Abstract
Malaria, caused by Plasmodium sp, results in almost one million deaths and over 200 million new infections annually. The World Health Organization has recommended that artemisinin-based combination therapies be used for treatment of malaria. Artemisinin is a sesquiterpene lactone isolated from the plant Artemisia annua. However, the supply and price of artemisinin fluctuate greatly, and an alternative production method would be valuable to increase availability. We describe progress toward the goal of developing a supply of semisynthetic artemisinin based on production of the artemisinin precursor amorpha-4,11-diene by fermentation from engineered Saccharomyces cerevisiae, and its chemical conversion to dihydroartemisinic acid, which can be subsequently converted to artemisinin. Previous efforts to produce artemisinin precursors used S. cerevisiae S288C overexpressing selected genes of the mevalonate pathway [Ro et al. (2006) Nature 440:940–943]. We have now overexpressed every enzyme of the mevalonate pathway to ERG20 in S. cerevisiae CEN.PK2, and compared production to CEN.PK2 engineered identically to the previously engineered S288C strain. Overexpressing every enzyme of the mevalonate pathway doubled artemisinic acid production, however, amorpha-4,11-diene production was 10-fold higher than artemisinic acid. We therefore focused on amorpha-4,11-diene production. Development of fermentation processes for the reengineered CEN.PK2 amorpha-4,11-diene strain led to production of > 40 g/L product. A chemical process was developed to convert amorpha-4,11-diene to dihydroartemisinic acid, which could subsequently be converted to artemisinin. The strains and procedures described represent a complete process for production of semisynthetic artemisinin.
Malaria, caused primarily by the Plasmodium falciparum and Plasmodium vivax parasites, is a disease overwhelmingly affecting areas of poverty in the developing world. There were over 200 million new infections and an estimated 781,000 deaths in 2009, predominantly affecting children under five years of age (1). P. falciparum, which causes the most virulent form of malaria, has become resistant to all inexpensive antimalarial drugs readily available in the developing world (2, 3). An antimalarial agent to which widespread resistance has not been observed is artemisinin, a sesquiterpene lactone peroxide extracted from the shrub Artemisia annua. The World Health Organization (WHO) recommends the use of artemisinin derivatives in combination with another effective schizontocidal drug, a treatment known as artemisinin-based combination therapy (ACT), for the first-line treatment of malaria (3). In recent years there have been large fluctuations in the price and availability of ACTs (4), and in 2007–2008 less than 15% of children under five years of age with fever received ACTs in 11 out of 13 countries surveyed (5). There is an urgent need to increase the supply of artemisinin to both stabilize the price and increase the availability of ACTs in the developing world, but a significant increase in the cultivation of A. annua would be needed to satisfy projected global demand (6). Chemical synthesis of artemisinin is not economically feasible because of the complexity and low yield of the process (7), however semisynthesis of artemisinin following microbial production of a precursor molecule may be feasible (6). This report describes progress toward the production of semisynthetic artemisinin.
Heterologous production of amorpha-4,11-diene (Fig. 5, compound 1) was first described from Escherichia coli which had been engineered to express the mevalonate pathway from Saccharomyces cerevisiae and amorpha-4,11-diene synthase (ADS) from A. annua (8). Development of a two-phase partitioning bioreactor allowed production of 0.5 g/L of amorpha-4,11-diene (9). Analysis of transcriptional responses and pathway metabolites showed that 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase limited production, leading to perturbation of fatty acid biosynthesis and generalized membrane stress (10, 11). Subsequent replacement of the S. cerevisiae HMG-CoA reductase and HMG-CoA synthase with equivalent enzymes from Staphylococcus aureus, and concomitant fermentation development, allowed production of 27 g/L of amorpha-4,11-diene (12).
Fig. 5.
Synthesis of dihydroartemisinic acid from amorpha-4,11-diene. Only the R isomer is shown.
Production of amorpha-4,11-diene from S. cerevisiae was initially developed as a tool to aid the cloning of the cytochrome P450 believed to be responsible for the production of artemisinic acid, a late-stage oxidized precursor of artemisinin (13). A series of engineering steps, namely high-level expression of ADS, overexpression of farnesyl diphosphate synthase and the catalytic domain of HMG-CoA reductase, reduced expression of squalene synthase, and generalized increased expression of the mevalonate pathway by overexpression of an activated allele of the UPC2 transcription factor, UPC2-1, allowed production of 153 mg/L of amorpha-4,11-diene in shake-flasks. This strain was used for functional expression of CYP71AV1 [also known as amorphadiene oxidase (AMO)], the cytochrome P450 responsible for the three-step oxidation of amorpha-4,11-diene, along with its cognate reductase, AaCPR (A. annua cytochrome P450 reductase), producing > 100 mg/L artemisinic acid (13). Incorporation of all A. annua-derived genes (ADS, CYP71AV1, and AaCPR) on a single expression plasmid allowed production of 2.5 g/L of artemisinic acid by development of a galactose-based fermentation process (14), though plasmid stability was shown to be lower in a strain producing artemisinic acid compared to a strain producing amorpha-4,11-diene, and production of artemisinic acid led to induction of pleiotropic drug resistance genes (15). We now describe the creation of new S. cerevisiae strains and processes for the production of amorpha-4,11-diene, culminating in a 250-fold increase in production of amorpha-4,11-diene to 40 g/L concentration, and a process for its efficient chemical conversion to the artemisinin precursor dihydroartemisinic acid.
Results
Eliminating Utilization of Galactose.
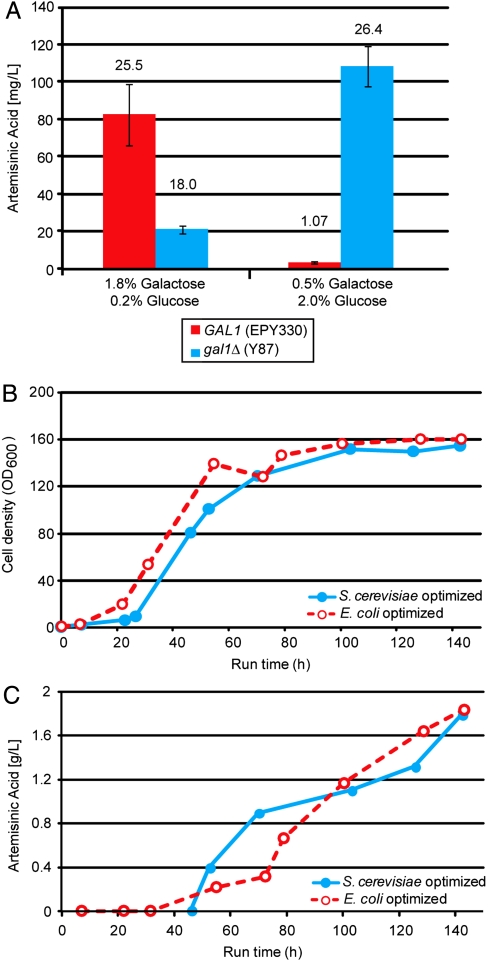
Any process involving fermentation to economically produce precursors for the semisynthesis of artemisinin for the developing world would be unable to use galactose as a sole carbon source as it is too expensive. Strains of S. cerevisiae lacking the gal1 gene are unable to metabolize galactose, but galactose still acts as a gratuitous inducer (16) for all galactose-regulated genes in the artemisinic acid-producing strain EPY330 (15). Fig. 1A shows that deleting GAL1 in EPY330 (= Y87; Table 1) allows production of artemisinic acid using glucose as a carbon source and galactose as a gratuitous inducer. Transcript levels were not analyzed, but given that the GAL regulon is still subject to glucose repression, it is likely that induction occurred following the diauxic shift and ethanol was used for artemisinic acid biosynthesis. Specific production of artemisinic acid by the gal1Δ strain grown in a high concentration of glucose (2.0%) is similar to the GAL1 strain grown in 1.8% galactose (Fig. 1A).
