Abstract
Expression of the homeobox protein CDX1 is lost or reduced in a significant proportion of colorectal carcinomas (CRCs) but the underlying mechanism for this is unclear. We have demonstrated absence of CDX1 mRNA expression in 7 of 37 CRC cell lines and shown that all 7 cell lines have a methylated CDX1 promoter. Twenty-five cell lines showed both CDX1 mRNA expression and an unmethylated CDX1 promoter. The five remaining cell lines had a partially methylated CDX1 promoter and all expressed CDX1 mRNA; when treated with the demethylating agent, 5-aza-2′-deoxycytidine, these five cell lines all showed increased CDX1 expression. No mutations were found in the promoter and coding regions of CDX1. One polymorphism was demonstrated in each of the promoter, 5′ UTR, and coding region of exon 1 of CDX1, but there were no associations between CDX1 mRNA expression and different polymorphic genotypes. Similarly, there was no association between CDX1 mRNA expression and loss of heterozygosity at the CDX1 locus. In conclusion, absence or reduction of CDX1 expression in CRC seems to be primarily regulated by promoter methylation and is probably selected for because of its impact on the differentiation of colonocytes.
CDX1 is a homeobox protein involved in regulating normal development of intestinal epithelium in utero (1). In adulthood, CDX1 expression is maintained in small and large intestinal epithelium (1, 2) although its exact physiological role at this time remains uncertain. That the protein has an oncosuppressor function is supported by in vitro and in vivo evidence of loss of CDX1 expression in colorectal carcinogenesis (3, 4). The mechanism underlying this loss is, however, unclear. Possible roles for genetic and/or epigenetic controls must therefore be considered. There is an ever growing list of oncosuppressor genes whose expressions are silenced through specific methylation of gene promoter CpG islands (CGIs). These genes all show 5′ regions that are rich in CGIs (5). In sharing this feature, CDX1 represents a prime candidate for promoter methylation control. Indeed, half way through our study, Suh et al. published preliminary data supporting this hypothesis (6).
The most comprehensive way of analyzing a gene for evidence of promoter methylation control is by direct sequencing after bisulfite modification of target DNA (so called “bisulfite sequencing”) (7). Ideally, this analysis should be accompanied by looking for reexpression of the silenced gene after treatment with the demethylating agent, 5-azacytidine or the less toxic 5-aza-2′-deoxycytidine (5aza2). Suh et al. (6) showed CDX1 promoter methylation status to explain CDX1 mRNA expression among eight colorectal carcinoma (CRC) cell lines and also successfully used 5aza2 treatment to induce or increase CDX1 mRNA expression in six of the cell lines. Nonetheless, two important questions regarding control of CDX1 expression in CRC remain unanswered. First, possible contributions of gene mutation and/or loss of heterozygosity (LOH) have not yet been excluded. Second, it is unclear which of the CGIs in the CDX1 promoter are most important to the control of gene expression. To help answer these questions, we have studied a significantly larger panel of CRC cell lines by using a mixture of promoter methylation analyses and gene mutation and LOH studies.
Methods
Cells and Tissues Studied. Thirty-seven CRC cell lines and the normal lymphoblastoid cell line Bristol 8 were studied. DNA and RNA were extracted from cell pellets and DNA from dissected normal colonic mucosa (dissected from paraffin-embedded, formalin-fixed normal colonic tissue) by using the DNeasy (Qiagen, Crawley, U.K.) or RNeasy kits (Qiagen). Unless stated otherwise, all cell pellets were prepared from cell lines at 50–80% confluence.
RT-PCR. cDNA was synthesized from 2 μg of denatured RNA (65°C for 5 min) through incubation at 37°C for 60 min with final quantities/concentrations of 1 μM of oligo(dT)s, 4 units of Omniscript reverse transcriptase (Qiagen), 10 units of RNase inhibitor (Ambion, Huntingdon, U.K.), and 0.5 mM each dNTP. The primers used for the CDX1 and β-actin RT-PCRs are shown in Table 1, and the PCR protocols are given below.
