Abstract
MicroRNAs (miRNAs) are small noncoding RNAs that regulate eukaryotic gene expression by binding to regions of imperfect complementarity in mRNAs, typically in the 3′ UTR, recruiting an Argonaute (Ago) protein complex that usually results in translational repression or destabilization of the target RNA. The translation and decay of mRNAs are closely linked, competing processes, and whether the miRNA-induced silencing complex (RISC) acts primarily to reduce translation or stability of the mRNA remains controversial. miR-122 is an abundant, liver-specific miRNA that is an unusual host factor for hepatitis C virus (HCV), an important cause of liver disease in humans. Prior studies show that it binds the 5′ UTR of the messenger-sense HCV RNA genome, stimulating translation and promoting genome replication by an unknown mechanism. Here we show that miR-122 binds HCV RNA in association with Ago2 and that this slows decay of the viral genome in infected cells. The stabilizing action of miR-122 does not require the viral RNA to be translationally active nor engaged in replication, and can be functionally substituted by a nonmethylated 5′ cap. Our data demonstrate that a RISC-like complex mediates the stability of HCV RNA and suggest that Ago2 and miR-122 act coordinately to protect the viral genome from 5′ exonuclease activity of the host mRNA decay machinery. miR-122 thus acts in an unconventional fashion to stabilize HCV RNA and slow its decay, expanding the repertoire of mechanisms by which miRNAs modulate gene expression.
Keywords: RNA decay, viral host factor
MicroRNAs (miRNAs) typically regulate eukaryotic gene expression by binding to regions of imperfect complementarity in the 3′ UTR of mRNAs, recruiting an Argonaute (Ago) protein complex that results in translational repression or destabilization of the target RNA (1). Although miRNAs regulate a majority of genes, an unresolved question is whether the miRNA-induced silencing complex (RISC) acts primarily to reduce translation or enhance decay of the mRNA, two closely linked, competing processes (2–4). miR-122 is an abundant, liver-specific miRNA, comprising >50% of mature miRNAs in human hepatocytes and regulating the expression of numerous hepatic genes, including those involved in fatty acid and cholesterol metabolism (5, 6). It is also a very unusual host factor required for replication of hepatitis C virus (HCV), an important cause of liver disease in humans (7, 8). Prior studies show that miR-122 binds the 5′ UTR of the positive-strand HCV RNA genome (7, 9), stimulating viral protein expression and promoting viral replication by a poorly understood mechanism (10, 11).
Although miR-122 does not directly stimulate HCV RNA synthesis (12, 13), its ability to promote genome amplification is independent of its regulation of hepatic metabolism (12) and requires the binding of its “seed sequence” (nucleotides 2–8) to two conserved sites (S1 and S2) in the viral 5′ UTR (7, 9). Additional “supplementary” base-pairing between miR-122 and HCV RNA sequences upstream of S1 and S2 has also been recognized recently and shown to be essential for promotion of genome amplification (14, 15). The miR-122 binding sites are near the 5′ end of the RNA and immediately upstream of an internal ribosome entry site (IRES) that has high affinity for the 40S ribosome subunit (16). The unusual ability of miR-122 to stimulate viral protein translation (10, 11) is dependent on where it binds, because miR-122 suppresses expression of capped reporter mRNAs that contain the HCV target sequence in the 3′ UTR (9). Translation enhancement only partially explains the role of miR-122 in the HCV life cycle, however, because mutant viral RNAs that are deficient in miR-122 binding are far more handicapped in their ability to replicate than viral RNAs with mutations in the IRES that result in quantitatively comparable defects in translation (11).
Although there has been speculation that miR-122 might promote genome amplification and viral protein expression by physically stabilizing HCV RNA (14, 17), previous experimental results suggest this is not the case and that miR-122 does not enhance RNA stability (7, 10). Here we present a contrasting view and show that binding of miR-122 to the 5′ terminus of HCV RNA in association with Ago2 significantly slows decay of the viral RNA genome in infected cells. miR-122 thus acts in an unconventional fashion to stabilize HCV RNA, expanding the repertoire of mechanisms by which miRNAs modulate gene expression.
