Google Scholar]
- 37.Hideshima, T., Chauhan, D., Schlossman, R., Richardson, P. & Anderson, K. C. (2001) Oncogene 20, 4519–4527. [DOI] [PubMed] [Google Scholar]
- 38.Neri, A., Murphy, J. P., Cro, L., Ferrero, D., Tarella, C., Baldini, L. & Dalla-Favera, R. (1989) J. Exp. Med. 170, 1715–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shtivelman, E., Lifshitz, B., Gale, R. P. & Canaani, E. (1985) Nature 315, 550–554. [DOI] [PubMed] [Google Scholar]
- 40.Uddin, S., Yetter, A., Katzav, S., Hofmann, C., White, M. F. & Platanias, L. C. (1996) Exp. Hematol. 24, 622–627. [PubMed] [Google Scholar]
- 41.Aalto, Y., El-Rifa, W., Vilpo, L., Ollila, J., Nagy, B., Vihinen, M., Vilpo, J. & Knuutila, S. (2001) Leukemia 15, 1721–1728. [DOI] [PubMed] [Google Scholar]
- 42.Gorre, M. E., Mohammed, M., Ellwood, K., Hsu, N., Paquette, R., Rao, P. N. & Sawyers, C. L. (2001) Science 293, 876–880. [DOI] [PubMed] [Google Scholar]
- 43.Rossow, K. L. & Janknecht, R. (2001) Cancer Res. 61, 2690–2695. [PubMed] [Google Scholar]
- 44.Sakabe, T., Shinomiya, T., Mori, T., Ariyama, Y., Fukuda, Y., Fujiwara, T., Nakamura, Y. & Inazawa, J. (1999) Cancer Res. 59, 511–515. [PubMed] [Google Scholar]
- 45.Lee, M. P. & Feinberg, A. P. (1998) Cancer Res. 58, 1052–1056. [PubMed] [Google Scholar]
- 46.Li, Y., DeFatta, R., Anthony, C., Sunavala, G. & De Benedetti, A. (2001) Oncogene 20, 726–738. [DOI] [PubMed] [Google Scholar]
- 47.Chauhan, D., Li, G., Auclair, D., Hideshima, T., Richardson, P., Podar, K., Mitsiades, N., Mitsiades, C., Li, C., Kim, R. S., et al. (2003) Blood 101, 3606–3614. [DOI] [PubMed] [Google Scholar]
- 48.Chauhan, D., Hideshima, T., Rosen, S., Reed, J. C., Kharbanda, S. & Anderson, K. C. (2001) J. Biol. Chem. 276, 24453–24456. [DOI] [PubMed] [Google Scholar]
- 49.Latres, E., Chiarle, R., Schulman, B. A., Pavletich, N. P., Pellicer, A., Inghirami, G. & Pagano, M. (2001) Proc. Natl. Acad. Sci. USA 98, 2515–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nevins, J. R. (2001) Hum. Mol. Genet. 10, 699–703. [DOI] [PubMed] [Google Scholar]
- 51.Kalitsis, P., Earle, E., Fowler, K. J. & Choo, K. H. (2000) Genes Dev. 14, 2277–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Strahl, B. D. & Allis, C. D. (2000) Nature 403, 41–45. [DOI] [PubMed] [Google Scholar]
- 53.Schreiber, S. L. & Bernstein, B. E. (2002) Cell 111, 771–778. [DOI] [PubMed] [Google Scholar]
- 54.Druker, B. J., Sawyers, C. L., Kantarjian, H., Resta, D. J., Reese, S. F., Ford, J. M., Capdeville, R. & Talpaz, M. (2001) N. Engl. J. Med. 344, 1038–1042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.