Abstract
ATP synthases are the primary source of ATP in all living cells. To catalyze ATP synthesis, these membrane-associated complexes use a rotary mechanism powered by the transmembrane diffusion of ions down a concentration gradient. ATP synthases are assumed to be driven either by H+ or Na+, reflecting distinct structural motifs in their membrane domains, and distinct metabolisms of the host organisms. Here, we study the methanogenic archaeon Methanosarcina acetivorans using assays of ATP hydrolysis and ion transport in inverted membrane vesicles, and experimentally demonstrate that the rotary mechanism of its ATP synthase is coupled to the concurrent translocation of both H+ and Na+ across the membrane under physiological conditions. Using free-energy molecular simulations, we explain this unprecedented observation in terms of the ion selectivity of the binding sites in the membrane rotor, which appears to have been tuned via amino acid substitutions so that ATP synthesis in M. acetivorans can be driven by the H+ and Na+ gradients resulting from methanogenesis. We propose that this promiscuity is a molecular mechanism of adaptation to life at the thermodynamic limit.
Keywords: A1Ao ATPase, energy conservation, ion specificity, methanogens, energetic limit
From archaea to man, the common enzyme in cellular bioenergetics is the ATP synthase. This membrane-bound macromolecular complex produces ATP, the primary energy source in the cell, at the expense of transmembrane ion gradients established by diverse enzymes, which vary across organisms and organelles. ATP synthases and related ATPases arose from a common ancestor and evolved into three distinct classes: the V1V0 ATPase present in eukarya; the F1F0 ATP synthase found in bacteria, mitochondria, and chloroplasts; and the A1A0 ATP synthase present in archaea (1, 2). All of these synthases are rotary machines that comprise two coupled units, one residing in the cytoplasm (F1/A1/V1), and another (Fo/Ao/Vo) embedded in the membrane. The former catalyzes ATP synthesis (or hydrolysis), and the latter is coupled to the transmembrane electrochemical potential (3–8).
The ion specificity of ATP synthases/ATPases is encoded by the membrane-embedded domain, and in particular in the rotor ring. This ring is an oligomeric assembly of multiple copies of subunit c, each of which carries at least one ion binding site. The architecture of the membrane domain enables the rotation of the c ring around its central axis in a manner that is tightly coupled to ion flow across the membrane (9–12). The rotation of the c ring is also mechanically transmitted to the cytoplasmic domain, where it sustains a conformational cycle conducive to ATP synthesis (13, 14).
Most F1F0 ATP synthases and V1V0 ATPases use protons as the coupling ion, but some use sodium ions instead (15–17). However, Na+-driven F1F0 ATP synthases have so far been found only in anaerobic bacteria, whose metabolism leads to the generation of a primary Na+ gradient, rather than a H+ gradient (16, 17). Na+ is clearly preferred over H+ in these ATP synthases (18, 19). This finding is also true at acidic pH, whereby ATP hydrolysis activity increases with increasing Na+ concentration in a typical Michaelis–Menten kinetic, indicating very high Na+ specificity under any physiological condition. Thus, conventional wisdom holds that ATP synthases are either H+- or Na+-coupled but do not use both cations in vivo.
Methanogenic archaea are one of the very few life forms that produce both H+ and Na+ primary electrochemical potentials simultaneously. Methanogenesis is believed to be one of the first biochemical pathways to sustain life on earth, despite its low energetic output (20). All methanogens investigated so far have a sodium-motive methyl-tetrahydromethanopterin:coenzyme M methyltransferase that is also able to generate a sodium ion potential during methanogenesis. The free-energy gain from this reaction allows for the translocation of only around 1.7 mol Na+ per mole of substrate. Evolutionary advanced methanogens, such as strains of the genus Methanosarcina, have cytochromes and a membrane-integral ion-carrier, methanophenazine, the H+ transport mechanism of which is similar to that of ubiquinol. This reaction energizes the translocation of 2 mol of H+. Because at least 3 mol of ions are required for synthesis of 1 mol of ATP (21), it appears that both the Na+ and the H+ gradient must be somehow used for survival.
Thus, the question that arises is how these gradients are used for the synthesis of ATP. A possibility discussed in the literature is the presence of two distinct ATP synthases, one coupled to H+ and another to Na+ (20). However, this notion is not supported by inspection of genome sequences. It is more probable that a Na+/H+ antiporter converts the Na+ gradient into a H+ gradient, or vice versa. It is also conceivable, however, that ATP synthesis may in some cases be driven directly by both Na+ and H+ gradients (20). This phenomenon has never been reported for any ATP synthase in physiological conditions, but such promiscuity has been observed in other biological systems for example, some cation/solute symporters (22, 23) and motility channels (24).
Here, we address this long-standing question using energetically intact inside-out membrane vesicles (IMV) derived from Methanosarcina acetivorans. Our results demonstrate the operation of an ATPase that concurrently drives the translocation of both Na+ and H+ under physiological conditions. This observation, not previously reported for any life form, strongly indicates that both Na+ and H+ gradients are used simultaneously for ATP synthesis in this organism. Using molecular models and computer simulations, we uncover the structural basis for the observed promiscuity of this ATP synthase, in terms of the intrinsic ion selectivity of its c ring rotor. In summary, this study sheds light on a molecular mechanism of adaptation to life at the thermodynamic limit, and substantiates an emerging paradigm in which the ion specificity of ATP synthases is seen as a spectrum, rather than as two opposing extremes, reflecting the natural diversity of environmental constraints.
Results
IMVs Are Bioenergetically Intact.
To assess the possibility of Na+ and H+ transport, we established a system to prepare IMVs from M. acetivorans (SI Materials and Methods). To demonstrate bioenergetic coupling under physiological conditions, we tested whether metabolism leads to ATP synthesis by IMVs. Upon addition of methanol, methanogenesis started, and a simultaneous increase in ATP content was detected (Fig. S1). The ATPase inhibitor dicyclohexylcarbodiimide (DCCD) prevented ATP synthesis, indicating the generation of an ion gradient that in turn can drive ATP synthesis.
