Abstract
Cyanobacteria are a prolific source of secondary metabolites, including compounds with toxic and enzyme-inhibiting activities. Microcystins and nodularins are the end products of a secondary metabolic pathway comprised of mixed polyketide synthases and nonribosomal peptide synthetases. Both peptides are potent natural toxins produced by distantly related genera of cyanobacteria. Horizontal gene transfer is thought to play a role in the sporadic distribution of microcystin producers among cyanobacteria. Our phylogenetic analyses indicate a coevolution of housekeeping genes and microcystin synthetase genes for the entire evolutionary history of the toxin. Hence they do not corroborate horizontal transfer of genes for microcystin biosynthesis between the genera. The sporadic distribution of microcystin synthetase genes in modern cyanobacteria suggests that the ability to produce the toxin has been lost repeatedly in the more derived lineages of cyanobacteria. The data we present here strongly suggest that the genes encoding nodularin synthetase are recently derived from those encoding microcystin synthetase.
Toxic cyanobacterial blooms have been reported from fresh and brackish water bodies for >100 years (1, 2). Toxic blooms have led to the deaths of wild and domestic animals all over the world (see a list in ref. 3) and are a health risk for human beings via recreational or drinking water (4). For example, cyanobacterial hepatotoxins, microcystins, have been held responsible for the deaths of 60 patients after renal dialysis with water contaminated with the toxin in Brazil (5). Chronic low-level exposure to microcystins has been suspected to cause human hepatocellular carcinoma in China (4). The World Health Organization has set a provisional guideline value of 1.0 μg·liter–1 for this toxin (6).
Microcystins, cyclic heptapeptide hepatotoxins, are by far the most prevalent of the cyanobacterial toxins and are produced by strains of the distantly related cyanobacterial genera Microcystis, Anabaena, Planktothrix, and more rarely Anabaenopsis, Haplosiphon, and Nostoc (2). In brackish waters, the cyclic pentapeptide hepatotoxin, nodularin, is produced by strains of the genus Nodularia (2). Microcystins and nodularin are potent inhibitors of eukaryotic protein phosphatases type 1 and 2A (7, 8). In addition to these peptide toxins, cyanobacteria have been found to produce a wide variety of linear (e.g., aeruginosins and microginins) and cyclic peptides (e.g., anabaenopeptins, nostopeptolides, and anabaenopeptilides), which may not be acutely toxic but have other bioactivities, such as serine protease inhibition (9, 10).
Most small cyanobacterial peptides contain unusual, nonproteinogenic amino acids and are synthesized by nonribosomal enzyme complexes (11–13). The enzyme complex responsible for microcystin biosynthesis is encoded by the microcystin synthetase gene cluster. It spans ≈55 kb and includes genes for peptide synthetases, polyketide synthases, mixed peptide synthetase and polyketide synthases, and tailoring enzymes (14–16). Insertional inactivation of a microcystin synthetase gene abolished the production of microcystin (17). All genera with microcystin-producing strains also contain related strains that lack the ability to produce this toxin. It is now thought that the difference between microcystin-producing (toxic) and nonproducing (nontoxic) strains of cyanobacteria lies primarily in the presence or absence of microcystin synthetase gene cluster (14, 18).
Horizontal gene transfer is now widely acknowledged as an important force shaping the genome structure of bacteria. Recent lateral gene transfer or a series of gene losses both have been proposed to explain the sporadic distribution of toxic strains of Microcystis (19–22). This hypothesis remains uncorroborated by phylogenetic analyses to date. It is not clear how distantly related genera of cyanobacteria gained the ability to produce microcystins. However, the presence of similar secondary metabolites in distantly related groups of organisms is usually attributed to horizontal gene transfer or convergent evolution. Therefore, the possibility that the microcystin synthetase genes in the different genera are ancient and related to one another through common descent is of particular interest.
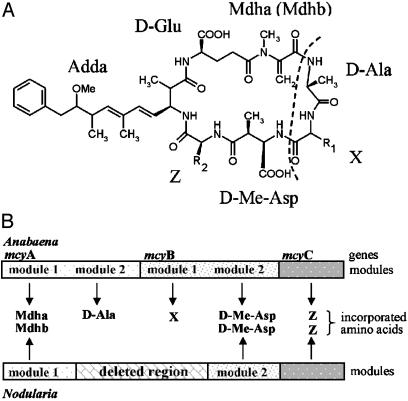
Nodularins are produced solely by strains of the genus Nodularia (2). Similarities in the chemical structures (Fig. 1) and biological action of microcystins and nodularins indicate that these compounds are closely related (2). The exact relationship between nodularins and microcystins is ambiguous (23), and it is not clear whether the structural similarity between nodularins and microcystins is reflected at the genetic level. However, a similar biosynthetic pathway is anticipated to produce both peptides based on these structural similarities (23). Recently, it has been suggested that microcystin synthetase genes are derived from nodularin synthetase genes (16).
