Abstract
Skeletal muscle cells have served as a paradigm for understanding mechanisms leading to cellular differentiation. The proliferation and differentiation of muscle precursor cells require the concerted activity of myogenic regulatory factors including MyoD. In addition, chromatin modifiers mediate dynamic modifications of histone tails that are vital to reprogramming cells toward terminal differentiation. Here, we provide evidence for a unique dimension to epigenetic regulation of skeletal myogenesis. We demonstrate that the lysine methyltransferase G9a is dynamically expressed in myoblasts and impedes differentiation in a methyltransferase activity-dependent manner. In addition to mediating histone H3 lysine-9 di-methylation (H3K9me2) on MyoD target promoters, endogenous G9a interacts with MyoD in precursor cells and directly methylates it at lysine 104 (K104) to constrain its transcriptional activity. Mutation of K104 renders MyoD refractory to inhibition by G9a and enhances its myogenic activity. Interestingly, MyoD methylation is critical for G9a-mediated inhibition of myogenesis. These findings provide evidence of an unanticipated role for methyltransferases in cellular differentiation states by direct posttranslational modification of a transcription factor.
Skeletal muscle differentiation is controlled by the MyoD family of myogenic regulatory factors and chromatin modifying enzymes that reconfigure chromatin at muscle-specific promoters (1–6). Epigenetic modifications constitute a complex regulatory layer to restrict or facilitate gene expression. In undifferentiated cells, marks of transcriptional repression including H3K9 and H3K27 methylation are apparent on early and late muscle gene promoters, along with reduced acetylation of histone tails. Many enzymes that regulate these histone modifications have been studied, including histone deacetylases (HDACs) and histone methyltransferases (HMTs). Protein complexes of the polycomb (PcG) group catalyze trimethylation of H3K27 (H3K27me3), and HMTs from the Suv39h family mediate H3K9 methylation (7–12). The onset of differentiation requires extensive reprogamming at muscle promoters. Several coactivator proteins including CBP/p300, P/CAF, the arginine methyltransferases Carm1/Prmt4 and Prmt5, and the SWI/SNF remodeling complexes are recruited, which substitute the repressive chromatin configuration in undifferentiated cells with transcriptional activation marks. Despite extensive investigations, however, it remains unclear how MyoD, which is expressed in undifferentiated proliferating myoblasts, remains transcriptionally inert and unable to activate muscle-specific genes until appropriate differentiation cues are established (13–16).
A key modification that is apparent on the early MyoD target gene myogenin in undifferentiated muscle cells is H3K9me2 (9, 10). This transcriptional repressive mark is mediated mostly by the SET domain containing Suv39 family members that include EHMT2/G9a and Suv39h1 (17). Although both G9a and Suv39h1 mediate H3K9 methylation, their activities are distinct. G9a is mainly responsible for mono- and dimethylation of H3K9 (H3K9me1 and H3K9me2, respectively), which is scattered in euchromatin, whereas Suv39h1, the principal enzyme responsible for accumulation of H3K9me3, is enriched in heterochromatin. More recent studies have demonstrated that in addition to mediating H3K9me2, G9a can methylate nonhistone substrates, including CDYL1, WIZ, ACINUS, and C/EBPβ (18), and also associates with de novo DNA methyltransferases 3a and 3b to repress transcription (19). Loss of G9a in mice leads to early embryonic lethality, which has precluded the understanding of its function in specific cellular differentiation pathways (20).
In this study, we demonstrate that G9a is a unique inhibitor of skeletal muscle differentiation that plays a dominant role in MyoD activation. G9a-mediated inhibition of the muscle differentiation program depends on its methyltransferase activity, because pharmacological blockage of G9a activity, or expression of a catalytically inactive mutant, fail to impact myogenesis. Interestingly, G9a interacts with MyoD in undifferentiated cells, and methylates it at K104, which inhibits its activity. Mutation of K104 renders MyoD refractory to inhibition by G9a and enhances its myogenic potential. Our data identify MyoD methylation as a distinct epigenetic modification that plays a prominent role in restricting MyoD activity in myoblasts and, consequently, skeletal muscle differentiation.
Results
G9a Inhibits Skeletal Muscle Differentiation in a Methyltransferase Activity-Dependent Manner.
