Abstract
The suprachiasmatic nucleus (SCN) of the hypothalamus is the neural locus of the circadian clock. To explore the organization of the SCN, two strains of transgenic mice, each bearing a jellyfish green fluorescent protein (GFP) reporter, were used. In one, GFP was driven by the promoter region of the mouse Period1 gene (mPer1) (Per1::GFP mouse), whereas in the other, GFP was inserted in the promoter region of calbindin-D28K-bacterial artificial chromosome (CalB::GFP mouse). In the latter mouse, GFP-containing SCN cells are immunopositive for gastrin-releasing peptide. In both mouse lines, light-induced Per1 mRNA and Fos are localized to the SCN subregion containing gastrin-releasing peptide. Double-label immunohistochemistry reveals that most gastrin-releasing peptide cells (∼70%) contain Fos after a brief light pulse. To determine the properties of SCN cells in this light-responsive region, we examined the expression of rhythmic Period genes and proteins. Gastrin-releasing peptide-containing cells do not express detectable rhythms in these key components of the molecular circadian clock. The results support the view that the mammalian SCN is composed of functionally distinct cell groups, of which some are light induced and others are rhythmic with respect to clock gene expression. Furthermore, the findings suggest that gastrin-releasing peptide is a potential mediator of intercellular communication between light-induced and oscillator cells within the SCN.
Keywords: circadian rhythms, SCN, GFP reporter, transgenic, GRP, mPer1, clock gene
Introduction
Daily rhythms in physiology and behavior persist in the absence of temporal information from the environment. Importantly, these self-sustained rhythms are synchronized, or entrained, to the environment by appropriately timed external cues. The neural locus controlling endogenous circadian rhythmicity lies in the suprachiasmatic nucleus (SCN) of the hypothalamus (Klein, 1991), with each nucleus containing 8000-10,000 cells in rodents (Van den Pol, 1980; Abrahamson and Moore, 2001; Moore et al., 2002). In mammals, photic cues from the day/night cycle reach the SCN via the retinohypothalamic tract (RHT) (Morin, 1994). It is not well understood how the individual cells that constitute this nucleus perform the dual functions of oscillating and resetting.
Circadian oscillation is a property of individual SCN cells (Welsh et al., 1995). At the molecular level, the core circadian rhythm-generating mechanism involves an autoregulatory transcription-translation feedback loop. In mammals, the positive transcriptional arm of the loop is driven by the proteins Clock and Bmal1, whereas the protein products of the transcribed clock genes Period (Per1, Per2, and Per3) and Cryptochrome (Cry1 and Cry2) comprise the negative arm (Lowrey and Takahashi, 2000; Shearman et al., 2000; Young and Kay, 2001). Entrainment to local environmental time is thought to result from adjustments of various steps in these oscillating feedback loops by light and other stimuli.
In mammals, the photosensitive elements necessary for resetting the SCN lie in the eye (Panda et al., 2003). Direct photic input from the RHT reaches only a fraction of SCN cells (Groos and Mason, 1980; Meijer et al., 1986; Cui and Dyball, 1996). The intercellular mechanisms for communication of phase resetting by photic stimuli from retinorecipient cells to the rest of the SCN are not known. Additionally, it is unclear whether all SCN cells are oscillators (Reppert and Weaver, 2001, 2002). For example, in the hamster, SCN regions marked by vasopressin (VP)-containing cells are characterized by rhythmic expression of Per1 and Per2 mRNA. In contrast, in the caudal SCN, a region marked by calbindin-expressing cells, light-induced but not rhythmic expression of these genes is detected (Hamada et al., 2001). Electrophysiological studies demonstrate a lack of rhythmicity in calbindin cells and support the view that some SCN cells are not oscillators (Jobst and Allen, 2002). It is possible that the hamster SCN, with its discrete population of densely packed calbindin-expressing cells, is unique among mammals. Alternatively, it may be that two distinct populations of cells, one rhythmic and the other nonrhythmic with regard to clock gene expression, are a general feature of mammalian SCN organization.
The present results reveal that in mice, some SCN cells express Per1 rhythmically, whereas others do not. In addition, there is a population of cells containing gastrin-releasing peptide that is light induced but does not express detectable rhythms in Per genes.
Materials and Methods
Animals. Two strains of transgenic mice, each bearing a jellyfish green fluorescent protein (GFP) reporter, were used. The mPer1::d2EGFP transgenic mouse (termed here Per1::GFP; a gift from Dr. D. McMahon, Vanderbilt University, Nashville, TN) was made with the B6C3F1 hybrid mouse as described previously (Kuhlman et al., 2000). The mice used in the present studies were hemizygous for the d2EGFP transgene, which is a degradable form of GFP.
The calbindin-D28K-bacterial artificial chromosome (BAC)::GFP transgenic mouse (termed here CalB::GFP; a gift from Dr. N. Heintz, Rockefeller University, New York, NY) was made by insertion of enhanced GFP (EGFP) (Yang et al., 1997) in the calbindin BAC (Research Genetics/Invitrogen, Carlsbad, CA) by homologous recombination. The homologous region was upstream of the start codon of the calbindin gene. The first stretch of the sequence included the calbindin-D28K promoter followed by the first linker sequence and then the EGFP construct followed by the second linker, with the tail sequence containing more of the calbindin gene, including the start codon (at position 1879).
There are several advantages to using these transgenic mice. First, the Per1-driven GFP is a more sensitive marker than the Per protein revealed by an anti-Per antibody (Witkovsky et al., 2003). Second, our results focus on the role of gastrin-releasing peptide-containing cells of the SCN. Gastrin-releasing peptide cells are difficult to detect unless the animal is treated with colchicine, because a dense fiber plexus obscures cell bodies. In the present study, we found that CalB::GFP could be used as a reporter for gastrin-releasing peptide (see below), thereby eliminating these technical concerns. Finally, in both cases, the GFP transgene permits the living neuron to be visualized, making it available for future electrophysiological and imaging studies.
Experimental animals were housed in translucent propylene cages (29 × 19 × 12.5 cm) in a 12 hr light/dark (LD) cycle (n = 24) or were transferred to constant darkness (DD; n = 24) for at least 2 d before experimentation. The rooms were maintained at 21 ± 1°C. For animals in DD, a white-noise generator (91 sound pressure level) masked environmental noise and a dim red light (0.5-1 lux) allowed for animal maintenance. All animals were provided with food and water ad libitum and cared for in accordance with the Columbia University Institutional Animal Care and Use Committee and Animal Welfare regulations.