Results
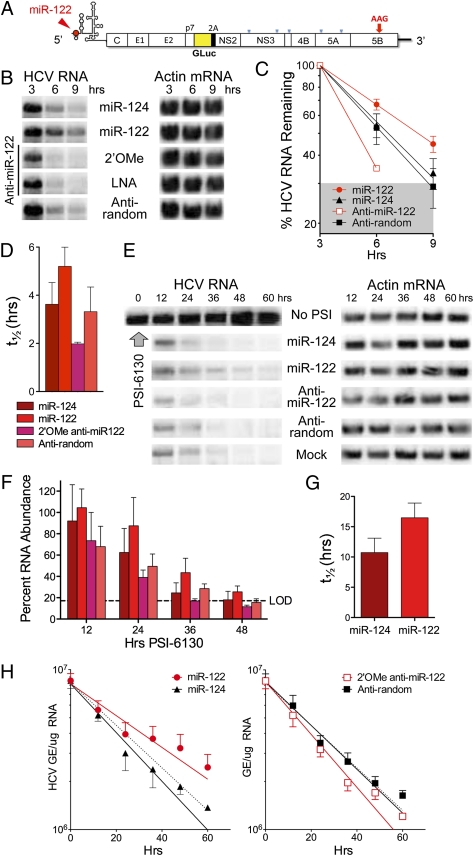
We studied how miR-122 influences the stability of synthetic HCV RNA transfected into human hepatoma cells. Northern blots demonstrated significant increases in the abundance of a replication-defective viral RNA (H77S/GLuc2A-AAG, that contains a lethal mutation in its RNA polymerase) (Fig. 1A) when it was electroporated into cells together with duplex miR-122 (Fig. 1B). miR-124, a brain-specific miRNA that does not bind HCV RNA, had no such effect. Conversely, cotransfection of viral RNA with 2'O-methyl-modified (2'OMe) or locked nucleic acid antisense oligoribonucleotides capable of sequestering miR-122 reduced the abundance of HCV RNA at 3, 6, and 9 h after electroporation. These differences were reproducible in multiple experiments and observed with HCV RNAs containing 5′ UTR sequence from either genotype 1 (H77) or genotype 2 (JFH1) virus. We estimated the rate of viral RNA decay by PhosphorImager analysis of Northern blots, normalizing HCV RNA abundance (HCV RNA/actin mRNA) to that present at 3 h after electroporation to compare rates of decay between 3 and 9 h under different conditions (Fig. 1C). This allowed for recovery of the cells after electroporation, and when the data were fit to a one-phase decay model, indicated that the half-life (t1/2) of HCV RNA in cells cotransfected with miR-122 was 5.2 h vs. 3.60 h for cells cotransfected with miR-124 (P = 0.0035 by the extra sum-of-squares F test) and 3.3 vs. 2.0 h for cells transfected with anti-random vs. the anti–miR-122 antagomir (P = 0.0016) (Fig. 1D). Similar differences in rates of decay were observed when HCV RNA was assayed by quantitative RT-PCR (qRT-PCR) (Fig. S1A). Differences in RNA decay were matched by differences in viral protein expression (Fig. S1B), monitored by measuring Gaussia luciferase (GLuc) encoded by sequence inserted into the viral genome (Fig. 1A). Taken together, these results indicate that miR-122 positively regulates the stability of transfected HCV RNA. The corresponding increase in protein expression provides a logical explanation for the enhanced HCV translation reported previously (10, 11).
Fig. 1.
miR-122 stabilizes the HCV RNA genome. (A) Organization of H77S/GLuc2A-AAG RNA that expresses GLuc as part of the HCV polyprotein (31). The AAG mutation in NS5B ablates genome amplification. The position at which miR-122 binds the 5′ UTR is shown. (B) Northern blot of H77S/GLuc2A-AAG and actin RNA (loading control) in Huh-7.5 cells after electroporation of viral RNA together with duplex miRNAs or antisense oligoribonucleotides. (C) Quantitation of HCV RNA relative to actin mRNA by phosphorimaging of Northern blots from five independent electroporation experiments. Data shown represent the mean percentage RNA remaining (±SEM) relative to that present at 3 h after electroporation under each condition. Data were fit to a one-phase decay model (R2 = 0.87–0.99). The shaded area represents the approximate limit of detection (LOD; 9 h posttransfection data from anti-miR-122–transfected cells were below the LOD and excluded from analysis). (D) Mean HCV RNA t1/2 estimated by fitting the Northern blot data to a one-phase decay model, as shown in C. Error bars indicate 95% confidence intervals. (E) Northern blot of HCV RNA in persistently infected (>3 wk) cells treated with PSI-6130 (10 μM or >10-fold the EC50) and transfected with miRNAs or 2'OMe oligoribonucleotides. (F) PhosphorImager quantitation of Northern blots in two replicate experiments involving PSI-6130 treatment. Results were normalized to RNA abundance in mock-transfected cells at 12 h. See D for the key. (G) Mean HCV RNA t1/2 ± SD in cells supplemented with miR-122 vs. miR-124 after PSI-6130 treatment, estimated by fitting PhosphorImager data from four independent experiments to a one-phase decay model. P = 0.007 by two-sided paired t test. (H) qRT-PCR determination of HCV RNA decay in infected cells treated with PSI-6130 and supplemented with miRNAs and antisense oligoribonucleotides as in E. Data are from three replicate infected cultures, and represent HCV genome equivalents (GE) per μg total RNA ± SD (left) HCV RNA in cells supplemented with miR-122 or miR-124. Data were fit to a one-phase decay model (R2 = 0.86–0.95): HCV RNA t1/2 =30.3 h for miR-122 vs. 19.8 h for miR-124 (P = 0.018). Right: Cells treated with 2’-OMe anti-miR-122 or anti-random (R2 = 0.95–0.96): t1/2 =18.2 h for anti-miR-122 vs. 21.9 h for anti-random (P = 0.023). The dashed line in both panels represents the one-phase decay curve in mock-treated cells.