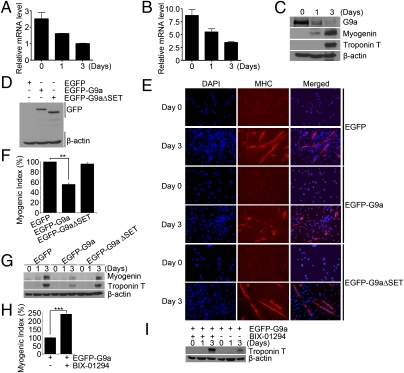
To examine whether G9a plays a role in skeletal myogenesis, we first investigated its expression in undifferentiated and differentiated cells. G9a mRNA was expressed at high levels in C2C12 cells and in undifferentiated primary myoblasts, and it was down-regulated during differentiation (Fig. 1 A and B). Consistently, G9a protein levels also declined upon differentiation and inversely correlated with the expression of MyoD target genes myogenin and troponin T (Fig. 1C). G9a was expressed in the nucleus in undifferentiated myoblasts and remained nuclear, albeit at much lower expression, in MHC+ differentiated cells (Fig. S1). To investigate whether G9a regulates myogenic differentiation, we performed gain-of-function studies. C2C12 cells expressing full-length G9a (EGFP-G9a), or a deletion mutant lacking the SET domain (EGFP-G9aΔSET), were analyzed for their ability to differentiate relative to control vector (EGFP) expressing cells. Both EGFP-G9a and EGFP-G9aΔSET were expressed at similar levels (Fig. 1D). In contrast to control and EGFP-G9aΔSET cells, EGFP-G9a cells exhibited a significant reduction in differentiation as evidenced by reduced myogenic index (Fig. 1 E and F). Moreover, myogenin and troponin T levels were reduced in EGFP-G9a cells (Fig. 1G), indicating that G9a-mediated inhibition of myogenesis depends on its methyltransferase activity. To confirm these findings, BIX-01294, a inhibitor of G9a activity (21) was used. G9a-overexpressing cells were left untreated, or treated with BIX-01294 for 3 d, and the extent of differentiation examined. Inhibition of G9a activity in EGFP-G9a–overexpressing cells led to a significant rescue of myogenic differentiation (Fig. 1H) and troponin T expression (Fig. 1I).
Fig. 1.
G9a inhibits differentiation in a methyltransferase activity-dependent manner. (A and B) G9a mRNA was examined in undifferentiated (Day 0) and differentiated (Days 1 and 3) in C2C12 cells (A), as well as primary myoblasts (B) by Q-PCR. (C) G9a, myogenin, and troponin T protein levels were analyzed in C2C12 cells by Western blot. (D) C2C12 overexpressing EGFP-G9a, EGFP-G9aΔSET, or EGFP (vector) were analyzed for G9a expression by Western blot. (E) Control and EGFP-G9a or EGFP-G9aΔSET expressing cells were immunostained with α-MHC antibody. (F) A significant decrease in myogenic index was apparent in EGFP-G9a cells compared with control and EGFP-G9aΔSET cells. (G) Control, EGFP-G9a, and EGFP-G9aΔSET cells were analyzed for myogenin and troponin T expression by Western blot. (H and I) EGFP-G9a–overexpressing cells were left untreated or treated with BIX-01294 for 3 d. A significant rescue of the myogenic index (H) and troponin T levels (I) was apparent upon inhibition of G9a activity. Error bars indicate mean ± SD. **P < 0.01; ***P < 0.001.
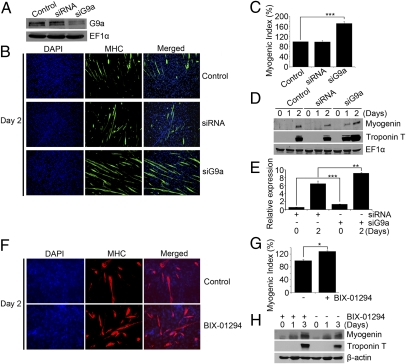
To confirm that endogenous G9a plays a role in myogenic differentiation, we used two approaches. In one, G9a expression was inhibited with siRNA, and in the other, its activity was targeted by using BIX-01294. C2C12 cells were transfected with G9siRNA (siG9a), or as a control, with scrambled siRNA (siRNA). Densitometric analysis of Western blots revealed a 71% down-regulation of G9a expression in siG9a cells compared with untransfected cells, or those expressing siRNA (Fig. 2A). Compared with controls, siG9a cells exhibited markedly higher myogenic differentiation (Fig. 2 B and C), and an earlier onset of myogenin and troponin T expression (Fig. 2D). Moreover, myogenin mRNA was up-regulated in siG9a cells cultured in growth and differentiation medium (Fig. 2E). Consistently, inhibition of endogenous G9a activity in control C2C12 cells resulted in a significant increase in myogenic differentiation (Fig. 2 F and G), as well as myogenin and troponin T levels (Fig. 2H).