Colchicine injections. CalB::GFP mice maintained in a 12 hr LD cycle (n = 4) were anesthetized intraperitoneally with 60 mg/kg ketamine and 5 mg/kg xylazine, placed into the stereotaxic apparatus (David Kopf Instruments, Tujunga, CA), and prepared for aseptic surgery. A 10 μl Hamilton syringe (Hamilton, Reno, NV) was used to inject 2 μl of colchicine (10 μg/μl in 0.9% saline; Sigma, St. Louis, MO) in the lateral ventricle (stereotaxic coordinates relative to bregma are as follows: flat skull, anteroposterior, +1 mm; mediolateral, +0.7 mm; dorsoventral, -3.0 mm from the top of the skull). Animals were killed 36 hr later. Anterior hypothalamic sections from these animals were used to visualize gastrin-releasing peptide-containing cell bodies.
Retinal tract tracing. Animals of each strain were maintained in a 12 hr LD cycle (n = 4) and anesthetized and injected monocularly with 0.05 μg of cholera toxin-β (CTβ; List Biological Laboratories, Campbell, CA) in 2 μl of 1.8% NaCl. After 24 hr, animals were perfused at zeitgeber time 10 (ZT10). Brain sections were used for CTβ-GFP double label.
Detection of rhythmic responses. To confirm in our CalB::GFP mice the previously reported pattern of rhythmic Per2 expression, animals were housed in a 12 hr LD cycle and killed at ZT14 and ZT2 (n = 2 per group), times of peak and trough expression (Field et al., 2000). For circadian studies of Per1-GFP, hemizygous Per1::GFP mice housed in DD were killed at the projected circadian time 2 (CT2), CT6, CT10, CT14, CT18, and CT22 (n = 2 per time point), as reported previously for Per1::GFP mice (LeSauter et al., 2003).
Detection of light-induced responses. In initial studies, we assessed whether the transgenic mice respond normally to a light pulse. For light-induced Per1 mRNA studies, mice (n = 3) were maintained in DD for 2 d, exposed to a 30 min light pulse (1000 lux at cage level) starting at projected CT13, and killed at CT13:30 (Shigeyoshi et al., 1997). For light-induced Fos, Per1::GFP (n = 2) and CalB::GFP (n = 3) animals received a 30 min light pulse at projected CT14 and were killed 1 hr after the end of the light pulse. Control animals for light-induced Per1 mRNA and Fos were also maintained in DD for 2 d, not exposed to light, and killed at the same circadian time as experimental animals (n = 2 per group).
Perfusion. Animals were deeply anesthetized (200 mg/kg pentobarbital, i.p.). Animals killed in the dark were anesthetized under the dim red light with their heads covered with a light-proof hood until they were perfused. Mice were perfused intracardially with 50 ml of saline followed by 100 ml of 4% paraformaldehyde in 0.1 m phosphate buffer (PB), pH 7.3. Brains were postfixed for 18-24 hr at 4°C and cryoprotected in 20% sucrose in 0.1 m PB overnight. RNase-free buffer was used for in situ hybridization at both the perfusion and sectioning steps. For immunohistochemistry, 35 μm cryostat sections were processed free floating; for Per1 mRNA in situ hybridization and GFP immunocytochemistry, alternate sections (30 μm) were processed free floating.
Immunohistochemistry. Single-labeled sections were processed with a modified avidin-biotin-immunoperoxidase technique (LeSauter and Silver, 1999) using diaminobenzidine (DAB) (Sigma) as the chromogen. Double-labeled sections were incubated in donkey serum for 1 hr, then in the two primaries made in different host species for 48 hr, and then in the appropriate donkey secondary conjugated to the Cy2 and Cy3 fluorescent chromogens (1:200; Jackson ImmunoResearch, West Grove, PA) for 2 hr. Sections were mounted and coverslipped with Permount (Fisher Scientific, Houston, TX) for DAB or Krystalon (EMD Chemicals, Gibbstown, NJ) for Cy2 and Cy3.
For Fos-GFP double-label immunocytochemistry (ICC), in which both primaries are raised in rabbits, sections were incubated for 1 hr in normal goat serum and then in GFP for 48 hr at 4°C, in biotinylated goat anti-rabbit for 1 hr, and in avidin-biotin complex (Elite kit; Vector Laboratories, Burlingame, CA) for 1 hr; they were then amplified for 30 min with biotinylated tyramide (60 μl/10 ml 0.1 m PBS) with 1:1000 H2O2 and incubated with avidin-Cy2 for 2 hr at room temperature. Sections were then washed in phosphate buffer. Next, sections were incubated for 1 hr in normal donkey serum, in Fos overnight at 4°C, and then in donkey anti-rabbit-Cy3 for 2 hr at room temperature.
To test for specificity, primary antibodies were omitted in some runs. To minimize potential inter-run variability in staining, all sections were run simultaneously.
Antibodies. Rabbit polyclonal GFP (1:40,000; Molecular Probes, Eugene, OR) and monoclonal mouse calbindin (1:20,000; Sigma) were used as single-label antibodies. The following antibodies for double-label ICC were used: rabbit polyclonal GFP antibody (1:800,000) and a rabbit polyclonal gastrin-releasing peptide antibody (1:3000; Diasorin, Stillwater, MN) for gastrin-releasing peptide-GFP, GFP (1:40,000) and mouse monoclonal Per2 (1:5000; Alpha Diagnostic, San Antonio, TX) for Per2-GFP, rabbit polyclonal GFP (1:20,000) and choleragenoid (1:10,000; List Biological Laboratories) for CTβ-GFP, rabbit polyclonal GFP (1:800,000) and rabbit polyclonal anti-Fos (1:40,000; Santa Cruz Biotechnology, Santa Cruz, CA) for Fos-GFP, and rabbit polyclonal GFP (1:20,000) and guinea pig polyclonal arginine vasopressin (AVP) (1:20,000; Diasorin) for GFP-AVP.