Because transfected RNA is likely to be subject to different decay pathways than replicating viral genomes in infected cells, we determined whether miR-122 also slows degradation of viral RNA in infected cells treated with PSI-6130, a potent and specific nucleoside inhibitor that arrests new viral RNA synthesis (18). Under these conditions, as expected, viral RNA degraded more slowly than after electroporation (compare Fig. 1 B and E). However, its rate of decay was reduced when miR-122 was transfected simultaneously with PSI-6130 treatment (Fig. 1E). When fit to a one-phase decay model, PhosphorImager data from replicate experiments (Fig. 1F) indicated a significant difference in the rate constant for HCV RNA decay, k (k = ln(2)/t1/2), in cells supplemented with miR-122 vs. miR-124 (P = 0.048 by the extra sum-of-squares F test). The difference in the t1/2 was highly significant statistically (P = 0.007 by two-sided paired t test) (Fig. 1G). Likewise, increases in the decay rate in cells transfected with anti-miR-122 vs. the control anti-random oligonucleotide were also significant (P = 0.014 by F test), whereas decay rate constants were similar in cells receiving anti-miR-124, anti-random, or mock treatment (P > 0.05). Similar results were observed when miR-122 was transfected into cells 8 h after the addition of PSI-6130, which would allow for any potential delay in suppression of viral RNA synthesis due to the need for phosphorylation of the inhibitor (Fig. S1 C and D).
In a completely independent set of experiments, we used qRT-PCR to quantify HCV RNA in infected cells treated with PSI-6310. The results suggested a longer t1/2 for HCV RNA (≈19 h vs. ≈10 h) in miR-124–treated cells than that determined by Northern analysis. This is likely to reflect the small size of the RNA segment detected in the RT-PCR assay (221 bases vs. the 9.7-kb RNA genome detected in Northern blots) and the inability of the RT-PCR assay to discriminate between intact and partially degraded RNAs. However, we again observed significant differences in HCV RNA decay rates in cells supplemented with miR-122 vs. miR-124, or anti-miR-122 vs. anti-random (Fig. 1H). Although the magnitude of this effect is relatively small (not unlike the impact of miRNAs on cellular mRNA translation), these data show collectively that miR-122 reproducibly stabilizes the viral RNA genome in infected cells.
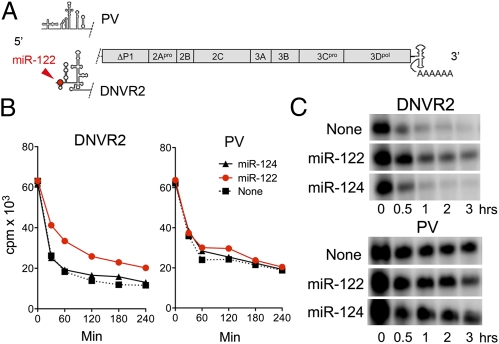
We next determined whether miR-122 could directly stabilize RNA in a cell-free system. For this, we compared poliovirus (PV) RNA and a related RNA (DNVR2) in which the PV 5′ UTR was replaced with the HCV 5′ UTR (Fig. 2A). The stabilities of these RNAs have been compared previously in S10 translation mixtures prepared from HeLa cells (19, 20), providing a useful context for these experiments. PV RNA is stabilized in these extracts by poly(rC) binding protein 1 (hnRNP1-E1), which associates with a 5′-terminal cloverleaf RNA structure, and decays more slowly than DNVR2 RNA (20). However, DNVR2 RNA stability was increased and approximated that of PV RNA when duplex miR-122, but not miR-124, was added to S10 reactions before the viral RNA (Fig. 2 B and C). The stabilization of DNVR2 RNA by miR-122 was reproducible and statistically significant (Fig. 2 legend). In contrast, neither miRNA enhanced stability of PV RNA lacking HCV sequence. Thus, miR-122 directly regulates stability of RNA containing the HCV 5′ UTR and does not accomplish this indirectly by modulating cellular gene expression.
Fig. 2.
miR-122 stabilizes synthetic RNA containing the HCV 5′ UTR. (A) Structure of DNVR2 and PV RNAs (19), which differ only in 5′ UTR sequence. (B) miR-122 slows decay of DNVR2 RNA in HeLa S10 lysate. Data shown represent acid-precipitable α-[32P]-CTP-labeled RNA in HeLa S10 reaction mixtures (19) containing 1 μM of duplex miRNA. The DNVR2 decay constant in miR-122– vs. miR-124–supplemented mixtures, estimated by fitting the data to a one-phase decay model (R2 = 0.972–0.989), differed significantly (P = 0.002). (C) RNA extracted from HeLa S10 reaction mixtures and fractionated by electrophoresis in 0.8% agarose.