Fig. 2.
Myogenic differentiation is enhanced by loss of G9a. (A) C2C12 cells were transfected with G9a siRNA (siG9a), scrambled siRNA (siRNA), or left untransfected (control). Lysates were analyzed for endogenous G9a protein levels by Western blot. (B) Cells were subjected to differentiation assays and stained with α-MHC antibody (green) on Day 2 after differentiation. Nuclei were stained with DAPI. (C) Quantification of myogenic index on Day 2 after differentiation revealed significantly enhanced MHC+ cells in siG9a cells. (D) Myogenin and troponin T expression was determined by Western blot at Days 0, 1, and 2 after differentiation. (E) Myogenin mRNA levels were measured in control and siG9a cells at D0 and D2 after differentiation by Q-PCR. Error bars indicate mean ± SD. (F and G) Endogenous G9a activity was blocked in C2C12 cells with BIX-01294. MHC+ cells (F) and myogenic index (G) were determined in cells left untreated (-), or after treatment (+) with BIX-01294 for 2 d. (H) Myogenin and troponin T expression was increased in cells treated with BIX-01294 compared with control untreated cells. *P < 0.05; **P < 0.01; ***P < 0.001.
G9a Interacts with and Inhibits MyoD Activity.
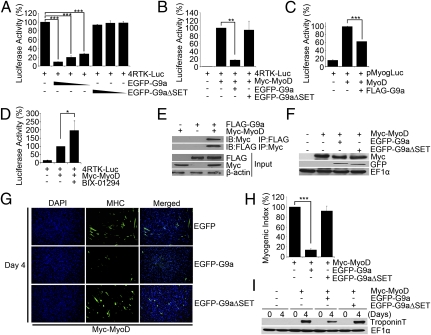
Because G9a is expressed in myoblasts and inhibits genes that are induced during differentiation, we tested whether it regulates myogenic regulatory factor (MRF) activity. C2C12 myoblasts were transfected with the MRF reporter 4RTK-Luc in the absence and presence of EGFP-G9a or EGFP-G9aΔSET. Interestingly, EGFP-G9a potently repressed the reporter activity in a SET-domain–dependent manner (Fig. 3A), suggesting that G9a may inhibit endogenous MyoD and/or Myf5 transcriptional activity. To examine the effect of G9a specifically on MyoD activity, C3H10T1/2 fibroblast cells were used that do not express endogenous MyoD, but readily undergo myogenic conversion upon exogenous MyoD expression. Cells were transfected with 4RTK-Luc and MyoD in the absence and presence of EGFP-G9a and EGFP-G9aΔSET. MyoD activated the reporter, whereas coexpression of EGFP-G9a, but not EGFP-G9aΔSET, blocked this effect (Fig. 3B). Similarly, MyoD-mediated activation of the myogenin promoter reporter (22) was inhibited by G9a (Fig. 3C). Consistent with these findings, MyoD-induced reporter activity was augmented in the presence of BIX-01294 (Fig. 3D). To investigate the mechanisms underlying the inhibition of its activity, we tested whether G9a associates with MyoD. Immunoprecipitation of either G9a or MyoD revealed an interaction between the two proteins (Fig. 3E), and colocalization of endogenous G9a and MyoD was apparent in undifferentiated cells (Fig. S2). To examine the impact of G9a on MyoD function, myogenic conversion assays were performed. 10T1/2 cells were transfected with MyoD alone, or together with equivalent amounts of EGFP-G9a and EGFP-G9aΔSET (Fig. 3F). EGFP-G9a, but not EGFP-G9aΔSET, inhibited MyoD-dependent myogenic conversion as seen by a significant reduction in the presence of myosin heavy chain (MHC+) myotubes (Fig. 3G), myogenic index (Fig. 3H), and troponin T expression (Fig. 3I).
Fig. 3.
G9a inhibits MyoD activity. (A) C2C12 cells were transfected with 4RTK-Luc (50 ng) in the absence and presence (50, 25, and 10 ng) of EGFP-G9a or EGFP-G9aΔSET. Data are means ± SD. (B and C) 10T1/2 cells were transfected with 4RTK-Luc and MyoD along with EGFP-G9a, and EGFP-G9aΔSET (B), or with pMyogLuc (80 ng) and MyoD (50 ng), without and with Flag-G9a (50 ng) (C). (D) 10T1/2 cells were transfected with 4RTK-Luc and MyoD in the absence and presence of BIX-01294. (E) 293 cells were transfected with Flag-G9a and Myc-MyoD. Proteins were immunoprecipitated with α-Flag or α-Myc agarose beads and immunoblotted with α-Myc or α-Flag antibody, respectively. Lysates (input) were analyzed for G9a, and MyoD expression was analyzed by Western blot. (F–I) 10T1/2 cells were transfected with MyoD, EGFP-G9a and EGFP-G9aΔSET and analyzed by Western blot (F). Transfected cells were subjected to myogenic conversion assays. Cells were stained with α-MHC (green) antibody (G). The percentage of MHC+ cells was quantified relative to MyoD alone, which was given a value of 100%. A significant decrease in myogenic index was apparent in cells overexpressing EGFP-G9a and MyoD (H). Troponin T levels analyzed by Western blot were reduced in cells coexpressing MyoD and G9a compared with controls (I). Data are mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001.