In situ hybridization. Brains were marked for identification, and experimental and control sections were run in the same wells. To detect mPer1 or calbindin mRNA, in situ hybridization histochemistry was performed as described previously (Yan and Silver, 2002). In brief, tissue sections were processed with proteinase K (1 mg/ml, 0.1 m Tris buffer, pH 8.0, 50 mm EDTA) for 10 min at 37°C and 0.25% acetic anhydride in 0.1 m triethanolamine for 10 min. Sections were incubated in hybridization buffer (60% formide, 10% dextran sulfate, 10 mm Tris-HCl, pH 8.0, 1 mm EDTA, pH 8.0, 0.6 m NaCl, 0.2% N-laurylsarcosine, 500 mg/ml, 200 mg/ml tRNA, 1× Denhardt's solution, 0.25% SDS, and 10 mm dithiothreitol) containing digoxigenin-labeled mPer1 or calbindin antisense cRNA probes for 16 hr at 60°C. After a high-stringency posthybridization wash, sections were treated with RNase A and were then further processed for immunodetection with a nucleic acid detection kit (Boehringer Mannheim, Indianapolis, IN). Sections were incubated in 1.0% of a blocking reagent in buffer 1 (100 mm Tris-HCl buffer, 150 mm NaCl, pH 7.5) for 1 hr at room temperature and then incubated at 4°C in an alkaline phosphatase-conjugated digoxigenin antibody diluted 1:5000 in buffer 1 for 3 d. On the following day, sections were washed in buffer 1 twice (5 min each) and incubated in buffer 3 (100 mm Tris-HCl buffer, pH 9.5, containing 100 mm NaCl and 50 mm MgCl2) for 5 min. They were then incubated in a solution containing nitroblue tetrazolium salt (0.34 mg/ml) and 5-bromo-4-chloro-3-indolyl phosphate toluidinium salt (0.18 mg/ml) (Roche, Indianapolis, IN) for 8 hr. The colorimetric reaction was halted by immersing the sections in buffer 4 (10 mm Tris-HCl containing 1 mm EDTA, pH 8.0). Sections were mounted on slides, and coverslips were applied with Permount (Fisher Scientific). Alternate sections were matched with their adjacent sections from the ICC runs for analysis.
Microscopy and quantification. Sections were examined on an Eclipse E800 microscope (Nikon, Melville, NY). Images were captured with a cooled CCD camera (SPOT; Morrell, Melville, NY) and stored on a personal computer for subsequent analysis. For DAB-stained tissue, images were captured in an 8 bit grayscale. Fluorescently stained sections were excited using filters for GFP (480 ± 20 nm) and Texas Red (560 ± 40 nm) with each channel acquired independently and then combined digitally with the SPOT software. Images were processed using Photoshop 7 (Adobe Systems, Mountain View, CA).
To assess regional distribution of Per1 mRNA and GFP, images of digoxigenin sections were merged with the adjacent DAB sections by aligning of landmarks around the SCN (optic chiasm, ventricles, and vasculature). To determine the number of double-labeled cells, each color channel was examined separately. Cells in which there was a Fos-positive signal and GFP-positive cytoplasm were scored as double labeled. Because both gastrin-releasing peptide and GFP staining are cytoplasmic, cells positive for each peptide under each color channel were scored as double labeled. This analysis was confirmed using confocal microscopy.
Confocal microscopy was used to examine the colocalization of GFP and gastrin-releasing peptide and the contacts between retinal fibers and GFP cells in both strains of mice using an Axiovert 100TV fluorescence microscope (Zeiss, Thornwood, NY) with a Zeiss LSM 410 scanning confocal attachment. Sections were excited sequentially with an argon-krypton laser using the standard excitation wavelength for Cy2 and Cy3. Stacked images were collected as 1 μm multitract optical sections. LSM 3.95 software (Zeiss) was used to superimpose red and green images of the sections. Each section of SCN was examined in its entirety in 1 μm steps.
The number of Fos and gastrin-releasing peptide cells was determined using the Abercrombie (1946) correction factor. The diameter of the gastrin-releasing peptide cells and of the Fos nuclei was derived from their perimeters measured using NIH Image (version 1.61). The perimeters of 30 gastrin-releasing peptide cells and 30 Fos nuclei were measured in each animal, and diameters were calculated. There were no differences in cell diameters between strains, and the data were combined. Gastrin-releasing peptide-immunoreactive (IR) cell diameter was 8.4 ± 0.2 μm (mean ± SEM), and Fos-IR nuclei diameter was 5.6 ± 0.1 μm.
Results
Identification and localization of GFP in the CalB::GFP mouse
CalB::GFP mouse: GFP cells contain gastrin-releasing peptide
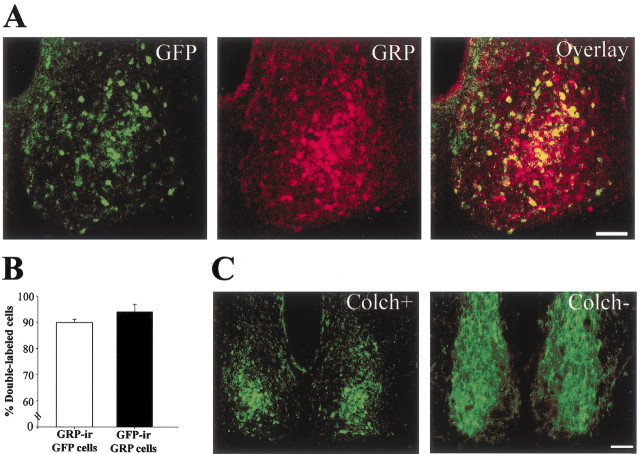
In the SCN of the CalB::GFP mouse, GFP-containing cells are concentrated in an SCN region bearing two noteworthy features: first, it is rich in gastrin-releasing peptide cells (Moore and Silver, 1998); second, it contains cells that lack rhythmic expression of Per1 mRNA or Per1 or Per2 protein (LeSauter et al., 2003). We now show that GFP-IR in the SCN of the CalB::GFP mouse is expressed in gastrin-releasing peptide-containing cells (Fig. 1A). In animals treated with colchicine, gastrin-releasing peptide-containing cell bodies can be resolved. In untreated animals, a dense gastrin-releasing peptide fiber plexus extends into the medial and dorsal SCN, making it difficult to delineate gastrin-releasing peptide cell bodies (Fig. 1C). Using conventional fluorescence microscopy techniques, quantitative analysis of sections labeled for GFP and gastrin-releasing peptide in colchicine-treated CalB::GFP mice indicates that 89.8 ± 1.3% (mean ± SEM) of GFP cells contain gastrin-releasing peptide, and that 94.0 ± 2.1% of gastrin-releasing peptide cells contain GFP (Fig. 1B). These results were confirmed at the confocal level, with 86.4 ± 1.0% of GFP cells containing gastrin-releasing peptide and 91.8 ± 4.1% of gastrin-releasing peptide cells containing GFP. In summary, the data indicate that in this transgenic line, GFP is a marker for the gastrin-releasing peptide cells.
Figure 1.