Fig. 1.
Production of artemisinic acid in Gen 1.0 yeast strains. (A) Production of artemisinic acid in shake-flask cultures of EPY330 (GAL1) and Y87 (gal1Δ) in defined media containing either low or high concentrations of glucose (± 1 SD of three flasks). Numbers above the bars show specific production (mg/L/OD600). (B) Growth and (C) artemisinic acid production in fed-batch glucose-limited fermentors of Y87 containing a high-copy plasmid expressing E. coli-optimized ADS/CYP71AV1/AaCPR and Y137 containing pAM322, expressing S. cerevisiae-optimized ADS/CYP71AV1/AaCPR.
Table 1.
Strain genotypes and products
| Strain | Base strain | Genotype and [plasmid] | Source | Produces |
| Gen 1.0 | ||||
| EPY330 | S288C | MATα his3Δ1 leu2Δ0 PGAL1-tHMG1 ∷ δ1 PGAL1-UPC2-1 ∷ δ2 erg9 ∷ PMET3-ERG9 ∷ HIS3 PGAL1-ERG20 ∷ δ3 PGAL1-tHMG1 ∷ δ4 [pESC-leu2d-ADS/AMO/CPR] | (15) | AA |
| Y87 | S288C | EPY330 gal1Δ∷K. lactis URA3 lys2Δ0 | This work | AA |
| Y135 | CEN.PK2 | MATα erg9 ∷ Kanr-PMET3-ERG9 trp1 ∷ TRP1-PGAL1-ERG20 his3 ∷ HIS-PGAL1-tHMG1 leu2 ∷ HIS-PGAL1-tHMG1 ura3 ∷ URA3-PGAL1-upc2-1 [pAM322: 2μ-leu2d PGAL1-ADSPGAL10-CYP71AV1 PGAL1-AaCPR (codon opt)] | This work | AA |
| Y137 | S288C | As Y87, but plasmid is pAM322 | This work | AA |
| Y151 | CEN.PK2 | As Y135, but plasmid is pAM426 [2μ-leu2d PGAL1-ADS (codon opt)] | This work | Amorph |
| Gen 2.0 | ||||
| Y224 | CEN.PK2 | 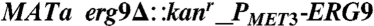 , leu2-3, , leu2-3,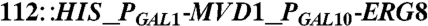 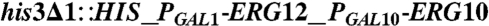 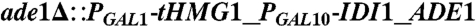 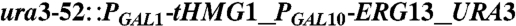 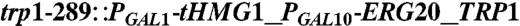 [pAM322] [pAM322] |
This work | AA |
| Y227 | CEN.PK2 | As Y224, but plasmid is pAM426 | This work | Amorph |
| Y293 | CEN.PK2 | Y227 gal80Δ∷natr | This work | Amorph |
| Y337 | CEN.PK2 | Y227 gal1Δ gal7Δ gal10Δ∷HphA | This work | Amorph |
Amorph, amorpha-4,11-diene; AA, artemisinic acid. A complete list of strains, including progenitor strains, is listed in Table S1.
Codon Optimization of Heterologous Genes.
We investigated whether S. cerevisiae codon optimization of heterologous genes would increase production of amorpha-4,11-diene or artemisinic acid. For production of artemisinic acid the plasmid containing all three A. annua genes, pESC-Leu2d∷ADS/AMO/CPR (15) was resynthesized with S. cerevisiae codon-optimized A. annua genes, and the orientation of the PGAL1-ADS-TPGK1 expression cassette reversed. The reversal of the GAL1 promoters ensures that recombination cannot occur [which would lead to elimination of CYP71AV1 (AMO) and ADS in pESC-Leu2d∷ADS/AMO/CPR] to generate pAM322 (Fig. S1). We compared production of artemisinic acid by strains containing either pESC-Leu2d∷ADS/AMO/CPR (strain Y87) or pAM322 (strain Y137). Y137 grew (Fig. 1B) and produced artemisinic acid (Fig. 1C) to similar levels as Y87, suggesting that codon optimization did not improve production. However, given the elimination of the possibility for intergenic recombination to occur, all subsequent strains producing artemisinic acid contained pAM322. A plasmid expressing only ADS (pAM426) was generated by recircularization of pAM322 following digestion by PvuII, eliminating the AMO/AaCPR expression cassette (Fig. S1). To allow fair comparison between artemisinic acid and amorpha-4,11-diene production, pAM426 was used in all subsequent amorpha-4,11-diene-producing strains.
Reconstruction of S. cerevisiae Production Strains.
