Abstract
Binding of the talin-1 FERM (4.1/ezrin/radixin/moesin) domain to the β3 cytosolic tail causes activation of the integrin αIIbβ3. The FERM domain also binds to acidic phospholipids. Although much is known about the interaction of talin-1 with integrins and lipids, the relative contribution of each interaction to integrin regulation and possible synergy between them remain to be clarified. Here, we examined the thermodynamic interplay between FERM domain binding to phospholipid bilayers and to its binding sites in the β3 tail. We found that although both the F0F1 and F2F3 subdomains of the talin-1 FERM domain bind acidic bilayers, the full-length FERM domain binds with an affinity similar to F2F3, indicating that F0F1 contributes little to the overall interaction. When free in solution, the β3 tail has weak affinity for the FERM domain. However, appending the tail to acidic phospholipids increased its affinity for the FERM domain by three orders of magnitude. Nonetheless, the affinity of the FERM for the appended tail was similar to its affinity for binding to bilayers alone. Thus, talin-1 binding to the β3 tail is a ternary interaction dominated by a favorable surface interaction with phospholipid bilayers and set by lipid composition. Nonetheless, interactions between the FERM domain, the β3 tail, and lipid bilayers are not optimized for a high-affinity synergistic interaction, even at the membrane surface. Instead, the interactions appear to be tuned in such a way that the equilibrium between inactive and active integrin conformations can be readily regulated.
Keywords: cytoskeleton, plasma membrane, platelet aggregation
The integrin αIIbβ3 resides on the platelet surface in equilibrium between resting and active conformations (1–5). On circulating platelets, αIIbβ3 is constrained in its inactive conformation to prevent spontaneous platelet aggregation. Similarly, the cytoskeletal protein talin-1, whose binding to the β3 cytosolic tail (CT) stabilizes the active conformation of αIIbβ3 (6–8), is sequestered away from the β3 CT (9). Besides interacting with integrins, talin-1 interacts with negatively charged phospholipids (10–12) and phosphoinositides (13–16). Stimuli generated at sites of vascular damage recruit talin-1 to the platelet plasma membrane, thereby promoting αIIbβ3 activation (17–19). Much is known about the interaction of talin-1 with integrins and lipids, but the relative contribution of each interaction to integrin regulation and possible synergy between them remain to be clarified. Elucidating these interactions is important for understanding events leading to and controlling the formation of integrin complexes and for potential pharmacologic modulation of integrin signaling.
Talin-1 is a 250-kD protein containing a 45-kD N-terminal head domain attached via a flexible linker to a 200-kD C-terminal rod domain (7). Because the head domain is packed against the rod domain in its inactive state (20), talin-1 recruitment to the membrane and to integrin β CTs requires disruption of this interaction (14). At integrin-mediated cellular adhesions, the orientation of talin-1 is highly polarized, with its head domain associated with the plasma membrane and its rod domain making numerous cytoskeletal contacts (21).
The talin-1 head domain is a member of the FERM (4.1/ezrin/radixin/moesin) domain family (22, 23). Canonical FERM domains, such as that of radixin, consist of F1, F2, and F3 subdomains adopting ubiquitin, acyl-CoA-binding protein, and phosphotyrosine-binding (PTB)/pleckstrin homology (PH) domain folds, respectively, and are assembled into a trefoil arrangement (Fig. S1 A and B). The talin-1 FERM domain contains a fourth subdomain, F0, preceding F1 that also adopts a ubiquitin-like fold (12). The four talin-1 subdomains have an extended, nearly linear, configuration (Protein Data Bank ID code 3IVF; Fig. S1C) (24). Whereas radixin binds its targets, such as the CD44 CT, in a groove formed by helix α1 and strand β5 of its F3 subdomain in an antiparallel β–β fashion (Fig S1A), the talin-β integrin CT complex contains a second interface with the N-terminal helical portion of the CT (Fig. S1D) (25, 26). This positions talin to modulate interaction between membrane-proximal cytosolic and transmembrane segments of integrin α and β subunits (25–27).
Here, we examine the thermodynamic interplay between phospholipid bilayers and β CT binding sites in the talin-1 FERM domain. The impetus for this study arose, in part, from conflicting findings regarding the ability of the talin-1 FERM domain to interact with the β3 CT. In solution, the affinity of talin-1 for β integrin CTs is nearly 1,000-fold weaker than the affinity of radixin for CD44 (28) or of talin-1 for the C terminus of PIP5KI-γ (25). However, talin-1 binds to the β3 CT with high affinity when the CT is immobilized on a carboxymethylated dextran surface (29). This suggests that the interaction of talin-1 with integrins is optimal when the β subunit CT is presented on a hydrophilic negatively charged surface such as the inner leaflet of plasma membranes.
To assess the interplay between membrane and β CT binding interactions, we tested whether the talin-1 FERM subdomains F0F1 and F2F3 (Fig. S2 B and C), each of which can interact with membranes (12), bind cooperatively or independently to a variety of negatively charged lipids and compared their binding to the complete FERM domain (F0-F3; Fig. S2A). We then measured the effect of the membrane environment on the affinity of FERM domain binding to the β3 CT. We found that although the membrane environment increased the overall affinity of the FERM domain for β3, the interactions between the FERM domain, the β3 CT, and lipid bilayers were not optimized for a high-affinity synergistic interaction, even at the membrane surface. Instead, the interactions appeared to be tuned in such a way that the equilibrium between inactive and active integrin conformations can be readily regulated.
Results
The Talin-1 FERM Domain Contains an Extended Positively Charged Surface.