Identification of GFP-IR cells in a CalB::GFP mouse. A, Confocal images (1 μm) of the SCN labeled for GFP (green) and gastrin-releasing peptide (GRP) (red). The overlay shows that most cells colocalize both peptides, quantitatively shown in the bar graph (B). C, Photomicrographs of the caudal SCN showing gastrin-releasing peptide-IR in colchicine-treated (Colch+) and untreated (Colch-) animals. Without colchicine treatment, the dense gastrin-releasing peptide fiber plexus obscures the gastrin-releasing peptide cells. Scale bars, 100 μm.
To identify calbindin cells in the CalB::GFP mouse, we stained alternate sections for calbindin protein and mRNA. The distribution of calbindin mRNA corresponds to that of its protein and confirmed that calbindin-containing cells are scattered throughout the SCN (Moore and Silver, 1998). However, in the SCN of the CalB::GFP mouse, the GFP reporter is not expressed in cells containing calbindin. Instead it occurs in gastrin-releasing peptide cells of the SCN (vide supra). Restated, in this transgenic line, GFP is not contained in calbindin-expressing cells of the SCN but is a marker for the gastrin-releasing peptide-containing cells. Similar discrepancies are seen in other brain regions. For example, GFP but not calbindin is seen in the dorsolateral septum and medial habenula, the dorsolateral aspect of the paraventricular nucleus. Conversely, calbindin but not GFP is seen in the medial preoptic area, the medial aspect of the paraventricular nucleus. Finally, in some areas, including cerebellar Purkinje cells and the caudate-putamen, GFP, calbindin mRNA, and protein occur in the same cells. Similar mismatches have been reported in other transgenic lines (DiLeone et al., 2000).
CalB::GFP mouse: localization of GFP in relation to the AVP cell population
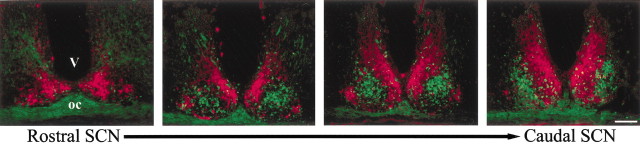
We delineated the topography of gastrin-releasing peptide cells in relation to the distribution of the well characterized SCN peptide AVP. It can clearly be seen in the CalB::GFP mouse that the gastrin-releasing peptide cells (reported by GFP) are densely packed in the central core of the SCN (Fig. 2). Caudally, GFP cells are concentrated more laterally. GFP neurons are localized to SCN regions mainly devoid of AVP-containing cells.
Figure 2.
Photomicrographs showing GFP (green) and AVP (red) immunostaining through the extent of the nucleus in a Calb::GFP mouse. V, Third ventricle; oc, optic chiasm. Scale bar, 100 μm.
Localization of light-responsive cells
CalB::GFP mouse: gastrin-releasing peptide cells (marked by GFP) are light responsive
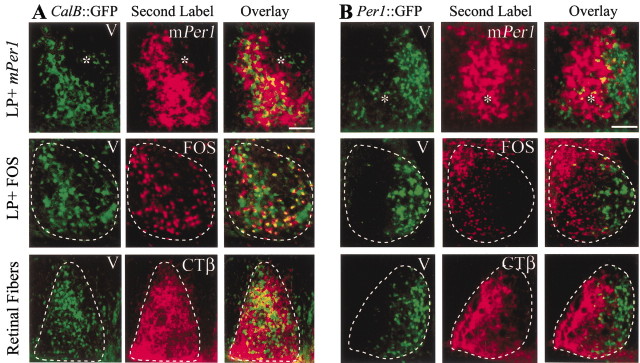
After a 30 min light pulse at CT13, Per1 mRNA is induced in the same regions in which GFP-IR is expressed. This can be seen in Figure 3A (top row), where both mPer1 and GFP staining occur in the laterocaudal SCN region.
Figure 3.
A, Light-induced responses (mPer1 and Fos) and retinal input to SCN in relation to GFP-containing cells in a CalB::GFP mouse. In this mouse, GFP-IR is a marker for gastrin-releasing peptide cells (Fig. 1). The top panels are alternate sections labeled for GFP and mPer1 and the overlay. The middle panels show sections double labeled for GFP and Fos. The bottom panels depict GFP, retinal fibers, and their overlap in double-labeled sections. B, Light-induced responses (mPer1 and Fos) and retinal input to SCN in relation to GFP-containing cells in a Per1::GFP mouse. Top panels are alternate sections labeled for GFP and mPer1 and the overlay. Middle panels show sections double labeled for GFP and Fos. Bottom panels depict GFP, retinal fibers, and their overlap. Dashed lines delineate the boundaries of the SCN. V, Third ventricle; LP, light pulse. The asterisk indicates a blood vessel used for orientation. Scale bars, 100 μm.
A light pulse at CT14 induces Fos in a region of the SCN in which GFP is expressed (Fig. 3A, middle row). Quantification of double-label immunocytochemistry indicates that there are 990 ± 11.4 Fos-positive nuclei in each SCN 1 hr after a CT14 light pulse. Within the caudal half of the SCN (the region rich in gastrin-releasing peptide cells), GFP cells account for 44.1 ± 2.9% of the total population of Fos-positive cells and 71.9 ± 3.9% of GFP-IR cells contain Fos, whereas in the entire nucleus, Fos-positive GFP-IR cells account for 33.0 ± 0.4% of the total amount of Fos-IR.
Tract-tracing studies using CTβ injected into the retina confirm that this laterocaudal SCN region of GFP-IR cells receives dense retinal input (Fig. 3A, bottom row). Confocal microscopic examination of CTβ-labeled retinal appositions on GFP cells demonstrates that 77.2 ± 3.8% of GFP cells are contacted by retinal fibers.
Per1::GFP mouse: GFP cells (expressing Per1 mRNA) are not light responsive In the Per1::GFP mouse, light-induced Per1 mRNA (Fig. 3B, top row), Fos (Fig. 3B, middle row), and dense retinal input (Fig. 3B, bottom row) occur in the ventral and lateral aspects of the caudal SCN. These light-induced responses occur outside the area of GFP-IR that delineates rhythmic Per1 gene-expressing cells of the SCN. It should be noted that GFP protein expression peaks at 3.5 hr after a light pulse (Kuhlman et al., 2000, 2003); consequently, at 1.5 hr after the light pulse, light-induced GFP is not detected. This allowed us to distinguish between endogenously rhythmic and light-induced cell populations. Quantification of double-label immunochemistry indicates that 1 hr after a CT14 light pulse, only 9.2 ± 3.4% of Per1::GFP cells contain Fos, and that the population of Fos-positive GFP cells accounts for 4.8 ± 1.1% of Fos-IR cells within the region.