Table 1. Primers, amplicon size, and specific protocol details for PCRs used.
| Protocol details
|
|||||
|---|---|---|---|---|---|
| PCR | Forward (F) and reverse (R) primers (5′ to 3′) | Size, bp | T° | Time, s | No. of cycles |
| CDX1 bisulfite sequencing | F:TTGTGTGAAGTTGGTTTAGAATTT | 505 | 56 | 45 | 35 |
| R:ACACATAACCCACATACATAATAAC | |||||
| CDX1 methylated | F:GTTGGGGTCGCGGGCGGTTC | 185 | 63 | 45 | 35 |
| R:AATCCTTATCCAACACATAACCCACATA | |||||
| CDX1 unmethylated | F:TTTGGTTGGGGTTGTGGGTGGTTT | 189 | 63 | 45 | 35 |
| R:AATCCTTATCCAACACATAACCCACATA | |||||
| CDX1 promoter sequencing | F:GAAGGACAAGGTGTTCAGGCCG | 421 | 62 | 45 | 35 |
| R:GTCCAGCACATAGCCCACATACATG | |||||
| CDX1 exon 1a sequencing | F:CGAGTTCAGGTGAGCGGTTGC | 250 | 60 | 30 | 35 |
| R:CCGGCTCCACGTGAGAGTAGC | |||||
| CDX1 exon 1b sequencing | F:CCGCAGTACCCCGACTTCTCCA | 350 | 60 | 30 | 35 |
| R:GGCGAGGGAAGGGTCCTTAC | |||||
| CDX1 exon 2 WAVE | F:CACCTTCCTTCCCGGCTCCTTCTG | 222 | 65 | 30 | 35 |
| R:GTCCTCGCACTGCTCCTATGGC | |||||
| CDX1 exon 3 WAVE | F:GCTGCCCACTCCATCTCTGTCC | 450 | 55 | 60 | 35 |
| R:CCCAGCACCCGAGTCCCCAG | |||||
| CDX1 RT-PCR | F:AGGACAAGTACCGCGTGGTCTA | 553 | 66 | 60 | 30 |
| R:CCTCTGAACGTATGGAGGAGGA | |||||
| Beta-actin RT-PCR | F:ACACCTTCTACAATGAGC | 600 | 56 | 60 | 25 |
| R:ACGTCACACTTCATGATG | |||||
T°, annealing temperature.
5aza2 Treatment. Cells grown to 60–70% confluence were incubated for 72 h in culture media containing a final 5aza2 (Sigma) concentration of 5 μM. After 72 h, the cells were washed and reincubated with fresh culture medium (without 5aza2) for a further 24 h before harvesting.
Bisulfite Sequencing. DNA (1 μg of DNA per sample) was modified with the CpGenome DNA modification kit (Intergen, Purchase, NY). A 450-bp region upstream of exon 1 of CDX1 was amplified by using the “CDX1 bisulfite sequencing PCR” primer pair shown in Table 1 and the protocol given below. This primer pair was designed to bind sequences lacking any CGIs, therefore avoiding any preferential amplification of methylated or unmethylated DNA strands. Forty microliters of the PCR products were purified (QIAquick kit, Qiagen), and 4 μl of the purified product was used for cloning (Topo TA Cloning kit, Invitrogen) and direct sequencing by using the Big Dye Terminator Cycle Sequencing kit (Applied Biosystems) and the fluorescent sequencer ABI 377 (Applied Biosystems).
Methylation-Specific PCR (MS-PCR). A common reverse primer was used for both the methylated and unmethylated PCRs (Fig. 1 and Table 1). The forward primers for these PCRs were designed to anneal to the same stretch of four CGIs but to show differential annealing according to the methylation status of the CGIs (Fig. 1). “Partial methylation” of the CDX1 promoter was defined as presence of amplified products with both methylated and unmethylated PCRs.
Fig. 1.
MS-PCR for CDX1. (A and B) Primer-binding sites for both CDX1 methylated PCR (M-PCR) and unmethylated PCR (UM-PCR) are shown. Examples of bisulfite-modified CDX1 DNA sequences are given showing completely methylated or unmethylated CGIs. CGIs are highlighted in bold, and base positions relative to the CDX1 transcription start site are shown at the end of each sense strand. Only the bisulfite-modified sense strand is amplified by either the M-PCR and UM-PCR. The common reverse primer binds to this sense strand, and the complementary strand thus produced forms the target site for the forward primers.(C) Examples of M-PCR and UM-PCR using bisulfite-modified C10 (C), LOVO (LO), and LS174T (LS) DNA and genomic LOVO DNA (G) as templates. Water blanks (–) and 100-bp ladders (bright band represents 500 bp) are also shown. These data accurately reflected the bisulfite sequencing results for the three CRC cell lines, as seen in Figs. 4 and 5, whereas no bands were produced from amplifying genomic, unmodified DNA.