Na+ Stimulates ATP Hydrolysis by IMVs of M. acetivorans.
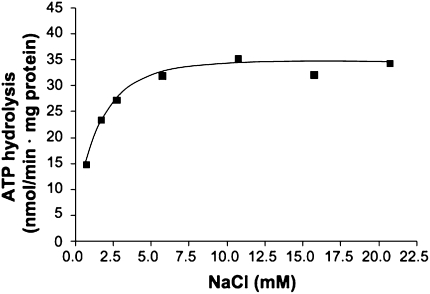
IMVs of M. acetivorans (SI Materials and Methods) hydrolyzed ATP with a rate of 29–60 mU/mg protein. ATP hydrolysis was optimal at pH 7, in a buffer containing 25 mM Pipes-KOH, 25 mM MgSO4⋅7H2O, 10 mM KHSO3, and 0.4 M sucrose (SI Materials and Methods). The Na+ concentration in the buffer was 0.75 mM. At that Na+ concentration, the rate of ATP hydrolysis was only 14.7 nmol⋅min-1 · mg-1 protein, but it could be stimulated by addition of NaCl. Maximal ATPase activity was observed at 6 mM NaCl, with half maximal activity at 0.92 ± 0.37 mM (Fig. 1). Addition of K+ did not stimulate ATP hydrolysis.
Fig. 1.
Na+ stimulation of ATP hydrolysis. Washed membrane vesicles (final concentration 0.7 mg/mL) were incubated in a total volume of 1 mL in a 25 mM Pipes-KOH buffer, pH 7, containing 25 mM MgSO4⋅7H2O, 0.4 M sucrose, 17 μM valinomycin, 10 mM KHSO3, 40 mM KCl, and increasing Na+ concentrations. The Na+ concentration of the assay without additional Na+ was 0.75 mM.
Na+ Transport Is Coupled to ATP Hydrolysis.
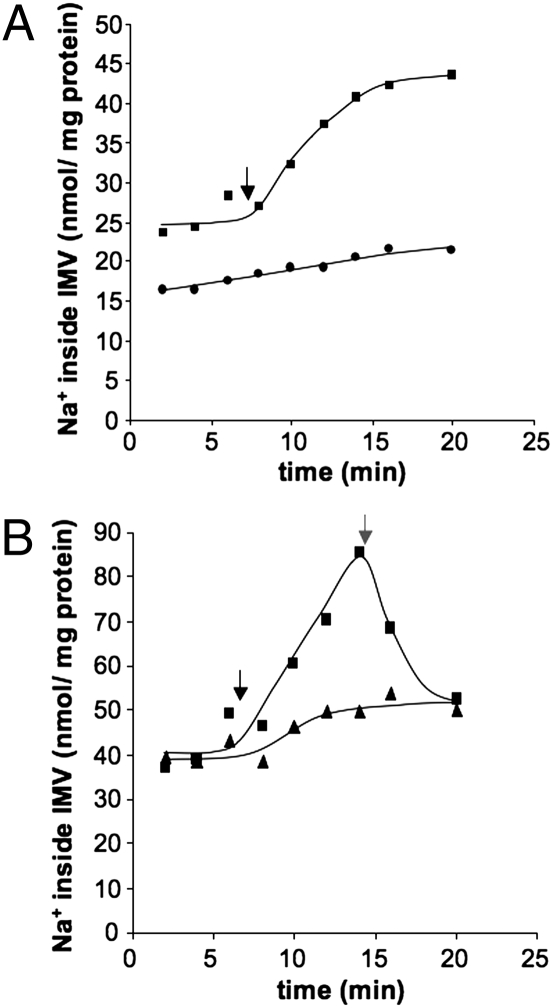
To determine whether ATP hydrolysis is not only stimulated by Na+ but also accompanied by Na+ transport, IMVs were incubated in the presence of 10 mM 22Na+ (SI Materials and Methods). Upon addition of ATP, 22Na+ was accumulated in the lumen of the IMVs (Fig. 2A). The rate of 22Na+ transport was accelerated by addition of Na+ (Fig. S2). Maximal Na+ transport was observed at 6 mM, half maximal at 1.1 ± 0.4 mM, which fits to the Km value determined for ATP hydrolysis. The Vmax was dependent on the vesicle preparation and ranged between 2.3 and 7.7 nmol Na+ per min⋅mg protein. Na+ transport was inhibited by DCCD (70% at a concentration of 750 μM) and diethylstilbestrol (DES) (62% at a concentration of 250 μM), two well-known ATP synthase inhibitors. This finding is consistent with the hypothesis that the A1A0 ATP synthase present in the IMVs catalyses ATP-driven Na+ translocation.
Fig. 2.
ATP-dependent, primary Na+ transport. Washed membrane vesicles were diluted in 1 mL 25 mM Pipes-KOH buffer, pH 7, containing 25 mM MgSO4⋅7H2O and 0.4 M sucrose. The assay was supplemented with 17 μM valinomycin, 10 mM NaHSO3, and 50 mM KCl. The reaction was started by addition of 5 mM of K2-ATP (black arrow). In A, one assay (●) did not receive ATP. In B, one assay (▲) was preincubated with ETH157; the other received ETH157, as indicated by the gray arrow (■).
Na+ Transport Is Primary and Electrogenic.
As a result of Na+ translocation, not only a chemical substance is transported but also a positive charge; thus, a transmembrane electrical potential (ΔΨ) is established that is positive on the inside. This ΔΨ should in turn slow down further charge translocation. Conversely, charge compensation should enhance Na+ transport by diminishing the magnitude of ΔΨ. This result was indeed observed. The K+ ionophore valinomycin (17 μM) in the presence of 50 mM KCl stimulated Na+ transport by 20%, indicating electrogenic Na+ movement.