Localization of rhythmic cell populations
CalB::GFP mouse: Per2 protein is not rhythmic in gastrin-releasing peptide cells (marked by GFP)
Per1 and Per2 proteins are expressed rhythmically in the SCN. We next determined whether rhythmic expression of Per proteins occurs in gastrin-releasing peptide cells of the CalB::GFP mouse using GFP as a reporter for gastrin-releasing peptide cells. Because Per1 and Per2 are expressed in the same SCN region and likely in the same cells (LeSauter et al., 2003; our unpublished data), and because commercially produced Per2 antibodies are available, we examined Per2 and gastrin-releasing peptide coexpression. Per2 is high in the rostral and mediocaudal SCN and low or absent in GFP cells at both peak and trough expression times (Fig. 4A, ZT14, ZT2).
Figure 4.
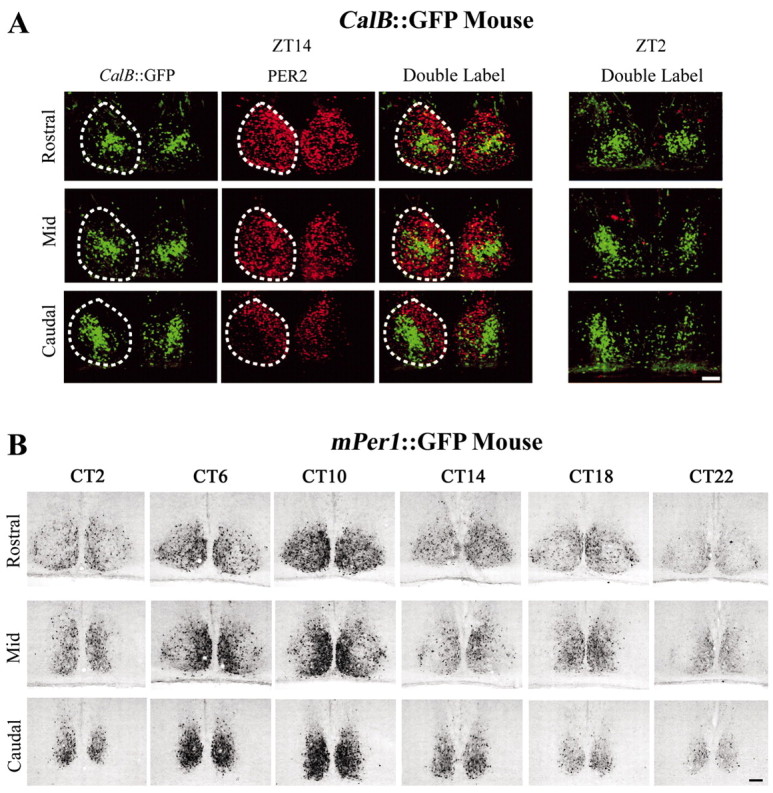
A, CalB::GFP mouse. Pictured are photomicrographs showing mPer2 at peak (ZT14) and trough (ZT2) expression times in relation to gastrin-releasing peptide-containing cells marked by GFP. Sections depict rostral, mid-caudal (Mid), and caudal (top to bottom) levels of the SCN photographed for GFP, Per2, and the overlay of both peptides at ZT14 (left to right). Note that Per2 is sparse in the region of CalB::GFP cells, even at the time of peak Per2 expression. Dashed lines delineate the boundaries of the SCN. B, Per1::GFP mouse. Shown is circadian expression of GFP throughout the SCN at 4 hr intervals. At peak expression times, rhythmic expression of Per1::GFP (CT10) occurs in the same region as Per2 (ZT14) and is absent from the regions expressing CalB::GFP in A. Scale bars, 100 μm.
Per1::GFP mouse: GFP (reporting Per1 mRNA) is rhythmically expressed
In the Per1::GFP mouse, GFP-IR shows a circadian expression pattern with a peak at CT10 and a trough at CT22. GFP staining is restricted to the rostral and medial aspects of the mid-SCN and caudal SCN (Fig. 4B).
Discussion
An enigma in understanding SCN function is how this nucleus, comprised of thousands of independent circadian oscillators, works as a whole to express a coherent daily rhythm in electrical and metabolic activity and organizes coherent circadian responses in the body. An early view of the mammalian SCN posited a linear signaling pathway into and out of the SCN with oscillator cells receiving afferent input and sending signals to target sites (Eskin, 1979). More recently, it has been suggested that all SCN neurons are oscillators (Reppert and Weaver, 2001, 2002). A variation on this theme is that ventral and lateral SCN neurons oscillate with low amplitude, whereas dorsal and medial neurons exhibit high-amplitude oscillation, with retinal input reaching only the former group (Shibata et al., 1984; Moore, 1996; Nakamura et al., 2001; Yan and Okamura, 2002). The present results suggest a third possibility: that the SCN is comprised of two fundamentally different cell types. One population, including gastrin-releasing peptide-containing cells, expresses Per mRNA in response to a light pulse but does not have a detectable rhythm in endogenous Per expression. The second population oscillates on a circadian basis with respect to clock genes but does not express Per genes immediately after a light pulse. This result is consistent with previous findings in mice (Yan and Silver, 2002) and hamsters (Hamada et al., 2001; Jobst and Allen, 2002).
A model of how this two-compartment SCN can sustain rhythmicity has been suggested (Antle et al., 2003). In this view, rhythmic cells are conceptualized as limit cycle-Van den Pol oscillators, which themselves are reset by “gate cells.” The gate cells provide a coordinating master signal for synchronization to maintain coherence among a population of independent oscillators expressing different periods. The proposed model neither excludes nor requires interoscillator coupling. Restated, the present results indicate that the coordinated action of two distinct SCN cell types forms the anatomical and functional basis for the integrated rhythmicity of SCN tissue suggested by this model.
A universal feature of the mammalian SCN is retinal input to the SCN via the RHT (Morin, 1994). Knowing the mechanism responsible for resetting SCN pacemakers by light is fundamental to understanding entrainment and phase shifting. In all species, only some SCN cells receive direct retinal input, with species differences in RHT termination sites. In rats, most retinal input terminates in the ventral region, with little or no retinal input to the dorsomedial SCN (Moore, 1996), whereas in hamsters, retinal input is concentrated in the ventrolateral aspect of the SCN (Johnson et al., 1988). RHT input to the mouse SCN is very dense in the ventral and lateral aspects of the nucleus and sparse in the dorsal and medial regions (Abrahamson and Moore, 2001) (Figs. 2, 3). In a pattern consistent with the anatomical connections of the retina, the response of immediate early genes (IEGs) in the SCN after photic stimulation is also topographically and temporally restricted. Light pulses in the subjective night cause rapid induction of mPer1 and mPer2 in the ventral and lateral aspects of the mouse SCN, with a time-dependent “spread” to the dorsal and medial portions of the nucleus (Yan and Silver, 2002).