Fig. 1.
Comparison of the chemical structures of microcystins and nodularins and the hypothetical events leading to nodularin synthetase gene cluster. (A) The general structure of microcystins and nodularin. Microcystin is a cyclic peptide containing seven amino acids, d-Ala-X-d-MeAsp-Z-Adda-d-Glu-Mdha, where X and Z represent variable l-amino acids, d-Me-Asp is d-erythro-β-methylaspartic acid, Mdha is N-methyldehydroalanine, and Adda is β-amino acid 3-amino-9-methoxy-2,6,8-trimethyl-10-phenyldeca-4,6-dienoic acid. Nodularin differs from microcystins by lacking the amino acids d-Ala and X and having N-methyldehydrobutyrine (Mdhb) in place of Mdha. The dashed line indicates the two amino acids absent in nodularins. (B) A schematic diagram illustrating the peptide synthetase region (mcyA-mcyC)of Anabaena and the proposed organization of the homologous region of the gene cluster for nodularin biosynthesis in Nodularia if evolved by a mutation changing the substrate specificity of the first module of mcyA and the deletion of the last module of mcyA and the first module of mcyB (see text).
The construction and comparison of phylogenetic trees are perhaps the best ways to assess the contribution of horizontal gene transfer to the evolutionary history of a gene family (24). Incongruence is taken to indicate a role for horizontal gene transfer, whereas congruence is consistent with descent through common ancestry. Therefore, to resolve the relationship between microcystin and nodularin synthetase genes and the role of horizontal gene transfer in the evolutionary history of these two compounds, we undertook a molecular phylogenetic study. We compared two data sets comprised of genes involved in primary metabolism and genes involved directly in the synthesis of microcystins and nodularins. These sequences were analyzed and tested for congruence.
Materials and Methods
Taxon Sampling. We obtained strains representative of known producers of microcystins and nodularins available in public culture collections. Genomic DNA was extracted from 36 strains of the filamentous heterocystous genera Anabaena, Nodularia, and Nostoc, the filamentous genus Planktothrix, and the unicellular genus Microcystis (Table 1, which is published as supporting information on the PNAS web site). Just two strains of the genus Nostoc are known to produce microcystins (25, 26), one of which was available for this study.
Amplification and Sequencing. To study the evolution of this biosynthetic system in cyanobacteria we chose three regions of the microcystin synthetase (mcy) gene cluster. A fragment of 291–297 bp from the mcyA gene was amplified with mcyA-Cd 1R (5′-aaaagtgttttattagcggctcat-3′) and mcyA-Cd 1F (5′-aaaattaaaagccgtatcaaa-3′) primers (27). This part of the mcyA gene encodes part of the condensation domain, which catalyzes a condensation reaction to form a peptide bond between the growing peptide and d-alanine. An 818-bp region of the mcyD gene was amplified with mcyDF (5′-gatccgattgaattagaaag-3′) and mcyDR (5′-gtattccccaagattgcc-3′) primers. This fragment of the mcyD gene encodes parts of both the β-ketoacyl synthase and the acyltransferase domains. An 809-to 812-bp region of the mcyE gene was amplified with the mcyE-F2 (5′-gaaatttgtgtagaaggtgc-3′) and mcyE-R4 (5′-aattctaaagcccaaagacg-3′) primers. The mcyE gene fragment codes for a partial adenylation domain and a phosphopantetheine-binding site, the region, which activates glutamic acid.
To investigate the role of horizontal gene transfer in the evolution of the microcystin biosynthetic gene cluster, we amplified and sequenced the 16S rRNA and rpoC1 genes. The rpoC1 gene fragment of 731 bp was amplified with degenerate primers RF (5′-tgggghgaaagnacaytncctaa-3′) and RR (5′-gcaaancgtccnccatcyaaytgba-3′). PCRs for mcyD, mcyE, and rpoC1 were performed in a 20-μl final volume containing 1 μl of DNA, 1× DynaZyme II PCR buffer, 250 μM of each deoxynucleotide, 0.5 μM of both PCR primers, and 0.5 unit of DynaZyme II DNA polymerase (Finnzymes, Espoo, Finland). The following protocol was used: 95°C, 3 min; 30× (94°C, 30 sec; 56°C, 30 sec; 72°C, 1 min); and 72°C, 10 min. The mcyE PCR products of Nodularia sp. strains were cloned with the TOPO TA cloning kit (Invitrogen) according to the manufacturer's instructions. A region containing the 16S rRNA gene and the internal transcribed spacer 1 was amplified by using primers and conditions as described (28) from strains for which the 16S rRNA sequence data were not available.