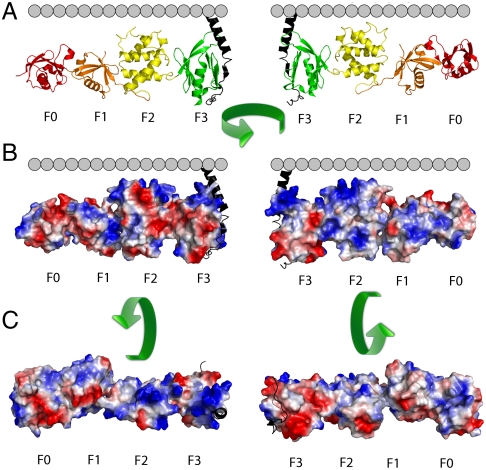
Projection of an electrostatic surface on the talin-1 FERM domain revealed that it contains an extended positively charged surface (Fig. 1) (24). This is especially pronounced for the F2 and F3 subdomains. The F0 and F1 subdomains also contain positively charged surfaces, and F1 contains a flexible loop that readily binds to membranes but was removed for crystallization (20). The positively charged surfaces on F0F1 are not completely aligned with those on F2F3. Although this could reflect a strong bias for maintaining the surfaces in an unaligned state in solution or the selection of a single conformation from a flexible ensemble due to crystal packing, X-ray scattering studies argue against a flexible ensemble of structures (24). Thus, if the solution conformation is maintained, F0F1 should make only modest contributions toward the affinity of the complete FERM domain over that of the F2F3 alone. Alternatively, if F0F1 and F2F3 change their mutual orientation away from the minimum energy solution conformation, this would attenuate the affinity relative to the value expected if they were preorganized to interact favorably with the surface.
Fig. 1.
Orientation of the talin-1 FERM domain with respect to the inner leaflet of the plasma membrane. (A) The interaction of talin-1 with the β integrin CT (black) places its FERM domain in proximity to the inner plasma membrane leaflet. (B) Projection of an electrostatic surface onto the talin-1 FERM domain. Red denotes negative- and blue denotes positive-charge density. (C) Talin-1 FERM domain rotated 90° showing the electrostatic surface as viewed from the membrane (Left) and cytosol (Right).
Binding of the Talin-1 FERM Domain Fragment F2F3 to Negatively Charged Phospholipids.
Before studying the interaction of the talin-1 FERM domain with the β3 CT tethered to membrane, we investigated whether the F2F3 subdomain, which makes the most direct contact with the β3 CT, differentially recognizes negatively charged phospholipids.
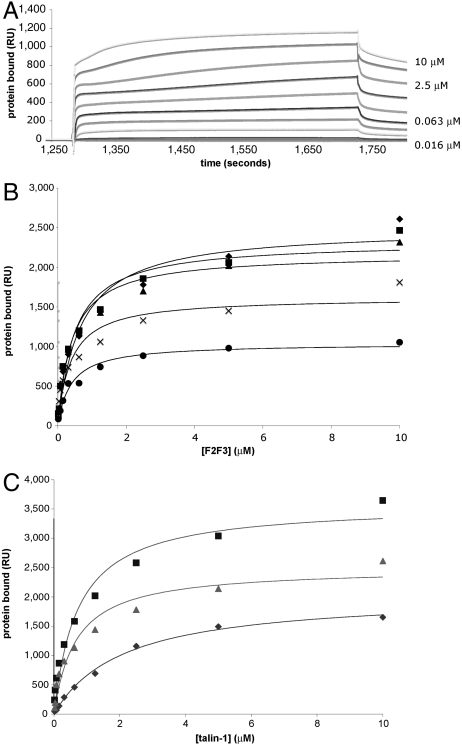
We used surface plasmon resonance (SPR) to measure the interaction of F2F3 with phospholipid bilayers immobilized on a hydrophobically modified Biacore L1 SPR chip (30). All measurements were made relative to a surface containing 100% phosphatidylcholine (PC). F2F3 binding to 20% 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L serine (POPS) and 80% 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) was measured at steady state (Fig. 2A) and conformed to a single-site binding model as a function of F2F3 concentration. The dissociation constant (Kd) for F2F3 binding to 20% phosphatidylserine (PS)-containing bilayers was 0.92 μM (Table 1). In comparison, the Kd for bilayers containing 10% PtdIns(4,5)P2 was 0.67 μM, suggesting that F2F3 may have a slight preference for PtdIns(4,5)P2 over PS. Nonetheless, F2F3 bound to bilayers containing the diphosphoinositides PtdIns(3,5)P2 and PtdIns(3,4)P2 and the monophosphoinositide PtdIns(5)P with affinities similar to PtdIns(4,5)P2, indicating that it does not strongly differentiate between negatively charged head groups (Fig. 2B and Table 1).
Fig. 2.
Steady-state binding of the talin-1 FERM domain and its subdomains to phospholipid bilayers. (A) SPR sensorgrams of F2F3 binding to a bilayer composed of 20% POPS and 80% POPC immobilized on a Biacore L1 SPR chip. F2F3 was injected in 2-fold increments up to a concentration of 10 μM. (B) Steady-state isotherms for F2F3 binding to negatively charged phospholipids. Diamonds, 10% PtdIns(4,5)P2; squares, 10% PtdIns(3,5)P2; triangles, 10% PtdIns(3,4)P2; Xs, PtdIns(5)P; circles, 20% phosphatidylserine. (C) Steady-state isotherms for F0-F3 (squares), F2F3 (triangles), and F0F1 (diamonds) binding to bilayers containing 10% PtdIns(4,5)P2. Data from B and C were fit to a one-site binding model using BIAevaluation software.
Table 1.
Steady-state SPR measurements of talin-1 F2F3 subdomain binding to phospholipids
| Lipid, % | Kd, μM | Maximum binding, RU |
| PS (20%) | 0.92 ± 0.260 | 1,210 ± 66 |
| PtdIns(4,5)P2 (10%) | 0.67 ± 0.07 | 2,490 ± 225 |
| PtdIns(3,4)P2 (10%) | 0.42 ± 0.09 | 2,170 ± 146 |
| PtdIns(3,5)P2 (10%) | 0.50 ± 0.053 | 2,330 ± 193 |
| PtdIns(5)P (10%) | 0.37 ± 0.029 | 1,770 ± 146 |
Binding to the indicated phospholipids was fit to a one-site binding model using BIAevaluation. The data shown represent the mean and standard deviation of triplicate injections.