To test whether the observed Na+ accumulation inside the IMVs is caused by a primary translocation of Na+ and not the result of a secondary Na+ gradient, for example built up by a Na+/H+ antiporter, we determined the effect of the protonophores 3,5-di-tert-butyl-hydroxybenzylidenemalonitrile (SF6847) and carbonyl cyanide m-chlorophenylhydrazone (CCCP). If the Na+ gradient was secondary, and because of a primary H+ gradient, the protonophore should abolish the Na+ accumulation. However, Na+ accumulation was not affected by 10 μM SF6847 or CCCP. In contrast, the addition of the sodium ionophore N,N,N,N′-tetracyclohexyl-1,2-phenylendioxydiacetamide (ETH157) (20 μM) completely prevented Na+ accumulation (Fig. 2B). In summary, the ionophore experiments clearly demonstrate that the ATP-dependent Na+ transport is indeed primary and electrogenic.
Evidence for Concurrent Transport of Na+ and H+.
In bacterial Na+-coupled F1Fo ATP synthases, ATP hydrolysis is impaired in the absence of Na+. At low (5.3) and high (9.0) pH values ATP hydrolysis is restored by the addition of Na+, showing typical Michaelis–Menten kinetics (15, 19, 25). These observations indicate that these ATP synthases are unequivocally Na+-specific under physiological conditions and not driven by H+. This finding is consistent with the fact that the organisms known to have Na+ F1Fo ATP synthases have only sodium-motive enzymes (decarboxylases, Rnf complex) to couple substrate oxidation to membrane energization (16, 17, 26).
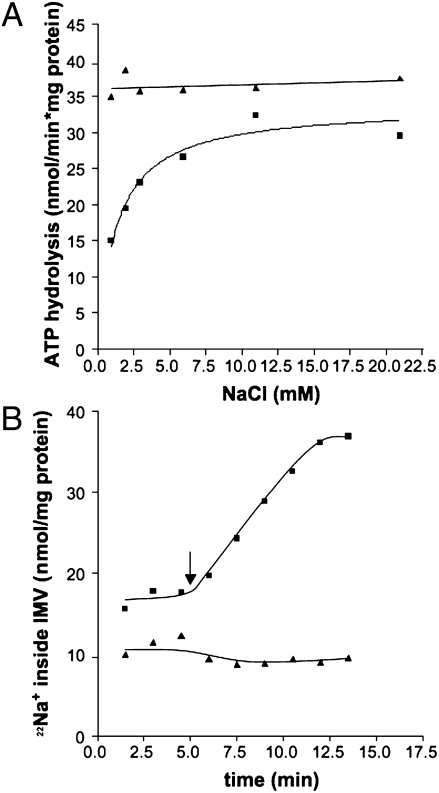
To assess whether the A1Ao ATP synthase from M. acetivorans shares these features, the dependence of ATP hydrolysis on Na+ concentration was analyzed at two different pH values (SI Materials and Methods). Most surprisingly, and in sharp contrast to the known Na+ F1Fo ATP synthases, ATP hydrolysis at pH 5 was not stimulated by Na+ (Fig. 3A). Moreover, the ATPase activity at pH 5 was even higher than that at pH 7 and saturating Na+ concentrations. This finding strongly indicates that under these conditions, full activity is achieved with protons as the coupling ion. This indication was corroborated by the fact that Na+ transport was not observed at pH 5 and 10 mM Na+ (Fig. 3B).
Fig. 3.
Effect of pH on Na+-dependence of ATP hydrolysis (A) and Na+ transport (B). Washed membrane vesicles [final concentration 0.73 mg/mL (A) or 0.13 mg/mL (B)] were incubated in a 25 mM Pipes-KOH/Mes-HCl buffer containing 25 mM MgSO4⋅7H2O and 0.4 M sucrose. One milliliter of assay was supplemented with 17 μM valinomycin, 10 mM KHSO3, 40 mM KCl, and increasing Na+ concentrations. The pH was adjusted to 7 (■) or 5 (▲). The Na+ concentration of the assay without additional Na+ was 0.95 mM. The Na+ concentration in B was 10 mM.
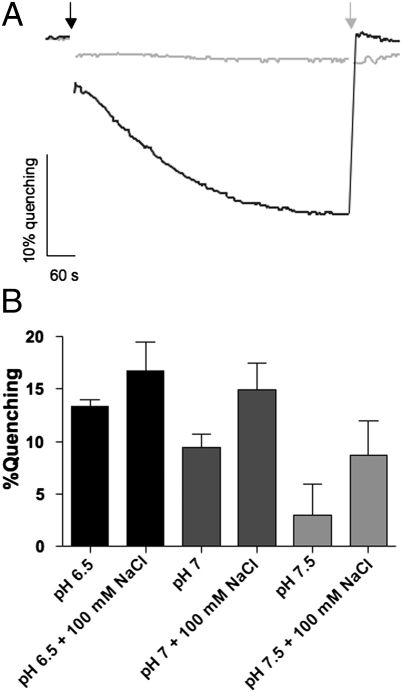
To establish whether H+ is translocated under the same conditions at which active Na+ transport occurs, we monitored the development of a pH gradient across the vesicle membranes using 9-amino-6-chloro-2-methoxyacridine (ACMA) (SI Materials and Methods). Upon addition of ATP, the fluorescence of ACMA was quenched, demonstrating proton transport into the vesicle. Subsequent addition of (NH4)2SO4, which dissipates the H+ gradient across the membrane, restored the fluorescence signal to the level before ATP addition. Preincubation with DCCD (or the protonophores SF6847 or CCCP) prevented fluorescence quenching (Fig. 4A), indicating that the pH gradient (inside acidic) is generated by the ATP synthase, working in hydrolysis mode. As expected, inward H+ transport was stimulated by high H+ concentrations, and decreasing H+ concentrations reduced transport rates (Fig. 4B). ATP hydrolysis, however, was approximately constant under these conditions. Importantly, H+ transport occurred in the presence of Na+ (Fig. 4B), and specifically under the conditions of Na+ translocation, at pH 7. Na+ stimulated H+ transport (Fig. 4B), which is consistent with observed stimulation of ATP hydrolysis under these conditions. In support of this notion is the observed increase in stimulation with decreasing H+ concentration. Taken together, this data demonstrates that under physiological pH and Na+ concentrations, H+ and Na+ concurrently bind to and are transported by the c ring of the methanogenic ATP synthase.
