Abstract
The ΔF508 mutation in the cystic fibrosis transmembrane conductance regulator (CFTR) gene is the most common cause of cystic fibrosis. The mutation disrupts biosynthetic processing, reduces channel opening rate, and decreases protein lifetime. In contrast to human CFTR (hCFTR)-ΔF508, mouse CFTR-ΔF508 is partially processed to the cell surface, although it exhibits a functional defect similar to hCFTR-ΔF508. To explore ΔF508 abnormalities, we generated human–mouse chimeric channels. Substituting mouse nucleotide-binding domain-1 (mNBD1) into hCFTR partially rescued the ΔF508-induced maturation defect, and substituting mouse membrane-spanning domain-2 or its intracellular loops (ICLs) into hCFTR prevented further ΔF508-induced gating defects. The protective effect of the mouse ICLs was reverted by inserting mouse NBDs. Our results indicate that the ΔF508 mutation affects maturation and gating via distinct regions of the protein; maturation of CFTR-ΔF508 depends on NBD1, and the ΔF508-induced gating defect depends on the interaction between the membrane-spanning domain-2 ICLs and the NBDs. These appear to be distinct processes, because none of the chimeras repaired both defects. This distinction was exemplified by the I539T mutation, which improved CFTR-ΔF508 processing but worsened the gating defect. Our results, together with previous studies, suggest that many different NBD1 modifications improve CFTR-ΔF508 maturation and that the effect of modifications can be additive. Thus, it might be possible to enhance processing by targeting several different regions of the domain or by targeting a network of CFTR-associated proteins. Because no one modification corrected both maturation and gating, perhaps more than a single agent will be required to correct all CFTR-ΔF508 defects.
Keywords: anion channel, protein biosynthesis
Mutations in the gene encoding the cystic fibrosis transmembrane conductance regulator (CFTR) anion channel cause cystic fibrosis (CF) (1, 2). The most common CF mutation deletes Phe508 (ΔF508, also called F508del). CFTR-ΔF508 is misprocessed; it fails to escape the endoplasmic reticulum (ER) and is degraded rather than trafficking to the cell membrane (3–6). Reducing the incubation temperature allows CFTR-ΔF508 channels to escape ER retention; however, compared with wild-type (WT) CFTR, they have a reduced open state probability (Po) and a shorter lifetime at the cell membrane (7–10). Crystal structures locate F508 on the solvent-exposed surface of nucleotide-binding domain (NBD)-1 and away from the NBD1/NBD2 dimer interface (11–13). Based on crystal structures of bacterial ATP-binding cassette (ABC) transporters and cross-linking studies, this exposed position may contact the intracellular loops (ICLs) of the membrane-spanning domains (MSDs) (14–16).
A recent cross-species comparison showed that mouse CFTR (mCFTR)-ΔF508 is partially processed like its wild-type counterpart (17). Partial processing occurred in cell types from several species, suggesting that it was not likely attributable to the presence or absence of a specific chaperone or other cellular protein. Given the similarities between mCFTR and human CFTR (hCTFR), these data also suggested that small sequence differences might significantly influence CFTR-ΔF508 biosynthesis. Differences between hCFTR-ΔF508 and mCFTR-ΔF508 provided an opportunity to learn more about the structural determinants of CFTR-ΔF508 processing, as well as its gating. Therefore, we hypothesized that substituting sequences of mCFTR into hCFTR might prevent the ΔF508 defects. To test this hypothesis, we generated human–mouse CFTR (hmCFTR) chimeras with and without the ΔF508 mutation and examined the effects on biosynthetic processing and gating.
Results
Inserting mNBD1 into hCFTR Partially Rescues the ΔF508 Processing Defect.
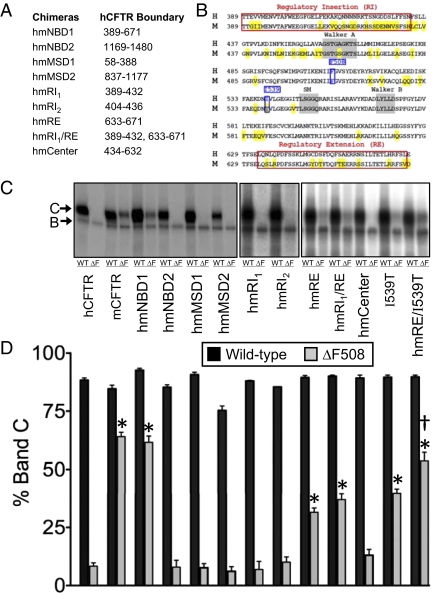
We constructed chimeras with an hCFTR backbone (Fig. 1 A and B) and assessed processing by examining CFTR glycosylation. CFTR undergoes core glycosylation in the ER and migrates as “band B” on electrophoresis; after reaching the Golgi complex, it is more extensively glycosylated and migrates as “band C” (3–5).