Mutations in S1 and S2 that ablate miR-122 binding are lethal to replication of HCV that has been adapted to growth in cell culture (HJ3-5 virus) (11). Similarly, Northern blots revealed that an HCV mutant defective in miR-122 binding at both sites (S1-S2-p6m; Fig. 3A) (11), as well as polymerase function (GDD to GND substitution in NS5B), was not stabilized by miR-122 when transfected into hepatoma cells (Fig. 3B, compare lanes 1–3 vs. 4–6). In contrast, the complementary miR-122 mutant (miR-122p6) (Fig. 3A) did stabilize S1-S2-p6m RNA (Fig. 3B, lanes 1–3 vs. 7–9) but not viral RNA with WT S1 and S2 sequences. The differences in HCV RNA abundance apparent in these blots were reproduced in multiple experiments, statistically significant (Fig. S2A), and mirrored by differences in GLuc expressed from these nonreplicating viral RNAs (Fig. S2B). Collectively, these data indicate that HCV RNA is physically stabilized as a result of miR-122 binding to its 5′ UTR, a unique action for a miRNA.
Fig. 3.
Stabilization of HCV RNA requires miR-122 binding and is independent of translation. (A) Upper: miR-122 and mutant miR-122p6 guide-strand sequences. Lower: 5′ terminal sequence of HCV (HJ3-5/GLuc2A virus), with S1 and S2 binding sites shown in red. Point mutations (underlined) in the related S1-S2-p6m-GND mutant (11) are shown above. SL-1 and SL-2 are putative stem-loop structures in the 5′ UTR. (B) Northern blots of HJ3-5/GLuc2A-GND and the related S1-S2-p6m-GND mutant RNA after transfection into Huh-7.5 cells with RNA oligoribonucleotides as in Fig. 1B. (C) Putative secondary structure of the HCV 5′ UTR, showing the location of stem-loop IIId and the G(266-8)C IRES mutation that ablates translation. (D) Northern blot showing HCV RNA abundance in MEFs transfected with HCV RNA (H77S/GLuc2A-AAG) containing (Upper) the WT 5′ UTR vs. (Lower) the translationally inactive G(266-8)C mutant, and supplemented with the indicated miRNAs. (E) PhosphorImager quantitation of HCV RNA in Northern blots of 6-h cell lysates from two independent experiments carried out as shown in D.
Because mRNA translation and decay are closely coupled processes (2), we considered the possibility that miR-122 could stabilize the RNA by promoting its translation. We thus evaluated its ability to slow decay of an RNA containing three consecutive base substitutions in an RNA loop within the HCV IRES [mutant G(266-8)C; Fig. 3C]. These base changes eliminate IRES affinity for the 40S ribosome particle and ablate translation (11, 16), even in cells supplemented with miR-122 (Fig. S3A). Consistent with the notion that translation and decay are intrinsically linked (2), Northern blots of cells transfected with equivalent amounts of WT and G(266-8)C RNA (both containing an NS5B mutation ablating RNA replication) consistently showed a lower abundance of the G(266-8)C mutant, suggesting that it was less stable than RNA with a WT 5′ UTR (Fig. 3 D and E). Nonetheless, decay of the translationally inactive mutant was significantly slowed when the cells were supplemented with miR-122 (Fig. 3 D and E). These experiments were carried out in murine embryonic fibroblasts (MEFs) that do not express detectable endogenous miR-122. Similar results were obtained both in MEFs and hepatoma cells with another IRES mutant, G(267)C, that retains affinity for the 40S subunit but is also translationally dead (11, 16) (Fig. S3 B–E). Collectively these results show that miR-122 does not stabilize HCV RNA by promoting its translation or enhancing its association with ribosomes. Active engagement in translation and miR-122 promote stability of the RNA genome independently, and possibly additively.
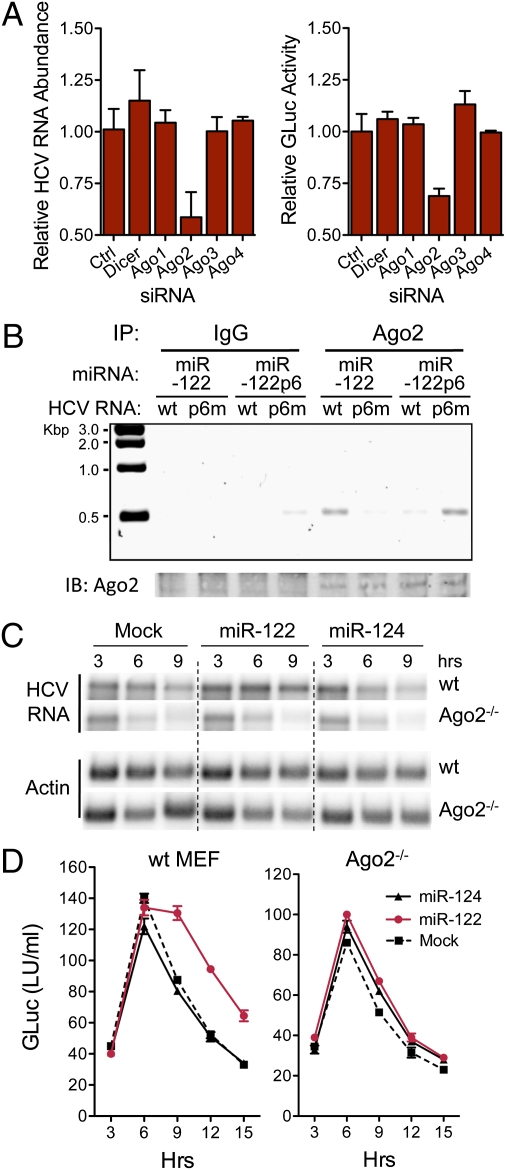
Randall et al. (21) reported previously that RNAi-mediated depletion of any of the Ago proteins (particularly Ago4) or Dicer inhibited the ability of HCV to infect cells. In contrast, we found that only Ago2 depletion (Fig. S4A) inhibited HCV RNA replication in hepatoma cells with previously established infection (Fig. 4A). Ago2 depletion had no effect on cell growth (Fig. S4B) but reduced both HCV RNA abundance and protein expression (Fig. 4A). This was not observed with depletion of other Ago proteins or Dicer. Although we are uncertain why our results differ from those of Randall et al. (21), they suggest that Ago2 plays a special role in HCV replication. Two recent studies are consistent with this: they show Ago2 to be required for miR-122 to promote HCV genome amplification (15, 22). However, a third study found no impairment in miR-122 modulation of HCV in Ago2-depleted cells (14).