Although there was no substantial difference in the affinity of F2F3 for bilayers containing various diphosphoinositides, the total amount of bound F2F3 depended on the net negative charge of the membrane, implying that electrostatics plays a role in the interaction. PtdIns(4,5)P2, PtdIns(3,5)P2, and PtdIns(3,4)P2 each have a charge of −4 per head group, whereas PtdIns(5)P has a charge of −3, and PS has a charge of −1. If F2F3 binding is affected by charge, the amount of protein bound to 10% PtdIns(5)P would be expected to be roughly 75% of the amount bound to 10% diphosphoinositides. Similarly, 50% as much F2F3 would be expected to bind to 20% PS as to 10% diphosphoinositides. The maximum resonance units (RU) of F2F3 bound to 10% PtdIns(4,5)P2, 10% PtdIns(3,5)P2, and 10% PtdIns(3,4)P2 were similar at 2,490, 2,330, and 2,170 RU, respectively, but we found approximately 75% as much F2F3 bound to 10% PtdIns(5)P (1,770 RU) and 50% as much F2F3 bound to 20% PS (1,200 RU), confirming that protein binding depends on the charge of the membrane.
Binding of Talin-1 FERM Domain and Its Subdomains to PtdIns(4,5)P2.
Next, we compared the affinity of the complete talin-1 FERM domain (F0-F3) for PtdIns(4,5)P2 with that of F0F1 and F2F3. PtdIns(4,5)P2 was chosen as the lipid substrate for these measurements because F2F3 does not differentiate between various phosphoinositides.
Phospholipid bilayers containing 10% PtdIns(4,5)P2 were immobilized on a Biacore L1 chip, and all measurements were made relative to a surface containing 100% PC. We found that F0-F3 bound to 10% PtdIns(4,5)P2 with a Kd of 980 nM, comparable to the Kd for F2F3 of 670 nM, whereas the Kd for F0F1 was 2.6 μM (Table 2). Thus, both F0F1 and F2F3 readily interact with PtdIns(4,5)P2, but when combined, the affinity for bilayers was not enhanced. The relative stoichiometries of F0F1, F2F3, and F0-F3 also suggest that the subdomains do not form a cooperative membrane binding surface. The mass ratios for F0-F3, F2F3, and F0F1 are 2∶1.1∶1.0. F0-F3 has twice the mass of F2F3 and nearly twice the resonance units of F0-F3 (3,960 RU) bound to the sensor chip as F2F3 (2,490 RU) (Fig. 2C, Table 2). Thus, F0-F3 and F2F3 have similar stoichiometries for the phospholipid surface. A similar amount of F0F1 bound to the sensor chip (2,290 RU) as F2F3, but with lower affinity. Thus, although F0F1 can interact with PtdIns(4,5)P2, the affinity and stoichiometry of the full FERM domain for PtdIns(4,5)P2 is similar to that of F2F3, indicating that F0F1 does not substantially cooperate with F2F3 in FERM domain binding to membranes.
Table 2.
Steady-state SPR measurements of talin FERM domain binding to phospholipid bilayers containing 10% PtdIns(4,5)P2
| FERM domain construct | Kd, μM | Maximum binding, RU |
| F0-F3 | 0.98 ± 0.10 | 3,960 ± 258 |
| F2F3 | 0.67 ± 0.07 | 2,490 ± 220 |
| F0F1 | 2.63 ± 0.84 | 2,290 ± 258 |
Binding results were fit to a one-site binding model using BIAevaluation. The data shown represent the mean and standard deviation of triplicate injections.
Enthalpy of Talin-1 FERM Domain Binding to PTdIns(4,5)2.
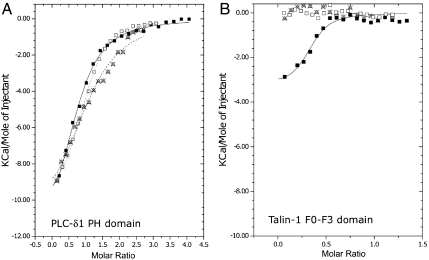
It has been reported that talin FERM domains bind to negatively charged vesicles in solution (12, 13, 24, 31, 32). To confirm these observations, we used isothermal titration calorimetry (ITC) to measure the enthalpy of F0-F3 binding to large unilamellar vesicles (LUVs) composed of 90% POPC and 10% PtdIns(4,5)P2. As a positive control, we measured binding of the PH domain of phospholipase C-δ1 (PLC-δ1) to PtdIns(4,5)P2 (33).
Titration of 100 μM PLC-δ1 PH domain into a LUV suspension containing 1%, 3%, and 10% PtdIns(4,5)P2 generated binding isotherms with molar enthalpies of −11.7, −10.2, and -12.8 kcal/mol and dissociation constants of 2.8, 2.1, and 4.2 μM, respectively (Fig. 3A and Table S1). These results are comparable to a previously published binding enthalpy of -11.9 kcal/mol and Kd of 1.1 μM for PLC-δ1 PH domain binding to 1% PtdIns(4,5)P2 bilayers (33).
Fig. 3.
Comparison of PLC-δ1 PH domain and talin-1 FERM domain binding to PtdIns(4,5)P2 and Ins(1,4,5)P2. (A) ITC titration of 100 μM PLC-δ1 PH domain into LUVs containing 1% (Xs), 3% (open squares), and 10% (closed squares) PtdIns(4,5)P2. The titrations were globally fit to a OneSite binding model using MicroCal/Origin. (B) ITC titration of 190 μM talin-1 FERM domain (F0-F3) into LUVs containing 1% (Xs), 3% (open squares), and 10% (closed squares) PtdIns(4,5)P2 No enthalpy was generated for 1% or 3% LUVs. Titration into 10% PtdIns(4,5)P2 was globally fit to a OneSite binding model using MicroCal/Origin.
