Abstract
Passive transfer of broadly neutralizing human antibodies against HIV-1 protects macaques against infection. However, HIV-1 uses several strategies to escape antibody neutralization, including mutation of the gp160 viral surface spike, a glycan shield to block antibody access to the spike, and expression of a limited number of viral surface spikes, which interferes with bivalent antibody binding. The latter is thought to decrease antibody apparent affinity or avidity, thereby interfering with neutralizing activity. To test the idea that increasing apparent affinity might enhance neutralizing activity, we engineered bispecific anti–HIV-1 antibodies (BiAbs) that can bind bivalently by virtue of one scFv arm that binds to gp120 and a second arm to the gp41 subunit of gp160. The individual arms of the BiAbs preserved the binding specificities of the original anti-HIV IgG antibodies and together bound simultaneously to gp120 and gp41. Heterotypic bivalent binding enhanced neutralization compared with the parental antibodies. We conclude that antibody recognition and viral neutralization of HIV can be improved by heteroligation.
Keywords: bispecific antibodies, HIV gp160, heterotypic binding
The human serologic response to HIV-1 is polyclonal and targets both internal and viral surface proteins, but only antibodies directed against the HIV envelope spike, gp160, mediate viral neutralization (1, 2). Although antibodies that neutralize autologous viruses are common, only a fraction of the patients infected with HIV-1 develop broadly neutralizing serologic activity, and only 2 to 3 y after infection (3–9). Several broadly neutralizing antibodies (bNAbs) to HIV-1 gp160 have been isolated, including a group that binds to gp120 (10–18) and a group that is specific for gp41 (19–21). Importantly, passive transfer of bNAbs to monkeys effectively protects them against simian-human immunodeficiency virus infection (22–31), and it has therefore been proposed that vaccines that elicit such antibodies may be protective against infection in humans (1, 2, 32–34).
Broad and potent anti-HIV antibodies are rare in part because there are numerous features of the HIV envelope protein that make it a poor target. These include (i) rapid mutation of variable regions followed by a selection of neutralization escape mutants (35–41), (ii) carbohydrate shielding (36, 42), (iii) conformation masking (43), (iv) steric occlusion (44, 45), (v) transient epitope exposure (46), and (vi) nonfunctional envelope spikes (47, 48). In addition to these well-established “defense” mechanisms, it has been proposed that the low density of functional HIV gp160 on the viral surface may render the anti-HIV antibodies less efficient for viral neutralization by impeding their bivalent binding to the virus (49–51).
Because the functional properties of an antibody are strongly influenced by its binding activity, an increased affinity or avidity for a critical epitope often results in higher potency (50, 52–58); consequently, bivalent binding of specific antibodies to HIV should enhance neutralization. One directly testable prediction of this hypothesis is that anti-HIV antibodies that bind bivalently to their target show increased neutralizing potency. To test this idea, we artificially engineered anti-HIV gp120/41 bispecific antibodies (BiAbs) bearing one antigen-binding site directed against a nonneutralizing gp41 epitope and a second to one of a number of different neutralizing gp120 epitopes. Three different anti-gp120/41 BiAbs were generated that showed simultaneous binding to gp120 and gp41 antigens. When tested for viral neutralization, we found enhanced neutralization by the anti-HIV gp120/41 BiAbs compared with the original anti-gp120 IgG antibodies. Our results show that heteroligation by bivalent antibody binding to two different epitopes on gp160 can lead to more efficient viral neutralization.
Results
Engineering Anti-HIV gp120/41 BiAbs.
To produce antibodies that bind simultaneously to both gp120 and gp41 subunits of HIV-1 gp160, we engineered scFv-Fc IgG-like molecules, also called immunoadhesins, bearing two distinct antigen-binding sites using one of three different anti-gp120s and one anti-gp41 (Fig. 1A). The scFv fragments were produced by overlapping PCR using variable heavy- and light-chain domain (VH and VL) genes encoding human anti-gp120, anti-gp41, or a control antibody (mGO53), which does not bind to gp160 and is not polyreactive (Table S1) (59–61). Specific primers were used to introduce a flexible (G3S)4 linker between VH and VL domains (Fig. S1A). To efficiently produce antibody heterodimers, specific scFv fragments were cloned into a γ1-expression vector modified to introduce a “knob into hole” that favors heterodimer formation (62) and a unique HIS or FLAG tag at the C terminus of each scFv-Fc arm (Fig. 1A and Fig. S1 B and C).
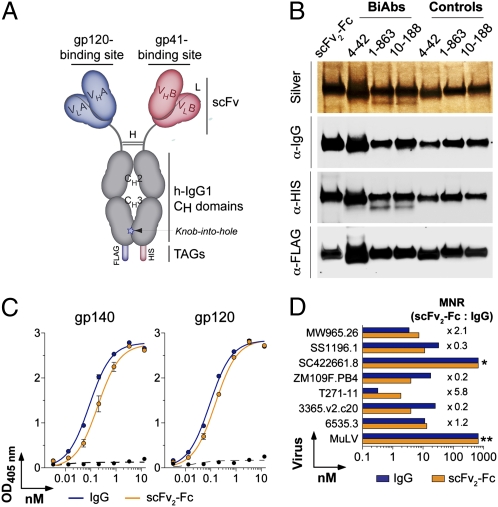
Fig. 1.