Fig. 4.
ATP-dependent H+ transport. Washed membrane vesicles (final concentration 0.32 mg/mL) were incubated in 1 mL of 5 mM Pipes, 5 mM Tricine, 25 mM MgSO4, 150 mM KCl, and 4 μM ACMA. (A) Effect of DCCD on H+ transport at pH 7. ATP (2 mM) was added as indicated (black arrow). The quenching was abolished by 20 μL saturated (NH4)2SO4 (gray arrow). The reaction was inhibited by DCCD (750 μM) (gray line). The Na+ concentration in the assay was 1 mM. (B) Effect of NaCl and pH on H+ transport. The pH and NaCl concentration were as indicated. The quenching percentage is plotted as a function of assay conditions.
Structural Models of Subunit c in the Na+- and H+-Bound States.
As mentioned, the ring of c subunits in the membrane domain of the ATP synthase carries a series of ion binding sites, approximately halfway through the membrane (27). These sites are sequentially loaded and unloaded as the ring rotates around its axis and those sites exit the lipid environment and enter the interface with the subunit a, which is adjacent to the c ring in the membrane (28). It is thus logical to assume that the ion-specificity of the ATP synthase is defined to a large extent by the rotor ring.
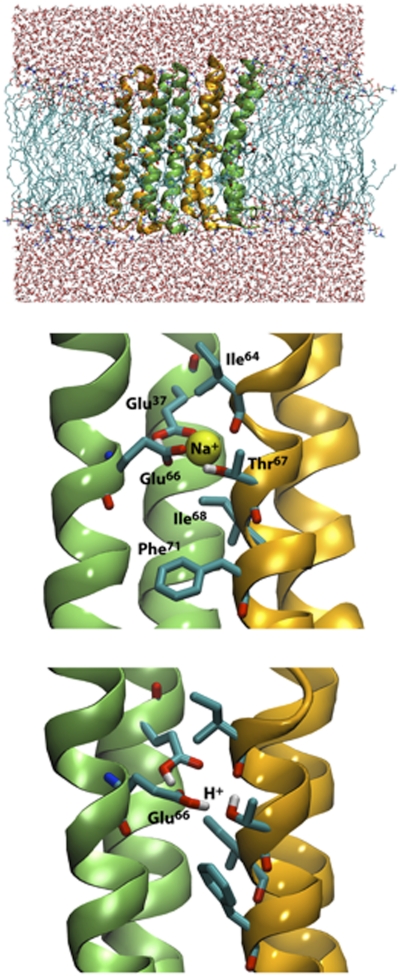
To rationalize the apparent ion promiscuity of the M. acetivorans ATP synthase, we analyzed the structure and selectivity of its c ring using computer modeling and simulation methods (SI Materials and Methods). First, structural models of a c4 oligomer were created in both the Na+- and H+-bound states, taking advantage of the sequence homology between subunit c of M. acetivorans and those of Enterococcus hirae, Ilyobacter tartaricus, and Spirulina platensis, for which atomic structures are known (29–31) (Fig. S3). Molecular dynamics simulations of these c4 oligomers embedded in a phospholipid membrane were then carried out to refine the models in a realistic environment. Finally, these models were used to analyze the thermodynamic selectivity for H+ or Na+ of the M. acetivorans membrane rotor through free-energy calculations.
Representative snapshots of the molecular simulations of the Na+- and H+-bound c4 constructs are shown in Fig. 5. In the Na+-bound state, the ion is coordinated by the conserved Glu66 side-chain in one c subunit and a backbone carbonyl group (from Ile64) in the adjacent one, as seen in, for example, the rotor ring of I. tartaricus (30). Two additional ion-coordinating side-chains in that structure, namely a serine and a glutamine, are replaced in M. acetivorans by Thr67 and Glu37; by analogy, we expect the latter to be constitutively protonated. Thr67 and Glu37 form identical interactions as their counterparts in I. tartaricus, both with the bound Na+ and with the conserved Glu66, which accepts a hydrogen-bond from each. Nevertheless, two important interactions that favor binding of Na+ over H+ in I. tartaricus are missing in M. acetivorans. Ile68 replaces a threonine, and leaves no room for a key structural water (30), and Phe71 replaces a tyrosine (32). In the H+-bound state of the M. acetivorans c subunit, Glu66 becomes protonated and donates a hydrogen bond to Thr67. The interaction of Glu66 with the protonated Glu37 remains, but the hydroxyl group of Thr67 reorients to hydrogen-bond with the carbonyl group in Ile64, which was previously engaging the bound Na+. This exact interaction pattern has not been observed experimentally as yet, but it is remarkably similar to that predicted in theoretical studies of H+ coordination in other H+-coupled rotors (32, 33).
Fig. 5.
Hypothetical ion-coordination structure in the ATP synthase c ring from M. acetivorans. (Top) Snapshot of a molecular dynamics simulation of a model c4 oligomer in a phospholipid membrane. (Middle) Close-up view of the ion-binding site, in the Na+-bound state, in the model based on the c11 ring from I. tartaricus. (Bottom) Close-up view of the ion-binding site, in the H+-bound state, in the model based on the c15 ring from S. platensis.