MyoD Is Methylated by G9a.
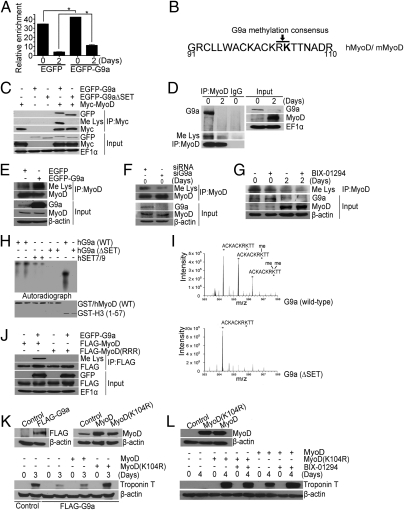
To examine the mechanisms underlying inhibition of MyoD activity, we first examined by chromatin immunoprecipitation (ChIP) assays enrichment of H3K9me2, a transcriptional repression mark mediated by G9a. Consistent with previous reports (9), H3K9me2 was apparent on the myogenin promoter in undifferentiated EGFP-expressing C2C12 cells and declined upon differentiation. In G9a-overexpressing cells, increased H3K9me2 was apparent in both undifferentiated and differentiated cells (Fig. 4A). Because G9a complexes with MyoD and is known to methylate nonhistone proteins, we wondered whether MyoD could be a target for G9a-mediated methylation. Analysis of the human and mouse MyoD cDNA sequence revealed a single G9a-methylation consensus RK at K104 (Fig. 4B). To test this possibility, MyoD was coexpressed with EGFP-G9a, or EGFP-G9aΔSET, and its methylation was examined after immunoprecipitation. Both EGFP-G9a and EGFP-G9aΔSET interacted with MyoD. However, MyoD was methylated only in presence of EGFP-G9a (Fig. 4C). Consistently, endogenous MyoD immunoprecipitated with G9a in myoblasts and was methylated (Fig. 4D). To examine whether G9a is involved in MyoD methylation, we modulated its levels and examined the impact on MyoD methylation. Overexpression of G9a resulted in enhanced MyoD methylation (Fig. 4E) and, conversely, abrogated it in siG9a cells (Fig. 4F). Inhibition of endogenous G9a activity resulted in reduced MyoD methylation in both undifferentiated and differentiated cells (Fig. 4G). Moreover, deletion of the ankyrin repeats in G9a (G9aΔANK) disrupted the association with MyoD and, correspondingly, its methylation, indicating that interaction with G9a is required for MyoD methylation (Fig. S3A).
Fig. 4.