The mcyD and mcyE gene products were sequenced directly with primers used for amplification except for the cloned mcyE sequences of Nodularia sp. strains, which were sequenced with primers anchored in the pCR2.1-TOPO vector, M13F (–20) and M13R. At least three clones of the mcyE PCR product were sequenced from each strain of Nodularia to identify potential errors introduced by nucleotide misincorporation by the DynaZyme II enzyme. The rpoC1 gene products were sequenced with the amplification primers and two additional internal sequencing primers RintF (5′-gatatgcccctgcgggatgt-3′) and RintR (5′-acatcccgcaggggcatatc-3′). The 16S rRNA region of the amplified PCR products was sequenced directly by using sets of internal primers (29). Sequencing of the mcyD, mcyE, and 16S rRNA genes was performed by Genome Express (Meylan, France). The rpoC1 products were sequenced with Applied Biosystems PRISM 310 Genetic Analyzer. The mcyA sequences were assembled as described by Hisbergues et al. (27). The chromatograms of mcyD, mcyE, rpoC1, and 16S rRNA gene sequences were checked and edited with the chromas 2.2 program (Technelysium Pty, Tewantin, Australia). Contig assembly and alignment of the sequences were performed with bioedit sequence alignment editor (30).
Sequence Divergence. We calculated the number of nonsynonymous substitutions per nonsynonymous site (KA) and the number of synonymous substitutions per synonymous site (KS) by using mega 2.1 (31). A KA/KS ratio >1 indicates positive selection for advantageous mutations, whereas a KA/KS ratio >1 indicates purifying selection to prevent the spread of detrimental mutations (32).
Phylogenetic Analyses. Phylogenetic analyses were conducted by using paup* (33). We investigated competing hypotheses on the sporadic distribution of microcystin synthetase genes and the evolutionary relationship between nodularins and microcystins by reconstructing the evolutionary history of microcystin and nodularin synthetase genes. A 1,799-bp data set comprised of two peptide synthetase genes and one polyketide synthetase gene was assembled. The aligned data sets were based on sequences with the following lengths: mcyA, 250 bp; mcyD, 780 bp; and mcyE, 769 bp. These sequences were combined with the sequence available from Microcystis aeruginosa PCC 7806 (15) and Planktothrix agardhii NIVA-CYA 126/8 (16). Primer sequences and ambiguous regions of the alignments were excluded.
Outgroups for the mcyA, mcyE, and mcyD gene fragments were obtained by identifying homologues in blast searches (Table 2, which is published as supporting information on the PNAS web site). A χ2 test was performed to test for base frequency compositional bias. Outgroup sequences, which passed the χ2 test, belonged to the nostopeptolide gene cluster from Nostoc sp. GSV224 (34), the anabaenopeptilide gene cluster from Anabaena sp. 90 (35), and putative polyketide synthase and peptide synthetase gene clusters from the complete genomes of Nostoc punctiforme PCC 73102 (www.jgi.doe.gov) and Nostoc sp. PCC 7120 (36). Only conserved and reliably aligned sequence regions from the outgroup sequences were used to minimize potential phylogenetic reconstruction artifacts derived from the use of distant outgroups (37). Concordance of the genes used to construct the data sets was evaluated with the conservative partition homogeneity test (38) implemented with paup* (33). We used random taxon addition (10 replicates), tree bisection-reconnection branch-swapping, and heuristic searches with 1,000 repartitions of the data. The partition homogeneity test indicated that these three genes could be combined (P = 0.119). The data from all three genes were concatenated to increase the amount of information available in phylogenetic analyses.
To investigate the role of horizontal gene transfer in the distribution of microcystin synthetase genes among cyanobacteria we assembled a 2,186-bp data set comprised of 16S rRNA (1,455 bp) and rpoC1 (731 bp) sequences from the same set of taxa. These genes are conserved and widely used as tools for phylogenetic classification. The 16S rRNA and rpoC1 gene sequences of the early branching cyanobacteria Gloeobacter violaceus PCC 7421 and Thermosynechococcus elongatus BP-1 were used as outgroups. No incongruence between the 16S rRNA and rpoC1 topologies could be found in the partition homogeneity test (38), so the data from these two genes were combined (P = 0.9980). Phylogenetic trees were inferred by using minimum-evolution, maximum-parsimony, and maximum-likelihood optimization criteria. To test the stability of monophyletic groups 2,000 bootstrap replicates were analyzed. In distance analyses, the minimum-evolution method using LogDet distances (39) with 10 heuristic searches, random additionsequence starting trees, and tree bisection and reconnection branch arrangements was used. Maximum-likelihood and maximum-parsimony analyses were performed with the same heuristic search procedure as minimum evolution. However, in maximum likelihood, bootstrap analyses were performed without the 10 random sequence addition replicates. The GTR model of DNA substitution with a γ distribution of rates and constant sites removed in proportion to base frequencies was used in maximum-likelihood analyses.