M. acetivorans c Ring Is Chemically H+-Selective, but also Binds Excess Na+.
The structural insights discussed above indicate that Na+ coordination is possible in the M. acetivorans ATP synthase, but that it is to some degree suboptimal relative to the canonical Na+-coupled systems from, for example, I. tartaricus or its homolog Propionigenium modestum. In contrast, the mode of H+ coordination in M. acetivorans seems to be very much comparable to those in, for example, H+-coupled cyanobacterial rings. We therefore reasoned that the specificity of the M. acetivorans c ring may be somewhere in between these extremes. To assess this hypothesis more quantitatively, we used the so-called “alchemical perturbation” method to extract the free energy of Na+/H+ selectivity from additional molecular dynamics simulations (SI Materials and Methods).
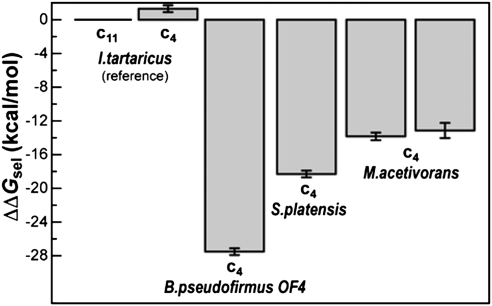
The result of these calculations for our models of the M. acetivorans c ring binding site are summarized in Fig. 6, with reference to analogous data obtained for I. tartaricus, S. platensis, and Bacillus pseudofirmus OF4. This analysis unequivocally shows that the M. acetivorans c ring is strongly H+-selective; specifically, its H+ selectivity is eight to nine orders-of-magnitude greater than the I. tartaricus c ring, which is weakly H+-selective and physiologically Na+-coupled (32). Nevertheless, the M. acetivorans c ring favors H+ to a significantly lesser degree than that of S. platensis, a cyanobacterium for which Na+ binding has not been detected. In particular, the M. acetivorans ring is about 1,000 times more likely to bind Na+. We attribute this difference largely to the presence of Thr67 in the Na+-coordination sphere, instead of a hydrophobic alanine in S. platensis. The contrast is even more pronounced with the alkalophile B. pseudofirmus OF4 (34), in which the c ring binding sites are more hydrophobic and thus extremely H+ selective.
Fig. 6.
Computed free energy of H+/Na+ selectivity of the c subunit binding sites in M. acetivorans, relative to that of the I. tartaricus c11 ring. More negative values indicate greater selectivity for H+ and against Na+. All values are given in reference to analogous calculations for the complete c11 ring from I. tartaricus, previously reported (32). Selectivity values are shown for two alternative models of a M. acetivorans c4 oligomer, one based on the architecture of the I. tartaricus c11 ring (Left), and another based on the c15 ring from S. platensis (Right). The computed selectivity of analogous c4 constructs from I. tartaricus, S. platensis, and B. pseudofirmus OF4 is also given.
Discussion
Based on experimental assays of ATP hydrolysis and ion transport in bioenergetically active inverted membrane vesicles from the marine archaeon M. acetivorans, we have obtained several lines of evidence that demonstrate that the rotary mechanism of its A1A0 ATP synthase is coupled to the concurrent transmembrane translocation of both Na+ and H+ under physiological conditions. Our interpretation of this experimental data rests on a comparative theoretical analysis of the ion selectivity the c-ring rotor, based on computer simulations. It is therefore assumed that the specificity of the enzyme is conferred primarily by the ion binding sites in this transmembrane structure. In principle, it is plausible that other subunits within the membrane domain contribute as well; in particular, subunit a is believed to provide aqueous access pathways for the transported ions into the c ring binding sites (35–37), and could therefore confer additional specificity to the enzyme. However, to our knowledge no experimental evidence or bioinformatic analysis to date has indicated that this subunit is ion-specific.
In contrast, the thermodynamic principle of ion selectivity of the c subunit rotor has been recently delineated by theoretical analyses of Na+- and H+-driven rotors (32), produced either by X-ray diffraction, computer modeling, or a combination thereof (30, 31, 33, 38). In this view, all c rings are seen as intrinsically H+ selective, as a result of the presence of a conserved carboxylate side-chain (Glu/Asp) in the binding sites. This baseline H+ selectivity is, however, insufficient to ensure coupling to the proton motive force in physiological settings, because Na+ is typically in great excess over H+. On the other hand, the intrinsic H+ selectivity that the Glu/Asp confers is too large to ensure strict coupling to Na+ gradients. Thus, ATP synthase c rings that are exclusively H+- or Na+-coupled have had to either boost or weaken this intrinsic H+ selectivity. To accomplish the required specificify, it appears that the key adaptation is the amino acid composition of the binding sites (rather than for example, ring stoichiometry, or c subunit arrangement). To enhance H+ binding over Na+, the sites must become more hydrophobic; to favor Na+ binding, they must increase the number of polar side-chains capable of cation-coordination (32).
The ion specificity of the ATP synthase should, therefore, be seen as a discrete spectrum. In alkalophilic organisms, such as B. pseudofirmus, the only polar side-chain in the c subunit binding site is the conserved Glu, and its predicted H+-selectivity is extreme. At the other end of the spectrum, the binding sites in E. hirae or I. tartaricus include three possible Na+ coordinating interactions (in addition to the carboxylate and a backbone contact), and are therefore only marginally H+-selective, and thus Na+-coupled at physiological ion concentrations. Two of these polar side-chain interactions are preserved in the M. acetivorans c ring, shifting the H+-specificity of the structure upwards, so that concurrent coupling to H+ and Na+ ions (simultaneously bound to different sites in the rotor) may take place in physiological or in vitro settings where Na+ is in sufficient excess.