Fig. 4.
Ago2 binds HCV RNA in association with miR-122 and is required for stabilization. (A) RNAi depletion of Ago2 impairs HCV genome amplification in persistently infected (HJ3-5/GLuc2A virus) cells. Left: HCV RNA abundance relative to that in cells transfected with nontargeting siRNA (Ctrl) 72 h after siRNA transfection. RNA was assayed by qRT-PCR; data are mean ± range of paired cultures. Right: GLuc activity secreted into media between 48 and 72 h, relative to si-Ctrl-transfected cultures (mean ± range). (B) miR-122 binds HCV RNA as a complex with Ago2. Lysates were prepared from WT MEFs 6 h after electroporation with HJ3-5/GND or S1-S2-p6m-GND RNA (Fig. 3A), mixed with miR-122 or miR-122p6, and immunoprecipitated with anti-Ago2 antibody. After extensive washing, RNA was extracted from the precipitates and subjected to a one-step HCV-specific RT-PCR (30 cycles). HCV RNA was enriched in precipitates from HJ3-5/GND–transfected cells supplemented with miR-122, or S1-S2-p6m-GND cells supplemented with miR-122p6. (C) Northern blots showing miR-122 does not stabilize HCV RNA in Ago2−/− MEFs. Cells were electroporated with HCV RNA (H77S/GLuc2A-AAG) together with miR-122, miR-124, or no miRNA (Mock), then lysed at 3-h intervals and assayed for HCV RNA abundance. miR-122 stabilized HCV RNA only in WT MEFs and was without effect in Ago2−/− cells. (D) GLuc activity in supernatant fluids from MEFs cotransfected with HCV RNA and the indicated miRNA (mean ± range). Data shown are representative of two or more independent experiments.
Consistent with the stabilization of HCV RNA requiring interaction with an miR-122–associated RISC complex, we observed no stabilizing or translation-enhancing effect in hepatoma cells transfected with single-stranded miR-122 (guide strand only, rather than the duplex miRNA transfected in previous experiments) (Fig. S4C). To determine whether miR-122 binds as a complex with Ago2, we lysed MEFs shortly after electroporation with HCV RNA together with duplex miR-122 and used RT-PCR to interrogate Ago2 immunoprecipitates for the presence of viral RNA. HCV RNA with WT S1 and S2 sequence coimmunoprecipitated with Ago2 (Fig. 4B). However, RNA with point mutations in S1 and S2 that ablate miR-122 binding (Fig. 3A) coimmunoprecipitated with Ago2 only when cells were supplemented with the complementary miR-122 mutant, miR-122p6 (Fig. 4B). Transfected HCV RNA was also enriched in anti-Flag immunoprecipitates from hepatoma cells ectopically expressing Flag-Ago2 vs. Flag-Ago1 (Fig. S5). Thus, miR-122 binds the HCV 5′ UTR in association with Ago2. Lesser amounts of Ago1 may also be present in the complex.