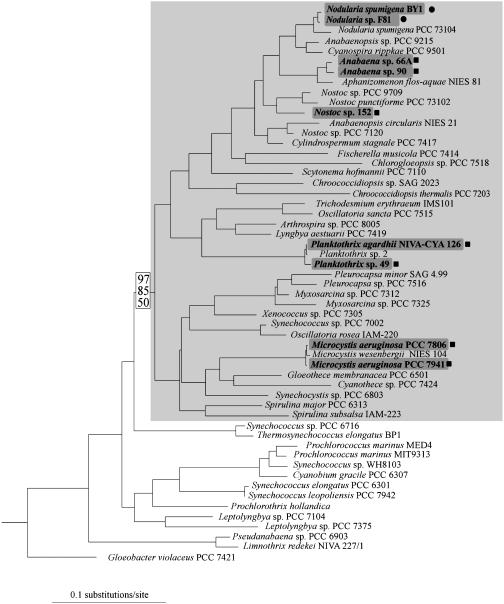
To illustrate the sporadic distribution of microcystin and nodularin-producing cyanobacteria, we constructed a maximum-likelihood tree based on the 16S rRNA gene sequences from 55 cyanobacterial strains and three outgroup species. The distribution of known microcystin and nodularin producers was mapped to this tree. The bootstrap support for the monophyly of the lineages containing known microcystin and nodularin producers was assessed by using maximum parsimony and minimum evolution.
Templeton's test (40) was used to compare alternative phylogenetic hypothesis concerning the relative positions of nodularin and microcystin synthetase genes (16). Templeton's test was done in paup* by using the conservative two-tailed Wilcoxon rank sum test.
Results
We analyzed the 16S rRNA and rpoC1 data set and the microcystin synthetase gene data set separately with distance, maximum-parsimony, and maximum-likelihood optimization criteria. The maximum-likelihood topologies generated from each data set were perfectly congruent (Fig. 2). This finding was also true of analyses performed separately with the mcyA (26 taxa), mcyD (18 taxa), and mcyE (30 taxa) genes in both rooted and unrooted analysis (data not shown). Bootstrap analyses were conducted to measure the stability of the observed phylogenetic patterns and revealed two well-supported topologies (Fig. 2). The conservative parsimony-based partition homogeneity test (38) found no support for incongruence between these two data sets (P = 1.000). The sequence divergence between the taxa included in this analysis from the 16S rRNA gene and rpoC1 data set and the microcystin synthetase gene data set were comparable (data not shown). Sequence divergences in the microcystin synthetase gene data set were much higher than expected in an evolutionary scenario, favoring recent horizontal gene transfer as a mechanism to explain the sporadic distribution of microcystin producers among cyanobacteria. To determine whether the microcystin synthetase genes are under positive or negative selection pressure, we compared the number of nonsynonymous substitutions per nonsynonymous site (KA) to the number of synonymous substitutions per synonymous site (KS). The KA/KS ratio was well below 1 in pairwise comparisons from representative strains of each genus. A low KA/KS ratio is indicative of purifying selection in which deleterious mutations affecting the protein sequence are selected against and is consistent with an ancient origin of the microcystin synthetase genes.
Fig. 2.
Congruence between the 16S rRNA and rpoC1 data set and the microcystin synthetase gene data set. (A) A maximum-likelihood tree based on the 16S rRNA and rpoC1 data set (–lnL = 8004.26493). (B) A maximum-likelihood tree based on the mcyA, mcyD, and mcyE data set (–lnL = 8781.50660). In both trees, branch lengths are proportional to sequence change. Minimum-evolution, maximum-parsimony, and maximum-likelihood bootstrap values from 2,000 bootstrap replicates are at the nodes. Outgroup taxa used in the construction of these trees are not shown.
Our data also resolve the relationship between nodularin and microcystin synthetase genes. The topological congruence we observe suggests that nodularin synthetase genes are derived from microcystin synthetase genes (Fig. 2). Rearranging the ingroup topology to make the nodularin and microcystin synthetase genes reciprocally monophyletic resulted in significantly worse trees with the Templeton test (P = 0.02). This finding suggests that nodularin synthetase genes are derived from microcystin synthetase genes (Fig. 2).
The 16S rRNA gene was used to reconstruct the distribution of known microcystin and nodularin producers and revealed a robust maximum-likelihood tree (Fig. 3 and Table 3, which is published as supporting information on the PNAS web site). Known microcystin- and nodularin-producing taxa fell into a clade containing the majority of living cyanobacteria including representatives from all five orders of cyanobacteria and some of the more speciose lineages of cyanobacteria. Bootstrap support for this clade was high in distance but low in parsimony analyses (Fig. 3).
Fig. 3.
A maximum-likelihood tree based on the 16S rRNA gene illustrating the sporadic distribution of cyanobacterial genera known to produce microcystins (–lnL = 17594.65327). Strains of the genera Planktothrix, Microcystis, Anabaena, and Nostoc produce microcystins (▪), whereas strains of the genus Nodularia produce nodularins (•). All strains in the lineage of cyanobacteria, which originally possessed microcystin synthetase genes, are contained within a light gray box. Support for this clade is given as follows: minimum evolution with LogDet distances, minimum evolution with maximum-likelihood distances, and maximum-parsimony bootstrap values. Outgroup taxa used in the construction of this tree are not shown.