Our structural analysis of the M. acetivorans rotor assumes first that the specificity of the binding sites in a c4 oligomer is comparable to that of the complete c ring, the stoichiometry of which is to date unknown. In this regard, we note that the computed selectivity of the I. tartaricus c4 construct differs from that of the c11 ring by less than one order-of-magnitude (i.e., < 1.4 kcal/mol) (Fig. 5). By analogy, we expect that the calculated selectivity of the c4 oligomer from M. acetivorans will be a good estimate of that of the complete ring. A second assumption is that the essential factors contributing to ion specificity can be captured in models based on known crystal structures. To address this theory, we created two independent models based on c rings of noticeably different architecture, namely those of I. tartaricus and S. platensis. These rings differ in the stagger of the outer and inner helices, as well as in the bend angle of the outer helix and in their stoichiometry. Moreover, one model was created in the Na+ bound state, and in the other a bound H+ was assumed. That the computations for these different models yield the same selectivity values (Fig. 6) indicates that these models capture the key factors controlling the specificity of the c ring.
Finally, it is worth stressing that this analysis is focused on the thermodynamic component of the ion selectivity of the c ring. This analysis is therefore not dependent on factors such as the electrochemical gradient, the nature of the interface with subunit a, or the lipid composition of the membrane. These factors likely influence the rotary mechanism and transport kinetics of the enzyme, and therefore might modulate its ion selectivity. However, we postulate that the relative binding affinity of Na+ and H+ to the c ring is the primary cause for the physiological specificity of the ATP synthase.
In conclusion, we have experimentally demonstrated that the rotary mechanism of the A1A0 ATP synthase in M. acetivorans is coupled to the translocation of both Na+ and H+ across the membrane under physiological conditions. Using molecular modeling and computer simulations, we have rationalized this unprecedented observation in terms of the structure of the c-subunit rotor and the adaptations of its amino acid sequence. Taken together, these data strongly suggest that ATP synthesis in M. acetivorans is concurrently driven by physiological gradients of Na+ and H+. Given the low free-energy gain resulting from methanogenesis (20, 39), the simultaneous use of Na+ and H+ gradients for ATP production appears to be a convenient adaptation to the environmental challenges facing M. acetivorans. It will be of interest to investigate whether or not this adaptation has occurred in nonmarine, cytochrome-containing methanogens as well (e.g., Methanosarcina mazei).
Materials and Methods
Preparation of Washed Membrane Vesicles.
M. acetivorans cultivation and crude vesicle preparation was carried out as described in SI Materials and Methods. For the preparation of washed membrane vesicles, M. acetivorans cells were grown in 20-L fermenters with 120 mM acetate as sole carbon and energy source. The cells were harvested aerobically at an A578 of 0.4–0.5 and protoplasts were prepared as described in SI Materials and Methods. Therefore, DTE and resazurin were omitted. For cell disruption, the cells were suspended in a 25 mM Pipes-KOH buffer, pH 7, containing 25 mM MgSO4 and 0.4 M sucrose. Before disruption, 100 μM PMSF as well as a few crystals of DNase were added. The protoplasts were passed through a French pressure cell at 400 PsiG, which resulted in the formation of IMVs. The remaining protoplast as well as the cell debris was removed by centrifugation (4,000 × g, 10 min, 4 °C). The supernatant was centrifuged at 135,000 × g for 1 h at 4 °C to collect membrane vesicles. The membrane fraction was washed and afterward resuspended in the same Pipes buffer to a final protein concentration of 10–15 mg/mL. The protein concentrations were determined by the method described by Bradford (40).
To determine whether the IMVs are bioenergetically coupled, energization with an artificial ammonium gradient or with growth substrate was tested, as described in SI Materials and Methods.
Measurement of Na+ Translocation.
The experiments were performed under aerobic conditions in 25 mM Pipes-KOH buffer, pH 7, containing 25 mM MgSO4⋅7H2O and 0.4 M sucrose at 37 °C in a water bath in a 3.5 mL glass vial. One milliliter of buffer was supplemented with 17 μM valinomycin and 10 mM KHSO3 or NaHSO3, 40 or 50 mM KCl (depending on the cation of the sulfite used) and NaCl as indicated. Then, 20 μM ETH157, 10 μM SF6847, 10 μM CCCP, 250 μM DES, or 750 μM DCCD were added as indicated. 22NaCl (final activity 0.5 μCi/mL carrier-free) was added and incubated for 40 min to ensure Na+ equilibrium before the reaction was started. After 6 min, 5 mM K2-ATP was added. The Na+ concentration of the assay was determined with a Na+-selective electrode (model 720A; Orion). When the effect of the pH had to be analyzed, a buffer containing 25 mM Pipes-KOH/Mes-HCl, 25 mM MgSO4⋅7H2O, 0.4 M sucrose, 17 μM valinomycin and 10 mM NaHSO3 at pH 5 or 7 was used. SF6837, DCCD, DES, ETH157, and valinomycin were added as ethanolic solutions. CCCP was solved in DMSO. Controls received the solvent only. The samples were taken and radioactivity was measured as described previously (41).
Measurement of H+ Translocation.
The experiments were performed under aerobic conditions in 5 mM Pipes-KOH, 5 mM Tricine-HCl buffer, containing 25 mM MgSO4⋅7H2O, 150 mM KCl and 4 μM ACMA at 37 °C. Then, 100 mM NaCl, 10 μM SF6847, 10 μM CCCP, or 750 μM DCCD were added as indicated. After equilibration of fluorescence 2 mM K2-ATP was added. The quench was abolished by the addition of 20 μL of saturated (NH4)2SO4 solution. Fluorescence was measured in a cuvettete in a fluorescence spectrophotometer (F-4500; Hitachi) with excitation at 410 nm and emission at 490 nm.
Structural Modeling and Simulation.
A description of the methodology for modeling the structure of Na+- and H+-bound c4 oligomers is provided in SI Materials and Methods, along with details on the molecular dynamics simulations and the calculations of the free energy of selectivity.