G9a-mediated methylation of MyoD at K104 impairs its myogenic activity. (A) ChIP assays were performed at Day 0 and Day 2 on the myogenin promoter. Increased H3K9me2 was apparent in EGFP-G9a cells at both stages. Error bars indicate mean ± SD. (B) Schematic representation of human (h)/mouse(m) MyoD cDNA sequence. A G9a methylation consensus (RK) at K104 is highlighted. (C) 293 cells were transfected with EGFP-G9a, EGFP-G9aΔSET, and Myc-MyoD. MyoD was immunoprecipitated and analyzed for its association with G9a, and methylation, using α-GFP and α-methyl lysine (Me Lys) antibodies. (D) Endogenous MyoD was immunoprecipitated from C2C12 cells on Day 0 and 2 after differentiation, followed by immunoblotting with α-G9a, α-Me Lys, and α-MyoD antibodies. Input shows expression of G9a and MyoD in the lysates. (E). C2C12 cells were transfected with EGFP and EGFP-G9a. Endogenous MyoD was immunoprecipitated from myoblasts and analyzed by α-Me Lys and α-MyoD antibodies. Input shows G9a and MyoD expression. (F) Endogenous MyoD was immunoprecipitated from myoblasts expressing siG9a and control siRNA, and analyzed with α-Me Lys and α-MyoD antibodies. Lysates were analyzed for MyoD and G9a expression. (G) C2C12 cells were differentiated in the absence and presence of BIX-01294 for 2 d. Endogenous MyoD was immunoprecipitated followed by Western blotting with α-Me Lys antibody. (H) In vitro methylation of recombinant GST-tagged human MyoD by G9a and Set7/9 using purified recombinant G9a, G9aΔSET, and Set7/9. GST-H3 was used as a positive control. (I) LC-MS of tryptic peptides of MyoD after treatment with the catalytically active and inactive recombinant G9a methyltransferase. (J) Flag-MyoD and Flag-MyoD(RRR) were transfected with EGFP-G9a in 293 cells. Flag-MyoD was immunoprecipitated, and immunoblotted with α-Me Lys and α-Flag antibodies. Lysates were analyzed for G9a and MyoD levels. (K) Flag-G9a–overexpressing cells (Left) were transfected with MyoD and MyoD(K104R). Vector transfected cells were used as control (Right). Troponin T levels were analyzed at 0 and 3 d after differentiation (Lower). (L) C3H10T1/2 cells were transfected with MyoD and MyoD(K104R) (Upper) and differentiated in the absence and presence of BIX-01294. Troponin T was analyzed at 0 and 4 d after differentiation (Lower). *P < 0.05.
We then examined whether G9a directly methylates MyoD in vitro by using GST-MyoD. GST-histone H3 (GST-H3) was used as a control. G9a methylated MyoD, albeit at a lower efficiency compared with H3 (Fig. 4H), whereas no methylation was apparent with the SET mutant (hG9aΔSET). In addition, SET7/9 also weakly methylated MyoD, suggesting that additional methyltransferases may also modify MyoD. Liquid chromatography-mass spectrometry (LC-MS) of the recombinant MyoD polypeptide treated with wild-type or mutant (ΔSET) G9a identified K104 as the site for methylation (Fig. 4I), consistent with a G9a-methylation consensus at this residue (Fig. 4B). To validate these findings, we transfected 293 cells with wild-type MyoD and MyoD(RRR), in which K99, K102, and K104 are mutated to arginine (RRR) (14) along with EGFP-G9a. MyoD was immunoprecipitated and probed for methylation by using methyl-lysine antibody. In contrast to MyoD, MyoD(RRR) was not methylated in presence of G9a (Fig. 4J).
To address the significance of H3K9me2 versus MyoD methylation in G9a-mediated inhibition of myogenesis, we examined the ability of MyoD and MyoD(K104R) (where K104 is mutated to arginine; ref. 14) to rescue the differentiation block imposed by G9a. In G9a-overexpressing cells (Fig. 4K, Upper Left), equivalent levels of each MyoD construct was expressed (Fig. 4K, Upper Right). Interestingly, MyoD(K104R) was more efficient than MyoD at rescuing myogenic differentiation (Fig. S3 B and C) and troponin T levels (Fig. 4K, Lower), likely due to its inability to be methylated by G9a. To validate these findings, we examined the myogenic potential of MyoD and MyoD(K104R) in myogenic conversion assays (Fig. 4L, Upper). As expected, expression of MyoD alone resulted in troponin T expression and myogenic differentiation, which was augmented by BIX-01294 (Fig. 4L, Lower and Fig. S3D). In contrast, expression of MyoD(K104R) at similar levels resulted in higher troponin T expression and myogenic differentiation, which, however, was unresponsive to G9a inhibition by BIX-01294. Consistently, MyoD(K104R) showed increased transactivation of the myogenin promoter (Fig. S3E), although the levels of H3K9me2 were unaltered in the presence of MyoD and MyoDK104R (Fig. S3F). Together, these results support a dominant role for MyoD K104 methylation in the inhibition of differentiation by G9a.
Discussion
The transition of cells from an undifferentiated to a differentiated state provide a powerful system for investigating epigenetic mechanisms that influence cellular fate. The results of this study demonstrate an unconventional epigenetic mechanism that connects the lysine methyltransferase G9a to maintenance of an undifferentiated fate by methylation of MyoD.