Discussion
The high degree of congruence between the microcystin synthetase gene data set and the 16S rRNA and rpoC1 data set and comparable levels of sequence divergence between the two data sets are consistent with an ancient origin of microcystins. This finding indicates that the phylogenetic marker genes and the microcystin synthetase genes have coevolved for the entire evolutionary history of this toxin. Our results show that microcystin synthetase genes were present in the last common ancestor of a large number of cyanobacteria. Therefore, the sporadic distribution of microcystin synthetase genes in modern cyanobacteria suggests that the ability to produce the toxin has been lost repeatedly in the more derived lineages of cyanobacteria. On the other hand, strains of cyanobacteria not previously suspected of producing microcystins may retain the genes necessary for their synthesis. Because microcystins and nodularins are highly toxic to humans and pose a serious health risk to water users, more taxa should be carefully checked for the presence of these toxins. This would be particularly important for strains of genera like Spirulina, Arthrospira, and Aphanizomenon that are commonly used in health food supplements (41).
It is possible that the congruence we observed is a phylogenetic artifact. Unexpectedly high levels of sequence divergence could be caused by accelerated evolution with positive selection pressure promoting amino acid divergence (42). However, low levels of nonsynonymous substitutions suggest that this is not the case. Phylogenetic artifacts are also known to result from the use of distant outgroups, long-branch attraction, heterogeneous base composition, and site-specific rate variation (37). However, in this study midpoint rooting also resulted in congruent trees in distance, maximum parsimony, and maximum likelihood. Congruence between maximum parsimony and methods thought to be more robust to long-branch attraction such as distance and maximum likelihood suggests that long-branch attraction does not play a role in the observed topologies (Fig. 2). The results of the LogDet method, which addresses potential concerns with heterogeneity of base composition (37), agree with results for maximum likelihood and maximum parsimony (Fig. 2). Maximum-likelihood analyses accounted for site-specific rate variation with a γ distribution of rates and resulted in a tree that was congruent with the trees generated from distance and maximum-parsimony methods (Fig. 2). Limited taxon sampling may play a role in the congruence we observe. The same topology we have found (Fig. 2.) is found in 16S rRNA trees with larger taxon sampling. However, we have sampled all available producers of microcystins and nodularins, but confirmation of this will have to await the discovery of additional genera capable of making microcystins.
Previous studies have suggested that horizontal gene transfer may play a role in the distribution of hepatotoxic strains within the genus Microcystis (19–22). However, there is no clear experimental evidence, which favors horizontal gene transfer over gene loss as a mechanism to explain the sporadic distribution of toxin-producing strains in the genus Microcystis. Our results suggest that the microcystin synthetase genes were originally present and that nontoxic strains of Microcystis have lost the ability to produce microcystins. Our results also indicate that microcystin biosynthesis is a very ancient secondary metabolic pathway and that there is no lateral transfer of mcy gene clusters between the genera. However, they do not rule out the possibility that parts of the microcystin synthetase gene cluster are of more recent origin and might be laterally transferred between strains within a genus. Indeed, the existence of many microcystin variants could imply a rapid evolution of certain gene domains. Recombination between mcyC and the first module of mcyB is thought to contribute to at least some of the variation observed in peptide structure in strains of the genus Microcystis (22). However, our results imply that the microcystin synthetase genes themselves are ancient and, therefore, it is likely that the recombination event is relatively recent.
Our study showed that nodularin synthetase genes clustered closely with microcystin synthetase genes of other filamentous heterocystous cyanobacteria (Anabaena and Nostoc) (Fig. 2). It is anticipated here that nodularin synthetase genes were formed from the ancestral microcystin synthetase gene set, through a relatively recent deletion of the two peptide synthetase modules that catalyze the incorporation of d-Ala and X into microcystin (the last mcyA module and the first mcyB module) and by mutation changing the substrate specificity coded by the first module of mcyA (Fig. 1B). There are >65 reported structural variants of microcystins (2). Our study suggests that nodularins can be regarded as the most extreme structural variants of microcystins. This recent origin of nodularins from microcystins proposed here is consistent with the production of nodularins by a single cyanobacterial genus and the limited structural variation of nodularins in comparison to microcystins (2).