Next, we titrated 190 μM F0-F3 into a suspension of LUVs composed of 90% POPC and 10% PtdIns(4,5)P2. After subtracting the heat of dilution, F0-F3 binding was slightly exothermic (approximately -3 kcal/mol) with a stoichiometry of approximately 1 F0-F3 to 3 PtdIns(4,5)P2 phospholipids (Fig. 3B). Titration of higher F0-F3 concentrations resulted in precipitation of the mixture in the calorimetry cell—confirming the interaction of F0-F3 with membranes, but precluding thermodynamic analysis. Surprisingly, similar experiments using LUVs containing 1% and 3% PtdIns(4,5)P2 did not generate measurable enthalpy (Fig. 3B). Therefore, to confirm that F0-F3 actually binds to bilayers containing 1% and 3% PtdIns(4,5)P2, we measured the interaction by SPR and found that F0-F3 bound with high affinity and roughly 1/10th the resonance units as 10% bilayers (Fig. S3).
Lastly, we attempted to measure the interaction F0-F3 with the inositol headgroup of PtdIns(4,5)P2 [Ins(1,4,5)P3] free in solution (Fig. S4). Two mM Ins(1,4,5)P3 was titrated into a 150 μM F0-F3 solution, but no binding enthalpy was generated. By contrast, titration of 120 μM Ins(1,4,5)P3 into a 6 μM solution of the PLC-δ1 PH domain generated an exothermic binding curve with thermodynamic parameters comparable to previously published results (Table S1) (33).
Talin-1 FERM Domain Binding to the β3 CT Immobilized on Negatively Charged Carboxymethylated Dextran.
Previous measurements of talin-1 FERM domain binding to the β3 CT used a β3 construct beginning at Lys716 and included a linker for attaching the construct to an SPR chip (29). Our β3 CT construct, designed to mimic the β3 CT as it exits the membrane, was attached via Cys719 to a CM-5 SPR sensor chip functionalized with N-[ϵ-maleimidocaproic acid]-hydrazide to generate thiol-reactive groups (Fig. S1D). F0-F3 was then flowed over the chip at concentrations up to 1.0 μM. The resulting sensorgrams (Fig. S5) could be fit to two binding events with dissociation constants of 155 nM and 3.5 μM. The biphasic behavior could have resulted from heterogeneity of β3 on the chip surface or heterogeneity of the interaction between F0-F3 and the carboxymethylated dextran (34). Regardless, the interaction between the talin-1 FERM domain and the β3 CT may be sensitive to such heterogeneity because we hypothesize that the FERM domain binds to the β3 CT and the inner membrane surface simultaneously.
Binding of the Talin-1 FERM Domain to the β3 CT Domain Tethered to Negatively Charged Bilayer.
To address whether the talin-1 FERM domain binds the β3 CT and a negatively charged surface in a concerted fashion, we first used fluorescence polarization (FP) and ITC to measure the affinity of talin-1 FERM domain for the β3 CT when both are free in solution.
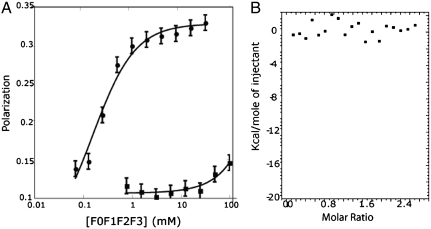
For FP, the β3 CT was labeled with maleimide-functionalized BODIPY-TMR at Cys719. When F0-F3, at concentrations up to 100 μM, was titrated into a 20 nM solution of labeled β3 CT, the FP increased from 0.1 to 0.16 (Fig. 4A). These results were fit to a binding isotherm with a Kd of approximately 600 μM, confirming that the talin-1 FERM domain and the β3 CT do not interact strongly in solution (26, 35). As a positive control, we titrated F0-F3 into a 20 nM solution of a fluorescein isothiocyanate (FITC)-labeled peptide corresponding to the C terminus of PIP5KIγ (Fig. 4B). The resulting change in FP was fit to an isotherm with a Kd of 160 M ± 50 M, consistent with reported affinities of 170 nM to 1.4 μM for other PIP5KIγ constructs with slightly different sequences (36, 37).
Fig. 4.
Talin-1 FERM domain binding to the β3 CT immobilized on negatively charged surfaces or free in solution. (A) FP of F0-F3 titrated into 20 nM BODIPY-TMR β3. FP was measured at 575 nm after excitation at 530 nm (squares). F0-F3 was also titrated into a 20 nM FITC-labeled PIP5KI-γ (black circles). FP was measured at 528 nm after excitation at 485 nm. (B) ITC of 140 μM β3 CT titrated into 10.5 μM F0-F3 revealed no significant enthalpy above the heat of dilution.
We also used ITC to confirm that the β3 CT and F0-F3 do not interact strongly in solution. There was no significant enthalpy above background heats of dilution when F0-F3, up to a final concentration of approximately 30 μM protein, was titrated into 10 μM peptide (Fig. 4B). This contrasts with a recent report in which talin-2 and the β1D CT interacted with a Kd of approximately 20 μM and a molar enthalpy of approximately -17 kcal/mol (26). This difference is associated with several subtle but significant differences between the β1 and β3 CT sequences.