Design and production of HIV-gp120/41 BiAbs. (A) Schematic diagram shows the gp120/41 BiAbs made as scFv2-Fc IgG-like molecules bearing one antibody binding site to gp120 and the other to gp41. scFv, single-chain variable fragment; VH, heavy-chain variable domain; VL, light-chain variable domain; H, hinge; L, (G4S)3 linker; CH, heavy-chain constant domain; h-IgG1, human IgG1. (B) Silver staining and Western blot analyses of gp120/41 BiAbs and BiAb controls (anti-gp120/mGO53 heterodimers). (C) Binding analyses of 10-188 IgG antibody and scFv2-Fc control to gp120 and gp140 antigens measured by ELISA. The x axis shows the antibody concentration (nM) required to obtain the ELISA values (OD405 nm) indicated on the y axis. The dotted lines show mGO53 negative control (61). (D) Neutralizing activity of 10-188 IgG and scFv2-Fc control, measured by TZM-bl assay. The x axis shows the antibody concentration (nM) required to achieve IC50 for each virus indicated on the y axis. The MNR values comparing the IC50 concentrations of 10-188 scFv2-Fc and 10-188 IgG are given for each tested virus. All experiments were performed at least in duplicate. Error bars indicate SEM. *Not neutralized; **negative control.
Bivalent scFv-Fc Recapitulates Binding and Neutralizing Properties of Anti-HIV IgG Antibodies.
To determine whether the antigen-binding and neutralizing properties of the original IgG antibodies can be preserved in scFv-Fcs, we produced homodimeric scFvs (scFv2-Fc) using an anti-V3 antibody, 10-188 (59). Purified 10-188 scFv2-Fc recognized artificial YU-2 gp140 trimers and YU-2 gp120 by ELISA with the same binding profiles as 10-188 IgG antibody (Fig. 1 B and C). Moreover, the in vitro neutralizing activity of the 10-188 scFv2-Fc in TZM-bl cells was similar to the native antibody (Fig.1D and Table S2). Thus, scFv2-Fc can preserve the binding and neutralizing properties of anti-HIV antibodies.
HIV gp120/41 BiAbs Exhibit Dual Binding to gp120 and gp41.
We selected three different human anti-gp120 antibodies with modest neutralizing activity and one anti-gp41 antibody with no neutralizing activity for BiAb production. Antibody 10-188 binds to a linear epitope in the variable loop V3 (gp120V3) (59), and the other two anti-gp120 antibodies recognize conformational epitopes in the CD4 binding site (CD4bs) and CD4-induced coreceptor binding site (CD4i) (1-863 and 4-42, respectively) (Table S1) (60). Anti-gp120 antibodies were paired with anti-gp41 antibody 5-25, which is directed against the immunodominant linear epitope of gp41 (gp41ID) (63) (Fig. S2 A and B) or with the nonspecific mGO53 control (61). Three gp120/41 BiAbs (10-188/5-25, 1-863/5-25, and 4-42/5-25) and their corresponding gp120/mGO53 control heterodimers (10-188/mGO53, 1-863/mGO53, and 4-42/mGO53) were produced and purified to ≈90% purity using successive affinity chromatography (Fig. S1 C and D). The anti-gp41 5-25/mGO53 heterodimer was also produced as additional control. Coexpression of both arms of the BiAbs was confirmed by anti-HIS or -FLAG Western blotting (Fig. 1B).
First, we analyzed the binding of the anti-gp120/41 BiAbs and controls to gp140, gp120, or gp41 using surface plasmon resonance (SPR) and ELISAs. Similar to the parental anti-HIV IgGs, all anti-gp120/41 BiAbs and BiAb controls bound to gp140 trimers (Fig. 2A and Fig. S3A). In addition, anti-gp120/41 BiAbs recognized both gp120 and gp41 ligands, whereas the gp120/mGO53 and gp41/mGO53 BiAb controls only bound to gp120 and gp41, respectively (Fig. 2A and Fig. S3B). To further show that anti-gp120/41 BiAbs and BiAb controls bind to gp140 with the same specificity as the original anti-gp120 and anti-gp41 IgG antibodies, we performed ELISAs using gp120V3 and gp41ID peptides, as well as two mutant proteins: gp120(D368R), which interferes with the binding of antibodies to the CD4bs (64), and gp120(I420R), which interferes with the binding of antibodies to the CD4i (65). As expected the 10-188–derived BiAbs recognized the gp120V3 peptide and both of the gp120 mutant proteins, whereas the 1-863– and 4-42–derived BiAbs showed reduced binding to gp120(D368R) and gp120(I420R), respectively (Fig. S4). In addition, only anti-gp120/41 BiAbs and anti-gp41 5-25/mGO53 heterodimer reacted against gp41ID peptide by ELISA (Fig. S2).