Microcystins are one of the few known natural examples of combined polyketide synthase and peptide synthetase systems (15). The rapamycin (43), bleomycin (44), yersiniabactin (45), and mycobactin (46) biosynthetic gene clusters are known to contain both polyketide synthase and peptide synthetase modules. More recently, a small group of hybrid biosynthetic gene clusters have been identified, in which polyketide synthase and peptide synthetase modules exist within the same ORF. These hybrid systems occur in the mycosubtilin (47), myxothiazol (48), iturin A (49), and microcystin synthetases (15). Little is known about the evolution of these true hybrid polyketide/peptide synthetases. Studies have suggested that the ketosynthase domain in mixed polyketide and peptide synthetase genes have a separate evolutionary origin to type I polyketide synthases (50, 51). However, it is unclear whether the combination of these two systems is of recent origin. Congruence between the polyketide synthase and peptide synthetase portions of the mcy gene cluster and the 16S rRNA and rpoC1 data set demonstrates that the combination of these two systems is an ancient collaboration in the production of microcystins.
The function of nodularins and microcystins is currently unresolved. Microcystins are widely believed to have evolved in response to grazing pressure by zooplankton (52). Metazoans such as copepods and cladocerans are often envisaged as the target organisms of microcystins (52). Molecular clocks set a divergence time of 1,576 million years ago for the crown eukaryotic lineages (53). However, the oldest fossils of filamentous akinete-forming cyanobacteria are dated to 1,500 million to 2,000 million years ago (54, 55). There is controversy whether such microfossils, in particular the oldest known fossils, are truly representatives of modern cyanobacteria (56). However, these fossils of akinete-forming cyanobacteria suggest that the Anabaena, Nostoc, and Nodularia genera and thus, the common ancestor of microcystin-producing cyanobacteria, are at least this old and that microcystin production may predate the metazoan lineage. Thus, microcystins and nodularins may have evolved to serve other functions such as siderophoric scavenging of trace metals (57) or signaling and gene regulation (58). In the context of function, it remains puzzling why closely related strains may differ with respect to their ability to produce microcystins and nodularins. A possible explanation may arise from the fact that strains that do not harbor genes for the biosynthesis of microcystins or nodularins in their genomes do contain genes for the synthesis of other nonribosomal peptides. These other peptides may serve the same or at least similar functions as microcystins and nodularins, respectively. Any model that attempts to explain why cyanobacteria make these peptides must now account for the age of the toxin synthetase genes. We anticipate here that the high levels of sequence divergence between microcystins synthetase genes from the distantly related genera of microcystin-producing cyanobacteria will allow novel approaches to the molecular ecology of bloomforming cyanobacteria and may provide answers to the longstanding debate over the role of microcystins and nodularins.
Supplementary Material
Acknowledgments
We thank Stefano Ventura for providing internal 16S rRNA primer sequences. This work was supported by grants from the European Union TOPIC (FMRX-CT98-0246), MIDI-CHIP (EVK2-CT-1999-00026), and CYANOTOX (ENV4-CT98-0802) projects and Academy of Finland Grant 46812.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. AJ515451–AJ515466, AJ515468–AJ515475 (mcyA), AY382530–AY382556 (mcyE), AY424980–AY424994 (mcyD), AY424995–AY4225003 (rpoC1), and AY439281–AY439283 (16S rRNA)].
References
- 1.Francis, G. (1878) Nature 18, 11–12. [Google Scholar]
- 2.Sivonen, K. & Jones, G. (1999) in Toxic Cyanobacteria in Water: A Guide to Their Public Health Consequences, Monitoring, and Management, eds. Chorus, I. & Bartram, J. (Spoon, London), pp. 41–111.
- 3.Ressom, R., Soong, F. S., Fitzgerald, J., Turczynowicz, L., El Saadi, O., Roder, D., Maynard, T. & Falconer, I. (1994) Health Effects of Toxic Cyanobacteria (Blue-Green Algae) (National Health and Medical Research Council, Australian Government Publishing Service, Canberra), p. 108.
- 4.Kuiper-Goodman, T., Falconer, I. & Fitzgerald, J. (1999) in Toxic Cyanobacteria in Water: A Guide to Their Public Health Consequences, Monitoring, and Management, eds. Chorus, I. & Bartram, J. (Spoon, London), pp. 113–153.
- 5.Jochimsen, E. M., Carmichael, W. W., An, J., Cardo, D. M., Cookson, S. T., Holmes, C. E. M., Antunes, M. B., de Melo Filho, D. A., Lyra, T. M., Barret, V. S. T., et al. (1998) N. Engl. J. Med. 338, 873–878. [DOI] [PubMed] [Google Scholar]
- 6.World Health Organization (1998) Guidelines for Drinking Water Quality: Addendum to Volume 2, Health Criteria and Other Supporting Information (W.H.O., Geneva), 2nd Ed.