Resetting of the SCN oscillators is thought to occur rapidly after a light pulse (Best et al., 1999; Watanabe et al., 2001), and significant effort has been devoted to identifying the phenotype of retinorecipient SCN cells. Estimates of the size and identity of this population of light-responsive cells range from 20 to 40% of SCN cells and vary depending on the assay used [electrophysiology (Sawaki, 1979; Meijer et al., 1986; Aggelopoulos and Meissl, 2000) or IEG expression (Earnest et al., 1993; Silver et al., 1996; Castel et al., 1997; Aioun et al., 1998)]. The present study shows that 1 hr after a brief light pulse, most GFP cells (in the CalB::GFP mouse) in the SCN express Fos. Consistent with our results in mice, retinal terminals contact gastrin-releasing peptide cells in both rats and hamsters (Tanaka et al., 1997; Aioun et al., 1998). VIP cells of the SCN are also known to receive direct retinal input. In rats, 0-10% of VIP cells, which lie in the ventrolateral retinorecipient SCN region, express Fos content after a light pulse (Earnest et al., 1993; Mikkelsen et al., 1994; Romijn et al., 1996). Retinal terminals on VIP cells have been observed in the rat but not in the hamster (Tanaka et al., 1993; Aioun et al., 1998). VIP, like gastrin-releasing peptide, is rhythmic in LD but not in DD, although the former peaks at night and the latter in the day (Shinohara et al., 1993). The precise distinct roles of these peptides remain to be determined. To summarize, although only 20-30% of SCN cells are directly light responsive (using c-fos IEG expression as an assay), the present study indicates that most gastrin-releasing peptide-containing cells lie in this group.
The relationship of peptidergic phenotype and afferent and efferent connections of the SCN to its function has been a long-standing curiosity. Previous work in numerous species indicates the existence of two anatomical divisions, designated core and shell (or ventrolateral and dorsomedial) on the basis of chemoarchitecture and connections (Card and Moore, 1984; Moore, 1992, 1996). With regard to chemoarchitecture, SCN cells are heterogeneous in their peptidergic content and distribution (Van den Pol, 1980; Van den Pol and Tsujimoto, 1985). AVP is found primarily in the dorsomedial SCN, whereas gastrin-releasing peptide and VIP cells lie in the ventrolateral SCN (Moore and Silver, 1998; Silver et al., 1999; Abrahamson and Moore, 2001; Moore et al., 2002). Of the calcium-binding proteins, calbindin is localized to the ventrolateral region of both the rat and hamster SCN and is sparsely distributed through the mouse SCN, whereas calretinin is found in the ventral mouse SCN and the lateral rat SCN and is sparsely distributed in the hamster SCN (Moore and Silver, 1998; Silver et al., 1999; Arvanitogiannis et al., 2000). The present study indicates that clock gene expression can be mapped to the anatomical and phenotypic characteristics of the SCN. There is a region of mouse SCN in which rhythmic expression of mPer1 mRNA and Per1 and Per2 proteins is not detected (present results) (LeSauter et al., 2003). In mice, this region includes cells that are immunopositive for gastrin-releasing peptide and express both light-induced mPer1 and Fos.
Numerous lines of evidence indicate an important role for gastrin-releasing peptide in light-induced phase shifts. Injections of gastrin-releasing peptide or a mixture of peptides, of which gastrin-releasing peptide is one component, produce photic-like phase shifts in behavior and electrical activity during subjective night (Piggins and Rusak, 1993; Piggins et al., 1994, 1995; Albers et al., 1995; McArthur et al., 2000). It has also been shown that the phase-shifting effects of a gastrin-releasing peptide injection can be blocked by the application of the gastrin-releasing peptide antagonist [d-F5Phe6,d-Ala11]BN(6-13)OMe (Piggins et al., 1995). In mice, gastrin-releasing peptide injections induce mPer1 gene expression in dorsal SCN neurons, and gastrin-releasing peptide receptor-deficient mice show both blunted mPer1 and behavioral phase-shifting responses (Aida et al., 2002). Together, the data indicate that gastrin-releasing peptide contributes to phase resetting and has effects similar to photic stimuli.
These studies reveal that the SCN is composed of both rhythmic and light-induced nonrhythmic components. Photic information from the environment reaches the SCN via the RHT and impacts a subset of SCN cells, which are light induced but not rhythmic with respect to clock gene expression. Gastrin-releasing peptide cells fall into this category. The present results, characterizing clock gene expression within this peptidergically and functionally identified SCN cell type, provide the foundation for understanding intercellular mechanisms of phase setting in SCN tissue that enable it to generate coherent circadian rhythms in the entire animal.
Footnotes
This work was supported by National Institute of Neurological Disorders and Stroke Grant 37919 and National Institute of Mental Health Grant 63341 to R.S. and a Natural Sciences and Engineering Research Council of Canada fellowship to I.K. We thank Drs. Michael Lehman and Lance Kriegsfeld for helpful comments on a previous draft of this manuscript.
Correspondence should be addressed to Dr. Rae Silver, Department of Psychology, 406 Schermerhorn Hall, MC5501, 1190 Amsterdam Avenue, New York, NY 10027. E-mail: qr@columbia.edu.