Fig. 1.
Processing of hmCFTR chimeras. (A) Boundaries of the NBD and MSD chimeras. (B) Amino acid comparison of hNBD1 and mNBD1. Mouse residues that differ from human residues are highlighted in yellow. Walker A, Walker B, and signature motifs (SMs) are shaded in gray. F508 and I539 are shown in blue boxes. RI (RI1 389–432 and RI2 404–436) and RE (633–671) boundaries refer to the regions identified in the NBD1 crystal structure (11) and are denoted in red boxes. (C) Autoradiographs of immunoprecipitated and in vitro-phosphorylated human, mouse, and human-mouse chimeras. Bands B and C are indicated by arrows. (D) Quantitation of band C relative to the total of bands B and C. Statistical significance was tested with one-way ANOVA with a Dunnett multiple comparison posttest. *Difference compared with hCFTR-ΔF508 (P < 0.05). †Difference in hmRE/I539T compared with hmRE and I539T (P < 0.05) (hCFTR, n = 15; mCFTR, n = 13; hmNBD1, n = 11; hmNBD2, n = 6; hmMSD1, n = 3; hmMSD2, n = 7; hmRI1, n = 3; hmRI2, n = 2; hmRE, n = 7; hmRI1/RE, n = 5; human–mouse Center, n = 6; hI539T, n = 4; hmRE/I539T, n = 4).
Wild-type hCFTR, mCFTR, and all the NBD and MSD chimeras generated band C (Fig. 1 C and D). Consistent with previous reports, the ΔF508 mutation prevented processing of hCFTR, whereas mCFTR-ΔF508 generated substantial band C (17). Interestingly, when we introduced mNBD1-ΔF508 into hCFTR (hmNBD1-ΔF508), the chimera generated band C at levels similar to those of mCFTR-ΔF508 (Fig. 1 C and D), indicating partial rescue of the processing defect. In contrast, mNBD2, mMSD1, and mMSD2 substitutions did not rescue the misprocessing. These results suggest that the “permissive” properties that allow mCFTR-ΔF508 processing are largely confined to mNBD1. In addition, pulse-chase experiments showed that the degradation rate of hmNBD1-ΔF508 did not substantially differ from that of hmNBD1 (Fig. S1).
mNBD1 Regulatory Extension Improves hCFTR-ΔF508 Processing.
To identify portions of mNBD1 responsible for improved hmNBD1-ΔF508 processing, we replaced smaller regions of hNBD1, choosing regions based on hNBD1 and mNBD1 crystal structures (11, 12) and differences between human and mouse primary sequences (Fig. 1 A and B). CFTR NBD1 contains two regions not present in other ABC transporters: an N-terminal sequence called the regulatory insertion (RI) and a C-terminal sequence called the regulatory extension (RE) (11).
Substituting portions of mNBD1 did not alter the glycosylation pattern of WT hCFTR (Fig. 1 C and D). However, mouse RE (mRE) alone and together with mouse RI1 (mRI1) increased hCFTR-ΔF508 band C production toward that of mCFTR-ΔF508. In contrast, mRI1, mRI2, and mouse Center failed to improve CFTR-ΔF508 processing (Fig. 1 C and D). We also individually mutated eight residues that differ between mRE and human RE (hRE), but no one mutation improved hCFTR-ΔF508 processing (Fig. S2). Thus, partial rescue depended on more than one difference between the mRE and hRE.
hNBD1 has an Ile at residue 539, whereas mNBD1 has a Thr (Fig. 1B). Previous reports indicated that an I539T mutation partially improved hCFTR-ΔF508 processing (18–20). We found the same (Fig. 1 C and D). In addition, we found that combining I539T with human–mouse RE (hmRE) caused hCFTR-ΔF508 to produce more band C than either substitution alone. Moreover, the proportion of band C in the chimera containing hmRE plus I539T was similar to that obtained when the entire mNBD1-ΔF508 replaced the human domain. Interestingly, mouse Center (433–632) contains T539 but did not rescue CFTR-ΔF508 processing, suggesting that the surrounding context is also important for rescue (Fig. 1 C and D).
Thus, the mNBD1 sequence is sufficient to correct hCFTR-ΔF508 processing partially, and more than one region of NBD1 is involved.
mMSD2 Prevents ΔF508 from Increasing Interburst Interval.