Mutation Analyses. A 400-bp region upstream of exon 1 of CDX1 was amplified (see Table 1 for primers), and the products were directly sequenced, as above. Exon 1 of CDX1 was amplified by two sets of overlapping primers (“exon 1a” primers for upstream half and “exon 1b” primers for downstream half; see Table 1) and directly sequenced, as above. The protocols used for all these PCRs are given below. Exon 2 and the coding region of exon 3 of CDX1 were amplified by using the primers shown in Table 1 and the protocols given below. The PCR products were denatured for 4 min at 95°C and then allowed to hybridize by using 42 cycles (of 1-min length each) starting at 95°C and dropping by 1.6°C per cycle to 27.8°C. These products were then run on a denaturing HPLC machine (WAVE, Transgenomic, Crewe, U.K.).
LOH Studies. The 2-bp repeat microsatellite loci D5S436 (5q33.1), D5S410 (5q33.2), and D5S422 (5q34) flank the CDX1 gene locus (5q33.1). These three highly informative microsatellite markers (heterozygosity scores for D5S436, D5S410 and D5S422: 0.83, 0.79, and 0.84, respectively) were studied through amplification by using commercial fluorescently labeled PCR primer pairs (ABI PRISM Linkage Mapping Set version 2.5, Applied Biosystems). Genomic DNA (10 ng) from each of the 37 CRC cell lines was used for each PCR. The amplified products were analyzed by using the fluorescent sequencer ABI 377 (Applied Biosystems) and the computer software genotyper v3.7 (Applied Biosystems). Because no corresponding normal tissues were available for analysis, LOH of CDX1 was defined as hemizygosity/homozygosity seen for all three markers (corresponding to P = 0.0057).
PCRs. For the CDX1 and β-actin RT-PCRs, the CDX1 bisulfite sequencing PCR, the CDX1 exon 2 and exon 3 WAVE PCRs, and for both methylated and unmethylated PCRs, a 50-μl PCR volume was used comprising final quantities/concentrations of 0.2 μM each primer, 1 unit of AmpliTaq Gold (Perkin–Elmer) polymerase, 1.5 mM MgCl2, 200 μM each dNTP and 200 ng of cDNA (for CDX1 RT-PCR), 100 ng of cDNA (for β-actin RT-PCR), 100 ng of DNA (CDX1 exon 2 and exon 3 WAVE PCRs), or 2 μl of bisulfite-modified DNA (for bisulfite sequencing PCR and MS-PCRs). The cycling conditions used for these PCRs were a denaturation step of 95°C for 10 min, 25–35 cycles of 95°C for 45 s, annealing temperature for 45 s, and 72°C for 30–60 s (see Table 1 for specific details) and a final extension step of 72°C for 10 min. Because of the high GC content of the CDX1 promoter and exon 1, a different PCR protocol using “Q” solution (Qiagen) was required to optimize amplification. For these reactions, a 50-μl PCR volume was used comprising final quantities/concentrations of 0.2 μM of each primer, 6 μl of “Q” solution, 1 unit of Hotstar Taq polymerase (Qiagen), 1.5 mM MgCl2, 200 μM each dNTP, and 100 ng of DNA. The cycling conditions used for these PCRs were a denaturation step of 95°C for 12 min, 35 cycles of 95°C for 30 s, annealing temperature (see Table 1 for details) for 30 s, and 72°C for 45 s, and a final extension step of 72°C for 10 min. All PCR products were visualized by electrophoresis and ethidium bromide staining in 2% agarose gels.