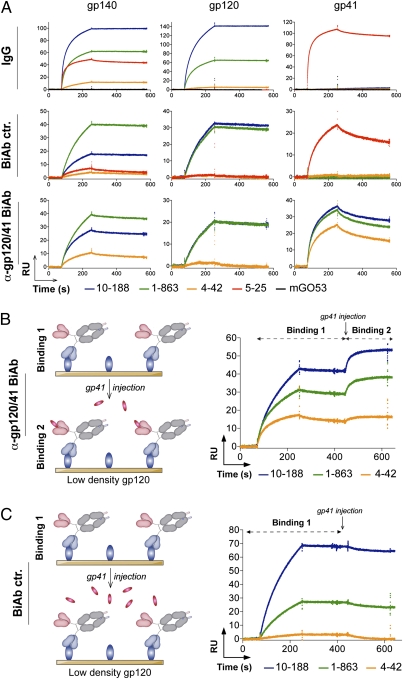
Fig. 2.
Dual antigen binding of HIV-gp120/41 BiAbs. (A) SPR analyses of the interaction of the anti-gp120/41 BiAbs, IgG, and BiAb controls (BiAb ctr.) with the gp140, gp120, and gp41 ligands immobilized at low density on the sensor chips (100 RU). Graphs show SPR sensorgrams over time for the binding of the selected antibodies. (B) Graph shows SPR analysis of the interaction of the anti-gp120/41 BiAbs monovalently bound to gp120 immobilized on the low-density chip (100 RU) (binding 1) with in solution-injected gp41 (binding 2), as illustrated by the schematic diagram (Left). (C) SPR analyses show no interaction of the BiAb controls monovalently bound to gp120 immobilized on the low-density chip (100 RU) (binding 1) with in solution-injected gp41, as illustrated by the schematic diagram (Left). All experiments were performed at least in duplicate. Representative data are shown.
Taken together, these results indicate that anti-gp120/41 BiAbs are capable of dual antigen recognition of the HIV envelope protein owing to the conservation of the original antibody binding specificities against gp120 and gp41.
HIV gp120/41 BiAbs React Simultaneously with gp120 and gp41.
To determine whether the anti-gp120/41 BiAbs are capable of binding simultaneously to gp120 and gp41, we performed SPR “sandwich” experiments. Soluble gp41 was injected in solution over anti-gp120/41 or control BiAbs bound to gp120 immobilized on SPR chips (Fig. 2 B and C). In contrast to the anti-gp120 BiAb controls, which failed to bind to gp41, anti-gp120/41 BiAbs showed reactivity to soluble gp41 that was consistent with high-affinity antigen capture (Fig. 2 B and C). Thus, anti-HIV BiAbs are able to bind simultaneously to gp120 and gp41, without the specific binding of one interfering with the binding of the other.
HIV-1 Neutralization Potency Is Enhanced by Heteroligation.
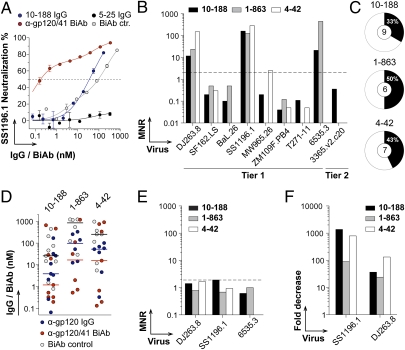
To evaluate the in vitro neutralizing activity of the anti-gp120/41 BiAbs, we measured their capacity to inhibit TZM-bl cell infection by pseudovirus variants (Table S2). To determine whether they display increased neutralization potency and/or breadth, we compared them with BiAb controls and parental anti-gp120 antibodies. The results are expressed as molar neutralization ratio (MNR), to account for any differences in the number of binding sites per antibody. First, it is noteworthy that the anti-gp120/41 BiAbs were in most cases far more potent than their respective BiAb controls to neutralize the selected HIV variants (Fig. S5 and Tables S2 and S3). Most importantly, the anti-gp120/41 BiAbs showed an increased neutralizing potency compared with the parental anti-gp120 IgG antibodies for many of the viruses tested (Fig. 3 A–D and Tables S2 and S3). For instance, 1-863 BiAb showed a 24-, 128-, and >455-fold increased neutralization for DJ263.8, SS1196.1, and 6535.3, respectively, compared with native 1-863 (Fig. 3B, Fig. S5, and Table S2). Overall, the anti-gp120/41 BiAbs enhanced the neutralization of 33–50% of the selected HIV variants compared with their parental IgG counterparts (Fig. 3C). Furthermore, the anti-gp120/41 BiAbs showed lower median IC50 concentrations for the neutralization of those virus strains than the IgG and BiAb controls (Fig. 3D).
Fig. 3.
Neutralizing activity of HIV-gp120/41 BiAbs. (A) Graph shows neutralization curves of SS1196.1 pseudovirus for 10-188 BiAb and controls. The x axis shows the antibody concentration (nM) required to achieve 50% neutralization (IC50), indicated by the dashed line. (B) Bar graph shows the MNR values comparing the IC50 concentrations of the anti-gp120/41 BiAbs and parental anti-gp120 IgGs for each tested virus. (C) Pie charts show the frequency of virus variants showing enhanced neutralization with the anti-gp120/41 BiAbs compared with parental anti-gp120 IgG antibodies. (D) Dot graph shows the IC50 concentrations of anti-gp120/41 BiAbs compared with controls for the neutralization of the selected HIV viruses. Median IC50 values are indicated for each group by horizontal lines. (E) Bar graph shows the MNR values comparing the IC50 concentrations of the parental anti-gp120 IgGs mixed with 5-25 anti-gp41 IgG and anti-gp120 IgG mAbs alone for the selected viruses. (F) Bar graph shows the fold decreases for the neutralization activity of the anti-gp120/41 BiAbs in presence of gp41ID peptide against the selected pseudoviruses.