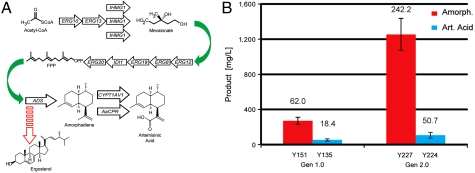
All prior strains, constructed in S. cerevisiae S288C, overexpressed selected mevalonate pathway genes from the galactose-regulated GAL1 promoter. Production of amorpha-4,11-diene and artemisinic acid were further increased by galactose-regulated (PGAL1) expression of the UPC2-1 transcription factor (13). UPC2-1 encodes an activated allele of UPC2 (17, 18), which increases transcription of most ergosterol pathway biosynthetic genes and is also a regulator of hypoxia-expressed genes. We decided to reconstruct the production strain so that every mevalonate pathway enzyme as far as ERG20 is transcribed from high-expression galactose-regulated promoters (PGAL1 or PGAL10). We elected to construct the new strains in S. cerevisiae CEN.PK2 rather than S288C. Whereas S288C has been much used for genetic investigation, and was the first strain to have its genome sequenced (19), little physiological information exists. CEN.PK2 has better characterized physiology, and its significantly higher sporulation efficiency (20, 21) simplifies strain construction using Mendelian genetics. CEN.PK2 was engineered in two ways: (i) the engineering scheme of the earlier S288C strains (13) was recapitulated to produce strains designated as Gen 1.0, and (ii) every enzyme of the mevalonate pathway as far as ERG20 was transcribed from the galactose-regulated divergent GAL1/ GAL10 promoters to produce Gen 2.0 strains (Fig. 2A, Fig. S2). Three copies of tHMG1 were integrated, as HMG-CoA reductase expression has been shown to be limiting (13). Table 1 and Table S1 summarize Gen 1.0 and Gen 2.0 strains that were constructed. Two Gen 2.0 strains were initially generated, Y224 containing pAM322 for production of artemisinic acid, and Y227 containing pAM426 (expressing only ADS; Fig. S1), for production of amorpha-4,11-diene. Production by Y224 and Y227 was compared to equivalent CEN.PK2 Gen 1.0 strains in galactose shake-flask cultures. Production of amorpha-4,11-diene by Y227 was approximately fivefold that of Y151, its Gen 1.0 equivalent, whereas production of artemisinic acid by Y224 was only approximately twofold that of Y135, its Gen 1.0 equivalent strain (Fig. 2B). Production of both amorpha-4,11-diene and artemisinic acid were, therefore, significantly increased by overexpression of the entire mevalonate pathway to ERG20 in Gen 2.0 strains, but the increase in production of amorpha-4,11-diene was more than double that of the increase in production of artemisinic acid. Most notably, production of amorpha-4,11-diene by Y227, the Gen 2.0 ADS-expressing strain, was over 10-fold higher than production of artemisinic acid by Y224, its equivalent Gen 2.0 artemisinic acid-producing strain. Interestingly, pAM322 was found to be very stable in Y224 [100% stable after 72 h growth in flasks, assessed by retention of the LEU2 plasmid selectable marker (15)], in contrast to the artemisinic acid-producing S288C Gen 1.0 strains containing pESC-Leu2d∷ADS/AMO/CPR (15), and CEN.PK2 Gen 1.0 strain bearing pAM322 (Y135; 70% stable after 72 h growth in flasks). Given the dramatically higher production of amorpha-4,11-diene compared to artemisinic acid in Gen 2.0 strains, or amorpha-4,11-diene in Gen 1.0 strains (Fig. 2B), we decided to pursue production of amorpha-4,11-diene in Gen 2.0 strains.
Fig. 2.
Construction of CEN.PK2 Gen 2.0 and comparison of production by Gen 1.0 and 2.0 strains. (A) Schematic of the mevalonate pathway showing genes overexpressed in Gen 2.0 strains from GAL1 or GAL10 promoters in block arrows. Dashed arrow represents the ergosterol pathway transcriptionally restricted at ERG9. (B) Production of amorpha-4,11-diene by Y151 (Gen 1.0) and Y227 (Gen 2.0), and artemisinic acid by Y135 (Gen 1.0) and Y224 (Gen 2.0) after 72 h growth in shake-flasks. Error bars show ± 1 SD of three flasks. Numbers above the bars show specific production (mg/L/OD600).
Fermentation Testing and Process Development.
Utilization of galactose was eliminated prior to testing production of amorpha-4,11-diene in fermentation by deleting the GAL1 GAL10 GAL7 gene cluster. The resulting strain, Y337 (= Y227 gal1Δ gal7Δ gal10Δ; Table 1), is unable to metabolize galactose, but still requires a low concentration of galactose for induction of the mevalonate pathway and ADS genes transcribed by galactose-regulated promoters. Y337 was grown in glucose-limited fed-batch fermentation with induction by galactose occurring at the end of the batch growth phase (SI Materials and Methods, Figs. S3–S5); under this fermentation condition Y337 produced 2.0 g/L amorpha-4,11-diene (Fig. 3B) with a yield of 1.11 Cmol % (molar percentage of substrate carbon added to the fermentation incorporated into amorpha-4,11-diene). We tested the effect of phosphate restriction on production in glucose-limited fed-batch fermentations (Fig. 3 A and B). A range of lower concentrations of phosphate in the feed, batch, or both were tested to identify phosphate delivery that would capture the increase in per cell productivity without disrupting carbon delivery in the fed-batch process to the extent the run would be less productive. Fig. S6 and Fig. S7 show that decreasing phosphate concentrations in the batch or feed media can indeed lead to higher production of amorph-4,11-diene. Highest production was achieved with 8 g/L KH2PO4 in the batch but no KH2PO4 in the feed, compared to 8 g/L KH2PO4 in the batch and 9 g/L KH2PO4 in the unrestricted process. Growth was somewhat restricted (Fig. 3A), but production of amorpha-4,11-diene was more than doubled, attaining 5.5 g/L after 95 h with a yield of 3.23 Cmol % (Fig. 3B). The use of ethanol has been described as a carbon source for the production of hydrocortisone (derived from the mevalonate pathway) in yeast (22). We also wished to see whether production could be increased when ethanol is provided as a carbon source, based on the notion that ethanol could increase the supply of cytosolic acetyl-CoA. To this end we tested mixed glucose/ethanol feeds following the glucose batch phase of growth. The mixed-feed strategy resulted in similar growth (Fig. 4A), but also gave a significant increase in production of amorpha-4,11-diene, attaining 16.5 g/L after 118 h with a yield of 6.03 Cmol % (Fig. 4B). It is also notable that production continued after maximum cell density had been achieved. Attempts to increase production of amorpha-4,11-diene by the use of phosphate restriction with mixed glucose/ethanol feeds gave variable results and were not pursued (Fig. S8, Fig. S9).
Fig. 3.
Development of fed-batch glucose-limited fermentations for the production of amorpha-4,11-diene. (A) Growth and (B) amorpha-4,11-diene production by Y337 in glucose-limited fermentations with and without phosphate limitation, and Y293 in glucose-limited fermentation. Production of amorpha-4,11-diene was induced in Y337 fermentations by addition of galactose (see SI Materials and Methods).
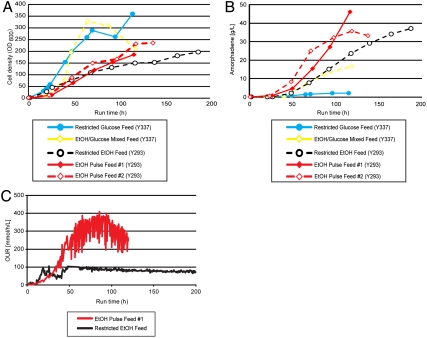
Fig. 4.