Results
Analysis of CGIs in the 5′-Flanking Region of CDX1. Of the 37 cell lines studied, 7 (19%) completely lacked expression of CDX1 mRNA on RT-PCR (Table 2). The lymphoblastoid cell line Bristol 8 also lacked CDX1 mRNA expression. Three of the CDX1 nonexpressing cell lines (C10, DLD1, and HCT116) were arbitrarily selected and treated with 5aza2 (Fig. 2). The strong reexpression of CDX1 mRNA induced by this treatment supported a role for promoter methylation in the regulation of CDX1 expression. We therefore analyzed the promoter region of CDX1 to identify CGIs whose methylation might be important to such regulation. The sequence of a 1-kb region upstream from exon 1 was derived from the University of California at Santa Cruz (UCSC) Genome Bioinformatics Site (www.genome.ucsc.edu). The first 400 bp upstream from the CDX1 transcription start site show a high density of CGIs, and, from –16 to –200 within this region, there is 93% homology with the mouse equivalent, as derived from the UCSC Genome Bioinformatics Site (Fig. 3). Further, this 200-bp 5′-flanking stretch contains a TATA box and numerous potential transcription factor-binding sites (Fig. 3).
Table 2. Results of CDX1 RT-PCR, methylated and unmethylated PCR (M-PCR and UM-PCR, respectively), polymorphism analyses (unless specified, the base shown represents homozygosity), and LOH studies for 37 colorectal carcinoma cell lines.
|
CDX1 polymorphism
|
|||||||
|---|---|---|---|---|---|---|---|
| CRC cell line | CDX1 mRNA | CDX1 M PCR | CDX1 UM PCR | Promoter | 5′ UTR | Exon 1 | LOH |
| C70 | + | - | + | C | G | G | Y |
| C75 | + | - | + | C | A | C | N |
| C84 | + | - | + | heteroC/T | heteroA/G | heteroC/G | N |
| C99 | + | - | + | C | A | C | N |
| C106 | + | - | + | C | A | C | Y |
| C125 | + | - | + | C | heteroA/G | heteroC/G | N |
| COLO206 | + | - | + | C | G | G | Y |
| HCA46 | + | - | + | C | heteroA/G | heteroC/G | N |
| HT55 | + | - | + | heteroC/T | heteroA/G | heteroC/G | N |
| JW | + | - | + | C | heteroA/G | heteroC/G | N |
| LS174T | + | - | + | C | A | C | N |
| LS 180 | + | - | + | C | A | C | N |
| LS1034 | + | - | + | C | G | G | N |
| LIM1863 | + | - | + | C | A | C | Y |
| SNU-C2B | + | - | + | C | heteroA/G | heteroC/G | N |
| SW403 | + | - | + | C | heteroA/G | heteroC/G | N |
| SW480 | + | - | + | C | G | G | Y |
| SW620 | + | - | + | C | G | G | Y |
| SW948 | + | - | + | C | A | C | N |
| T84 | + | - | + | T | G | G | Y |
| VACO4A | + | - | + | C | A | C | Y |
| VACO4S | + | - | + | C | A | C | Y |
| VACO 400 | + | - | + | C | A | C | N |
| VACO429 | + | - | + | T | G | G | Y |
| WIDR | + | - | + | C | A | C | Y |
| C10 | - | + | - | C | A | C | Y |
| CC20 | - | + | - | heteroC/T | heteroA/G | heteroC/G | N |
| DLD1 | - | + | - | C | G | G | N |
| HCT116 | - | + | - | C | heteroA/G | heteroC/G | N |
| LS 411 | - | + | - | C | A | C | N |
| NCI H716 | - | + | - | C | A | C | Y |
| RKO | - | + | - | heteroC/T | heteroA/G | heteroC/G | N |
| C32 | + | + | + | heteroC/T | G | G | N |
| CC07 | + | + | + | C | heteroA/G | heteroC/G | N |
| HCA7 | + | + | + | C | A | C | N |
| LOVO | + | + | + | C | A | C | N |
| NCI-747 | + | + | + | C | heteroA/G | heteroC/G | N |
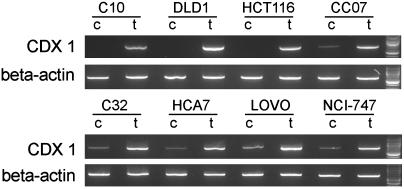
Fig. 2.
5aza2 treatment experiments on eight colorectal carcinoma cell lines. Treated (t) and corresponding control (c) samples are shown for each cell line. The gels show CDX1 mRNA expression as assessed with RT-PCR along with β-actin loading controls for each template. One hundred-base pair ladders (the Upper and Lower bright bands represent 1 kb and 500 bp, respectively) are also shown.
Fig. 3.