Interestingly, the same three viruses (DJ263.8, SS1196.1, and 6535.3) showed a higher sensitivity to the neutralization by the gp120/41 BiAbs compared with anti-gp120 IgG controls, irrespective of the specificity of the anti-gp120 arm (Fig. 3B, Fig. S5, and Tables S2 and S3).
HIV-1 Neutralization by gp120/41 BiAbs Requires Heteroligation.
To verify that the enhanced neutralization was not due to a synergetic effect caused by different BiAb molecules binding independently to gp41 and gp120, we tested the neutralization activity of equimolar mixtures of anti-gp120 and anti-gp41 IgGs and Fab fragments. As expected, neither IgG nor Fab mixtures led to enhanced neutralization of the sensitive HIV strains compared with parental anti-gp120 IgG (Fig. 3E, Fig. S6, and Table S2). In addition, to confirm that the enhanced neutralizing activity of the anti-gp120/41 BiAbs against the sensitive viruses was dependent on the binding of the gp41-specific arm, they were tested in a peptide competition neutralization assay wherein the gp41ID peptide was added to block the gp41 arm of the BiAb. The neutralizing activity of the gp120/41 BiAbs against the selected sensitive viruses was completely abrogated in the presence of gp41ID peptide (specific epitope of 5-25 anti-gp41 antibody) (Fig. 3F and Fig. S7). Indeed, the neutralization profiles of the anti-gp120/41 BiAbs in the presence of the peptide competitor resembled those of the corresponding anti-gp120 BiAb controls (Fig. S7B).
In summary, these experiments showed that the enhanced neutralization observed against the sensitive HIV viruses by the anti-gp120/41 BiAbs requires simultaneous binding to gp120 and gp41 epitopes.
Discussion
Homotypic bivalent antibody binding increases apparent affinity, or avidity, and thereby contributes to antibody-mediated neutralization of viruses that express high densities of surface spikes, such as respiratory syncytial and influenza viruses (50, 54–58). However, HIV has only a small number of surface gp160 trimers (49, 50). Cryoelectron microscopy tomography showed that mature viruses express 10–15 randomly distributed viral spikes, which would be spaced too far apart for a bivalent antibody to bridge (49–51). Nevertheless, bivalent binding could theoretically be achieved by altered antibody geometry, increased valency, or by intramolecular ligation within a trimeric envelope protein if it were compatible with the topographic distribution of the epitope on the trimer.
For example, polymeric 2F5 and 2G12 antibodies (IgA and IgM) showed greater neutralizing breadth and potency than their monomeric IgG counterparts (66). In addition, a dimeric form of 2G12 IgG bNAb also increased its neutralization potency (67). Cross-linking of HIV spikes has also been achieved by engineering artificial molecules composed of a CD4i-directed Fab or IgG fused to CD4, thereby producing a molecule that binds simultaneously to the CD4i site and the CD4bs on gp120. These artificial constructs were more potent neutralizers of selected HIV strains than the original IgG antibodies (68, 69). Finally, homotypic bivalent binding of 2F5 to its cognate epitope expressed on gp41 and to another 2F5-epitope artificially grafted into the gp120-variable loop 4 led to enhanced neutralizing potency (70).
Alternatively, bivalency can be achieved by heteroligation as documented for the 2F5 and 4E10 antibodies that bind to both the viral spike and envelope lipid (71–74). Although lipid reactivity is required for the neutralizing activity of these antibodies, the mechanism that enhances neutralization is not precisely defined (72, 74–76). One possibility is that neutralization is enhanced by bivalent heterotypic binding. Alternatively, lipid binding simply may increase the local concentration of the antibody on the membrane, effectively increasing the on rate and thereby the overall affinity (72). The anti-CD4i antibody, 21c, which binds to both CD4 and CD4i (when CD4 is bound to gp120), is an example of how heteroligation by a single antibody arm can increase antibody avidity to the HIV spike (77). Finally, the majority of anti-gp160 memory B-cell antibodies are polyreactive (59, 78) and capable of bivalent heterotypic binding or heteroligation between one high-affinity anti-HIV-gp160 combining site and a second low-affinity site on another molecular structure on HIV (78). This unusual form of bivalent binding, whereby one antibody arm binds to a specific ligand and the second to a hetero-ligand, also increases the apparent affinity of anti-HIV antibodies (78).
To directly assess the effect of heteroligation on neutralizing activity, we produced BiAbs composed of an anti-gp120 arm with neutralizing activity and a gp41 arm with no neutralizing activity. Anti-gp120/41 BiAbs preserved the specificity of the parental anti-HIV antibodies and bound to gp120 and gp41 simultaneously in SPR experiments. Addition of the gp41 arm but not nonspecific controls to the anti-gp120 in the BiAb increased neutralizing potency against some but not all HIV-1 strains compared with the parental anti-gp120 IgG antibodies. We hypothesize that the variability of the neutralizing activity of the anti-gp120/41 BiAbs could be explained by the heterogeneity between the various viral isolates tested, with respect to expression density of the HIV-1 envelope protein and conformational architecture of the gp160 trimer.
