The search for novel organic molecules with biological activity on human, animal, and plant physiological systems has passed through many phases over the centuries. From first steps testing single molecules on whole living systems to fully automated high-throughput screening (HTS) testing tens or hundreds of thousands of molecules per day on purified protein targets, the search has become ever more complex (1, 2). However, the increase in success has not been proportional to the effort and expense entailed. In particular, when considering the screening of “small molecules” (molecular mass <1,000 Da), the results of contemporary HTS have been plagued with problems of false positives, false negatives (3), and the abnormal behavior of certain molecules resulting from their physicochemical properties rather than their biological activity (4). In PNAS, Miller et al. (5) describe a significant evolution of current HTS technology that increases the confidence in the detection of truly active molecules by an order of magnitude.
High-potency, highly specific molecular ligands are of great importance to modern medicine and agriculture and can also be valuable research tools that significantly aid the elucidation of metabolic pathways and control mechanisms. In its infancy, searching—or “screening” as it is now called—for active molecules relied on the analysis of plant and animal extracts and was a laborious, slow, and time-consuming process. As technology progressed and more rational methods were elaborated in the 1980s, screening evolved to a “process” whereby series of novel synthetic molecules were tested systematically for activity on one or even several different “targets” or target systems. In the 1990s high-throughput robotic screening methods based on microtiter plates were developed that took advantage of industrial-scale automation and large-scale data processing. This has allowed modern-day drug screening laboratories to “process” several tens or even hundreds of thousands of molecules per day (1, 2).
The early promise of this technology has not, however, been fully realized (6), as is clearly evidenced by the ongoing efforts of (particularly) the pharmaceutical industry to identify novel molecules that are specific and potent ligands for novel targets. Indeed, the number of new drugs (termed new medical entities, or NMEs) is decreasing: in the period 2005–2010, 50% fewer NMEs were approved compared with the previous 5 y (7). In 2007, for example, only 19 NMEs were approved by the US Food and Drug Administration, the lowest number approved since 1983. Although the introduction and application of large-scale combinatorial chemistry methods, in silico virtual screening, X-ray crystallography, and sophisticated molecular modeling have helped to understand how small molecules bind to large proteins, such as enzymes and G protein-coupled receptors, HTS is still mostly based on random searching and often resembles a highly developed and very expensive search for the proverbial needle in a haystack.
There are several reasons for this limited success, and one of the most important is that modern HTS technologies usually only test molecules for activity at a single concentration and thus (at least initially) completely ignore the all-important “dose–response” relationship that underlies the basis of most molecular interactions in biological systems. This results in many false-positive and false-negative findings (3). Although HTS certainly reveals many “active” molecules, it also reveals numerous false positives and many molecules with bizarre dose–response relationships that on further examination render them useless as potential medicines or research tools (8). Indeed, the time, energy, and costs of the analysis of these false positives is one factor that currently restricts the discovery potential of HTS methods. The loss in economic value due to false negatives (molecules that are in reality active but are measured as inactive in HTS) is impossible to assess but has always been a recognized, omnipresent difficulty.
Miller et al. (5) describe a screening technique and strategy that represents a clear advance toward identifying in the first-pass screening campaign only those molecules that are truly acting in a reproducible and dose-dependent fashion. This system was used to screen a library of marketed drugs for inhibition of the enzyme protein tyrosine phosphatase 1B, a target for type 2 diabetes mellitus, obesity, and cancer, and the authors identified a number of unique inhibitors.
Their screening system is based on the use of droplets in a microfluidic system as independent microreactors, which play the same role as the wells of a microtiter plate. However, the reaction volume, which in conventional HTS microplate wells is a few microliters, is reduced to a few picolitres, a reduction of ≈1 million-fold. Droplet-based microfluidics is a rapidly developing technology (for reviews see refs. 9 and 10) that is already commercialized for targeted sequencing and digital PCR (see http://www.raindancetechnologies.com/ and http://www.bio-rad.com/).
The potential advantages of droplet-based microfluidics for the HTS of large biomolecules were graphically illustrated by its recent use to screen enzymes displayed on the surface of yeast (11). Directed evolution (a Darwinian system based on repeated rounds of mutation and selection in the laboratory) was used to improve the activity of horseradish peroxidase. For this experiment, ca. 108 individual enzyme reactions were screened in only 10 h, using <150 μL of total reagent volume. Compared with state-of-the-art robotic screening systems, the entire screen was performed with a 1,000-fold increase in speed and a significant reduction in cost (the total cost of the screen was only $2.50, compared with $15 million using microplates) (11).
The work of Miller et al. (5), however, provides a demonstration of the screening of a small-molecule library using droplet-based microfluidics, and the advantages that this can entail. The compounds to be tested are automatically injected one-by-one from microtiter plates into a continual stream of buffer, and the initial rectangular pulse of each compound is transformed into a concentration gradient using a simple system based on a microfluidic phenomenon first analyzed in the 1950s by Sir Geoffrey Taylor (12). As the compounds travel through a narrow capillary, because there is no turbulence in the microfluidic system, each compound is dispersed in an extremely predictable manner by a combination of diffusion and the parabolic flow profile in the capillary. The diluted compounds then enter a microfluidic chip where they are combined with the assay reagents (the target enzyme and a fluorogenic substrate) and segmented into droplets by two intersecting streams of inert fluorinated oil containing a surfactant. In this way thousands of independent microreactors are generated, each containing a slightly different concentration of compound but the same concentrations
Miller et al. describe a significant evolution of current HTS technology.