Sequences of the 5′-flanking regions for human and mouse CDX1. The base positions relative to the transcription start site for human CDX1 are shown above each line of sequence. CGIs are shown in bold. The TATA box is shown in purple, and highlighted in other colors are potential binding sites for the following transcription factors: SpI (blue), CCAAT/enhancer-binding protein (CEBP) (red), the zinc finger protein CTCF (green), and TCF/LEF (yellow).
The 400-bp, 5′-flanking region of CDX1 was amplified and sequenced from bisulfite-modified DNA prepared from two CDX1-expressing CRC cell lines (C70 and LS174T), a CDX1 nonexpressing CRC cell line (C10), Bristol 8, and from normal colonic mucosa (Fig. 4). This bisulfite sequencing showed, among the 3 CRC cell lines, a clear association between heavy methylation of CGIs in this 400-bp region and lack of CDX1 mRNA expression, and vice versa. Although not as densely methylated as C10, the CDX1 promoter CGIs were still predominantly methylated for Bristol 8. Although some of the sequenced clones from the normal colonic mucosa specimen may have originated from contaminating nonepithelial cells, we could still demonstrate that a majority of CDX1 promoter CGIs were unmethylated for this sample.
Fig. 4.
CDX1 promoter bisulfite sequencing. Sequencing data, together with CDX1 mRNA expression status, are shown for C70, LS174T, C10, Bristol 8, normal colonic mucosa, and CC07. For each case, relative location and methylation status of CDX1 promoter CGIs for 10 clones are shown. Each circle represents a CGI, and base positions relative to the CDX1 transcription start site are shown above each cluster of clones. Methylated CGIs are highlighted in gray. The horizontal solid lines encompass CGIs within the 200-bp length 5′-flanking region, which shows close homology to its mouse counterpart (see also Fig. 2). CGIs enclosed by the boxes (positions –53 to –65) represent the binding site of the methylation-specific forward primers (see also Fig. 1).
Earlier studies on the CRC cell line LOVO showed an increase in CDX1 mRNA expression with postconfluent growth, in keeping with recent similar findings of Qualtrough et al. (8). We hypothesized that this increase of expression might associate with apparent demethylation of CDX1 promoter CGIs, which are critical to regulating gene expression. After establishing equal sized subcultures of LOVO, one subculture was harvested after 4 days growth (at 40–50% confluence) whereas the second was harvested after 17 days growth (10 days postconfluence). The increase in CDX1 expression after postconfluent growth was associated with fewer clones showing methylation, particularly in the 200-bp 5′-flanking region (Fig. 5). Within this region, reanalyzing the methylation patterns of Bristol 8 and normal colonic mucosa showed the methylation status of four CGIs located at –53 to –65 (Fig. 4) to correlate closest with CDX1 mRNA expression. Further, the –53 to –65 CGI cluster contains a potential binding site for the transcription factor SpI (Fig. 3). This finding is in keeping with other genes such as MAGE-1(9) and p14ARF(10) whose critical promoter CGIs are located close to or within the binding sites of transcription factors.
Fig. 5.
Promoter bisulfite sequencing (presented as in Fig. 4) (A) and CDX1 RT-PCR (B) of LOVO harvested after 4 (D4) and 17 (D17) days' growth. The gel shows increased CDX1 mRNA expression with postconfluent growth. β-Actin loading controls, normal colonic mucosa serving as a positive control (+), water blanks (–), and 100-bp ladders (the Upper and Lower bright bands represent 1 kb and 500 bp, respectively) are also shown.
MS-PCR and Correlation with CDX1 mRNA Expression (Table 2). The forward primer for MS-PCR was therefore targeted at these 4 CGIs in this 200-bp 5′-flanking region (Fig. 1). Because there was no other, more distant cluster of CGIs whose methylation status correlated as closely with CDX1 expression, a common reverse primer was designed (for both the methylated and unmethylated PCRs) that annealed to a CpG-free region (Fig. 1). When tested on previously bisulfite-sequenced CRC cell lines, the MS-PCRs consistently reflected the methylation status of the –53 to –65 promoter CGIs: completely methylated for C10, partially methylated for LOVO, and completely unmethylated for LS174T (Figs. 1, 4, and 5). These MS-PCRs were therefore used to assess CDX1 promoter methylation status for the remaining 34 CRC cell lines.