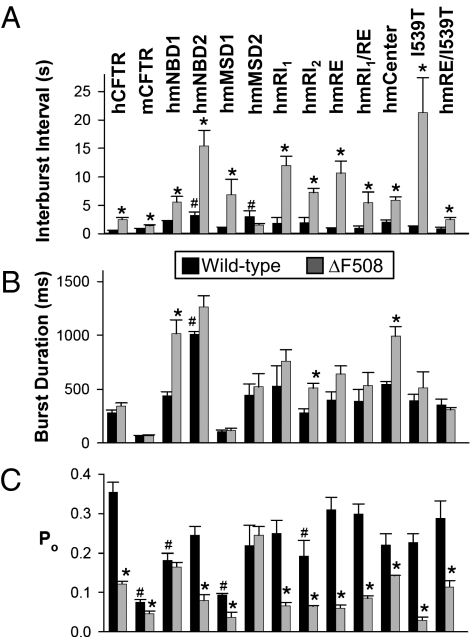
We asked whether the chimeras would prevent the ΔF508 gating defect, a prolonged interburst interval (8, 21). None of the mNBD1 or mMSD1 chimeras significantly altered the interburst interval of wild-type CFTR, whereas hmNBD2 and hmMSD2 prolonged it (Figs. 2A and 3). Of all the chimeras, only hmMSD2 prevented ΔF508 from prolonging the interburst interval. Thus, although substituting mMSD2 prolonged the interburst interval, it prevented ΔF508 from further increasing the interburst interval. Interestingly, the I539T mutation, which minimized the effect of ΔF508 on processing, actually accentuated the ΔF508-induced gating defect (Fig. 2A).
Fig. 2.
Single-channel kinetic properties of hmCFTR chimeras. Data are the mean ± SEM for interburst interval (A), burst duration (B), and Po (C) (n = 3–9 membrane patches for each construct). mCFTR has two open states (a subconductance opening and a full-conductance opening) (38). For mCFTR and mCFTR-ΔF508, we only counted full-conductance openings in the calculations. Neither hCFTR nor any of the chimeras had subconductance openings (Fig. 3), consistent with a previous report (22). Statistical significance was tested with one-way ANOVA with a Turkey posttest. *ΔF508 variant differed from its WT counterpart (P < 0.05). #Difference of non-ΔF508 variant compared with WT hCFTR (P < 0.05).
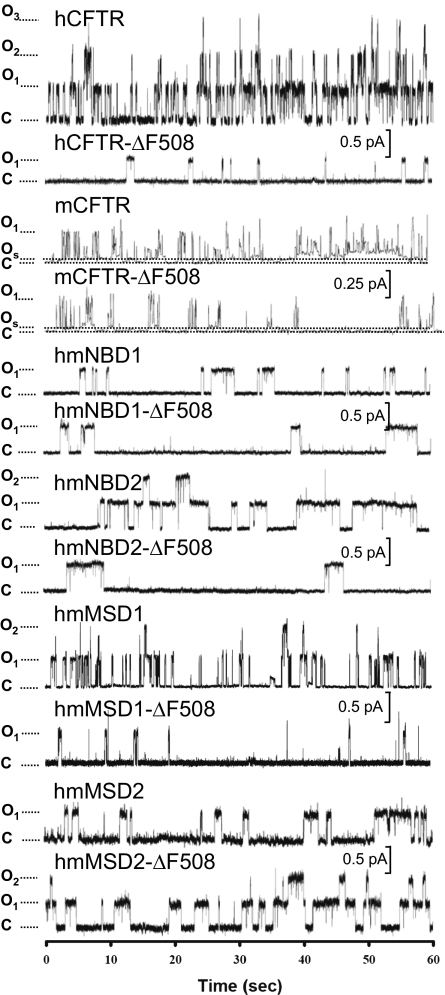
Fig. 3.
Representative traces of hCFTR and mCFTR and chimeras. Data are traces from excised, inside-out membrane patches containing one to three channels (n = 3–9 membrane patches for each construct). The cytosolic surfaces of the patches were continuously incubated in 75 nM protein kinase A and 1 mM ATP. The voltage was −50 mV for hCFTR-ΔF508, mCFTR, mCFTR-ΔF508, hmNBD1, and hmNBD1-ΔF508 or −80 mV for hCFTR, hmNBD2, hmNBD2-ΔF508, hmMSD1, hmMSD1-ΔF508, hmMSD2, and hmMSD2-ΔF508. The dotted line with “C” indicates the channel closed state. The dotted line with “Os” for mCFTR indicates the subconductance open state. The dotted line with “O” indicates the full open state. If a patch contained more than one channel, each open step is indicated by O1, O2, and O3. For purposes of illustration, traces were digitally filtered at 100 Hz with a Bessel filter, except for mCFTR, which was filtered at 10 Hz.
We also measured burst duration. With a wild-type hCFTR backbone, only the hmNBD2 chimera altered burst duration, and adding the ΔF508 mutation altered burst duration in hmNBD1, human–mouse Center, and hmRI2 (Fig. 2B). Of note, the prolonged burst duration in hmNBD1-ΔF508 generated a Po similar to that of hmNBD1, despite the increased interburst interval (Fig. 2 A–C).