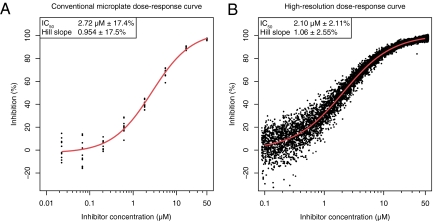
of the assay reagents. After generation, the droplets pass through an on-chip delay line and, after a suitable incubation period, the fluorescence of each droplet is analyzed. By premixing each compound with a near-infrared fluorescent dye before injection, it was possible to calculate the compound concentration in each droplet from its near-infrared fluorescence. In parallel, the degree of enzyme inhibition in the droplet was determined from the fluorescence of the product of the enzymatic reaction at a different wavelength. For each compound the data collected in just over 3 s was sufficient to generate a high-resolution dose–response profile containing ≈10,000 data points (compared with the usual 7 to 10 data points), allowing a determination of the IC50 (concentration of the test compound that inhibits by 50% the activity of the target) with a precision that has yet to be equaled using conventional microplate methods (Fig. 1). Comparison between the microplate and microfluidic methods shows that the microfluidic system generates IC50 values with a 95% confidence interval that is ≈10-fold smaller. Such high-resolution data should allow compounds with undesirable dose–response behavior to be eliminated as early as possible. For example, compounds for which inhibition rises more quickly with concentration than one would expect are normally unsuitable for further development (8).
Fig. 1.
Comparison of (A) a conventional dose–response curve produced using microplates (10 replicates at eight concentrations) and (B) a high-resolution dose–response curves produced using droplet-based microfluidics (data from 11,113 droplets). The curves are for inhibition of the enzyme β-galactosidase by 2-phenylethyl β-d-thiogalactoside. The 95% confidence limits for the fitted IC50 and Hill slope values (Inset in A and B) are approximately an order of magnitude more precise for the high-resolution dose–response curve (5).
The end result is a significant increase in the “confidence” in the data, and thus the task of identifying promising active molecules for further analysis is greatly simplified. Furthermore, by considerably increasing the number of measurements of molecule–target interaction, it can be expected that the false-negative and false-positive rates should be reduced to near zero.
The throughput of the current system is currently only one compound every 157 s. Hence, further work needs to be done to increase the throughput to allow the screening of 105 to 106 compounds, with a high-resolution dose–response curve for every compound, in a large primary screening campaign. Nevertheless, even at the current throughput, the approach should prove useful for focused or iterative drug screenings, which are dependent on data quality and rely on intelligent selection and refinement of chemical libraries rather than brute force (2). The precision with which the dose dependency can be measured is of extreme importance given the natural variation in the response of biological systems, and thus higher-quality measurements of the activity of test molecules will inevitably lead to a better understanding of structure–activity relationships and the underlying chemical biology. The savings in time and effort that could be achieved are yet to be fully assessed, but the method should at least increase the confidence in HTS data.
Footnotes
The author declares no conflict of interest.
See companion article on page 378 of issue 2 in volume 109.
References
- 1.Dove A. Drug screening—beyond the bottleneck. Nat Biotechnol. 1999;17:859–863. doi: 10.1038/12845. [DOI] [PubMed] [Google Scholar]
- 2.Mayr LM, Bojanic D. Novel trends in high-throughput screening. Curr Opin Pharmacol. 2009;9:580–588. doi: 10.1016/j.coph.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 3.Malo N, Hanley JA, Cerquozzi S, Pelletier J, Nadon R. Statistical practice in high-throughput screening data analysis. Nat Biotechnol. 2006;24:167–175. doi: 10.1038/nbt1186. [DOI] [PubMed] [Google Scholar]
- 4.Lipinski C, Hopkins A. Navigating chemical space for biology and medicine. Nature. 2004;432:855–861. doi: 10.1038/nature03193. [DOI] [PubMed] [Google Scholar]
- 5.Miller OJ, et al. High-resolution dose–response screening using droplet-based microfluidics. Proc Natl Acad Sci USA. 2011;109:378–383. doi: 10.1073/pnas.1113324109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell JB. Improving lead generation success through integrated methods: Transcending ‘drug discovery by numbers’. IDrugs. 2010;13:874–879. [PubMed] [Google Scholar]
- 7.Paul SM, et al. How to improve R&D productivity: The pharmaceutical industry's grand challenge. Nat Rev Drug Discov. 2010;9:203–214. doi: 10.1038/nrd3078. [DOI] [PubMed] [Google Scholar]
- 8.Shoichet BK. Interpreting steep dose-response curves in early inhibitor discovery. J Med Chem. 2006;49:7274–7277. doi: 10.1021/jm061103g. [DOI] [PubMed] [Google Scholar]
- 9.Teh SY, Lin R, Hung LH, Lee AP. Droplet microfluidics. Lab Chip. 2008;8:198–220. doi: 10.1039/b715524g. [DOI] [PubMed] [Google Scholar]
- 10.Theberge AB, et al. Microdroplets in microfluidics: An evolving platform for discoveries in chemistry and biology. Angew Chem Int Ed Engl. 2010;49:5846–5868. doi: 10.1002/anie.200906653. [DOI] [PubMed] [Google Scholar]
- 11.Agresti JJ, et al. Ultrahigh-throughput screening in drop-based microfluidics for directed evolution. Proc Natl Acad Sci USA. 2010;107:4004–4009. doi: 10.1073/pnas.0910781107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taylor G. Dispersion of soluble matter in solvent flowing slowly through a tube. Proc R Soc Lond A Math Phys Sci. 1953;219:186–203. [Google Scholar]