In agreement with an inhibitory role in skeletal muscle differentiaton, G9a is expressed at high levels in proliferating myoblasts and inversely correlated with MyoD transcriptional activity and the onset of myogenin expression during differentiation. Similar to G9a, other chromatin modifers are expressed in undifferentiated myoblasts including HDAC1 (7, 8) and Suv39h1, both of which interact with MyoD (12, 23). However, unlike these enzymes, which act locally to deacetylate and methylate histones resulting in chromatin configurations that are nonpermissive for MyoD transcriptional activity, G9a not only mediates H3K9me2, but also directly methylates MyoD at K104. Although G9a is known to mediate methylation of both histone and nonhistone substrates, the biological functions of nonhistone methylations are largely unclear. In this regard, our data provide unique insights into the function of methylation of a nonhistone substrate in the control of cellular differentiaion. Our data indicate that MyoD methylation, rather than alterations in H3K9me2, are significant in G9a-mediated inhibition of myogenesis. This inference is based on the strikingly differential abilities of wild-type MyoD and MyoD(K104R) in rescuing myogenic differentiation in G9a-overexpressing cells that have similar levels of H3K9me2. MyoD(K104R) exhibits higher myogenic potential in myogenic conversion assays, and in activation of the myogenin promoter, while being unresponsive to G9a. Based on the crystal structure of MyoD–DNA complex, K104 does not make contact with DNA and, correspondingly, its mutation does not impact MyoD DNA binding or nuclear localization (14, 24). Therefore, our data collectively argue that MyoD methylation is critical in regulation of its transcriptional activity, and the onset of MyoD-dependent gene expression during differentiation. Although methylation of MyoD has not been described in the literature, it is noteworthy that in vitro, MyoD is methylated not only by G9a, but also by Set7/9. It is unlikely that methylation of MyoD by G9a and Set7/9 occur on the same residue because they have distinct substrate requirements (25). MyoD is constitutively bound to several promoters in both myoblasts and myotubes. It is unclear whether G9a activity has a role in regulating MyoD-activated target genes in myoblasts. Nevertheless, G9a blocks the differentiation-inducing ability of MyoD through methylation of K104 in euchromatin of myoblasts. In addition to G9a, several other chromatin modifier complexes containing Suv39h1, HP1, HDAC1, and Ezh2 contribute toward the generation of a stable H3K9me3, H3K27me3, and deacetylated state, resulting in silencing of early and late myogenic promoters (11, 12, 23). During differentiation, the declining G9a levels would expectedly erase MyoD methylation. Moreover, Suv39h1, HDAC1, and Ezh2 also are down-regulated at the onset of differentiation. Thus, H3K9me2, H3K27me3, and histone deacetylation marks on MyoD target promoters are removed, paving the way for MyoD and histone activation that are permissive for myogenic differentiation.
Materials and Methods
Cell Culture and Plasmid Constructs.
C2C12, 293, C3H10T1/2, primary myoblasts, and Phoenix cells were cultured as described (26, 27). Plasmids Myc-MyoD, Flag-MyoD, Flag-MyoD(K104R), Flag-MyoD(RRR), 4RTK-Luc, pMyogLuc reporter harboring 1565 base pairs of upstream sequences from the myogenin gene, Flag-G9a, EGFP-G9a, and EGFP-G9aΔSET have been described (14, 22, 26–28). Primers used to generate Flag-G9aΔANK are provided in Table S1.
G9a Overexpression, siRNA, and Myogenic Differentiation.
For G9a overexpression, EGFP-G9a, EGFP-G9aΔSET, or Flag-G9a were transfected in C2C12 cells along with pBabe-puromycin. Cells were selected with 2 μg/mL puromycin (26) and differentiated in the absence or presence of 2.5 μM BIX-01294. For rescue experiments, Flag-G9a–expressing cells were transfected with MyoD or MyoD(K104R). For G9a knockdown, C2C12 cells were transfected with 100 nM control scrambled siRNA (Dharmacon; on-target plus control pool; nontargeting pool) or siRNA specific for G9a (Dharmacon; on-target plus smart pool, Mouse BAT 8; accession no. NM_147151; NM_145830) by using Lipofectamine RNAiMAX (Invitrogen). Myogenic conversion assays were done as described (27). Briefly, C3H10T1/2 cells were transfected with MyoD alone, or together with EGFP-G9a and EGFP-G9aΔSET. For rescue experiements, MyoD and MyoD(K104R) were transfected and differentiated in the absence and presence of 2.5 μM BIX-01294. Parental cells were transfected with empty vector. Differentiated cells were stained with α-MHC antibody (Sigma) and detected with Alexa Fluor-coupled secondary antibodies. Nuclei were stained with DAPI (Vector Laboratories) and visualized by fluorescence microscopy (Nikon Eclipse TE 2000-U) with 10× or 20× objective lens and MetaMorph software version 7.0r3. The percentage of MHC+ cells in controls were given a value of 100%. Myogenic index was calculated by quantifying the ratio of nuclei in MHC+ myotubes/total nuclei. At least 600 nuclei were counted.