Development of fed-batch glucose/ethanol and ethanol feed fermentations. (A) Growth and (B) amorpha-4,11-diene production by Y337 with restricted glucose feed and mixed glucose and ethanol feed, and Y293 with ethanol pulse feed (replicate fermentations) and restricted ethanol feed. (C) Oxygen uptake rate of Y293 ethanol pulse feed and restricted ethanol feed fermentors.
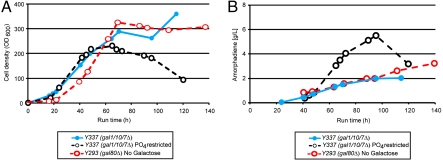
Y337 (= Y227 gal1Δ gal7Δ gal10Δ) does not metabolize galactose, but still requires addition of a low concentration of galactose to initiate production of amorpha-4,11-diene. It would be advantageous to eliminate the use of galactose altogether to further decrease the cost of amorpha-4,11-diene production. To this end, we deleted the GAL80 gene from Y227, the galactose-utilizing amorpha-4,11-diene producing Gen 2.0 strain. Gal80p is a negative regulator of the galactose regulon which acts by binding to the C-terminal transcriptional activation domain of Gal4p in nongalactose carbon sources; it is released from Gal4p upon addition of galactose, and in the absence of a repressing carbon source such as glucose the release of Gal80p from Gal4p allows induction of galactose-regulated genes (23). The amorpha-4,11-diene production strain with deletion of GAL80 was designated Y293 (= Y227 gal80Δ; Table 1). In glucose-limited fed-batch fermentation without addition of galactose, Y293 performed similarly to Y337, which required addition of galactose to induce production (Fig. 3). Given the similar behavior of Y293 and Y337 we elected to continue fermentation development with Y293, thus obviating the use of galactose altogether.
We wished to test whether production could be increased with the use of pure ethanol feeds. Two different feed strategies were tested, an ethanol pulse feed without ethanol restriction, and a restricted ethanol feed with lower oxygen uptake rate (OUR). The cell density achieved was appreciably lower than that achieved with either glucose or glucose/ethanol mixed feed (Fig. 4A). Production of amorpha-4,11-diene, however, was significantly higher than all earlier processes, attaining 41 g/L for the unrestricted ethanol pulse feed after 116 h (yield of 16.98 Cmol %); a replicate fermentation attained 36 g/L. A restricted ethanol feed produced 37 g/L amorpha-4,11-diene (yield of 18.67 Cmol %), albeit after a longer run time of 187 h (Fig. 4B). An important advantage of using a restricted ethanol feed process is that significantly lower OURs are required for production in comparison to the ethanol pulse feed process (Fig. 4C).
Chemical Conversion of Amorpha-4,11-diene to Dihydroartemisinic Acid.
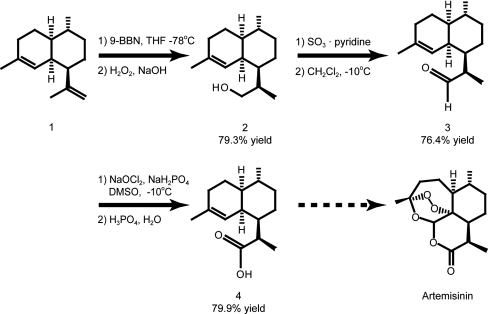
Amorpha-4,11-diene, compound 1, was converted to dihydroartemisinic acid, compound 4, in three steps beginning with selective hydroboration of the double bond at the 11-position (Fig. 5). Using the hindered borane 9-borabicyclo[3.3.1]nonane (9-BBN) a 79.3% yield of alcohol compound 2 was achieved as an 85∶15 mixture (by 1H - NMR) of epimers with the desired R-epimer predominating. Several methods reported to convert primary alcohols to carboxylic acids in one step (24, 25) produced complex mixtures from compound 2, presumably due to interaction with the olefinic linkage at the 4-position. Thus a two-step oxidation sequence was employed in which SO3 pyridine complex (26) oxidized compound 2 to aldehyde compound 3 (76.4%) followed by treatment of compound 3 with the oxidant NaOCl2 in DMSO (27, 28) to form compound 4 (79.9%). The use of DMSO as the solvent was crucial because the NaOCl formed in the reaction preferentially reacts with it, thus protecting the double bond at the 4-position. The overall yield of compound 4 from compound 1 by this route was 48.4%.
Discussion
Initial work showed that deletion of GAL1, a gene required for the utilization of galactose, allowed the use of galactose as a gratuitous inducer while eliminating its use as a carbon source. An attempt to increase production of artemisinic acid by codon optimization of the heterologous genes from A. annua was unsuccessful, perhaps reflecting the recent realization that expression optimization by simple codon usage bias is of limited value (29). However, the redesigned plasmid could not eliminate genes by recombination and was used for subsequent work.
We chose to reengineer CEN.PK2 rather than S288C to produce Gen 2.0 strains in which every enzyme of the mevalonate pathway leading to the production of farnesyl diphosphate was transcribed from high-expression galactose-regulated promoters. The enhanced sporulation efficiency of CEN.PK2 greatly simplified the construction of Gen 2.0 strains, and the better documented physiology of CEN.PK2 (20) suggested that this strain would be suitable for fermentative production. The use of the galactose-regulated GAL1,10 promoters to overexpress the mevalonate pathway genes allowed repression of expression during strain construction, thus avoiding buildup of any potentially toxic intermediates (8) which may have resulted in strain instability during construction. Intermediate buildup was not assayed in the final strains, and we did not investigate whether overexpression of all the mevalonate pathway genes was necessary. Regulated overexpression of all relevant mevalonate pathway genes in Gen 2.0 strains led to a fivefold increase in production of amorpha-4,11-diene in shake-flasks, but only a twofold increased production of artemisinic acid. Production of amorpha-4,11-diene by Y227 was over 10-fold higher than production of artemisinic acid by Y224, its equivalent Gen 2.0 artemisinic acid-producing strain (Fig. 2). This very significantly higher production led us to concentrate on the development of amorpha-4,11-diene production by fermentation. Low plasmid stability was not the reason for the poor production of artemisinic acid, as plasmid pAM322, expressing ADS, CYP71AV1, and AaCPR, was found to be 100% stable in Gen 2.0 strains, in contrast to Gen 1.0 strains where marked instability was observed (15). Presumably there is strong selective pressure for maintenance of pAM322 in Gen 2.0 strains, which is lacking in Gen 1.0 strains.