To determine whether Ago2 plays a functional role in stabilizing HCV RNA, we compared the ability of miR-122 to slow the decay of the RNA when electroporated with it into WT or Ago2-deficient (Ago2−/−) MEFs (23). This revealed a striking dependence on Ago2, because miR-122 had no effect in Ago2−/− MEFs, whereas it significantly stabilized the viral RNA and enhanced viral translation in matched WT cells (Fig. 4 C and D). Ectopic expression of human Flag-Ago2 in the Ago2−/− MEFs restored the ability of miR-122 to positively regulate protein expression from HCV RNA, further confirming the requirement for Ago2 (Fig. S6). To ascertain whether it is possible for other Ago proteins to functionally substitute for Ago2 in stabilizing HCV RNA, we also overexpressed human Flag-Ago1 in the Ago2−/− MEFs. This only partially rescued the ability of miR-122 to promote HCV protein expression (Fig. S6B), although immunoblots with anti-Flag antibody indicated that Flag-Ago1 was overexpressed at almost threefold the abundance of Flag-Ago2 and well above physiologically relevant levels (Fig. S6A). Thus, Ago2 is not unique in its ability to support miR-122 enhancement of HCV protein expression, but it seems to do this more efficiently than Ago1. This is consistent with the greater enrichment of HCV RNA we observed in Flag-Ago2 compared with Flag-Ago1 immunoprecipitates (Fig. S5). Additional experiments confirmed that Ago2 is the dominant Ago protein involved in miR-122 stabilization of HCV RNA. RNAi-mediated depletion of Ago2 reduced the ability of miR-122 to stabilize HCV RNA and promote its translation in HeLa cells (Fig. S7 A–D). Importantly, the magnitude of this effect mirrored the reduction in miR-122–mediated suppression of a reporter mRNA containing the HCV miR-122 binding sites within its 3′ UTR (9) (Fig. S7E).
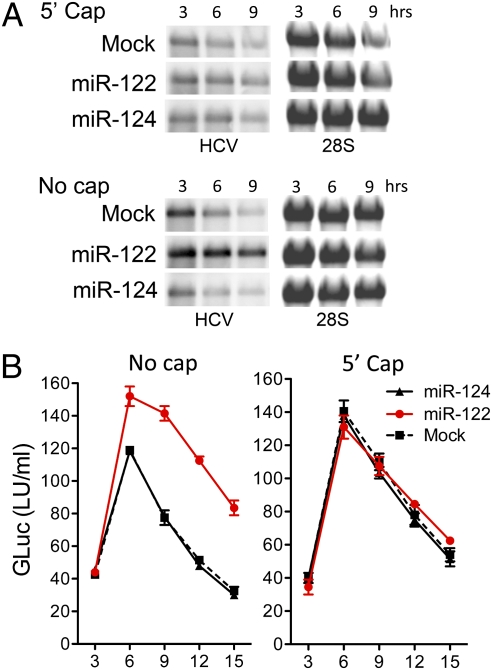
These results suggest a functional interaction of Ago2 with RNA decay machinery. Because HCV RNA lacks a 5′ m7G cap, one possibility is that miR-122 recruits an Ago2 RISC complex to the 5′ end of viral RNA that protects it from 5′ exonuclease activity. If so, this protective action should be rendered redundant by providing the RNA with a 5′ cap. To test this, we synthesized HCV RNAs with or without a 5′ nonmethylated guanosine cap analog and compared their stabilities after transfection into MEFs with or without miR-122. As anticipated, miR-122 had no effect on the rate of decay of the 5′ capped RNA, whereas it substantially stabilized the uncapped HCV RNA (Fig. 5A). The magnitude of protein expression (GLuc) from capped RNA was comparable to that from the uncapped RNA in cells supplemented with miR-122 and was not further increased by miR-122 (Fig. 5B). Thus, the 5′ cap enhanced stability and protein expression from HCV RNA, functionally substituting for miR-122 and providing strong evidence that the miR-122–Ago2 complex protects HCV RNA from 5′ exonuclease. Importantly, translation of the capped HCV RNA was IRES-dependent, because the nonmethylated 5′ cap lacked the ability to recruit eukaryotic initiation factors.
Fig. 5.
A nonmethylated 5′ guanosine cap functionally substitutes for miR-122 in stabilizing HCV RNA. (A) Northern blots of HCV RNA in lysates of MEFs after electroporation with HCV RNA (H77S/GLuc2A-AAG) (Fig. 1A) synthesized with or without a nonmethlyated G[5′]ppp[5′]G-RNA cap. HCV RNAs were transfected together with miR-122, miR-124, or no miRNA (Mock). 28S rRNA is shown as a loading control. (B) GLuc activity in supernatant fluids of MEFs transfected with capped or uncapped HCV RNAs (mean ± range of two replicate cultures). Data shown are representative of two or more independent experiments.
Discussion
Our results reveal a unique mechanism by which a miRNA in association with Ago2 regulates the expression of its target RNA, in this case the HCV genome. Although miR-10a enhances translation by binding the 5′ UTR of ribosomal protein mRNAs containing 5′ terminal oligopyrimidine (TOP) motifs (24), miRNAs have not been recognized to slow decay or up-regulate abundance of their RNA targets. miR-122 does this by recruiting an Ago2 RISC-like complex to the 5′ end of the HCV RNA genome. As evidenced by the IRES mutants (Fig. 3D and Fig. S3D), the stabilizing action of miR-122 does not require the target HCV RNA to be capable of translation and thus does not result from increased ribosomal loading. Stabilization also does not require the RNA to be replication competent (Fig. 1 B and C).