Copyright © 2004 Society for Neuroscience 0270-6474/04/240068-08$15.00/0
References
- Abercrombie M (1946) Estimation of nuclear populations from microtome sections. Anat Rec 94: 239-247. [DOI] [PubMed] [Google Scholar]
- Abrahamson EE, Moore RY (2001) Suprachiasmatic nucleus in the mouse: retinal innervation, intrinsic organization and efferent projections. Brain Res 916: 172-191. [DOI] [PubMed] [Google Scholar]
- Aggelopoulos NC, Meissl H (2000) Responses of neurones of the rat suprachiasmatic nucleus to retinal illumination under photopic and scotopic conditions. J Physiol (Lond) 523: 211-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida R, Moriya T, Araki M, Akiyama M, Wada K, Wada E, Shibata S (2002) Gastrin-releasing peptide mediates photic entrainable signals to dorsal subsets of suprachiasmatic nucleus via induction of period gene in mice. Mol Pharmacol 61: 26-34. [DOI] [PubMed] [Google Scholar]
- Aioun J, Chambille I, Peytevin J, Martinet L (1998) Neurons containing gastrin-releasing peptide and vasoactive intestinal polypeptide are involved in the reception of the photic signal in the suprachiasmatic nucleus of the Syrian hamster: an immunocytochemical ultrastructural study. Cell Tissue Res 291: 239-253. [DOI] [PubMed] [Google Scholar]
- Albers HE, Gillespie CF, Babagbemi TO, Huhman KL (1995) Analysis of the phase shifting effects of gastrin releasing peptide when microinjected into the suprachiasmatic region. Neurosci Lett 191: 63-66. [DOI] [PubMed] [Google Scholar]
- Antle MC, Foley DK, Foley NC, Silver R (2003) Gates and oscillators: a network model of the brain clock. J Biol Rhythms 18: 339-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvanitogiannis A, Robinson B, Beaule C, Amir S (2000) Calbindin-D28k immunoreactivity in the suprachiasmatic nucleus and the circadian response to constant light in the rat. Neuroscience 99: 397-401. [DOI] [PubMed] [Google Scholar]
- Best JD, Maywood ES, Smith KL, Hastings MH (1999) Rapid resetting of the mammalian circadian clock. J Neurosci 19: 828-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Card JP, Moore RY (1984) The suprachiasmatic nucleus of the golden hamster: immunohistochemical analysis of cell and fiber distribution. Neuroscience 13: 415-431. [DOI] [PubMed] [Google Scholar]
- Castel M, Belenky M, Cohen S, Wagner S, Schwartz WJ (1997) Light-induced c-Fos expression in the mouse suprachiasmatic nucleus: immunoelectron microscopy reveals co-localization in multiple cell types. Eur J Neurosci 9: 1950-1960. [DOI] [PubMed] [Google Scholar]
- Cui LN, Dyball RE (1996) Synaptic input from the retina to the suprachiasmatic nucleus changes with the light-dark cycle in the Syrian hamster. J Physiol (Lond) 497: 483-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiLeone RJ, Marcus GA, Johnson MD, Kingsley DM (2000) Efficient studies of long-distance Bmp5 gene regulation using bacterial artificial chromosomes. Proc Natl Acad Sci USA 97: 1612-1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnest DJ, DiGiorgio S, Olschowka JA (1993) Light induces expression of fos-related proteins within gastrin-releasing peptide neurons in the rat suprachiasmatic nucleus. Brain Res 627: 205-209. [DOI] [PubMed] [Google Scholar]
- Eskin A (1979) Identification and physiology of circadian pacemakers. Introduction. Fed Proc 38: 2570-2572. [PubMed] [Google Scholar]
- Field MD, Maywood ES, O'Brien JA, Weaver DR, Reppert SM, Hastings MH (2000) Analysis of clock proteins in mouse SCN demonstrates phylogenetic divergence of the circadian clockwork and resetting mechanisms. Neuron 25: 437-447. [DOI] [PubMed] [Google Scholar]
- Groos GA, Mason R (1980) The visual properties of rat and cat suprachiasmatic neurones. J Comp Neurol 349-356.
- Hamada T, LeSauter J, Venuti JM, Silver R (2001) Expression of period genes: rhythmic and nonrhythmic compartments of the suprachiasmatic nucleus pacemaker. J Neurosci 21: 7742-7750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobst EE, Allen CN (2002) Calbindin neurons in the hamster suprachiasmatic nucleus do not exhibit a circadian variation in spontaneous firing rate. Eur J Neurosci 16: 2469-2474. [DOI] [PubMed] [Google Scholar]
- Johnson RF, Morin LP, Moore RY (1988) Retinohypothalamic projections in the hamster and rat demonstrated using cholera toxin. Brain Res 462: 301-312. [DOI] [PubMed] [Google Scholar]
- Klien D, Moore R, Reppert S (1991) Suprachiasmatic nucleus: the mind's clock. Oxford: Oxford UP.
- Kuhlman SJ, Quintero JE, McMahon DG (2000) GFP fluorescence reports period 1 circadian gene regulation in the mammalian biological clock. NeuroReport 11: 1479-1482. [PubMed] [Google Scholar]
- Kuhlman SJ, Silver R, Le Sauter J, Bult-Ito A, McMahon DG (2003) Phase resetting light pulses induce Per1 and persistent spike activity in a subpopulation of biological clock neurons. J Neurosci 23: 1441-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeSauter J, Silver R (1999) Localization of a suprachiasmatic nucleus subregion regulating locomotor rhythmicity. J Neurosci 19: 5574-5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeSauter J, Yan L, Vishnubhotla B, Quintero JE, Kuhlman SJ, McMahon DG, Silver R (2003) A short half-life GFP mouse model for analysis of suprachiasmatic nucleus organization. Brain Res 964: 279-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowrey PL, Takahashi JS (2000) Genetics of the mammalian circadian system: photic entrainment, circadian pacemaker mechanisms, and posttranslational regulation. Annu Rev Genet 34: 533-562. [DOI] [PubMed] [Google Scholar]
- McArthur AJ, Coogan AN, Ajpru S, Sugden D, Biello SM, Piggins HD (2000) Gastrin-releasing peptide phase-shifts suprachiasmatic nuclei neuronal rhythms in vitro J Neurosci 20: 5496-5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer JH, Groos GA, Rusak B (1986) Luminance coding in a circadian pacemaker: the suprachiasmatic nucleus of the rat and the hamster. Brain Res 382: 109-118. [DOI] [PubMed] [Google Scholar]
- Mikkelsen JD, Larsen PJ, Sorensen GG, Woldbye D, Bolwig TG, Hastings MH, Ebling FJ (1994) A dual-immunocytochemical method to localize c-fos protein in specific neurons based on their content of neuropeptides and connectivity. Histochemistry 101: 245-251. [DOI] [PubMed] [Google Scholar]