Initial development of a fermentation process for the production of amorpha-4,11-diene used a derivative of the Gen 2.0 strain Y227 known as Y337 in which galactose utilization had been disabled by deletion of the GAL1 GAL10 GAL7 gene cluster. A glucose-limited fed-batch fermentation process in which amorpha-4,11-diene production was induced by the addition of a low concentration of galactose at the end of batch growth resulted in production of 2.0 g/L amorpha-4,11-diene. An initial attempt to increase production by phosphate limitation, on the rationale that restricting phosphate would limit growth and channel greater carbon flux to product, resulted in more than doubling of amorpha-4,11-diene production. The biggest increase in production was observed when ethanol was added to the feed, initially as a glucose/ethanol mixed feed following the glucose batch phase of growth. The rationale for adding ethanol to the feed was that direct supply of ethanol may increase the supply of cytosolic acetyl-CoA to the mevalonate pathway (30). An alternative explanation is that use of ethanol eliminated glucose repression, but this scenario is considered unlikely as glucose was still present in the feed, and the concentration of glucose in the feed stage of the fed-batch fermentations was very low, and mostly undetectable. However, transcript levels were not monitored during the different fermentation procedures, and it is not possible to unequivocally determine the reason for the increase in amorpha-4,11-diene production in a glucose/ethanol mixed-feed fermentation. The glucose/ethanol mixed-feed approach led to production of 16.5 g/L of amorpha-4,11-diene with significantly increased yield.
The final stage of fermentation development was conducted with Y293, an amorph-4,11-diene-producing strain in which expression of Gal80p (the negative-regulator of galactose-regulon gene expression) had been eliminated. The removal of Gal80p expression eliminated the need to add galactose for induction of amorpha-4,11-diene production. Y293 produced a similar concentration of amorph-4,11-diene to Y337 (which cannot metabolize galactose), but Y293 did not require addition of galactose to the fermentation. Two different strategies for feeding pure ethanol as feed carbon source were evaluated. Whereas an unrestricted ethanol feed (a pulse feed) produced the highest concentration of amorpha-4,11-diene, its high OUR makes it industrially infeasible. Conversely, the ethanol restricted-feed process, capable of producing 37 g/L of amorpha-4,11-diene, has a lower OUR that may be amenable to adaptation to industrial-scale fermentation. The use of S. cerevisiae for the production of amorpha-4,11-diene has considerable advantages over the earlier E. coli process (12), notably the absence of an expensive inducer such as IPTG, and the lower OUR required for the restricted-feed ethanol process. In large-scale fermentations, high oxygen uptake rates require high sparge rates, high agitation rates, and/or supplementation with pure oxygen (31). In addition, the heat generation resulting from rapid substrate and oxygen consumption requires increased cooling capacity to maintain temperature (32). Further process development is underway to implement practical solutions at large scale.
The overall aim of the strain construction and fermentation development was to create a process that could feasibly be developed for cost-effective production of artemisinin for the developing world. An earlier fermentation process for production of artemisinic acid used galactose for both growth and induction (14). A drawback with this process, other than the low titer of artemisinic acid produced (2.3 g/L), was the cost of galactose; galactose is approximately 100-fold more expensive than glucose (SI Materials and Methods). The advantage of eliminating catabolism of galactose by deletion of the GAL1 GAL10 GAL7 gene cluster is that galactose is required solely as an inducer, and the quantities added to the fermentor are much lower (10 g/L in the induction feed medium; SI Materials and Methods). The final strain used for fermentation development (Y293) contained a deletion of GAL80 and did not require addition of galactose for induction, thus providing a further cost saving. Ethanol is approximately twice the cost of glucose on a weight basis (SI Materials and Methods). However, addition of ethanol, initially as a mixed-feed then as pure ethanol feed, resulted in approximately 20-fold increased production of amorpha-4,11-diene [2.0 g/L with restricted glucose feed, compared to 41 g/L with unrestricted ethanol feed (Fig. 4)]. The yield of amorpha-4,11-diene production also increased significantly with ethanol feed, rising from 1.11 Cmol % with restricted glucose feed to 18.67 Cmol % with restricted ethanol feed; this increased yield demonstrates a dramatic increase in the conversion of substrate carbon into amorpha-4,11-diene. Given that the market price of artemisinin is over 200-fold higher than that of ethanol, and that the production of amorpha-4,11-diene is a significant fraction of the total production cost, it is economically reasonable to use ethanol as a fermentation carbon source. Nonetheless, efforts are underway to increase the production and yield of amorpha-4,11-diene from glucose.
Dihydroartemisinic acid (Fig. 5, compound 4) is a late-stage intermediate in a number of total syntheses of artemisinin (33) and its derivatives (34), and an efficient industrial-scale synthesis from amorpha-4,11-diene would greatly aid in efforts to produce large amounts of artemisinin-based antimalarial drugs. For small scale (submolar) synthesis scale we used 9-borabicyclo[3,3,1]nonane for the hydroboration of amorpha-4,11-diene to form dihydroartemisinic alcohol (Fig. 5, compound 2) because it did not affect the sensitive double bond at the 4-position. We elected to use a two-step oxidation process because common reagents for converting primary alcohols to carboxylic acids in one step are either chromium based or contain components that were likely to react with the sensitive double bond at the 4-position. The readily available and scalable sulfur trioxide-pyridine complex (35) smoothly converts compound 2 to dihydroartemisinic aldehyde (Fig. 5, compound 3), which is subsequently oxidized to dihydroartemisinic acid (Fig. 5, compound 4) with inexpensive inorganic reagents (26–28).
This work links together each of the key steps of a complete process for the production of semisynthetic artemisinin. The construction of a complete mevalonate pathway under strong induction, along with fermentation development, was crucial to obtaining high production of amorpha-4,11-diene. Combining high-level production of amorpha-4,11-diene with demonstration of successful chemical conversion to dihydroartemisinic acid represents a significant milestone toward the development of an economically viable process for the production of semisynthetic artemisinin. A major challenge remains that the Gen 2.0 reengineered strain of yeast did not produce significantly more artemisinic acid than the original Gen 1.0 strains (Fig. 2B). Production of artemisinic acid by S. cerevisiae at concentrations comparable to that of amorpha-4,11-diene by Y293 would be advantageous, as the chemical conversion of artemisinic acid to artemisinin would be simpler than production from amorpha-4,11-diene. To directly produce high levels of artemisinic acid from S. cerevisiae, the underlying cause of the lower production of artemisinic acid compared to amorpha-4,11-diene will need to be identified and remedied.