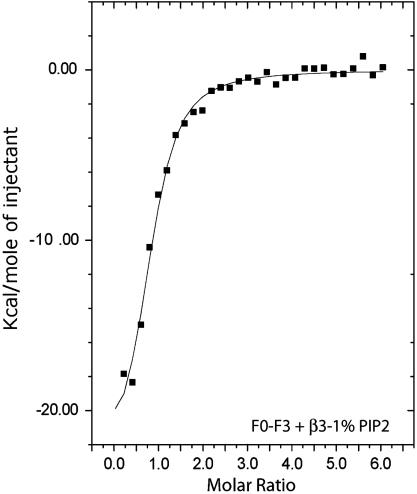
Because the interaction between the talin-1 FERM domain and PtdIns(4,5)P2 occurs with very low enthalpy, we were able to use ITC to measure talin-1 FERM domain binding to the β3 CT in the presence of PtdIns(4,5)P2. Accordingly, we immobilized the β3 CT on phospholipid bilayers by reacting a β3 CT construct containing Cys at residue 719 with maleimide-functionalized phosphatidylethanolamine on the surface of LUVs composed of POPC and either 1% PtdIns(4,5)P2, 20% PS, or 20% PS + 1% PtdIns(4,5)P2 (27). Titration of F0-F3 into a suspension of LUVs containing 1% PtdIns(4,5)P2 in the absence of immobilized β3 CT or into buffer containing the free β3 CT generated little or no enthalpy. By contrast, titration of 150 μM F0-F3 into a LUV suspension containing 5 μM immobilized β3 CT was significantly exothermic (Fig. 5). The titration was globally fit to a one-site binding isotherm, revealing a Kd of 860 nM and a molar enthalpy of -25.0 kcal/mol (Table S2). When the experiments were repeated with the β3 CT immobilized on bilayers containing 20% PS or 20% PS with 1% PtdIns(4,5)P2, nearly identical results were obtained (Fig. S6 A and B; Table S2). Thus, these experiments demonstrate that immobilizing the β3 CT on phospholipid bilayers increases its affinity for the talin-1 FERM domain by at least three orders of magnitude compared to the β3 CT free in solution, increasing its affinity from that of a very weak to a high-affinity interaction.
Fig. 5.
Talin-1 FERM domain binding to the β3 CT conjugated to negatively charged phospholipid bilayers. The talin-1 FERM domain (F0-F3, 150 μM) was titrated at 20 °C into a buffer-matched suspension of LUVs composed of 1% PtdIns(4,5)P2. The titration was globally fit to a OneSite binding model using MicroCal/Origin.
Discussion
Here, we found that presentation of the β3 CT in the context of an acidic bilayer enhances its affinity for the talin-1 FERM domain up to three orders of magnitude, depending on the phospholipid composition of the bilayer. However, FERM domain binding to the β3 CT contrasts with the stronger interaction of the FERM domain with PIP5K1γ and with the interaction of the talin-2 FERM domain and the β1D CT. Small changes in integrin β CT sequences have been shown to significantly increase their affinities for talin family FERM domains (26). This suggests that the interaction between the talin-1 FERM domain and the β3 CT has been evolutionarily tuned to be intrinsically weak, allowing it to be modulated by changes in phospholipid composition, posttranslational modifications, and additional protein–protein interactions.
To understand how these protein–protein and lipid–protein interactions modulate affinity, it is important to consider the various energetic components associated with the interaction of the talin-1 FERM domain and the β3 CT with lipids, as well as their interaction with each other. Previous studies reported on the interaction of the talin-1 FERM domain and the β3 CT with micelles and phospholipid vesicles (15, 27, 38–40). Here, we studied these interactions using a β3 CT construct tethered to phospholipid bilayers to understand the energetics of the overall process. Previously, we used this construct to study the dynamics of the β3 CT in the presence of bilayers by hydrogen–deuterium exchange (HDX). Although the construct lacks a transmembrane (TM) domain, it was designed computationally to attach via a residue predicted to be at the membrane interface and it contains all of the β3 residues necessary for interaction with talin-1. While it is possible that the TM domain could affect this interaction, our NMR and HDX studies indicate that the TM helix is broken by a kink as it exits the membrane, implying that the structure of the β3 CT distal to the helix disruption is unlikely to depend on residues buried within the membrane.
We can consider the overall free energy for the interactions of the bilayer-tethered β3 CT (RT ln(Kobs) = ΔGoverall) in terms of several components according to Eq. 1 below. It should be noted that this thermodynamic decomposition is path-independent and does not require kinetic ordering; nevertheless, all interactions here are rapid, on the second to millisecond time scale, indicating that they might rapidly interconvert on a physiologically relevant time scale.
| [1] |
where ΔGtalin–bilayer is the energy associated with talin head interaction with lipid, as determined directly from the titration of the FERM domains into lipid bilayers; ΔGtalin–peptide is the energy associated with talin head interaction with the β3 CT, as determined directly from the NMR or the fluorescent titrations reported above and which is sequence-specific (26) but only weakly favorable (Kdiss on the order of 0.2 to 0.6 mM); and ΔGreorganization is the free energy required to rearrange the conformations of isolated individual components and binary complexes to allow formation of a ternary complex (this term also includes statistical terms associated with the standard states). The electrostatic surfaces of F3, F2F3, and the full FERM domain (24, 41) suggest that F2F3 binds negatively charged bilayers in a manner highly conducive to forming a simultaneous interaction with the β3 CT; this finding has been supported by detailed coarse-grained molecular dynamics calculations (15). Thus, there is likely little reorganization energy associated with the interaction of these subdomains with bilayers. By contrast, the β3 CT, tethered to a bilayer, is in a rapid equilibrium that favors a conformation in which the peptide interacts with the membrane in a way that masks residues important for talin-1 binding and that would otherwise be located 10–30 Å into the cytoplasm (27). A second minor conformation is more exposed to the aqueous surroundings, presumably allowing rapid kinetic access to the FERM domain. HDX showed that the equilibrium favors the membrane-associated state by 100 to 1,000-fold relative to the exposed state, decreasing the overall affinity for binding cytosolic proteins by an equivalent amount. Thus, we expect that ΔGreorganization would be dominated by the rearrangement of the β3 CT within the bilayer.