Like our data, a previous study of wild-type chimeras reported that an hmNBD2 chimera had a prolonged interburst interval and burst duration compared with wild-type hCFTR (22). That study also reported increased interburst intervals and burst durations for an hmNBD1 chimera. Although our data showed a trend in that direction, the changes were not statistically significant; this difference might be attributable to different boundaries in the chimeras.
Inserting Mouse Sequences from ICL3 and ICL4 Prevents the ΔF508-Induced Gating Defect.
In MSD2, it is ICL3 and ICL4 that are predicted to interact with NBD1 (14–16, 23, 24). Therefore, in ICL3, we substituted the human 964LNT966 amino acid sequence with the mouse residues 964ISK966 (LNT964-966ISK) (Fig. 4A). Like hmMSD2, LNT964-966ISK prolonged the interburst interval, but, importantly, the F508 deletion failed to further increase it (Fig. 4B). Mutating the human sequence to alanine (LNT964-966AAA) also prevented ΔF508 from lengthening the interburst interval.
Fig. 4.
Single-channel kinetic properties of CFTR MSD2 chimeras. (A) Amino acid alignment of human and mouse ICL3 and ICL4. Residues that differ from human residues are highlighted in yellow. The red boxes denote the mutations that we studied. (B–D) Single-channel kinetic properties of indicated variants. Data are the mean ± SEM (n = 3–13 membrane patches for each construct). Details are provided in the legend for Fig. 2. *ΔF508 variant differed from its WT counterpart (P < 0.05). #Difference of non-ΔF508 variant compared with WT hCFTR (P < 0.05).
In ICL4, we found that substituting the human P1072 with the mouse T1072 (P1072T) (Fig. 4A) prevented ΔF508 from further increasing the interburst interval (Fig. 4B). The P1072A and G1069R variants failed to prevent a ΔF508 effect on interburst interval. These data identify ICL3 and ICL4 as important for the CFTR-ΔF508 gating defect.
Inserting Mouse NBDs Reverses the Protective Effect of mICL4s.
Our results suggest that substituting mouse ICL sequences might have altered the interactions with the human NBDs, thereby preventing ΔF508 from impairing gating. If that were the case, we reasoned that replacing the human NBDs with mouse NBDs would eliminate the protective effect. Indeed, an mNBD2 eliminated the protective effect of the ICL3 mutation (LNT964-966ISK), and an mNBD1 eliminated the protective effect of the ICL4 mutant P1072T (Fig. 4B). These effects were specific because mNBD2 did not revert the effect of the ICL4 mutant and mNBD1 did not revert the effect of the ICL3 mutant. These data further suggest that interactions between the NBDs and the MSD2 ICLs influence the effect of the ΔF508 mutation on gating.
Discussion
mNBD1 Sequences Partially Rescue ΔF508-Induced Processing Defects.
We found that mNBD1 was permissive for CFTR-ΔF508 processing, and much of the effect was recapitulated by the RE segment. However, mutating I539 to the mouse sequence also partially rescued processing, and the mouse RE and I539T were additive in their effects. The RE is predicted to lie on the solvent-exposed surface of the NBD1, away from F508, whereas I539 lies close to the predicted NBD1 homodimer interface and faces the ICLs (12).
Other variants can also improve CFTR-ΔF508 biosynthesis. (i) A genetic approach identified second-site suppressor mutations, including I539T, G550E, R553M/Q, and R555K (18–21, 25, 26). None of these lie in the RE. (ii) Other studies examined NBD1 mutations that improve its solubility for crystallization, including residues in the RE, in the RI, between the RI and RE, and around the LSGGQ motif (12). Double or triple combinations of some solubilizing mutations improved CFTR-ΔF508 processing (27). (iii) Mutating residues comprising an ER retention motif enhanced hCFTR-ΔF508 biosynthesis (26, 28–30). (iv) Deleting the NBD1 RI also restored hCFTR-ΔF508 maturation (31).
Our data, together with these other studies, point to the sequence and structure of NBD1 as critical for hCFTR-ΔF508 processing. Substitutions, single amino acid mutations, and deletions scattered throughout the domain and largely located on its surface improved CFTR-ΔF508 maturation. In addition, earlier studies indicated that the ΔF508 mutation altered NBD1 structure little other than locally modifying the surface in the region of F508 (12). These observations may allow speculation about the mechanisms by which NBD1 modifications improve hCFTR-ΔF508 processing. First, the variety of modifications, their disparate positions in NBD1, and their surface localizations suggest that enhanced maturation may not be caused by a markedly altered hmNBD1-ΔF508 structure. Second, previous studies suggested that the region of NBD1 around F508 interacts with MSD2 and that by disrupting that association, ΔF508 impairs folding (15, 16, 19). Although it is possible that some NBD1 modifications repair an NBD1/MSD2 functional interaction, this mechanism does not explain the effect of the mouse RE or other widely dispersed NBD1 modifications. Moreover, as we discuss further below, interventions that do appear to modify NBD1/MSD2 interactions did not rescue CFTR-ΔF508 biosynthesis. Third, although it is possible that modifying the solvent-exposed NBD1 surface changes contacts with other parts of CFTR, some of the regions modified are not predicted to touch other CFTR domains. Fourth, NBD1 modifications might change interactions with chaperones or associated proteins, and thereby facilitate CFTR-ΔF508 progression through the biosynthetic pathway. This possibility could explain how the broad varieties of NBD1 modifications located far away from F508 enhance CFTR-ΔF508 maturation.
mMSD2 Sequences Prevent ΔF508-Induced Gating Defects.