Materials and Methods
S. cerevisiae Strain Construction.
Five separate DNA integration constructs (Fig. S2) were made, all based on transcription of two mevalonate pathway genes from the divergent GAL1,10 promoter, as follows: S. cerevisiae genomic DNA covering several hundred nucleotides upstream of the desired integration site was amplified by PCR with a flanking PmeI restriction enzyme site. A selection marker was also PCR amplified from genomic DNA with 40 nt of overlap to the downstream terminus of the upstream integration site amplicon. The fragments were combined by overlap PCR (36). Several hundred nucleotides of genomic DNA downstream of the desired integration site were PCR amplified with a PmeI site at the downstream terminus. The two amplicons were joined by overlap PCR to yield a fragment (no. 1) of the following composition: (PmeI)-upstream integration fragment-selection marker-(XmaI)-downstream integration fragment-(PmeI). The resulting amplicon was ligated into the TOPO pCR2.1 vector (Invitrogen). The genomic locus encoding the open reading frame and transcriptional terminator of the first gene to be expressed was PCR amplified with an XmaI site at its upstream terminus. The PGAL1,10 promoter was PCR amplified with 40 nt of overlap to the upstream gene, and the promoter and gene combined by overlap PCR. The PGAL1,10 promoter also contained 40 nt of overlap to the downstream gene to be expressed. The genomic locus of the second gene to be expressed was PCR amplified, and the two amplicons combined by overlap extension to yield a fragment (no. 2) of the following composition: (XmaI)-upstream gene-PGAL1,10 promoter-downstream gene-(XmaI). The resulting amplicon was ligated into the TOPO ZERO Blunt II Cloning vector. Fragments no. 1 and no. 2 were combined by digesting the TOPO pCR2.1 vector containing fragment no. 1 with XmaI, and subsequent ligation with fragment no. 2 released from its TOPO ZERO Blunt II vector by XmaI digestion, to yield a fragment of the following composition: (PmeI)-upstream integration fragment-selection marker-(XmaI)-upstream gene-PGAL1,10 promoter-downstream gene-(XmaI)-downstream integration fragment-(PmeI). This combined fragment was released from the TOPO ZERO Blunt II Cloning vector by digestion with PmeI, purified, and used to transform S. cerevisiae CEN.PK2 derivatives. Full details of each construction are provided in SI Materials and Methods; Table S2 lists all oligonucleotide primers..
Media and Growth Conditions.
S. cerevisiae strains (Table 1) were grown on synthetic complete medium agar plates at 30 °C with 2% glucose as the carbon source unless otherwise indicated, lacking leucine where needed for selection of plasmids (37). Flask and fermentor media were based on that of van Hoek et al. (38) (SI Materials and Methods). Amorpha-4,11-diene produced in flask cultures was captured by addition of 20% vol/vol isopropyl myristate to the medium; amorpha-4,11-diene produced in fermentors was captured by the addition of 10% vol/vol methyl oleate. Fermentations took place in 2 L Sartorius Biostat B plus twins with gas-flow ratio controllers. Full details are provided in SI Materials and Methods.
Assay Methods.
Amorpha-4,11-diene.
Production of amorpha-4,11-diene was monitored by gas chromatography with flame-ionization detection (GC/FID). Fermentor samples were prepared by mixing 0.4 mL cell lysis reagent [two parts Novagen YeastBuster protein reagent (EMD Biosciences, P/N 71186-4) and one part 2 M HCl] with 0.1 mL whole broth and 1 mL ethyl acetate containing 10 mg/L transcaryophylene (surrogate; Sigma-Aldrich; ≥98.5% purity; P/N 22075) in a 2 mL glass vial. For determination of amorpha-4,11-diene from flask cultures the isopropyl myristate overlay was sampled, and diluted directly into ethyl acetate/transcaryophylene. The sample was mixed for 30 min on a vortex mixer. After phase separation 0.6 mL of the ethyl acetate layer was transferred to a GC vial for analysis. The ethyl acetate-extracted samples were analyzed using the GC/FID. Amorpha-4,11-diene peak areas were converted to concentration values from external standard calibrations using authentic compounds. A 1 μL sample was split 1∶20 and separated using a DB-WAX column (50 m × 200 μm × 0.2 μm; P/N 128-7052; Agilent) with hydrogen as the carrier gas at flow rate at 1.57 mL/ min. The temperature program for the analysis was as follows: the column was initially held at 150 °C for 3.0 min, followed by a temperature gradient of 5 °C/ min to a temperature of 250 °C, where the column was held at 250 °C for 5 min to elute all remaining components. Under these conditions transcaryophylene and amorpha-4,11-diene and elute at 4.95 and 5.77 min, respectively.
Artemisinic acid.
A 1 mL aliquot of well-mixed fermentation broth was diluted in 9 mL of methanol + 0.1% formic acid. The mixture is then mixed on a vortex mixer for 30 min and centrifuged at 16,000 × g for 5 min. One hundred microliters of the supernatant was diluted into 900 μL methanol + 0.1% formic acid. A 20 μL aliquot was injected on an Agilent 1200 HPLC with UV detection at 212 nm. A Supelco Discovery C8 column (4.6 mm × 100 mm × 5.0 μm, Supelco, P/N 569423-U) equipped with the appropriate guard column (4.0 mm × 20.0 mm, Supelco, P/N 59589-U) was used for separation, with the following gradient at a flow rate of 1 mL/ min (channel A, water + 0.1% formic acid; channel B, methanol + 0.1% formic acid): 0–0.5 min 70% B, gradually increasing to 97% B from 0.5 to 6.7 min, hold at 97% B until 7 min, decrease to 70% B from 7–7.5 min, and reequilibrate to 70% B from 7.5 to 9.5 min. The column was held at 25 °C during the separation. Under these conditions, artemisinic acid was found to elute at 6.3 min. Artemisinic acid peak areas were converted to concentrations from external standard calibrations of authentic compounds.
Chemistry.
General information.
NMR spectra were recorded in deuterated chloroform by Acorn NMR using a JEOL ECX-400 NMR spectrometer operating at 400 MHz for 1H and 100.5 MHz for 13C. Chemical shifts are reported in δ units relative to SiMe4 as an internal standard (δ = 0). GC/MS data were obtained with an Agilent 5975 inert mass selective detector coupled to an Agilent 6890 N network GC system using an Agilent 19091Z-005 50 m capillary column. Helium was used as the carrier gas. Operating conditions: inlet temperature 250 °C, initial temperature 50 °C for 0.5 min then ramp 5 °C/ min to 190 °C then ramp 60 °C/ min to 300 °C and hold for 1 min. Total run time 11.33 min. All reagents and solvents were obtained from Sigma-Aldrich and used as received. Thin layer chromatography (TLC) was performed on 2.5 × 7.5 cm silica gel 60 F254 glass plates from EMD Chemicals, Inc. using ethyl acetate/hexane mixtures as eluents. Column chromatography was conducted with EtOAc/hexane mixtures as eluents and Merck grade 9385 silica gel (230–400 mesh) obtained from Sigma-Aldrich.