Consideration of Eq. 1 shows that cytosolic protein binding to the β3 CT in a proper context depends on an energetic balance, which, in this case, includes a highly favorable interaction between talin-1 and the membrane. A modestly favorable free energy of interaction of the β3 CT with the talin-1 head is balanced by a modestly unfavorable free energy of rearrangement of the β3 CT within the bilayer. In net, the observed affinity for formation of the ternary complex remains similar to that for the binary interaction of the talin-1 head with acidic bilayers. Likewise, in the absence of integrin, the binary complex between the FERM domain and the membrane appears to energetically favor the F2F3 subunit, as F0F1 does not affect the overall affinity or stoichiometry of the interaction. This suggests that F0F1 might not interact favorably with the bilayer once it is tethered in place by the more stable F2F3 interaction. Alternatively, F0F1 might interact extensively when the FERM domain binds, but only after an energetically uphill conformational change that attenuates the overall affinity gain associated with F0F1-bilayer interaction. It is likely not accidental that the binary and ternary complexes are not optimized for cooperativity. Rather, they appear to be tuned to be readily regulated by changes in the lipid environment and by posttranslational modifications.
In summary, our studies reveal the rich interplay between structure, dynamics, kinetics, and thermodynamics that govern formation of the ternary complex between the β3 CT, phospholipid bilayers, and talin-1 FERM domain. This energetic dissection provides the basis for understanding how posttranslational modifications and changes in lipid composition might affect such processes. The situation becomes more complex as interactions with additional domains in integrin and talin, as well as other antagonistic or synergistic cytoplasmic proteins, are considered. Nevertheless, the present study illustrates the importance of considering the role of conformational changes, structural rearrangements, and conformational entropy in these processes, and provides a general approach to energetic decomposition that can form the basis for understanding regulation of even more complex systems.
Methods
Design of Recombinant Talin-1, β3 CT, and PIP5K1γ Constructs.
Talin-1, β3 CT, and PIP5K1γ constructs were cloned between the NdeI and XhoI restriction sites of the vector pet28a and contained an N-terminal 6xHis tag for purification, followed by a thrombin cleavage site for affinity tag removal. The talin-1 constructs (Fig. S2 A–C) consisted of the entire talin-1 FERM domain (F0-F3) extending from its N terminus through residue Ser425, the F0F1 subdomains extending from the N terminus through residue Asp201, and the F2F3 subdomains spanning residues Ser206 through Ser425.
The β3 CT construct (Fig. S2D), encompassing residues Thr720 through the C terminus at Thr762, was previously described (27). For the current experiments, residue Ile719 was replaced with Cys to enable the construct to be immobilized on the surface of bilayers or to be labeled via maleimide chemistry. A peptide corresponding to residues 646–654 of PIP5K1γ was also labeled with fluorescein isothiocyanate (FITC) (Fig. S2E).
cDNA in the vector pet28a were introduced into BL21-DE3 cells (Novagen) and expressed for 20 h at 20 °C in the presence of 0.5 mM IPTG. Cells were then lysed by sonication on ice in 20 mM imidazole buffer, pH 7.0, containing 500 mM NaCl, 1 mM EDTA, and a protease inhibitor cocktail (Sigma-Aldrich). After the lysates were cleared by centrifugation, the supernatants were loaded onto HiTrap Ni-NTA columns (GE Healthcare) and peptides were eluted using an imidazole gradient. Imidazole was dialyzed away overnight into 50 mM Tris buffer, pH 7.5 containing 100 mM NaCl and the 6xHis tag was removed using thrombin. The constructs were then purified by SP ion exchange chromatography.
Preparation of LUVs.
LUVs were generated by evaporating a mixture of POPC (Avanti Polar Lipids) and POPS (Avanti Polar Lipids) dissolved in chloroform under nitrogen; excess chloroform was removed under high vacuum overnight. Phosphoinositides (Cell Signaling) were dissolved in CHCl3/MeOH/0.1% HCl, mixed with POPC, and dried under nitrogen. Evaporated lipids were then resuspended in buffer, vortexed vigorously for 5 min, subjected to 3× freeze–thaw cycles, and extruded 20 to 30 times using 100-nm-diameter filters (Avanti Polar Lipids).
ITC.
ITC experiments were performed on a MicroCal ITC200 (MicroCal/GE). Samples were prepared in degassed 25 mM 3-(N-morpholino)propanesulfonic acid (MOPS) buffer, pH 6.8, containing 100 mM NaCl. Samples were loaded into cells after vigorous washing with buffer, and syringes were loaded with 50 μL of titrant. Each experiment consisted of 20 × 2.0 μL injections spaced at intervals of 2–5 min. A 0.2-μL injection preceded all experiments and was not included in fitting. Fitting was performed using the Origin software suite (MicroCal/Origin Systems) as single binding events. Heats of dilution were calculated by titration into the prepared diazylate and subtracted prior to fitting.
SPR.
All SPR experiments were performed on a Biacore 3000 (GE Healthcare). Each injection was done in triplicate. Kinetic experiments were performed in degassed 10 mM (4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES) buffer, pH 7.2, containing 100 mM NaCl. The CM5 chip was activated using an amine coupling kit (GE Healthcare). N-(2-Aminoethyl)maleimide (pH 7.0) was then injected and nonreacted activated groups were capped with 1 M ethanolamine (pH 7.0). The β3 CT was immobilized onto the CM5 chip via maleimide crosslinking to approximately 600 RU. Background maleimide was quenched with 2-mercaptoethanol. Talin-1 FERM domain constructs were injected at 50 μL/ min with a 2-min injection time and 400-s dissociation time. Binding data were fit to a double exponential using the BIAevaluation software.