We found that substituting mouse for human MSD2 and ICL sequences altered gating. These results are consistent with previous studies suggesting that ICL4 couples NBD activity to gating of the pore (16, 32). However, our data go further by showing that mouse ICL sequences prevented ΔF508 from further altering gating. In addition, substituting mouse for human NBDs reverted the effect; an mNBD2 eliminated the protective effect of an ICL3 mutation, and an mNBD1 eliminated the protective effect of an ICL4 mutation. These results suggest that the region around F508 links to the ICLs of MSD2. They also suggest that loss of F508 alters the interaction of both NBDs with MSD2.
Previous reports have also suggested a connection between the NBDs and the ICLs. The crystal structure of Sav1866 (a half transporter containing one MSD and one NBD) revealed that the MSD ICLs contact the NBDs of the opposite subunit (14). Cysteine cross-linking experiments in CFTR also predicted that NBD2 contacts ICL1 and ICL2 and that NBD1 contacts ICL3 and ICL4. Additional studies predicted that the region surrounding F508 interacts with residues in ICL4 (15, 16, 23, 24). Our findings, plus these observations, suggest that the two NBDs form a functional unit that interfaces with ICL3 and ICL4 to transmit conformational NBD changes to the channel gate. And the ΔF508 mutation disrupts that process.
mMSD2 Does Not Rescue CFTR-ΔF508 Processing, and mNBD1 Does Not Prevent the Gating Defects.
A striking finding of our study was the difference between the effect of mouse sequences on CFTR-ΔF508 gating and on processing. None of the substitutions modified both ΔF508-induced misprocessing and ΔF508-induced prolongation of the interburst interval. This distinction was exemplified by the I539T mutation. I539T improved processing; however, it not only failed to prevent the ΔF508 gating defect but actually further prolonged the interburst interval.
Other substitutions increased the Po of CFTR-ΔF508 by lengthening the burst duration rather than preventing ΔF508 from increasing the interburst interval. For example, hmNBD1, which partially improved processing, increased the Po with a prolonged burst duration instead of a shorter closed time. Other examples are the G550E and R555K mutations, which also partially rescued CFTR-ΔF508 processing and increased the Po by lengthening the burst duration. Although ΔF508 lengthened the interburst interval for both mutations, G550E reduced the magnitude of that increase (21, 26). In addition, a variant that combined G550E with R553M and R553K increased processing and current, although the effect on channel kinetics was not tested (33).
Our findings and consideration of earlier work suggest that the ΔF508 mutation affects two processes. First, it may impair NBD1 folding and/or stability, and that abnormality disrupts normal CFTR biosynthesis. This conclusion is consistent with studies on isolated NBD1 polypeptides (13, 20, 34). Second, it may impair interdomain interactions, and that abnormality disrupts normal CFTR gating. Thus, modifications of NBD1 or of MSD2 affect one or the other of these processes.
Implications for Therapeutics.
There is a substantial effort to identify compounds that improve CFTR-ΔF508 processing and/or function for therapeutic purposes. Our studies may have implications for such efforts. First, finding that I539T enhanced ΔF508-CFTR processing but reduced its channel activity suggests that a drug screening strategy that only detects cell surface CFTR-ΔF508 might miss adverse consequences for channel function. Second, the interface between NBD1 and ICL3 and ICL4 might be an important target for compounds that facilitate opening of ΔF508-CFTR. Third, because many different NBD1 modifications improved CFTR-ΔF508 maturation, it might be possible to target several different regions of NBD1 to enhance processing. In addition, finding that hmRE and I539T together improved CFTR-ΔF508 processing more than either alone suggests that simultaneously targeting more than one NBD1 site might be beneficial. Fourth, the diversity of NBD1 modifications that improve CFTR-ΔF508 biosynthesis suggested that modifications might alter interactions with chaperones and CFTR-associated proteins. Thus, targeting chaperones or a network of CFTR-associated proteins might be a reasonable therapeutic strategy. Fifth, because no one modification corrected both maturation and gating, perhaps more than a single agent will be required to correct all the CFTR-ΔF508 defects.
Materials and Methods
Human–Mouse Chimera Constructs.