Of the 37 cell lines, 7 showed CDX1 promoter methylation and all 7 did not express CDX1 mRNA. Twenty-five cell lines showed an unmethylated CDX1 promoter, and, in keeping with this result, all 25 lines showed CDX1 mRNA expression. The five remaining cell lines (C32, CC07, HCA7, LOVO, and NCI-747) showed partial CDX1 promoter methylation and all expressed CDX1 mRNA. Bisulfite sequencing of CC07 showed, as with LOVO, a variegated pattern of methylation of CGIs of the CDX1 promoter (Fig. 4). To test whether demethylation of the remaining methylated strands/cells could further increase CDX1 mRNA expression, C32, CC07, HCA7, LOVO, and NCI-747 were treated with 5aza2. In all five cases, this treatment led to an increase in CDX1 mRNA expression (Fig. 2).
CDX1 Mutation and LOH Analyses (Table 2). To exclude an additional genetic event contributing to loss of CDX1 expression and to try to explain the cases of partial promoter methylation, all 37 cell lines were analyzed for mutations in the CDX1 promoter and coding regions, and for LOH at the CDX1 gene locus. Denaturing HPLC (DHPLC) analysis failed to detect any mutations in exons 2 and 3 in any of the 37 CRC cell lines. The accuracy of DHPLC analysis is compromised when studying GC rich sequences. Therefore, we directly sequenced the promoter region and exon 1 of CDX1 for the 37 cell lines. No mutations were demonstrated in either region. However, these sequencing analyses did demonstrate a single-nucleotide polymorphism (SNP) each in the promoter region, 5′ UTR, and coding region of exon 1. The three SNPs showed complete linkage disequilibrium with one another, creating effectively three alleles. However, there were no obvious associations between the six possible genotypes generated by these three SNPs and CDX1 mRNA expression among the 37 CRC cell lines (χ2 test, P = 0.71). There was an excess of homozygotes for each of the three SNPs implying LOH. The expected frequency of LOH can be calculated by using the formula: (o–h)/(1 –h), where o and h are the observed and expected proportions of homozygotes in the study population (11). As o = 24/37 and h = 16.25/37, the expected frequency of LOH at CDX1 among the 37 cell lines was 37%. This compared favorably with the observed LOH frequency based on the three microsatellite markers studied, i.e., 13/37 cell lines, 35.1%. There was, however, no association between observed LOH at the CDX1 gene locus and CDX1 mRNA expression among the 37 cell lines (two-tailed Fisher exact test, P = 0.98).
Discussion
We have confirmed that CDX1 expression is absent in a significant proportion of CRC cell lines. The selection pressure for such loss of expression, however, remains uncertain. Although the loss of expression with carcinoma development (2–4) suggests a tumor suppressor role for CDX1, the protein has few known downstream effectors that would mediate such a role. An alternative explanation for loss of CDX1 expression in colorectal carcinogenesis relates to the protein's putative physiological role in mediating intestinal epithelial cell differentiation (1, 2). In keeping with this finding, postconfluent growth of CRC cell lines has been shown to induce functional differentiation (8) whereas we confirmed that such growth can increase expression of CDX1 mRNA. Loss of CDX1 expression may therefore be advantageous to CRC cells in maintaining a dedifferentiated state and so encouraging dysregulated growth.
We have shown that loss of CDX1 expression among CRC cell lines is attributable to promoter methylation and not mutations nor LOH of CDX1. Further, our use of several indirect approaches collectively suggested CGIs important in regulating CDX1 expression to be located between 53 to 63 bp upstream from the transcription start site. Our MS-PCR was therefore designed around these islands and should be useful in future studies for rapid representation of the functional methylation status of the CDX1 promoter.
Using the CDX1 MS-PCR, we showed a direct inverse relation between CDX1 promoter methylation and gene expression among 32 of 37 CRC cell lines. The remaining five cell lines showed partial methylation of the CDX1 promoter with CDX1 mRNA expression. In all five cases, completing the demethylation with 5aza2 treatment led to increased CDX1 expression. Bisulfite sequencing of two of these five cell lines confirmed that clones showed either predominant methylation or unmethylation of CGIs in the 200-bp 5′-flanking region of CDX1. It remains uncertain whether the differing clones/DNA strands originate from different cells or whether they represent different alleles within the same cell. We have shown in the cell line HCT116 that partial methylation of CGIs in the p16INK4a promoter relates to presence of a heterozygous downstream mutation: the unmethylated allele always contains the mutation whereas the methylated allele is always wild type (unpublished results). However, we could not demonstrate any mutations, polymorphic genotypes, or LOH patterns to similarly explain the partial CDX1 promoter methylation of the 5 aforementioned cell lines.