Dihydroartemisinic alcohol, compound 2.
Amorpha-4,11-diene (compound 1, 0.150 g, 0.734 mmol) was dissolved in tetrahydrofuran (THF; 5.0 mL, Aldrich Sure/Seal) and cooled to -78 °C under a blanket of N2. A 0.5 M solution of 9-BBN in THF (1.47 mL, 0.734 mol) was added via syringe drive over 19 min with magnetic stirring. The dry-ice acetone bath was allowed to come to room temperature and the reaction stirred for 16 h. The reaction was again cooled to -78 °C and a second portion of 9-BBN (1.47 mL, 0.734 mol) added via syringe drive over 4 min. The reaction was stirred for 16 h at which time TLC (20% EtOAC/hexane) revealed only a trace of compound 1 remaining. A solution of NaOH (0.294 g, 0.734 mol) in 2.5 mL of H2O was added, the mixture cooled to 0 °C with an ice bath and 0.75 mL of 30% H2O2 added. The ice bath was removed and after 1 h the THF was evaporated under reduced pressure. The residue was subjected to extractive workup with ether and H2O. The ether layer was dried (MgSO4) and evaporated under reduced pressure. Flash chromatography of the crude product on silica gel (20% EtOAc/hexane) provided 0.010 g of compound 1 and 0.119 g (79.3% based on recovered stating material) of compound 2 as a colorless oil consisting of a 85∶15 mixture (1H - NMR) of 11- R : 11 - S isomers of compound 1 that rapidly solidified. The 1H - NMR and MS spectra of the major isomer were identical to those previously published (39). For additional details, see Fig. 5.
Dihydroartemisinic aldehyde, compound 3.
Dihydroartemisinic alcohol (compound 2, 0.444 g, 2.00 mmol), triethylamine (1.12 mL, 0.808 g, 8.00 mmol), and 10.4 mL of 5∶1 CH2Cl2/DMSO were combined and cooled to -10 °C with an ice/NaCl bath. The mixture was magnetically stirred and SO3·py (0.796 g, 5.00 mmol) was added in three equal portions over 20 min. The ice bath was removed and the reaction stirred for 15 h at ambient temperature at which time GC/MS showed the reaction to be complete. The reaction mixture was poured into 10 mL of 10% aqueous citric acid solution and stirred for 10 min. The layers were separated and the organic phase washed with 10 mL of 10% aqueous citric acid solution, 10 mL of saturated NaHCO3 solution, 10 mL of NaCl solution, dried (MgSO4) and the CH2Cl2 removed under reduced pressure. The residue was passed through a 1 × 2 cm plug of silica gel with 20% EtOAc/hexane to afford 0.377 g (76.4%) of compound 3 with 1H - NMR and MS spectra identical to those previously published (40). For additional details, see Fig. 5.
Dihydroartemisinic acid, compound 4.
Dihydroartemisinic aldehyde (0.250 g, 1.13 mmol) was dissolved in 20 mL of DMSO with mechanical stirring followed by addition over 2 h of a solution of NaOCl2 (0.143 g, 1.58 mmol) and NaH2PO4 (0.938 g, 6.92 mmol) in 10.0 mL of H2O. After stirring at ambient temperature for an additional 2 h, 15 mL of aqeous NaHCO3 solution were added and the stirring continued for 15 h. The solution was acidified to pH 2 (Colorphast Strip) by the dropwise addition of concentration H3PO4, which resulted in the precipitation of compound 4 as fine white needles. Yield, 0.214 g (79.9%). The 1H - NMR and MS spectra of compound 4 were identical to those previously published (34). For additional details, see Fig. 5.
Supplementary Material
Acknowledgments.
We thank members of Jay Keasling’s laboratory for EPY330 and for many productive conversations, and also Jasper Rine and Hans van Dijken for advice and many fruitful discussions. We thank and acknowledge our friends and colleagues at Sanofi–Aventis, especially Denis Thibaut for valuable discussion around the use of ethanol as a carbon source, Bruno Dumas, Corinne Masson-Brocard, Paul Baduel, and Henri Farret. This research was conducted under the sponsorship of the Institute for OneWorld Health through generous support of the Bill and Melinda Gates Foundation.
Footnotes
Conflict of interest statement: All authors hold stock options or shares in Amyris, Inc.
This article is a PNAS Direct Submission.
See Author Summary on page 655.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1110740109/-/DCSupplemental.
References
- 1.Korenromp E, Miller J, Nahlen B, Wardlaw T, Young M. World Malaria Report 2005. Geneva: World Health Organization; 2005. [Google Scholar]
- 2.Bloland PB. Drug Resistance in Malaria. Geneva: World Health Organization; 2001. [Google Scholar]
- 3.Olumese P. Guidelines for the Treatment of Malaria. Geneva: World Health Organization; 2006. [Google Scholar]
- 4.Cohen J, Singh I, O’Brien M. Predicting global fund grant disbursements for procurement of artemisinin-based combination therapies. Malar J. 2008;7:200. doi: 10.1186/1475-2875-7-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aregawi M, Cibulskis R, Otten M, Williams R, Dye C. World Malaria Report 2008. Geneva: World Health Organization; 2008. [Google Scholar]
- 6.Hale V, Keasling JD, Renninger N, Diagana TT. Microbially derived artemisinin: A biotechnology solution to the global problem of access to affordable antimalarial drugs. Am J Trop Med Hyg. 2007;77(Suppl 6):198–202. [PubMed] [Google Scholar]
- 7.White NJ. Qinghaosu (artemisinin): The price of success. Science. 2008;320:330–334. doi: 10.1126/science.1155165. [DOI] [PubMed] [Google Scholar]
- 8.Martin VJ, Pitera DJ, Withers ST, Newman JD, Keasling JD. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat Biotechnol. 2003;21:796–802. doi: 10.1038/nbt833. [DOI] [PubMed] [Google Scholar]
- 9.Newman JD, et al. High-level production of amorpha-4,11-diene in a two-phase partitioning bioreactor of metabolically engineered Escherichia coli. Biotechnol Bioeng. 2006;95:684–691. doi: 10.1002/bit.21017. [DOI] [PubMed] [Google Scholar]