- Moore RY (1992) The fourth C.U. Ariens Kappers lecture. The organization of the human circadian timing system. Prog Brain Res 93: 99-115. [PubMed] [Google Scholar]
- Moore RY (1996) Entrainment pathways and the functional organization of the circadian system. Prog Brain Res 111: 103-119. [DOI] [PubMed] [Google Scholar]
- Moore RY, Silver R (1998) Suprachiasmatic nucleus organization. Chronobiol Int 15: 475-487. [DOI] [PubMed] [Google Scholar]
- Moore RY, Speh JC, Leak RK (2002) Suprachiasmatic nucleus organization. Cell Tissue Res 309: 89-98. [DOI] [PubMed] [Google Scholar]
- Morin LP (1994) The circadian visual system. Brain Res Brain Res Rev 19: 102-127. [DOI] [PubMed] [Google Scholar]
- Nakamura W, Honma S, Shirakawa T, Honma K (2001) Regional pacemakers composed of multiple oscillator neurons in the rat suprachiasmatic nucleus. Eur J Neurosci 14: 666-674. [DOI] [PubMed] [Google Scholar]
- Panda S, Provencio I, Tu DC, Pires SS, Rollag MD, Castrucci AM, Pletcher MT, Sato TK, Wiltshire T, Andahazy M, Kay SA, Van Gelder RN, Hogenesch JB (2003) Melanopsin is required for non-image-forming photic responses in blind mice. Science 301: 525-527. [DOI] [PubMed] [Google Scholar]
- Piggins HD, Rusak B (1993) Electrophysiological effects of pressure-ejected bombesin-like peptides on hamster suprachiasmatic nucleus neurons in vitro J Neuroendocrinol 5: 575-581. [DOI] [PubMed] [Google Scholar]
- Piggins HD, Cutler DJ, Rusak B (1994) Effects of ionophoretically applied bombesin-like peptides on hamster suprachiasmatic nucleus neurons in vitro Eur J Pharmacol 271: 413-419. [DOI] [PubMed] [Google Scholar]
- Piggins HD, Antle MC, Rusak B (1995) Neuropeptides phase shift the mammalian circadian pacemaker. J Neurosci 15: 5612-5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR (2001) Molecular analysis of mammalian circadian rhythms. Annu Rev Physiol 63: 647-676. [DOI] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR (2002) Coordination of circadian timing in mammals. Nature 418: 935-941. [DOI] [PubMed] [Google Scholar]
- Romijn HJ, Sluiter AA, Pool CW, Wortel J, Buijs RM (1996) Differences in colocalization between Fos and PHI, GRP, VIP and VP in neurons of the rat suprachiasmatic nucleus after a light stimulus during the phase delay versus the phase advance period of the night. J Comp Neurol 372: 1-8. [DOI] [PubMed] [Google Scholar]
- Sawaki Y (1979) Suprachiasmatic nucleus neurones: excitation and inhibition mediated by the direct retino-hypothalamic projection in female rats. Exp Brain Res 37: 127-138. [DOI] [PubMed] [Google Scholar]
- Shearman LP, Sriram S, Weaver DR, Maywood ES, Chaves I, Zheng B, Kume K, Lee CC, van der Horst GT, Hastings MH, Reppert SM (2000) Interacting molecular loops in the mammalian circadian clock. Science 288: 1013-1019. [DOI] [PubMed] [Google Scholar]
- Shibata S, Liou S, Ueki S, Oomura Y (1984) Influence of environmental light-dark cycle and enucleation on activity of suprachiasmatic neurons in slice preparations. Brain Res 302: 75-81. [DOI] [PubMed] [Google Scholar]
- Shigeyoshi Y, Taguchi K, Yamamoto S, Takekida S, Yan L, Tei H, Moriya T, Shibata S, Loros JJ, Dunlap JC, Okamura H (1997) Light-induced resetting of a mammalian circadian clock is associated with rapid induction of the mPer1 transcript. Cell 91: 1043-1053. [DOI] [PubMed] [Google Scholar]
- Shinohara K, Tominaga K, Isobe Y, Inouye ST (1993) Photic regulation of peptides located in the ventrolateral subdivision of the suprachiasmatic nucleus of the rat: daily variations of vasoactive intestinal polypeptide, gastrin-releasing peptide, and neuropeptide Y. J Neurosci 13: 793-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver R, Romero MT, Besmer HR, Leak R, Nunez JM, LeSauter J (1996) Calbindin-D28K cells in the hamster SCN express light-induced Fos. NeuroReport 7: 1224-1228. [DOI] [PubMed] [Google Scholar]
- Silver R, Sookhoo AI, LeSauter J, Stevens P, Jansen HT, Lehman MN (1999) Multiple regulatory elements result in regional specificity in circadian rhythms of neuropeptide expression in mouse SCN. NeuroReport 10: 3165-3174. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Ichitani Y, Okamura H, Tanaka Y, Ibata Y (1993) The direct retinal projection to VIP neuronal elements in the rat SCN. Brain Res Bull 31: 637-640. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Hayashi S, Tamada Y, Ikeda T, Hisa Y, Takamatsu T, Ibata Y (1997) Direct retinal projections to GRP neurons in the suprachiasmatic nucleus of the rat. NeuroReport 8: 2187-2191. [DOI] [PubMed] [Google Scholar]
- Van den Pol AN (1980) The hypothalamic suprachiasmatic nucleus of rat: intrinsic anatomy. J Comp Neurol 191: 661-702. [DOI] [PubMed] [Google Scholar]
- Van den Pol AN, Tsujimoto KL (1985) Neurotransmitters of the hypothalamic suprachiasmatic nucleus: immunocytochemical analysis of 25 neuronal antigens. Neuroscience 15: 1049-1086. [DOI] [PubMed] [Google Scholar]
- Watanabe K, Deboer T, Meijer JH (2001) Light-induced resetting of the circadian pacemaker: quantitative analysis of transient versus steady-state phase shifts. J Biol Rhythms 16: 564-573. [DOI] [PubMed] [Google Scholar]
- Welsh DK, Logothetis DE, Meister M, Reppert SM (1995) Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron 14: 697-706. [DOI] [PubMed] [Google Scholar]
- Witkovsky P, Veisenberger E, LeSauter J, Yan L, Johnson M, Zhang DQ, McMahon D, Silver R (2003) Cellular location and circadian rhythm of expression of the biological clock gene Period 1 in the mouse retina. J Neurosci 23: 7670-7676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Okamura H (2002) Gradients in the circadian expression of Per1 and Per2 genes in the rat suprachiasmatic nucleus. Eur J Neurosci 15: 1153-1162. [DOI] [PubMed] [Google Scholar]
- Yan L, Silver R (2002) Differential induction and localization of mPer1 and mPer2 during advancing and delaying phase shifts. Eur J Neurosci 16: 1531-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XW, Model P, Heintz N (1997) Homologous recombination based modification in Escherichia coli and germline transmission in transgenic mice of a bacterial artificial chromosome. Nat Biotechnol 15: 859-865. [DOI] [PubMed] [Google Scholar]
- Young MW, Kay SA (2001) Time zones: a comparative genetics of circadian clocks. Nat Rev Genet 2: 702-715. [DOI] [PubMed] [Google Scholar]