In summary, although the precise mode of bivalent binding achieved by the anti-gp120/41 BiAbs was not explored (i.e., intra- or interspikes interactions), the enhanced neutralization observed provides a proof of the concept that heteroligation, using HIV gp120/41 BiAbs, increases neutralization potency most likely via a gain in avidity.
Materials and Methods
scFv PCR and Expression-Vector Cloning.
scFv fragments were generated by classic overlapping PCR using VL and VH DNA fragments cloned into human Ig-expressing vectors (79) as templates, and encoding for anti-gp140 mAbs (59, 60) or mGO53 control mAb (61). Briefly, VL and VH DNA fragments were amplified using specific primers introducing a (G3S)4 linker sequence at the 3′ and 5′ ends of VL and VH DNA fragments, respectively (Fig. S1). PCR reactions were performed using 0.65 U of HotStart Taq polymerase (Promega) and comprised one cycle of 94 °C for 10 min, 35 cycles of 94 °C for 45 s, 59 °C for 45 s, and 72 °C for 1 min, and a final elongation step of 72 °C for 5 min. PCR products were purified using a NucleoSpin Extract II kit (Macherey-Nagel) and assembled by overlapping PCR using specific 5′AgeI VH and 3′ BstXI Jk primers (Fig. S1) and 0.32 U of Pfu Turbo DNA Polymerase (Agilent). PCR conditions comprised one cycle of 94 °C for 2 min, 35 cycles of 94 °C for 30 s, 59 °C for 45 s, and 72 °C for 1 min 30, and a final elongation step of 72 °C for 10 min. Purified scFv fragments were then cloned into a modified human IgG1-expressing vector as described below, using AgeI and BstXI restriction sites. Our standard cloning vector (79) was modified by PCR to introduce a FLAG or a Hexa-Histine (HIS) tag at the C terminus of the IgH constant domain 3 (CH3). The γ1-expressing vector was further modified by directed-site mutagenesis (QuikChange Site-Directed Mutagenesis Kit; Stratagene) to remove a BstXI site in the vector backbone and to introduce a C243A substitution. A “knob into hole” double mutation (a T366Y substitution on the FLAG-tagged arm and a Y407T substitution on the HIS-tagged arm) was also introduced in the γ1-expression vector to increase the production of heterodimers, as previously described (62) (Fig. 1A). Vectors containing scFv DNA fragment were isolated from transformed-DH10β bacteria using plasmid DNA purification kits (NucleoSpinPlasmid, Macherey-Nagel; or PureLink Plasmid Maxiprep Kit, Invitrogen), sequenced and compared with the original PCR-product sequences (MacVector).
Production and Purification of BiAbs.
Anti-HIV gp160 mAbs and BiAbs were produced by cotransfection of exponentially growing HEK 293T cells (ATCC, CRL-11268) using a polyethylenimine precipitation method as described previously (59). Equal amounts of scFvγ1_His- and scFvγ1_FLAG-expressing vectors (15 μg of each plasmid DNA per plate) were used for cotransfection. Cells were cultured for 4 d at 37 °C in a 5% CO2 air atmosphere before the harvesting of the supernatants. BiAbs were affinity purified using Protein G Sepharose beads (GE Healthcare) followed by HisPur cobalt-agarose (Pierce) according to the manufacturer's instructions. After dialysis in PBS, the proteins were separated by SDS/PAGE in 3–8% separating gels (Invitrogen) and were transferred onto nitrocellulose membranes followed by Western blot analysis with anti-FLAG (Sigma), anti-HIS (BD Biosciences), or anti-human IgG (BD Pharmingen) antibodies to monitor the heterodimer production. In parallel, gels were stained with Coomassie Blue G-250 or Silver stain (Thermo Scientific) to check the protein purity. Relative quantification of stained protein bands was performed using ImageJ 1.42q software (National Institutes of Health).
Fabs.
Fab fragments were produced from anti-gp140 IgG mAbs by papain digestion using Fab preparation kit (Pierce). Their purity was checked on G250 Coomassie blue-stained 4–12% NuPAGE gel (Invitrogen).
ELISAs.
High-binding 96-well ELISA plates (Costar) were coated overnight with 100 ng per well of purified antigens [YU-2 gp140, YU-2 gp120, YU-2 gp120(D368R), YU-2 gp120(I420R) (59), and gp41 (Acris)] in PBS. After washing, plates were blocked 2 h with 2% BSA, 1 μM EDTA, and 0.05% Tween-PBS (blocking buffer), and then incubated 2 h with IgG mAbs or BiAbs diluted at 26.7 nM and several consecutive 1:4 dilutions in PBS. After washings, the plates were revealed by incubation with goat HRP-conjugated anti-human IgG antibodies (Jackson ImmunoResearch) (at 0.8 μg/mL in blocking buffer) for 1 h and by adding 100 μL of HRP chromogenic substrate (ABTS solution; Invitrogen). Optical densities were measured at 405 nm (OD405nm) using an ELISA microplate reader (Molecular Devices). Background values given by incubation of PBS alone in coated wells were subtracted. To assay the antibody binding to the selected gp41 overlapping peptides (sequences shown in Fig. S2) and to gp120V3 peptide (RKSINIGPGGRALYTTGEII) (Proteomics Resource Center, The Rockefeller University) (60), the antibodies were tested using a previously described peptide-ELISA method (59). For competition ELISAs, high-binding ELISA plates (Costar) were coated with 100 ng per well of purified gp140 and gp41. Plates were blocked 2 h (with blocking buffer) and incubated for 2 h with anti-HIV gp140 IgG and BiAb antibodies at 26.7 nM in 1:2 serially diluted gp41589-608 peptide (gp41ID)-containing PBS (with a concentration ranging from 5.71 nM to 2.92 μM). Plates were developed as described above. All experiments were performed at least in duplicate.