Because a nonmethylated 5′ cap analog functionally substitutes for miR-122 (Fig. 5), the RISC-like complex recruited by miR-122 is likely to act by protecting the RNA from 5′ exonuclease. Whether this occurs simply as a result of physically masking the 5′ end of the viral RNA from 5′ exonuclease attack, or whether Ago2 plays a more complex role by influencing the association of HCV RNA with P bodies (25), sites of mRNA degradation and storage, remains to be determined. Binding of a RISC-like complex could also limit recognition of the 5′ triphosphate of HCV RNA by retinoic acid-inducible gene I (RIG-I), a ubiquitous innate immune pathogen recognition receptor (26) that is capable of inducing interferons and interferon-stimulated genes, including RNase L, an endonuclease, and ISG20, a 3′-5′ exonuclease (27). However, subversion of RIG-I–dependent viral RNA degradation played no role in the stabilization of HCV RNA in our experiments because the Huh-7.5 cells used are deficient in RIG-I signaling (28).
The mechanism by which miR-122 stabilizes HCV RNA is distinct from the up-regulation of translation by miRNAs that target AU-rich elements within the 3′ UTRs of some mRNAs, because the latter occurs only in quiescent cells arrested at the G0 phase of the cell cycle (29, 30). Moreover, miRNA-mediated stimulation of translation in quiescent cells has not been linked to stabilization of the target mRNA, as we show here for miR-122. Still to be determined is whether the protein composition of the RISC-like complex that is recruited to the 5′ UTR by miR-122 differs significantly from miRNA-induced RISC complexes involved in translational repression.
The stabilization of HCV RNA by miR-122 is likely to be responsible for the miR-122–induced enhancement of HCV translation reported previously (10, 11). Mutations in the viral RNA that prevent binding of miR-122 also ablate replication of infectious virus (11) and prevent stabilization of the genome by miR-122 (Fig. 3). Depletion of Ago2 has similar effects on replication, translation, and RNA stability (15, 22) (Fig. 4). This makes it difficult to distinguish between these consequences of miR-122 binding to the 5′ UTR or to determine the primary role played by miR-122 as a host factor for viral replication. However, stabilization of the genome is likely to be a key factor in the promotion of viral replication by miR-122. A recombinant HCV in which the U3 RNA sequence was inserted in lieu of the S1 binding site in the 5′ UTR was found recently to be less dependent upon miR-122 for replication (17), possibly because the U3 sequence stabilized the RNA, much as a 5′ cap did (Fig. 5). Nonetheless, it would not be surprising to find that miR-122 has other functions in the viral life cycle in addition to its role in stabilizing the viral RNA genome, perhaps in the initiation of viral RNA synthesis.
Methods
Viral RNA Stability in Transfected Cells.
RNA was transcribed in vitro (11) from pH77S/GLuc2A-AAG, which contains the complete genotype 1a HCV sequence with GLuc2A placed in-frame within the polyprotein-coding region (31) and a lethal GDD to AAG mutation in NS5B. Where indicated, a nonmethylated 5′ guanosine cap was added using the ScriptCap m7G Capping System (Epicentre Biotechnologies). Viral RNA (20 μg) and miRNA duplexes (11) or antisense oligoribonucleotides (1 μM) were mixed with 1 × 107 Huh-7.5 cells in a 4-mm cuvettete and pulsed once at 250 V, 950 μF, and 100 Ω in a Gene Pulser Xcell Total System (Bio-Rad). HeLa cells were electroporated at 300 V, 500 μF, and ∞ Ω, and MEFs at 400 V, 250 μF, and ∞ Ω. Cells were harvested at intervals and supernatant fluids assayed for GLuc activity (31) and HCV RNA abundance in cell lysates assessed by Northern blotting (11). Polyadenylated reporter RNAs encoding firefly or Cypridina luciferase were cotransfected to monitor transfection efficiency (11).
HCV-Infected Cells.
Synthetic HJ3-5 or HJ3-5/GLuc2A RNA (31) was transfected into 1 × 107 FT3-7 cells, which were passaged until >90% positive for core antigen in an immunofluorescence assay (11). siRNA pools targeting Ago1-4 or Dicer and control siRNA pools (Dharmacon) were transfected using siLentfect Lipid Reagent (Bio-Rad). miRNA duplexes or single-stranded oligoribonucleotides (50 nM) were transfected using Lipofectamine 2000 (Invitrogen).
Additional methods and associated references can be found in SI Methods.
Supplementary Material
Acknowledgments
We thank Lucinda Hensley for expert technical assistance; William F. Marzluff and Scott M. Hammond for helpful discussions; Alexander Tarakhovsky and Angela Santana, Rockefeller University, for mouse embryonic fibroblasts; Charles Rice, Rockefeller University, for Huh-7.5 cells; Thorleif Møller, Mirrx Therapeutics, for psi-Check2/Luc-3′HCV DNA; and Angela Lam and Phil Furman, Pharmasset, Inc., for PSI-6130. This study was supported in part by the University Cancer Research Fund and National Institutes of Health Grants RO1-AI095690, U19-AI040035, P20-CA150343 (to S.M.L.), and AI042189 (to D.J.B.). R.K.J. was supported by a James W. McLaughlin Fellowship.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1112263109/-/DCSupplemental.