For steady-state experiments, 100-nm LUVs were loaded onto an L1 chip. Talin-1 FERM domain constructs were injected for 10 min with a 120-s dissociation time and a 10 μL/ min flow rate. Between injections, new lipid surfaces were generated when full dissociation was not observed. Steady-state construct binding was compared to a reference channel containing 100% POPC. Binding was fit to a single component isotherm equation: KA ∗ [analyte] ∗ RUmax/(KA ∗ [analyte] ∗ n + 1) + background.
FP.
FP experiments were performed as previously described (42, 43). Briefly, FP was conducted in an ATF105 spectrofluorometer (Aviv Instrument, Inc) using a 0.3-cm path length cuvette. Spectra were measured using 1.0-nm slit widths at 25 °C. The β3 CT was labeled with maleimide-BODIPY-TMR at a cysteine introduced at residue 719. Talin-1 F0-F3 was titrated into 20 nM β3 BODIPY-TMR, and FP was measured at 575 nm after excitation at 530 nm. When F0-F3 was titrated into 20 nM FITC-labeled PIP5KI-γ, a dissociation constant and baseline parameters were derived from the fluorescence anisotropy signal/concentration isotherm using previously described methods (44).
Supplementary Material
Acknowledgments.
This work was supported by National Institutes of Health Grants HL40387(J.S.B. and W.F.D.), HL81012 (J.S.B. and W.F.D.), and EY18705 (K.B.B.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1117220108/-/DCSupplemental.
References
- 1.Luo BH, Carman CV, Springer TA. Structural basis of integrin regulation and signaling. Annu Rev Immunol. 2007;25:619–647. doi: 10.1146/annurev.immunol.25.022106.141618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banno A, Ginsberg MH. Integrin activation. Biochem Soc Trans. 2008;36:229–234. doi: 10.1042/BST0360229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett JS. Structure and function of the platelet integrin alphaIIbbeta3. J Clin Invest. 2005;115:3363–3369. doi: 10.1172/JCI26989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett JS, Berger BW, Billings PC. The structure and function of platelet integrins. J Thromb Haemost. 2009;7(Suppl):200–205. doi: 10.1111/j.1538-7836.2009.03378.x. [DOI] [PubMed] [Google Scholar]
- 5.Grigoryan G, Moore DT, Degrado WF. Transmembrane communication: General principles and lessons from the structure and function of the m2 proton channel, k(+) channels, and integrin receptors. Annu Rev Biochem. 2011;80:211–237. doi: 10.1146/annurev-biochem-091008-152423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harburger DS, Calderwood DA. Integrin signalling at a glance. J Cell Sci. 2009;122:159–163. doi: 10.1242/jcs.018093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Critchley DR. Biochemical and structural properties of the integrin-associated cytoskeletal protein talin. Annu Rev Biophys. 2009;38:235–254. doi: 10.1146/annurev.biophys.050708.133744. [DOI] [PubMed] [Google Scholar]
- 8.Kim C, Ye F, Ginsberg MH. Regulation of integrin activation. Annu Rev Cell Dev Biol. 2011;27:321–345. doi: 10.1146/annurev-cellbio-100109-104104. [DOI] [PubMed] [Google Scholar]
- 9.Beckerle MC, Miller DE, Bertagnolli ME, Locke SJ. Activation-dependent redistribution of the adhesion plaque protein, talin, in intact human platelets. J Cell Biol. 1989;109:3333–3346. doi: 10.1083/jcb.109.6.3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dietrich C, Goldmann WH, Sackmann E, Isenberg G. Interaction of NBD-talin with lipid monolayers. A film balance study. FEBS Lett. 1993;324:37–40. doi: 10.1016/0014-5793(93)81527-7. [DOI] [PubMed] [Google Scholar]
- 11.Goldmann WH, Senger R, Kaufmann S, Isenberg G. Determination of the affinity of talin and vinculin to charged lipid vesicles: A light scatter study. FEBS Lett. 1995;368:516–518. doi: 10.1016/0014-5793(95)00678-3. [DOI] [PubMed] [Google Scholar]
- 12.Goult BT, et al. Structure of a double ubiquitin-like domain in the talin head: A role in integrin activation. EMBO J . 2010;29:1069–1080. doi: 10.1038/emboj.2010.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Catimel B, et al. The PI(3,5)P2 and PI(4,5)P2 interactomes. J Proteome Res. 2008;7:5295–5313. doi: 10.1021/pr800540h. [DOI] [PubMed] [Google Scholar]
- 14.Goksoy E, et al. Structural basis for the autoinhibition of talin in regulating integrin activation. Mol Cell. 2008;31:124–133. doi: 10.1016/j.molcel.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalli AC, et al. The structure of the talin/integrin complex at a lipid bilayer: An NMR and MD simulation study. Structure. 2010;18:1280–1288. doi: 10.1016/j.str.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saltel F, et al. New PI(4,5)P2- and membrane proximal integrin-binding motifs in the talin head control beta3-integrin clustering. J Cell Biol. 2009;187:715–731. doi: 10.1083/jcb.200908134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abrams CS. Intracellular signaling in platelets. Curr Opin Hematol. 2005;12:401–405. doi: 10.1097/01.moh.0000176681.18710.e3. [DOI] [PubMed] [Google Scholar]
- 18.Moser M, Legate KR, Zent R, Fassler R. The tail of integrins, talin, and kindlins. Science. 2009;324:895–899. doi: 10.1126/science.1163865. [DOI] [PubMed] [Google Scholar]