Plasmids encoding human WT and ΔF508 CFTR have been described (21, 35). Mouse CFTR cDNA was kind gift from Christopher Boyd (University of Edinburgh, Edinburgh, Scotland) and Brandon Wainwright (University of Queensland, Brisbane, QLD, Australia). We subcloned human, mouse, and all chimera CFTR cDNAs into pcDNA3.1 (Invitrogen). Most hmCFTR chimera constructs were generated by ligating vector and insert fragments that were each products of blunt-end PCR (AccuPrime Pfx; Invitrogen). pcDNA3.1-hCFTR and pcDNA3.1-mCFTR were used as templates in all PCRs. All cDNAs were sequenced in their entirety to confirm proper sequence.
Vectors and Expression.
For protein-processing studies, 293T cells were transfected with pcDNA3.1-human, mouse, and the human–mouse chimeric CFTR with or without the ΔF508 mutation using Lipofectamine 2000 (Invitrogen). For patch-clamp studies, HeLa cells were transfected with CFTR and variant cDNA plasmids and Lipofectamine 2000 24 h after cells were seeded. Patch-clamp studies were done 24 h after transfection. Cells were cultured at 37 °C. For some of the CFTR-ΔF508 studies, cells were transferred to an incubator at 27 °C to correct the ΔF508-induced processing defect, which allowed us to do patch-clamp studies 24–48 h later.
Processing Studies.
293T cells were lysed 48 h after transfection, solubilized in lysis buffer [50 mM Tris (pH 7.4), 50 mM NaCl, 1% Nonidet P-40 and proteinase inhibitors, 2 μg/mL aprotinin, 7 μg/mL benzamidine⋅HCl, 1 μg/mL pepstatin A, and 2 μg/mL leupeptin), and centrifuged at 70,000 × g for 20 min at 4 °C. CFTR in the supernatant was immunoprecipitated with M3A7 and MM13-4 antibodies (Upstate Biotechnology), and then phosphorylated with γ-[32]ATP and the catalytic subunit of cAMP-dependent protein kinase (PKA; Promega) as described (35). Immunoprecipitates were electrophoresed on 6% SDS/PAGE, dried, and exposed to a phosphor screen for visualization (Fuji7000).
Patch-Clamp Studies.
We used excised, inside-out membrane patches. The pipette (extracellular) solution contained the following: 140 mM N-methyl-d-glucamine, 100 mM l-aspartic acid, 3 mM MgCl2, 5 mM CaCl2, and 10 mM Tricine (pH 7.3) with HCl (final Cl− concentration ∼51 mM). The bath (intracellular) solution contained the following: 140 mM N-methyl-d-glucamine, 3 mM MgCl2, 1 mM cesium EGTA, and 10 mM Tricine (pH 7.3) with HCl (final Cl− concentration 140 mM). Following patch excision, channels were activated with the catalytic subunit of cAMP-dependent protein kinase (PKA; Calbiochem, EMD Chemicals, Inc.) and ATP. Unless otherwise specified, PKA was present in all cytosolic solutions that contained ATP. All nucleotides were from Sigma–Aldrich. ATP was added as the Mg2+ salt. The holding voltage was −50 to −100 mV for single-channel experiments. Experiments were performed at room temperature (23–26 °C).
An Axopatch 200B amplifier (Axon Instruments, Inc.) was used for voltage clamping and current recording, and the pCLAMP software package (version 9.1; Axon Instruments, Inc.) was used for data acquisition and analysis. Data were digitized at 5 kHz. Current recordings were low-pass filtered at 500 Hz using an eight-pole Bessel filter (Model 900; Frequency Devices, Inc.) for analysis and at 100 Hz for display in figures (10 Hz for mCFTR display traces). Single-channel analysis was performed as previously described (36, 37) with a burst delimiter of 20 ms. Events ≤4 ms in duration were ignored. mCFTR subconductance opening was not taken into account in our studies (Fig. 3).
Supplementary Material
Acknowledgments
We thank Philip Karp, Pamela Hughes, Ping Tan, Huiyu Gong, and Theresa Mayhew for excellent technical assistance. We thank the In Vitro Models and Cell Culture Core for assistance, supported, in part, by National Heart, Lung, and Blood Institute Grants HL091842 and HL51670; Cystic Fibrosis Foundation Grants R458-CR02 and ENGLH9850; and National Institute of Diabetes and Digestive and Kidney Disease Grant DK54759. This work was also supported by National Institutes of Health Grant HL61234 and Cystic Fibrosis Foundation Grant OSTEDG06G0. M.J.W. is a Howard Hughes Medical Institute Investigator.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1120065109/-/DCSupplemental.