Surface Plasmon Resonance.
All experiments were performed with a Biacore T100 in HBS-EP+ running buffer (Biacore) at 25 °C as described previously (78). gp140, gp120, and gp41 proteins at 12.5 μg/mL were immobilized on CM5 chips (Biacore) by amine coupling at pH 4.5, resulting in an immobilization level of 100 response units (RUs). For binding analyses on the gp140-, gp120-, and gp41-derivatized chips, IgG mAbs and BiAbs were injected through flow cells at 500 nM and 1,000 nM, respectively, at flow rates of 35 μL/min with 3-min association and 5-min dissociation. The sensor surface was regenerated between each experiment with a 30-s injection of 10 mM glycine·HCl (pH 2.5) at a flow rate of 50 μL/min. Binding curves were plotted after subtraction of backgrounds (binding to control flow cells and signal of the HBS-EP+ running buffer) using Scrubber 2 software (Center for Biomolecular Interaction Analysis, University of Utah). For the analysis of the simultaneous binding of the anti-gp120/41 BiAb molecules to gp120 binding (binding 1) and to gp41 (binding 2), the anti-gp120/41 BiAbs and BiAb controls were injected through flow cells (at 700 nM) in HBS-EP+ running buffer at flow rates of 30 μL/min with 3-min association and 2-min dissociation. Purified gp41 (at a final concentration of 30 μg/mL) was then injected in HBS-EP+ running buffer at a flow rate of 20 μL/min with 2-min contact time. The sensor surface was regenerated as described above.
Neutralization Assays.
Virus neutralization was measured using a luciferase-based assay in TZM.bl cells, as previously described (80). For competition assays, purified anti-gp120/41 BiAbs were incubated with gp41ID peptide at a final concentration of 0.53 μM (in 0.5% DMSO PBS) before neutralization testing using the TZM.bl assay. Fold decreases for neutralization activity were calculated by dividing the IC50 values of the BiAbs in the presence of gp41ID peptide by the ones given by the controls (BiAbs at the same concentration in 0.5% DMSO PBS without peptide).
Supplementary Material
Acknowledgments
We thank Pamela J. Bjorkman and members of the M.C.N. laboratory for helpful discussions. This research was supported by The Rockefeller University, National Institutes of Health Grant PO1 AI081677, the International AIDS Vaccine Initiative, and Bill and Melinda Gates Foundation Comprehensive Antibody-Vaccine Immune Monitoring Consortium Grant 38619 (to M.S.S.). M.W. was supported by the German National Academic Foundation. M.C.N. is a Howard Hughes Medical Institute investigator.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1120059109/-/DCSupplemental.
References
- 1.Mascola JR, Montefiori DC. The role of antibodies in HIV vaccines. Annu Rev Immunol. 2010;28:413–444. doi: 10.1146/annurev-immunol-030409-101256. [DOI] [PubMed] [Google Scholar]
- 2.Zolla-Pazner S. Identifying epitopes of HIV-1 that induce protective antibodies. Nat Rev Immunol. 2004;4:199–210. doi: 10.1038/nri1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deeks SG, et al. Neutralizing antibody responses against autologous and heterologous viruses in acute versus chronic human immunodeficiency virus (HIV) infection: Evidence for a constraint on the ability of HIV to completely evade neutralizing antibody responses. J Virol. 2006;80:6155–6164. doi: 10.1128/JVI.00093-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sather DN, et al. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J Virol. 2009;83:757–769. doi: 10.1128/JVI.02036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doria-Rose NA, et al. Frequency and phenotype of human immunodeficiency virus envelope-specific B cells from patients with broadly cross-neutralizing antibodies. J Virol. 2009;83:188–199. doi: 10.1128/JVI.01583-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Binley JM, et al. Profiling the specificity of neutralizing antibodies in a large panel of plasmas from patients chronically infected with human immunodeficiency virus type 1 subtypes B and C. J Virol. 2008;82:11651–11668. doi: 10.1128/JVI.01762-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dhillon AK, et al. Dissecting the neutralizing antibody specificities of broadly neutralizing sera from human immunodeficiency virus type 1-infected donors. J Virol. 2007;81:6548–6562. doi: 10.1128/JVI.02749-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simek MD, et al. Human immunodeficiency virus type 1 elite neutralizers: Individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. J Virol. 2009;83:7337–7348. doi: 10.1128/JVI.00110-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gray ES, et al. Antibody specificities associated with neutralization breadth in plasma from human immunodeficiency virus type 1 subtype C-infected blood donors. J Virol. 2009;83:8925–8937. doi: 10.1128/JVI.00758-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corti D, et al. Analysis of memory B cell responses and isolation of novel monoclonal antibodies with neutralizing breadth from HIV-1-infected individuals. PLoS ONE. 2010;5:e8805. doi: 10.1371/journal.pone.0008805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burton DR, et al. A large array of human monoclonal antibodies to type 1 human immunodeficiency virus from combinatorial libraries of asymptomatic seropositive individuals. Proc Natl Acad Sci USA. 1991;88:10134–10137. doi: 10.1073/pnas.88.22.10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trkola A, et al. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol. 1996;70:1100–1108. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walker LM, et al. Protocol G Principal Investigators Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu X, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonsignori M, et al. Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. J Virol. 2011;85:9998–10009. doi: 10.1128/JVI.05045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scheid JF, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu X, et al. NISC Comparative Sequencing Program Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333:1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walker LM, et al. Protocol G Principal Investigators Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muster T, et al. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J Virol. 1993;67:6642–6647. doi: 10.1128/jvi.67.11.6642-6647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buchacher A, et al. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res Hum Retroviruses. 1994;10:359–369. doi: 10.1089/aid.1994.10.359. [DOI] [PubMed] [Google Scholar]
- 21.Zwick MB, et al. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J Virol. 2001;75:10892–10905. doi: 10.1128/JVI.75.22.10892-10905.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shibata R, et al. Neutralizing antibody directed against the HIV-1 envelope glycoprotein can completely block HIV-1/SIV chimeric virus infections of macaque monkeys. Nat Med. 1999;5:204–210. doi: 10.1038/5568. [DOI] [PubMed] [Google Scholar]
- 23.Mascola JR, et al. Protection of Macaques against pathogenic simian/human immunodeficiency virus 89.6PD by passive transfer of neutralizing antibodies. J Virol. 1999;73:4009–4018. doi: 10.1128/jvi.73.5.4009-4018.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trkola A, et al. Delay of HIV-1 rebound after cessation of antiretroviral therapy through passive transfer of human neutralizing antibodies. Nat Med. 2005;11:615–622. doi: 10.1038/nm1244. [DOI] [PubMed] [Google Scholar]
- 25.Hessell AJ, et al. Effective, low-titer antibody protection against low-dose repeated mucosal SHIV challenge in macaques. Nat Med. 2009;15:951–954. doi: 10.1038/nm.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parren PW, et al. Antibody protects macaques against vaginal challenge with a pathogenic R5 simian/human immunodeficiency virus at serum levels giving complete neutralization in vitro. J Virol. 2001;75:8340–8347. doi: 10.1128/JVI.75.17.8340-8347.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mascola JR, et al. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat Med. 2000;6:207–210. doi: 10.1038/72318. [DOI] [PubMed] [Google Scholar]
- 28.Hessell AJ, et al. Broadly neutralizing human anti-HIV antibody 2G12 is effective in protection against mucosal SHIV challenge even at low serum neutralizing titers. PLoS Pathog. 2009;5:e1000433. doi: 10.1371/journal.ppat.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hessell AJ, et al. Broadly neutralizing monoclonal antibodies 2F5 and 4E10 directed against the human immunodeficiency virus type 1 gp41 membrane-proximal external region protect against mucosal challenge by simian-human immunodeficiency virus SHIVBa-L. J Virol. 2010;84:1302–1313. doi: 10.1128/JVI.01272-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Veazey RS, et al. Prevention of virus transmission to macaque monkeys by a vaginally applied monoclonal antibody to HIV-1 gp120. Nat Med. 2003;9:343–346. doi: 10.1038/nm833. [DOI] [PubMed] [Google Scholar]
- 31.Ng CT, et al. Passive neutralizing antibody controls SHIV viremia and enhances B cell responses in infant macaques. Nat Med. 2010;16:1117–1119. doi: 10.1038/nm.2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mascola JR. HIV/AIDS: Allied responses. Nature. 2007;449:29–30. doi: 10.1038/449029a. [DOI] [PubMed] [Google Scholar]
- 33.Karlsson Hedestam GB, et al. The challenges of eliciting neutralizing antibodies to HIV-1 and to influenza virus. Nat Rev Microbiol. 2008;6:143–155. doi: 10.1038/nrmicro1819. [DOI] [PubMed] [Google Scholar]
- 34.Montefiori DC, Mascola JR. Neutralizing antibodies against HIV-1: Can we elicit them with vaccines and how much do we need? Curr Opin HIV AIDS. 2009;4:347–351. doi: 10.1097/COH.0b013e32832f4a4d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richman DD, Wrin T, Little SJ, Petropoulos CJ. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc Natl Acad Sci USA. 2003;100:4144–4149. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei X, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 37.Albert J, et al. Rapid development of isolate-specific neutralizing antibodies after primary HIV-1 infection and consequent emergence of virus variants which resist neutralization by autologous sera. AIDS. 1990;4:107–112. doi: 10.1097/00002030-199002000-00002. [DOI] [PubMed] [Google Scholar]
- 38.Arendrup M, et al. Autologous HIV-1 neutralizing antibodies: Emergence of neutralization-resistant escape virus and subsequent development of escape virus neutralizing antibodies. J Acquir Immune Defic Syndr. 1992;5:303–307. [PubMed] [Google Scholar]