Aside from variation of methylation pattern between clones, our bisulfite sequencing data also showed variation, between different cell lines, in the extent of CDX1 promoter CGI methylation along an individual DNA strand. In the cases of LS174T and C10, each clone studied showed either completely unmethylated or methylated CGIs. In the cases of LOVO, CC07, and Bristol 8, however, each clone studied showed a more “variegated” pattern of CGI methylation, with a general decrease in methylation in a 5′ to 3′ direction along the CDX1 promoter (Figs. 4 and 5). This finding would support the concept of DNA methylation spreading from upstream into a promoter region. (12) The actual variegated methylation pattern itself has been ascribed to positional shifts in this wave of methylation in relation to the binding of transcription factors (12). The partial methylation status of the five cell lines mentioned above may therefore merely represent different stages of methylation or demethylation in cell lines with less stably methylated CDX1 promoters. Why some cell lines should show a less stably methylated promoter will then require further investigation.
Acknowledgments
This work was funded by Cancer Research UK, the Digestive Disorders Foundation, and the Jean Shanks Foundation.
Abbreviations: 5aza2, 5-aza-2′-deoxycytidine; CGI, CpG island; CRC, colorectal carcinoma, MS-PCR: methylation-specific PCR; LOH, loss of heterozygosity; SNP, single-nucleotide polymorphism.
References
- 1.Freund, J. N., Domon-Dell, C., Kedinger, M. & Duluc, I. (1998) Biochem. Cell. Biol. 76, 957–969. [DOI] [PubMed] [Google Scholar]
- 2.Silberg, D. G., Furth, E. E., Taylor, J. K., Schuck, T., Chiou, T. & Traber, P. G. (1997) Gastroenterology 113, 478–486. [DOI] [PubMed] [Google Scholar]
- 3.Vider, B. Z., Zimber, A., Chastre, E., Gespach, C., Halperin, M., Mashiah, P., Yaniv, A. & Gazit, A. (2000) Biochem. Biophys. Res. Commun. 272, 513–518. [DOI] [PubMed] [Google Scholar]
- 4.Mallo, G. V., Rechreche, H., Frigerio, J. M., Rocha, D., Zweibaum, A., Lacasa, M., Jordan, B. R., Dusetti, N. J., Dagorn, J. C. & Iovanna, J. L. (1997) Int. J. Cancer 74, 35–44. [DOI] [PubMed] [Google Scholar]
- 5.Jubb, A. M., Bell, S. M. & Quirke, P. (2001) J. Pathol. 195, 111–134. [DOI] [PubMed] [Google Scholar]
- 6.Suh, E. R., Ha, C. S., Rankin, E. B., Toyota, M. & Traber, P. G. (2002) J. Biol. Chem. 277, 35795–35800. [DOI] [PubMed] [Google Scholar]
- 7.Frommer, M., McDonald, L. E., Millar, D. S., Collis, C. M., Watt, F., Grigg, G. W., Molloy, P. L. & Paul, C. L. (1992) Proc. Natl. Acad. Sci. USA 89, 1827–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qualtrough, D., Hinoi, T., Fearon, E. & Paraskeva, C. (2002) Gut 51, 184–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Smet, C., Lurquin, C., Lethe, B., Martelange, V. & Boon, T. (1999) Mol. Cell. Biol. 19, 7327–7335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng, S., Chen, P., McMillan, A., Lafuente, A., Lafuente, M. J., Ballesta, A., Trias, M. & Wiencke, J. K. (2000) Carcinogenesis 21, 2057–2064. [DOI] [PubMed] [Google Scholar]
- 11.Bodmer, W. F., Browning, M. J., Krausa, P., Rowan, A., Bicknell, D. C. & Bodmer, J. G. (1993) Ann. N.Y. Acad. Sci. 690, 42–49. [DOI] [PubMed] [Google Scholar]
- 12.Turker, M. S. (2002) Oncogene 21, 5388–5393. [DOI] [PubMed] [Google Scholar]