- 7.MacKintosh, C., Beattie, K. A., Klumpp, S., Cohen, P. & Codd, G. A. (1990) FEBS Lett. 264, 187–192. [DOI] [PubMed] [Google Scholar]
- 8.Goldberg, J., Huang, H.-B., Kwon, Y.-G., Greengard, P., Nairn, A. C. & Kuriyan, J. (1995) Nature 376, 745–753. [DOI] [PubMed] [Google Scholar]
- 9.Namikoshi, M. & Rinehart, K. L. (1996) J. Ind. Microbiol. Biotechnol. 17, 373–384. [Google Scholar]
- 10.Golakoti, T., Yoshida, W. Y., Chaganty, S. & Moore, R. E. (2000) Tetrahedron 56, 9093–9102. [Google Scholar]
- 11.Marahiel, M. A., Stachelhaus, T. & Mootz, H. D. (1997) Chem. Rev. 97, 2651–2673. [DOI] [PubMed] [Google Scholar]
- 12.von Döhren, H., Keller, U., Vater, J. & Zocher, R. (1997) Chem. Rev. 97, 2675–2705. [DOI] [PubMed] [Google Scholar]
- 13.Christiansen, G., Dittmann, E., Via Ordorica, L., Rippka, R., Herdman, M. & Börner, T. (2001) Arch. Microbiol. 176, 452–458. [DOI] [PubMed] [Google Scholar]
- 14.Nishizawa, T., Ueda, A., Asayama, M., Fujii, K., Harada, K.-I., Ochi, K. & Shirai, M. (2000) J. Biochem. 126, 779–789. [DOI] [PubMed] [Google Scholar]
- 15.Tillett, D., Dittmann, E., Erhard, M., von Döhren, H., Börner, T. & Neilan, B. A. (2000) Chem. Biol. 7, 753–764. [DOI] [PubMed] [Google Scholar]
- 16.Christiansen, G., Fastner, J., Erhard, M., Börner, T. & Dittmann, E. (2003) J. Bacteriol. 185, 564–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dittmann, E., Neilan, B. A., Erhard, M., von Döhren, H. & Börner, T. (1997) Mol. Microbiol. 26, 779–787. [DOI] [PubMed] [Google Scholar]
- 18.Meibner, K., Dittmann, E. & Börner, T. (1996) FEMS Microbiol. Lett. 135, 295–303. [DOI] [PubMed] [Google Scholar]
- 19.Otsuka, S., Suda, S., Li, R., Watanabe, M., Oyaizu, H., Matsumoto, S. & Watanabe, M. M. (1999) FEMS Microbiol. Lett. 172, 15–21. [DOI] [PubMed] [Google Scholar]
- 20.Neilan, B. A., Dittmann, E., Rouhiainen, L., Bass, R. A., Schaub, V., Sivonen, K. & Börner, T. (1999) J. Bacteriol. 181, 4089–4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tillett, D., Parker, D. L. & Neilan, B. A. (2001) Appl. Environ. Microbiol. 67, 2810–2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mikalsen, B., Boison, G., Skulberg, O. M., Fastner, J., Davies, W., Gabrielsen, T. M., Rudi, K. & Jakobsen, K. S. (2003) J. Bacteriol. 185, 2774–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moffitt, M. C. & Neilan, B. A. (2001) FEMS Microbiol. Lett. 196, 207–214. [DOI] [PubMed] [Google Scholar]
- 24.Ragan, M. A. (2001) Curr. Opin. Genet. Dev. 11, 620–626. [DOI] [PubMed] [Google Scholar]
- 25.Sivonen, K., Carmichael, W. W., Namikoshi, M., Rinehart, K. L., Dahlem, A. M. & Niemelä, S. I. (1990) Appl. Environ. Microb. 56, 2650–2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beattie, K. A., Kaya, K., Sano, T. & Codd, G. A. (1998) Phytochemistry 47, 1289–1292. [Google Scholar]
- 27.Hisbergues, M., Christiansen, G., Rouhiainen, L., Sivonen, K. & Börner, T. (2003) Arch Microbiol. 180, 402–410. [DOI] [PubMed] [Google Scholar]
- 28.Lepère, C., Wilmotte, A. & Meyer, B. (2000) Syst. Geogr. Pl. 70, 275–283. [Google Scholar]
- 29.Edwards, U., Rogall, T., Blöcker, H., Emde, M. & Böttger, E. C. (1989) Nucleic Acids Res. 17, 7843–7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hall, T. A. (1999) Nucleic Acids. Symp. Ser. 41, 95–98. [Google Scholar]
- 31.Kumar, S., Tamura, K., Jakobsen, I. B. & Nei, M. (2001) Bioinformatics 17, 1244–1245. [DOI] [PubMed] [Google Scholar]
- 32.Li, W. H. & Graur, D. (1991) Fundamentals of Molecular Evolution (Sinauer, Sunderland, MA).
- 33.Swofford, D. L. (2001) paup*: Phylogenetic Analysis Using Parsimony (* and Other Methods) (Sinauer, Sunderland, MA), version 4.0b8.