References
- 1.Riordan JR, et al. Identification of the cystic fibrosis gene: Cloning and characterization of complementary DNA. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- 2.Welsh MJ, Tsui LC, Boat TF, Beaudet AL. In: Cystic fibrosis. The Metabolic and Molecular Basis of Inherited Disease. 7th Ed. Scriver CR, Beaudet AL, Sly WS, Valle D, editors. New York: McGraw–Hill; 1995. pp. 3799–3876. [Google Scholar]
- 3.Cheng SH, et al. Defective intracellular transport and processing of CFTR is the molecular basis of most cystic fibrosis. Cell. 1990;63:827–834. doi: 10.1016/0092-8674(90)90148-8. [DOI] [PubMed] [Google Scholar]
- 4.Lukacs GL, et al. Conformational maturation of CFTR but not its mutant counterpart (delta F508) occurs in the endoplasmic reticulum and requires ATP. EMBO J. 1994;13:6076–6086. doi: 10.1002/j.1460-2075.1994.tb06954.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ward CL, Kopito RR. Intracellular turnover of cystic fibrosis transmembrane conductance regulator. Inefficient processing and rapid degradation of wild-type and mutant proteins. J Biol Chem. 1994;269:25710–25718. [PubMed] [Google Scholar]
- 6.Skach WR. Defects in processing and trafficking of the cystic fibrosis transmembrane conductance regulator. Kidney Int. 2000;57:825–831. doi: 10.1046/j.1523-1755.2000.00921.x. [DOI] [PubMed] [Google Scholar]
- 7.Denning GM, et al. Processing of mutant cystic fibrosis transmembrane conductance regulator is temperature-sensitive. Nature. 1992;358:761–764. doi: 10.1038/358761a0. [DOI] [PubMed] [Google Scholar]
- 8.Dalemans W, et al. Altered chloride ion channel kinetics associated with the delta F508 cystic fibrosis mutation. Nature. 1991;354:526–528. doi: 10.1038/354526a0. [DOI] [PubMed] [Google Scholar]
- 9.Lukacs GL, et al. The delta F508 mutation decreases the stability of cystic fibrosis transmembrane conductance regulator in the plasma membrane. Determination of functional half-lives on transfected cells. J Biol Chem. 1993;268:21592–21598. [PubMed] [Google Scholar]
- 10.Swiatecka-Urban A, et al. The short apical membrane half-life of rescued ΔF508-cystic fibrosis transmembrane conductance regulator (CFTR) results from accelerated endocytosis of ΔF508-CFTR in polarized human airway epithelial cells. J Biol Chem. 2005;280:36762–36772. doi: 10.1074/jbc.M508944200. [DOI] [PubMed] [Google Scholar]
- 11.Lewis HA, et al. Structure of nucleotide-binding domain 1 of the cystic fibrosis transmembrane conductance regulator. EMBO J. 2004;23:282–293. doi: 10.1038/sj.emboj.7600040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewis HA, et al. Impact of the deltaF508 mutation in first nucleotide-binding domain of human cystic fibrosis transmembrane conductance regulator on domain folding and structure. J Biol Chem. 2005;280:1346–1353. doi: 10.1074/jbc.M410968200. [DOI] [PubMed] [Google Scholar]
- 13.Thibodeau PH, Brautigam CA, Machius M, Thomas PJ. Side chain and backbone contributions of Phe508 to CFTR folding. Nat Struct Mol Biol. 2005;12:10–16. doi: 10.1038/nsmb881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dawson RJ, Locher KP. Structure of a bacterial multidrug ABC transporter. Nature. 2006;443:180–185. doi: 10.1038/nature05155. [DOI] [PubMed] [Google Scholar]
- 15.Loo TW, Bartlett MC, Clarke DM. Processing mutations disrupt interactions between the nucleotide binding and transmembrane domains of P-glycoprotein and the cystic fibrosis transmembrane conductance regulator (CFTR) J Biol Chem. 2008;283:28190–28197. doi: 10.1074/jbc.M805834200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Serohijos AW, et al. Phenylalanine-508 mediates a cytoplasmic-membrane domain contact in the CFTR 3D structure crucial to assembly and channel function. Proc Natl Acad Sci USA. 2008;105:3256–3261. doi: 10.1073/pnas.0800254105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ostedgaard LS, et al. Processing and function of CFTR-DeltaF508 are species-dependent. Proc Natl Acad Sci USA. 2007;104:15370–15375. doi: 10.1073/pnas.0706974104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeCarvalho AC, Gansheroff LJ, Teem JL. Mutations in the nucleotide binding domain 1 signature motif region rescue processing and functional defects of cystic fibrosis transmembrane conductance regulator delta f508. J Biol Chem. 2002;277:35896–35905. doi: 10.1074/jbc.M205644200. [DOI] [PubMed] [Google Scholar]