Supplementary Material
Acknowledgments
We thank Dr. Lucy Forrest for her suggestions on this manuscript and Alexander Krah for discussions. This work was funded by Deutsche Forschungsgemeinschaft SFB807; the Biodiversity and Climate Research Centre (BiK-F), Frankfurt am Main; the Ministry of Higher Education, Research, and the Arts (LOEWE); and the Cluster of Excellence “Macromolecular Complexes.” Computational resources were in part provided by the Juelich Supercomputing Centre.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1115796109/-/DCSupplemental.
References
- 1.Müller V, Grüber G. ATP synthases: Structure, function and evolution of unique energy converters. Cell Mol Life Sci. 2003;60:474–494. doi: 10.1007/s000180300040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grüber G, Wieczorek H, Harvey WR, Müller V. Structure-function relationships of A-, F- and V-ATPases. J Exp Biol. 2001;204:2597–2605. doi: 10.1242/jeb.204.15.2597. [DOI] [PubMed] [Google Scholar]
- 3.Abrahams JP, Leslie AGW, Lutter R, Walker JE. Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 4.Pänke O, Gumbiowski K, Junge W, Engelbrecht S. F-ATPase: Specific observation of the rotating c subunit oligomer of EFoEF1. FEBS Lett. 2000;472:34–38. doi: 10.1016/s0014-5793(00)01436-8. [DOI] [PubMed] [Google Scholar]
- 5.Sambongi Y, et al. Mechanical rotation of the c subunit oligomer in ATP synthase (FoF1): Direct observation. Science. 1999;286:1722–1724. doi: 10.1126/science.286.5445.1722. [DOI] [PubMed] [Google Scholar]
- 6.Stock D, Gibbons C, Arechaga I, Leslie AG, Walker JE. The rotary mechanism of ATP synthase. Curr Opin Struct Biol. 2000;10:672–679. doi: 10.1016/s0959-440x(00)00147-0. [DOI] [PubMed] [Google Scholar]
- 7.Yoshida M, Muneyuki E, Hisabori T. ATP synthase—A marvellous rotary engine of the cell. Nat Rev Mol Cell Biol. 2001;2:669–677. doi: 10.1038/35089509. [DOI] [PubMed] [Google Scholar]
- 8.Duncan TM, Zhou Y, Bulygin VV, Hutcheon ML, Cross RL. Probing interactions of the Escherichia coli FoF1 ATP synthase beta and gamma subunits with disulphide cross-links. Biochem Soc Trans. 1995;23:736–741. doi: 10.1042/bst0230736. [DOI] [PubMed] [Google Scholar]
- 9.Vik SB, Antonio BJ. A mechanism of proton translocation by F1Fo ATP synthases suggested by double mutants of the a subunit. J Biol Chem. 1994;269:30364–30369. [PubMed] [Google Scholar]
- 10.Feniouk BA, et al. The proton-driven rotor of ATP synthase: Ohmic conductance (10 fS), and absence of voltage gating. Biophys J. 2004;86:4094–4109. doi: 10.1529/biophysj.103.036962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Junge W, Sielaff H, Engelbrecht S. Torque generation and elastic power transmission in the rotary FoF1-ATPase. Nature. 2009;459:364–370. doi: 10.1038/nature08145. [DOI] [PubMed] [Google Scholar]
- 12.Dimroth P, von Ballmoos C, Meier T. Catalytic and mechanical cycles in F-ATP synthases. Fourth in the Cycles Review Series. EMBO Rep. 2006;7:276–282. doi: 10.1038/sj.embor.7400646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishmukhametov R, Hornung T, Spetzler D, Frasch WD. Direct observation of stepped proteolipid ring rotation in E. coli F0F1-ATP synthase. EMBO J. 2010;29:3911–3923. doi: 10.1038/emboj.2010.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yokoyama K, Nakano M, Imamura H, Yoshida M, Tamakoshi M. Rotation of the proteolipid ring in the V-ATPase. J Biol Chem. 2003;278:24255–24258. doi: 10.1074/jbc.M303104200. [DOI] [PubMed] [Google Scholar]
- 15.Murata T, Kawano M, Igarashi K, Yamato I, Kakinuma Y. Catalytic properties of Na+-translocating V-ATPase in Enterococcus hirae. Biochim Biophys Acta. 2001;1505:75–81. doi: 10.1016/s0005-2728(00)00278-4. [DOI] [PubMed] [Google Scholar]
- 16.Dimroth P. Bacterial sodium ion-coupled energetics. Antonie van Leeuwenhoek. 1994;65:381–395. doi: 10.1007/BF00872221. [DOI] [PubMed] [Google Scholar]
- 17.Schmidt S, Biegel E, Müller V. The ins and outs of Na+ bioenergetics in Acetobacterium woodii. Biochim Biophys Acta. 2009;1787:691–696. doi: 10.1016/j.bbabio.2008.12.015. [DOI] [PubMed] [Google Scholar]
- 18.Laubinger W, Dimroth P. The sodium ion translocating adenosinetriphosphatase of Propionigenium modestum pumps protons at low sodium ion concentrations. Biochemistry. 1989;28:7194–7198. doi: 10.1021/bi00444a010. [DOI] [PubMed] [Google Scholar]
- 19.Reidlinger J, Müller V. Purification of ATP synthase from Acetobacterium woodii and identification as a Na+-translocating F1Fo-type enzyme. Eur J Biochem. 1994;223:275–283. doi: 10.1111/j.1432-1033.1994.tb18992.x. [DOI] [PubMed] [Google Scholar]
- 20.Deppenmeier U, Müller V. Life close to the thermodynamic limit: How methanogenic archaea conserve energy. Results Probl Cell Differ. 2008;45:123–152. doi: 10.1007/400_2006_026. [DOI] [PubMed] [Google Scholar]
- 21.von Ballmoos C, Wiedenmann A, Dimroth P. Essentials for ATP synthesis by F1F0 ATP synthases. Annu Rev Biochem. 2009;78:649–672. doi: 10.1146/annurev.biochem.78.081307.104803. [DOI] [PubMed] [Google Scholar]