- 34.Hoffmann, D., Hevel, J. M., Moore, R. E. & Moore, B. S. (2003) Gene 311, 171–180. [DOI] [PubMed] [Google Scholar]
- 35.Rouhiainen, L., Paulin, L., Suomalainen, S., Hyytiäinen, H., Buikema, W., Haselkorn, R. & Sivonen, K. (2000) Mol. Microbiol. 37, 156–167. [DOI] [PubMed] [Google Scholar]
- 36.Kaneko, T., Nakamura, Y., Wolk, C. P., Kuritz, T., Sasamoto, S., Watanabe, A., Iriguchi, M., Ishikawa, A., Kawashima, K., Kimura, T., et al. (2001) DNA Res. 8, 205–213. [DOI] [PubMed] [Google Scholar]
- 37.Swofford, D. L., Olsen, G. J., Waddell, P. J. & Hillis, D. M. (1996) in Molecular Systematics, eds. Hillis, D. M., Moritz, C. & Mable, B. K. (Sinauer, Sunderland, MA), pp. 407–514.
- 38.Farris, J. S., Källersjö, M., Kluge, A. G. & Bult, C. (1994) Cladistics 10, 315–319. [DOI] [PubMed] [Google Scholar]
- 39.Lockhart, P. J., Steel, M. A., Hendy, H. D. & Penny, D. (1994) Mol. Biol. Evol. 11, 605–612. [DOI] [PubMed] [Google Scholar]
- 40.Templeton, A. R. (1983) Evolution 37, 221–244. [DOI] [PubMed] [Google Scholar]
- 41.Gilroy, D. J., Kauffman, K. W., Hall, R. A., Huang, X. & Chu, F. S. (2000) Environ. Health Perspect. 108, 435–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morrison, G. M., Semple, C. A., Kilanowski, F. M., Hill, R. E. & Dorin, J. R. (2003) Mol. Biol. Evol. 20, 460–470. [DOI] [PubMed] [Google Scholar]
- 43.Schwecke, T., Aparicio, J. F., Molnár, I., König, A., Khaw, L. E., Haydock, S. F., Oliynyk, M., Caffrey, P., Cortés, J., Lester, J. B., et al. (1995) Proc. Natl. Acad. Sci. USA 92, 7839–7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Du, L., Sánchez, C., Chen, M., Edwards, D. J. & Shen, B. (2000) Chem. Biol. 7, 623–642. [DOI] [PubMed] [Google Scholar]
- 45.Gehring, A. M., DeMoll, E., Fetherston, J. D., Mori, I., Mayhew, G. F., Blattner, F. R., Walsh, C. T. & Perry, R. D. (1998) Chem. Biol. 5, 573–586. [DOI] [PubMed] [Google Scholar]
- 46.Quadri, L. E., Sello, J, Keating, T. A., Weinreb, P. H. & Walsh, C. T. (1998) Chem. Biol. 5, 631–645. [DOI] [PubMed] [Google Scholar]
- 47.Duitman, E. H., Hamoen, L. W., Rembold, M., Venema, G., Seitz, H., Saenger, W., Bernhard, F., Reinhardt, R., Schmidt, M., Ullrich, C., et al. (1999) Proc. Natl. Acad. Sci. USA 96, 13294–13299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Silakowski, B., Schairer, H. U., Ehret, H., Kunze, B., Weinig, S., Nordsiek, G., Brandt, P., Blöcker, H., Höfle, G., Beyer, S., et al. (1999) J. Biol. Chem. 274, 37391–37399. [DOI] [PubMed] [Google Scholar]
- 49.Tsuge, K., Akiyama, T. & Shoda, M. (2001) J. Bacteriol. 183, 6265–6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Du, L., Sánchez, C. & Shen, B. (2001) Metab. Eng. 3, 78–95. [DOI] [PubMed] [Google Scholar]
- 51.Moffitt, M. C. & Neilan, B. A. (2003) J. Mol. Evol. 56, 446–457. [DOI] [PubMed] [Google Scholar]
- 52.DeMott, W. R. & Moxter, F. (1991) Ecology 72, 1820–1834. [Google Scholar]
- 53.Heckman, D. S., Geiser, D. M., Eidell, B. R., Stauffer, R. L., Kardos, N. L. & Hedges, S. B. (2001) Science 293, 1129–1133. [DOI] [PubMed] [Google Scholar]
- 54.Golubic, S., Sergeev, V. N. & Knoll, A. H. (1995) Lethaia 28, 285–298. [DOI] [PubMed] [Google Scholar]
- 55.Amard, B. & Bertrand-Sarfati, J. (1997) Precambrian Res. 81, 197–221. [Google Scholar]
- 56.Feng, D. F., Cho, G. & Doolittle, R. F. (1997) Proc. Natl. Acad. Sci. USA 94, 13028–13033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Utkilen, H. & Gjølme, N. (1995) Appl. Environ. Microbiol. 61, 797–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dittmann, E., Erhard, M., Kaebernick, M., Scheler, C., Neilan, B. A., von Döhren, H. & Börner, T. (2001) Microbiology 147, 3113–3119. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.