- 19.Shattil SJ, Kim C, Ginsberg MH. The final steps of integrin activation: The end game. Nat Rev Mol Cell Biol. 2010;11:288–300. doi: 10.1038/nrm2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goult BT, et al. The structure of an interdomain complex that regulates talin activity. J Biol Chem. 2009;284:15097–15106. doi: 10.1074/jbc.M900078200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanchanawong P, et al. Nanoscale architecture of integrin-based cell adhesions. Nature. 2010;468:580–584. doi: 10.1038/nature09621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fehon RG, McClatchey AI, Bretscher A. Organizing the cell cortex: The role of ERM proteins. Nat Rev Mol Cell Biol. 2010;11:276–287. doi: 10.1038/nrm2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frame MC, Patel H, Serrels B, Lietha D, Eck MJ. The FERM domain: Organizing the structure and function of FAK. Nat Rev Mol Cell Biol. 2010;11:802–814. doi: 10.1038/nrm2996. [DOI] [PubMed] [Google Scholar]
- 24.Elliott PR, et al. The structure of the talin head reveals a novel extended conformation of the FERM domain. Structure. 2010;18:1289–1299. doi: 10.1016/j.str.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wegener KL, et al. Structural basis of integrin activation by talin. Cell. 2007;128:171–182. doi: 10.1016/j.cell.2006.10.048. [DOI] [PubMed] [Google Scholar]
- 26.Anthis NJ, Wegener KL, Critchley DR, Campbell ID. Structural diversity in integrin/talin interactions. Structure. 2010;18:1654–1666. doi: 10.1016/j.str.2010.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Metcalf DG, et al. NMR analysis of the alphaIIb beta3 cytoplasmic interaction suggests a mechanism for integrin regulation. Proc Natl Acad Sci USA. 2010;107:22481–22486. doi: 10.1073/pnas.1015545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mori T, et al. Structural basis for CD44 recognition by ERM proteins. J Biol Chem. 2008;283:29602–29612. doi: 10.1074/jbc.M803606200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Calderwood DA, et al. The phosphotyrosine binding-like domain of talin activates integrins. J Biol Chem. 2002;277:21749–21758. doi: 10.1074/jbc.M111996200. [DOI] [PubMed] [Google Scholar]
- 30.Narayan K, Lemmon MA. Determining selectivity of phosphoinositide-binding domains. Methods. 2006;39:122–133. doi: 10.1016/j.ymeth.2006.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Isenberg G, Niggli V, Pieper U, Kaufmann S, Goldmann WH. Probing phosphatidylinositolphosphates and adenosinenucleotides on talin nucleated actin polymerization. FEBS Lett. 1996;397:316–320. doi: 10.1016/s0014-5793(96)01203-3. [DOI] [PubMed] [Google Scholar]
- 32.Martel V, et al. Conformation, localization, and integrin binding of talin depend on its interaction with phosphoinositides. J Biol Chem. 2001;276:21217–21227. doi: 10.1074/jbc.M102373200. [DOI] [PubMed] [Google Scholar]
- 33.Lemmon MA, Ferguson KM, O’Brien R, Sigler PB, Schlessinger J. Specific and high-affinity binding of inositol phosphates to an isolated pleckstrin homology domain. Proc Natl Acad Sci USA. 1995;92:10472–10476. doi: 10.1073/pnas.92.23.10472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schuck P, Zhao H. The role of mass transport limitation and surface heterogeneity in the biophysical characterization of macromolecular binding processes by SPR biosensing. Methods Mol Biol. 2010;627:15–54. doi: 10.1007/978-1-60761-670-2_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anthis NJ, et al. The structure of an integrin/talin complex reveals the basis of inside-out signal transduction. EMBO J . 2009;28:3623–3632. doi: 10.1038/emboj.2009.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Pereda JM, et al. Structural basis for phosphatidylinositol phosphate kinase type Igamma binding to talin at focal adhesions. J Biol Chem. 2005;280:8381–8386. doi: 10.1074/jbc.M413180200. [DOI] [PubMed] [Google Scholar]
- 37.Kong X, Wang X, Misra S, Qin J. Structural basis for the phosphorylation-regulated focal adhesion targeting of type Igamma phosphatidylinositol phosphate kinase (PIPKIgamma) by talin. J Mol Biol. 2006;359:47–54. doi: 10.1016/j.jmb.2006.02.048. [DOI] [PubMed] [Google Scholar]
- 38.Vinogradova O, et al. A structural mechanism of integrin alpha(IIb)beta(3) “inside-out” activation as regulated by its cytoplasmic face. Cell. 2002;110:587–597. doi: 10.1016/s0092-8674(02)00906-6. [DOI] [PubMed] [Google Scholar]
- 39.Vinogradova O, et al. Membrane-mediated structural transitions at the cytoplasmic face during integrin activation. Proc Natl Acad Sci USA. 2004;101:4094–4099. doi: 10.1073/pnas.0400742101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang J, et al. Structure of an integrin alphaIIb beta3 transmembrane-cytoplasmic heterocomplex provides insight into integrin activation. Proc Natl Acad Sci USA. 2009;106:17729–17734. doi: 10.1073/pnas.0909589106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garcia-Alvarez B, et al. Structural determinants of integrin recognition by talin. Mol Cell. 2003;11:49–58. doi: 10.1016/s1097-2765(02)00823-7. [DOI] [PubMed] [Google Scholar]
- 42.Yin H, et al. Activation of platelet alphaIIbbeta3 by an exogenous peptide corresponding to the transmembrane domain of alphaIIb. J Biol Chem. 2006;281:36732–36741. doi: 10.1074/jbc.M605877200. [DOI] [PubMed] [Google Scholar]
- 43.Caputo GA, et al. Computationally designed peptide inhibitors of protein-protein interactions in membranes. Biochemistry. 2008;47:8600–8606. doi: 10.1021/bi800687h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yin H, et al. Terephthalamide derivatives as mimetics of helical peptides: Disruption of the Bcl-x(L)/Bak interaction. J Am Chem Soc. 2005;127:5463–5468. doi: 10.1021/ja0446404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.