References
- 1.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 2.Balagopal V, Parker R. Polysomes, P bodies and stress granules: States and fates of eukaryotic mRNAs. Curr Opin Cell Biol. 2009;21:403–408. doi: 10.1016/j.ceb.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Djuranovic S, Nahvi A, Green R. A parsimonious model for gene regulation by miRNAs. Science. 2011;331:550–553. doi: 10.1126/science.1191138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang J, et al. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004;1:106–113. doi: 10.4161/rna.1.2.1066. [DOI] [PubMed] [Google Scholar]
- 6.Lanford RE, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 8.Chisari FV. Unscrambling hepatitis C virus-host interactions. Nature. 2005;436:930–932. doi: 10.1038/nature04076. [DOI] [PubMed] [Google Scholar]
- 9.Jopling CL, Schütz S, Sarnow P. Position-dependent function for a tandem microRNA miR-122-binding site located in the hepatitis C virus RNA genome. Cell Host Microbe. 2008;4:77–85. doi: 10.1016/j.chom.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henke JI, et al. microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J. 2008;27:3300–3310. doi: 10.1038/emboj.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jangra RK, Yi M, Lemon SM. miR-122 regulation of hepatitis C virus translation and infectious virus production. J Virol. 2010;84:6615–6625. doi: 10.1128/JVI.00417-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Norman KL, Sarnow P. Modulation of hepatitis C virus RNA abundance and the isoprenoid biosynthesis pathway by microRNA miR-122 involves distinct mechanisms. J Virol. 2010;84:666–670. doi: 10.1128/JVI.01156-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Villanueva RA, et al. miR-122 does not modulate the elongation phase of hepatitis C virus RNA synthesis in isolated replicase complexes. Antiviral Res. 2010;88:119–123. doi: 10.1016/j.antiviral.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Machlin ES, Sarnow P, Sagan SM. Masking the 5′ terminal nucleotides of the hepatitis C virus genome by an unconventional microRNA-target RNA complex. Proc Natl Acad Sci USA. 2011;108:3193–3198. doi: 10.1073/pnas.1012464108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roberts AP, Lewis AP, Jopling CL. miR-122 activates hepatitis C virus translation by a specialized mechanism requiring particular RNA components. Nucleic Acids Res. 2011;39:7716–7729. doi: 10.1093/nar/gkr426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kieft JS, Zhou K, Jubin R, Doudna JA. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA. 2001;7:194–206. doi: 10.1017/s1355838201001790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li YP, Gottwein JM, Scheel TK, Jensen TB, Bukh J. MicroRNA-122 antagonism against hepatitis C virus genotypes 1-6 and reduced efficacy by host RNA insertion or mutations in the HCV 5′ UTR. Proc Natl Acad Sci USA. 2011;108:4991–4996. doi: 10.1073/pnas.1016606108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma H, et al. Characterization of the metabolic activation of hepatitis C virus nucleoside inhibitor beta-D-2′-Deoxy-2′-fluoro-2′-C-methylcytidine (PSI-6130) and identification of a novel active 5′-triphosphate species. J Biol Chem. 2007;282:29812–29820. doi: 10.1074/jbc.M705274200. [DOI] [PubMed] [Google Scholar]
- 19.Lyons T, Murray KE, Roberts AW, Barton DJ. Poliovirus 5′-terminal cloverleaf RNA is required in cis for VPg uridylylation and the initiation of negative-strand RNA synthesis. J Virol. 2001;75:10696–10708. doi: 10.1128/JVI.75.22.10696-10708.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murray KE, Roberts AW, Barton DJ. Poly(rC) binding proteins mediate poliovirus mRNA stability. RNA. 2001;7:1126–1141. doi: 10.1017/s1355838201010044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Randall G, et al. Cellular cofactors affecting hepatitis C virus infection and replication. Proc Natl Acad Sci USA. 2007;104:12884–12889. doi: 10.1073/pnas.0704894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson JA, Zhang C, Huys A, Richardson CD. Human Ago2 is required for efficient miR-122 regulation of HCV RNA accumulation and translation. J Virol. 2011;85:2342–2350. doi: 10.1128/JVI.02046-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Carroll D, et al. A Slicer-independent role for Argonaute 2 in hematopoiesis and the microRNA pathway. Genes Dev. 2007;21:1999–2004. doi: 10.1101/gad.1565607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ørom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5'UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30:460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 25.Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 26.Hornung V, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 27.Zhou Z, et al. Antiviral activities of ISG20 in positive-strand RNA virus infections. Virology. 2011;409:175–188. doi: 10.1016/j.virol.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sumpter R, Jr, et al. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J Virol. 2005;79:2689–2699. doi: 10.1128/JVI.79.5.2689-2699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: MicroRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 30.Vasudevan S, Tong Y, Steitz JA. Cell-cycle control of microRNA-mediated translation regulation. Cell Cycle. 2008;7:1545–1549. doi: 10.4161/cc.7.11.6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shimakami T, et al. Protease inhibitor-resistant hepatitis C virus mutants with reduced fitness from impaired production of infectious virus. Gastroenterology. 2011;140:667–675. doi: 10.1053/j.gastro.2010.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.