Coimmunoprecipitation (Co-IP) Assays and Western Blots.
Co-IP assays were done as described (26). To detect endogenous interactions, 1mg of C2C12 lysate was incubated with 2μg of α-MyoD antibody. As a control, 2 μg of rabbit IgG was added to D0 lysate. The following antibodies were used in this study for Western blots: α-Flag, α-Myc, α-troponin T (Sigma); α-MyoD, α-GFP, α-myogenin (Santa Cruz Biotechnology); α-G9a (Cell Signaling, Upstate Biotechnology); and Me Lys (Abcam). α−β-actin or α-EF-1α was used for normalization.
Chromatin Immunoprecipitation (ChIP).
Assays were performed in C2C12 cells by using the ChIP assay kit (Upstate Biotechnology) as described (29) using 2 μg of H3K9me2 (Upstate Biotechnology) antibody. Primers for amplification of Myogenin and β-actin promoters are provided in Table S1.
Quantitative RT-PCR (Q-PCR).
Q-PCR was done as described (26). The cDNA was amplified with primers specific to G9a, myogenin, and GAPDH (Table S1).
In Vitro Methylation.
In vitro methylation reactions were conducted as described (28) by using 3 μg of purified poly-histidine–tagged recombinant proteins Set7/9 or G9a in a reaction buffer containing 50 mM Tris⋅HCl at pH 8.5, 5 mM MgCl2, 1 mM DTT, 1 μM [14C]-labeled S-adenosyl-l-methionine [14C]SAM (Perkin-Elmer), 50 mM SAM (Sigma), and 2 μg of GST-MyoD (43-224) or GST-histone H3 tail (1-57). Proteins were separated on a 15% SDS-polyacrylamide gel. Gels were visualized by Coomassie blue staining and fluorography. In parallel, the recombinant MyoD polypeptide, treated with wild-type or mutant (ΔSET) G9a, was digested with trypsin and analyzed by LC-MS analysis (Thermo Electron; Finnigan LTQ).
Statistical Analysis.
Values are reported as mean ± SD or SE. Statistical significance was determined by the Student t test, and P values of <0.05 were considered to be statistically significant (*P < 0.05; **P < 0.01; ***P < 0.001).
Supplementary Material
Acknowledgments
We thank D. Sassoon and G. Pavlath for reagents. V.S. is supported by the Intramural Research Program of the National Institute of Arthritis and Musculoskeletal and Skin Diseases/National Institutes of Health. This work was supported by grants from the National Medical Research Council, and Agency for Science, Technology, and Research Singapore Stem Cell Consortium (to R.T.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1111628109/-/DCSupplemental.
References
- 1.Sabourin LA, Rudnicki MA. The molecular regulation of myogenesis. Clin Genet. 2000;57:16–25. doi: 10.1034/j.1399-0004.2000.570103.x. [DOI] [PubMed] [Google Scholar]
- 2.Sartorelli V, Caretti G. Mechanisms underlying the transcriptional regulation of skeletal myogenesis. Curr Opin Genet Dev. 2005;15:528–535. doi: 10.1016/j.gde.2005.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tapscott SJ. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development. 2005;132:2685–2695. doi: 10.1242/dev.01874. [DOI] [PubMed] [Google Scholar]
- 4.McKinsey TA, Zhang CL, Olson EN. Control of muscle development by dueling HATs and HDACs. Curr Opin Genet Dev. 2001;11:497–504. doi: 10.1016/s0959-437x(00)00224-0. [DOI] [PubMed] [Google Scholar]
- 5.Albini S, Puri PL. SWI/SNF complexes, chromatin remodeling and skeletal myogenesis: It's time to exchange! Exp Cell Res. 2010;316:3073–3080. doi: 10.1016/j.yexcr.2010.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perdiguero E, Sousa-Victor P, Ballestar E, Muñoz-Cánoves P. Epigenetic regulation of myogenesis. Epigenetics. 2009;4:541–550. doi: 10.4161/epi.4.8.10258. [DOI] [PubMed] [Google Scholar]
- 7.Puri PL, et al. Class I histone deacetylases sequentially interact with MyoD and pRb during skeletal myogenesis. Mol Cell. 2001;8:885–897. doi: 10.1016/s1097-2765(01)00373-2. [DOI] [PubMed] [Google Scholar]