- 22.Pourcher T, Leclercq S, Brandolin G, Leblanc G. Melibiose permease of Escherichia coli: Large scale purification and evidence that H+, Na+, and Li+ sugar symport is catalyzed by a single polypeptide. Biochemistry. 1995;34:4412–4420. doi: 10.1021/bi00013a033. [DOI] [PubMed] [Google Scholar]
- 23.Tolner B, Ubbink-Kok T, Poolman B, Konings WN. Cation-selectivity of the L-glutamate transporters of Escherichia coli, Bacillus stearothermophilus and Bacillus caldotenax: Dependence on the environment in which the proteins are expressed. Mol Microbiol. 1995;18:123–133. doi: 10.1111/j.1365-2958.1995.mmi_18010123.x. [DOI] [PubMed] [Google Scholar]
- 24.Terahara N, Krulwich TA, Ito M. Mutations alter the sodium versus proton use of a Bacillus clausii flagellar motor and confer dual ion use on Bacillus subtilis motors. Proc Natl Acad Sci USA. 2008;105:14359–14364. doi: 10.1073/pnas.0802106105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kluge C, Dimroth P. Kinetics of inactivation of the F1Fo ATPase of Propionigenium modestum by dicyclohexylcarbodiimide in relationship to H+ and Na+ concentration: Probing the binding site for the coupling ions. Biochemistry. 1993;32:10378–10386. doi: 10.1021/bi00090a013. [DOI] [PubMed] [Google Scholar]
- 26.Biegel E, Schmidt S, González JM, Müller V. Biochemistry, evolution and physiological function of the Rnf complex, a novel ion-motive electron transport complex in prokaryotes. Cell Mol Life Sci. 2011;68:613–634. doi: 10.1007/s00018-010-0555-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meier T, Polzer P, Diederichs K, Welte W, Dimroth P. Structure of the rotor ring of F-Type Na+-ATPase from Ilyobacter tartaricus. Science. 2005;308:659–662. doi: 10.1126/science.1111199. [DOI] [PubMed] [Google Scholar]
- 28.Pogoryelov D, et al. Microscopic rotary mechanism of ion translocation in the Fo complex of ATP synthases. Nat Chem Biol. 2010;6:891–899. doi: 10.1038/nchembio.457. [DOI] [PubMed] [Google Scholar]
- 29.Murata T, Yamato I, Kakinuma Y, Leslie AG, Walker JE. Structure of the rotor of the V-Type Na+-ATPase from Enterococcus hirae. Science. 2005;308:654–659. doi: 10.1126/science.1110064. [DOI] [PubMed] [Google Scholar]
- 30.Meier T, et al. Complete ion-coordination structure in the rotor ring of Na+-dependent F-ATP synthases. J Mol Biol. 2009;391:498–507. doi: 10.1016/j.jmb.2009.05.082. [DOI] [PubMed] [Google Scholar]
- 31.Pogoryelov D, Yildiz O, Faraldo-Gómez JD, Meier T. High-resolution structure of the rotor ring of a proton-dependent ATP synthase. Nat Struct Mol Biol. 2009;16:1068–1073. doi: 10.1038/nsmb.1678. [DOI] [PubMed] [Google Scholar]
- 32.Krah A, et al. Structural and energetic basis for H+ versus Na+ binding selectivity in ATP synthase Fo rotors. Biochim Biophys Acta. 2010;1797:763–772. doi: 10.1016/j.bbabio.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 33.Leone V, Krah A, Faraldo-Gómez JD. On the question of hydronium binding to ATP-synthase membrane rotors. Biophys J. 2010;99:L53–L55. doi: 10.1016/j.bpj.2010.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Preiss L, Yildiz O, Hicks DB, Krulwich TA, Meier T. A new type of proton coordination in an F1Fo ATP synthase rotor ring. PLOS Biol. 2010;8:e1000443. doi: 10.1371/journal.pbio.1000443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Angevine CM, Herold KAG, Fillingame RH. Aqueous access pathways in subunit a of rotary ATP synthase extend to both sides of the membrane. Proc Natl Acad Sci USA. 2003;100:13179–13183. doi: 10.1073/pnas.2234364100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steed PR, Fillingame RH. Subunit a facilitates aqueous access to a membrane-embedded region of subunit c in Escherichia coli F1F0 ATP synthase. J Biol Chem. 2008;283:12365–12372. doi: 10.1074/jbc.M800901200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steed PR, Fillingame RH. Aqueous accessibility to the transmembrane regions of subunit c of the Escherichia coli F1F0 ATP synthase. J Biol Chem. 2009;284:23243–23250. doi: 10.1074/jbc.M109.002501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krah A, Pogoryelov D, Meier T, Faraldo-Gómez JD. On the structure of the proton-binding site in the Fo rotor of chloroplast ATP synthases. J Mol Biol. 2010;395:20–27. doi: 10.1016/j.jmb.2009.10.059. [DOI] [PubMed] [Google Scholar]
- 39.Thauer RK, Kaster AK, Seedorf H, Buckel W, Hedderich R. Methanogenic archaea: Ecologically relevant differences in energy conservation. Nat Rev Microbiol. 2008;6:579–591. doi: 10.1038/nrmicro1931. [DOI] [PubMed] [Google Scholar]
- 40.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 41.Heise R, Müller V, Gottschalk G. Presence of a sodium-translocating ATPase in membrane vesicles of the homoacetogenic bacterium Acetobacterium woodii. Eur J Biochem. 1992;206:553–557. doi: 10.1111/j.1432-1033.1992.tb16959.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.