- 10.Pitera DJ, Paddon CJ, Newman JD, Keasling JD. Balancing a heterologous mevalonate pathway for improved isoprenoid production in Escherichia coli. Metab Eng. 2007;9:193–207. doi: 10.1016/j.ymben.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 11.Kizer L, Pitera DJ, Pfleger BF, Keasling JD. Application of functional genomics to pathway optimization for increased isoprenoid production. Appl Environ Microbiol. 2008;74:3229–3241. doi: 10.1128/AEM.02750-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsuruta H, et al. High-level production of amorpha-4,11-diene, a precursor of the antimalarial agent artemisinin, in Escherichia coli. PLoS One. 2009;4:e4489. doi: 10.1371/journal.pone.0004489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ro DK, et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440:940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- 14.Lenihan JR, Tsuruta H, Diola D, Renninger NS, Regentin R. Developing an industrial artemisinic acid fermentation process to support the cost-effective production of antimalarial artemisinin-based combination therapies. Biotechnol Prog. 2008;24:1026–1032. doi: 10.1002/btpr.27. [DOI] [PubMed] [Google Scholar]
- 15.Ro DK, et al. Induction of multiple pleiotropic drug resistance genes in yeast engineered to produce an increased level of anti-malarial drug precursor, artemisinic acid. BMC Biotechnol. 2008;8:83. doi: 10.1186/1472-6750-8-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hovland P, Flick J, Johnston M, Sclafani RA. Galactose as a gratuitous inducer of GAL gene expression in yeasts growing on glucose. Gene. 1989;83:57–64. doi: 10.1016/0378-1119(89)90403-4. [DOI] [PubMed] [Google Scholar]
- 17.Davies BS, Wang HS, Rine J. Dual activators of the sterol biosynthetic pathway of Saccharomyces cerevisiae: Similar activation/regulatory domains but different response mechanisms. Mol Cell Biol. 2005;25:7375–7385. doi: 10.1128/MCB.25.16.7375-7385.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davies BS, Rine J. A role for sterol levels in oxygen sensing in Saccharomyces cerevisiae. Genetics. 2006;174:191–201. doi: 10.1534/genetics.106.059964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cherry JM, et al. Genetic and physical maps of Saccharomyces cerevisiae. Nature. 1997;387(Suppl 6632):67–73. [PMC free article] [PubMed] [Google Scholar]
- 20.van Dijken JP, et al. An interlaboratory comparison of physiological and genetic properties of four Saccharomyces cerevisiae strains. Enzyme Microb Technol. 2000;26:706–714. doi: 10.1016/s0141-0229(00)00162-9. [DOI] [PubMed] [Google Scholar]
- 21.Ben-Ari G, et al. Four linked genes participate in controlling sporulation efficiency in budding yeast. PLoS Genet. 2006;2:e195. doi: 10.1371/journal.pgen.0020195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szczebara FM, et al. Total biosynthesis of hydrocortisone from a simple carbon source in yeast. Nat Biotechnol. 2003;21:143–149. doi: 10.1038/nbt775. [DOI] [PubMed] [Google Scholar]
- 23.Lohr D, Venkov P, Zlatanova J. Transcriptional regulation in the yeast GAL gene family: A complex genetic network. FASEB J. 1995;9:777–787. doi: 10.1096/fasebj.9.9.7601342. [DOI] [PubMed] [Google Scholar]
- 24.Zhao M, Mano E, Song Z, Tschaen D. Oxidation of primary alcohols to carboxylic acids with sodium chlorite catalyzed by TEMPO and Bleach. Org Synth. 2004;81:195–201. [Google Scholar]
- 25.Yamaoka H, Moriya N, Ikunaka M. A practical RuCl3-catalyzed oxidation using trichloroisocyanuric acid as a stoichiometric oxidant under mild nonacidic conditions. Org Process Res Dev. 2004;8:931–938. [Google Scholar]
- 26.Krysan DJ, Haight AR, Menzia JA, Welch N. A stereoselective synthesis of the dihydroxyethylene dipeptide isostere, A-82768. Tetrahedron. 1994;50:6163–6172. [Google Scholar]
- 27.Dalcanale E, Montanari F. Selective oxidation of aldehydes to carboxylic acids with sodium chlorite-hydrogen peroxide. J Org Chem. 1986;51:567–569. [Google Scholar]
- 28.Bal BS, Childers WE, Pinnick HW. Oxidation of [alpha],[beta]-unsaturated aldehydes. Tetrahedron. 1981;37:2091–2096. [Google Scholar]
- 29.Welch M, Villalobos A, Gustafsson C, Minshull J. You’re one in a googol: Optimizing genes for protein expression. J R Soc Interface. 2009;6(Suppl 4):S467–476. doi: 10.1098/rsif.2008.0520.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Jong-Gubbels P, Vanrolleghem P, Heijnen S, van Dijken JP, Pronk JT. Regulation of carbon metabolism in chemostat cultures of Saccharomyces cerevisiae grown on mixtures of glucose and ethanol. Yeast. 1995;11:407–418. doi: 10.1002/yea.320110503. [DOI] [PubMed] [Google Scholar]
- 31.Stanbury PF, Whitaker A, Hall SJ. Principles of Fermentation Technology. Boston: Butterworth-Heinemann; [Google Scholar]
- 32.Bailey JE, Ollis DF. Biochemical Engineering Fundamentals. New York: McGraw-Hill; 1986. [Google Scholar]
- 33.Li Y, Huang H, Wu Y-L. Qinghaosu (artemisinin)—a fantastic antimalarial drug from a traditional Chinese medicine. In: Liang X-T, Fang W-S, editors. Medicinal Chemistry of Bioactive Natural Products. New York: Wiley; 2006. pp. 183–256. [Google Scholar]
- 34.Kim B, Sasaki T. Recent progress in the synthesis of artemisinin and its derivatives. Org Prep Proced Int. 2006;38:1–80. [Google Scholar]
- 35.Liu C, et al. Development of a large-scale process for an HIV protease inhibitor. Org Process Res Dev. 1997;1:45–54. [Google Scholar]
- 36.Horton RM. In vitro recombination and mutagenesis of DNA SOEing together tailor-made genes. Methods Mol Biol. 1997;67:141–149. doi: 10.1385/0-89603-483-6:141. [DOI] [PubMed] [Google Scholar]
- 37.Rose MD, Winston F, Hieter P. Methods in Yeast Genetics: A Laboratory Course Manual. Plainview, NY: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 38.van Hoek P, van Dijken JP, Pronk JT. Regulation of fermentative capacity and levels of glycolytic enzymes in chemostat cultures of Saccharomyces cerevisiae. Enzyme Microb Technol. 2000;26:724–736. doi: 10.1016/s0141-0229(00)00164-2. [DOI] [PubMed] [Google Scholar]
- 39.Sy L-K, Ngo K-S, Brown GD. Biomimetic synthesis of arteannuin h and the 3,2-rearrangement of allylic hydroperoxides. Tetrahedron. 1999;55:15127–15140. [Google Scholar]
- 40.Bertea CM, et al. Identification of intermediates and enzymes involved in the early steps of artemisinin biosynthesis in Artemisia annua. Planta Med. 2005;71:40–47. doi: 10.1055/s-2005-837749. [DOI] [PubMed] [Google Scholar]