- 8.Mal A, Sturniolo M, Schiltz RL, Ghosh MK, Harter ML. A role for histone deacetylase HDAC1 in modulating the transcriptional activity of MyoD: Inhibition of the myogenic program. EMBO J. 2001;20:1739–1753. doi: 10.1093/emboj/20.7.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang CL, McKinsey TA, Olson EN. Association of class II histone deacetylases with heterochromatin protein 1: Potential role for histone methylation in control of muscle differentiation. Mol Cell Biol. 2002;22:7302–7312. doi: 10.1128/MCB.22.20.7302-7312.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mal A, Harter ML. MyoD is functionally linked to the silencing of a muscle-specific regulatory gene prior to skeletal myogenesis. Proc Natl Acad Sci USA. 2003;100:1735–1739. doi: 10.1073/pnas.0437843100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caretti G, Di Padova M, Micales B, Lyons GE, Sartorelli V. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 2004;18:2627–2638. doi: 10.1101/gad.1241904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mal AK. Histone methyltransferase Suv39h1 represses MyoD-stimulated myogenic differentiation. EMBO J. 2006;25:3323–3334. doi: 10.1038/sj.emboj.7601229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Puri PL, et al. Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Mol Cell. 1997;1:35–45. doi: 10.1016/s1097-2765(00)80005-2. [DOI] [PubMed] [Google Scholar]
- 14.Sartorelli V, et al. Acetylation of MyoD directed by PCAF is necessary for the execution of the muscle program. Mol Cell. 1999;4:725–734. doi: 10.1016/s1097-2765(00)80383-4. [DOI] [PubMed] [Google Scholar]
- 15.Dilworth FJ, Seaver KJ, Fishburn AL, Htet SL, Tapscott SJ. In vitro transcription system delineates the distinct roles of the coactivators pCAF and p300 during MyoD/E47-dependent transactivation. Proc Natl Acad Sci USA. 2004;101:11593–11598. doi: 10.1073/pnas.0404192101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duquet A, et al. Acetylation is important for MyoD function in adult mice. EMBO Rep. 2006;7:1140–1146. doi: 10.1038/sj.embor.7400820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shinkai Y, Tachibana M. H3K9 methyltransferase G9a and the related molecule GLP. Genes Dev. 2011;25:781–788. doi: 10.1101/gad.2027411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rathert P, et al. Protein lysine methyltransferase G9a acts on non-histone targets. Nat Chem Biol. 2008;4:344–346. doi: 10.1038/nchembio.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tachibana M, Matsumura Y, Fukuda M, Kimura H, Shinkai Y. G9a/GLP complexes independently mediate H3K9 and DNA methylation to silence transcription. EMBO J. 2008;27:2681–2690. doi: 10.1038/emboj.2008.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tachibana M, et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16:1779–1791. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kubicek S, et al. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol Cell. 2007;25:473–481. doi: 10.1016/j.molcel.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 22.Friday BB, Horsley V, Pavlath GK. Calcineurin activity is required for the initiation of skeletal muscle differentiation. J Cell Biol. 2000;149:657–666. doi: 10.1083/jcb.149.3.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ait-Si-Ali S, et al. A Suv39h-dependent mechanism for silencing S-phase genes in differentiating but not in cycling cells. EMBO J. 2004;23:605–615. doi: 10.1038/sj.emboj.7600074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma PC, Rould MA, Weintraub H, Pabo CO. Crystal structure of MyoD bHLH domain-DNA complex: Perspectives on DNA recognition and implications for transcriptional activation. Cell. 1994;77:451–459. doi: 10.1016/0092-8674(94)90159-7. [DOI] [PubMed] [Google Scholar]
- 25.Qian C, Zhou MM. SET domain protein lysine methyltransferases: Structure, specificity and catalysis. Cell Mol Life Sci. 2006;63:2755–2763. doi: 10.1007/s00018-006-6274-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun H, et al. Stra13 regulates satellite cell activation by antagonizing Notch signaling. J Cell Biol. 2007;177:647–657. doi: 10.1083/jcb.200609007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Azmi S, Ozog A, Taneja R. Sharp-1/DEC2 inhibits skeletal muscle differentiation through repression of myogenic transcription factors. J Biol Chem. 2004;279:52643–52652. doi: 10.1074/jbc.M409188200. [DOI] [PubMed] [Google Scholar]
- 28.Nishio H, Walsh MJ. CCAAT displacement protein/cut homolog recruits G9a histone lysine methyltransferase to repress transcription. Proc Natl Acad Sci USA. 2004;101:11257–11262. doi: 10.1073/pnas.0401343101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gulbagci NT, et al. SHARP1/DEC2 inhibits adipogenic differentiation by regulating the activity of C/EBP. EMBO Rep. 2009;10:79–86. doi: 10.1038/embor.2008.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.