- 19.He L, et al. Restoration of domain folding and interdomain assembly by second-site suppressors of the DeltaF508 mutation in CFTR. FASEB J. 2010;24:3103–3112. doi: 10.1096/fj.09-141788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoelen H, et al. The primary folding defect and rescue of ΔF508 CFTR emerge during translation of the mutant domain. PLoS ONE. 2010;5:e15458. doi: 10.1371/journal.pone.0015458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Teem JL, Carson MR, Welsh MJ. Mutation of R555 in CFTR-ΔF508 enhances function and partially corrects defective processing. Receptors Channels. 1996;4:63–72. [PubMed] [Google Scholar]
- 22.Scott-Ward TS, et al. Chimeric constructs endow the human CFTR Cl- channel with the gating behavior of murine CFTR. Proc Natl Acad Sci USA. 2007;104:16365–16370. doi: 10.1073/pnas.0701562104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He L, et al. Multiple membrane-cytoplasmic domain contacts in the cystic fibrosis transmembrane conductance regulator (CFTR) mediate regulation of channel gating. J Biol Chem. 2008;283:26383–26390. doi: 10.1074/jbc.M803894200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mornon JP, Lehn P, Callebaut I. Atomic model of human cystic fibrosis transmembrane conductance regulator: Membrane-spanning domains and coupling interfaces. Cell Mol Life Sci. 2008;65:2594–2612. doi: 10.1007/s00018-008-8249-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teem JL, et al. Identification of revertants for the cystic fibrosis delta F508 mutation using STE6-CFTR chimeras in yeast. Cell. 1993;73:335–346. doi: 10.1016/0092-8674(93)90233-g. [DOI] [PubMed] [Google Scholar]
- 26.Roxo-Rosa M, et al. Revertant mutants G550E and 4RK rescue cystic fibrosis mutants in the first nucleotide-binding domain of CFTR by different mechanisms. Proc Natl Acad Sci USA. 2006;103:17891–17896. doi: 10.1073/pnas.0608312103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pissarra LS, et al. Solubilizing mutations used to crystallize one CFTR domain attenuate the trafficking and channel defects caused by the major cystic fibrosis mutation. Chem Biol. 2008;15:62–69. doi: 10.1016/j.chembiol.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 28.Hegedus T, et al. F508del CFTR with two altered RXR motifs escapes from ER quality control but its channel activity is thermally sensitive. Biochim Biophys Acta. 2006;1758:565–572. doi: 10.1016/j.bbamem.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 29.Roy G, Chalfin EM, Saxena A, Wang X. Interplay between ER exit code and domain conformation in CFTR misprocessing and rescue. Mol Biol Cell. 2010;21:597–609. doi: 10.1091/mbc.E09-05-0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim Chiaw P, et al. Functional rescue of DeltaF508-CFTR by peptides designed to mimic sorting motifs. Chem Biol. 2009;16:520–530. doi: 10.1016/j.chembiol.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 31.Aleksandrov AA, et al. Regulatory insertion removal restores maturation, stability and function of DeltaF508 CFTR. J Mol Biol. 2010;401:194–210. doi: 10.1016/j.jmb.2010.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cotten JF, Ostedgaard LS, Carson MR, Welsh MJ. Effect of cystic fibrosis-associated mutations in the fourth intracellular loop of cystic fibrosis transmembrane conductance regulator. J Biol Chem. 1996;271:21279–21284. doi: 10.1074/jbc.271.35.21279. [DOI] [PubMed] [Google Scholar]
- 33.Thibodeau PH, et al. The cystic fibrosis-causing mutation deltaF508 affects multiple steps in cystic fibrosis transmembrane conductance regulator biogenesis. J Biol Chem. 2010;285:35825–35835. doi: 10.1074/jbc.M110.131623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang C, et al. Integrated biophysical studies implicate partial unfolding of NBD1 of CFTR in the molecular pathogenesis of F508del cystic fibrosis. Protein Sci. 2010;19:1932–1947. doi: 10.1002/pro.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ostedgaard LS, Zeiher B, Welsh MJ. Processing of CFTR bearing the P574H mutation differs from wild-type and ΔF508-CFTR. J Cell Sci. 1999;112:2091–2098. doi: 10.1242/jcs.112.13.2091. [DOI] [PubMed] [Google Scholar]
- 36.Carson MR, Travis SM, Welsh MJ. The two nucleotide-binding domains of cystic fibrosis transmembrane conductance regulator (CFTR) have distinct functions in controlling channel activity. J Biol Chem. 1995;270:1711–1717. doi: 10.1074/jbc.270.4.1711. [DOI] [PubMed] [Google Scholar]
- 37.Cotten JF, Welsh MJ. Covalent modification of the nucleotide binding domains of cystic fibrosis transmembrane conductance regulator. J Biol Chem. 1998;273:31873–31879. doi: 10.1074/jbc.273.48.31873. [DOI] [PubMed] [Google Scholar]
- 38.Lansdell KA, Kidd JF, Delaney SJ, Wainwright BJ, Sheppard DN. Regulation of murine cystic fibrosis transmembrane conductance regulator Cl- channels expressed in Chinese hamster ovary cells. J Physiol. 1998;512:751–764. doi: 10.1111/j.1469-7793.1998.751bd.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.