- 39.Bunnik EM, Pisas L, van Nuenen AC, Schuitemaker H. Autologous neutralizing humoral immunity and evolution of the viral envelope in the course of subtype B human immunodeficiency virus type 1 infection. J Virol. 2008;82:7932–7941. doi: 10.1128/JVI.00757-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moore PL, et al. CAPRISA 002 Study; NIAID Center for HIV/AIDS Vaccine Immunology (CHAVI) Limited neutralizing antibody specificities drive neutralization escape in early HIV-1 subtype C infection. PLoS Pathog. 2009;5:e1000598. doi: 10.1371/journal.ppat.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rong R, et al. Escape from autologous neutralizing antibodies in acute/early subtype C HIV-1 infection requires multiple pathways. PLoS Pathog. 2009;5:e1000594. doi: 10.1371/journal.ppat.1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Binley JM, et al. Role of complex carbohydrates in human immunodeficiency virus type 1 infection and resistance to antibody neutralization. J Virol. 2010;84:5637–5655. doi: 10.1128/JVI.00105-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kwong PD, et al. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature. 2002;420:678–682. doi: 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- 44.Labrijn AF, et al. Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gp120 is sterically restricted on primary human immunodeficiency virus type 1. J Virol. 2003;77:10557–10565. doi: 10.1128/JVI.77.19.10557-10565.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schief WR, Ban YE, Stamatatos L. Challenges for structure-based HIV vaccine design. Curr Opin HIV AIDS. 2009;4:431–440. doi: 10.1097/COH.0b013e32832e6184. [DOI] [PubMed] [Google Scholar]
- 46.Frey G, et al. A fusion-intermediate state of HIV-1 gp41 targeted by broadly neutralizing antibodies. Proc Natl Acad Sci USA. 2008;105:3739–3744. doi: 10.1073/pnas.0800255105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moore PL, et al. Nature of nonfunctional envelope proteins on the surface of human immunodeficiency virus type 1. J Virol. 2006;80:2515-25–28. doi: 10.1128/JVI.80.5.2515-2528.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poignard P, et al. Heterogeneity of envelope molecules expressed on primary human immunodeficiency virus type 1 particles as probed by the binding of neutralizing and nonneutralizing antibodies. J Virol. 2003;77:353–365. doi: 10.1128/JVI.77.1.353-365.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu P, et al. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature. 2006;441:847–852. doi: 10.1038/nature04817. [DOI] [PubMed] [Google Scholar]
- 50.Klein JS, Bjorkman PJ. Few and far between: how HIV may be evading antibody avidity. PLoS Pathog. 2010;6:e1000908. doi: 10.1371/journal.ppat.1000908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Klein JS, et al. Examination of the contributions of size and avidity to the neutralization mechanisms of the anti-HIV antibodies b12 and 4E10. Proc Natl Acad Sci USA. 2009;106:7385–7390. doi: 10.1073/pnas.0811427106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barbas CF, 3rd, et al. In vitro evolution of a neutralizing human antibody to human immunodeficiency virus type 1 to enhance affinity and broaden strain cross-reactivity. Proc Natl Acad Sci USA. 1994;91:3809–3813. doi: 10.1073/pnas.91.9.3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roost HP, et al. Early high-affinity neutralizing anti-viral IgG responses without further overall improvements of affinity. Proc Natl Acad Sci USA. 1995;92:1257–1261. doi: 10.1073/pnas.92.5.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu H, et al. Ultra-potent antibodies against respiratory syncytial virus: Effects of binding kinetics and binding valence on viral neutralization. J Mol Biol. 2005;350:126–144. doi: 10.1016/j.jmb.2005.04.049. [DOI] [PubMed] [Google Scholar]
- 55.Edwards MJ, Dimmock NJ. Hemagglutinin 1-specific immunoglobulin G and Fab molecules mediate postattachment neutralization of influenza A virus by inhibition of an early fusion event. J Virol. 2001;75:10208–10218. doi: 10.1128/JVI.75.21.10208-10218.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Icenogle J, et al. Neutralization of poliovirus by a monoclonal antibody: Kinetics and stoichiometry. Virology. 1983;127:412–425. doi: 10.1016/0042-6822(83)90154-x. [DOI] [PubMed] [Google Scholar]
- 57.Schofield DJ, Stephenson JR, Dimmock NJ. Variations in the neutralizing and haemagglutination-inhibiting activities of five influenza A virus-specific IgGs and their antibody fragments. J Gen Virol. 1997;78:2431–2439. doi: 10.1099/0022-1317-78-10-2431. [DOI] [PubMed] [Google Scholar]
- 58.Smith TJ, Olson NH, Cheng RH, Chase ES, Baker TS. Structure of a human rhinovirus-bivalently bound antibody complex: Implications for viral neutralization and antibody flexibility. Proc Natl Acad Sci USA. 1993;90:7015–7018. doi: 10.1073/pnas.90.15.7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mouquet H, et al. Memory B cell antibodies to HIV-1 gp140 cloned from individuals infected with clade A and B viruses. PLoS ONE. 2011;6:e24078. doi: 10.1371/journal.pone.0024078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Scheid JF, et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature. 2009;458:636–640. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- 61.Wardemann H, et al. Predominant autoantibody production by early human B cell precursors. Science. 2003;301:1374–1377. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- 62.Ridgway JB, Presta LG, Carter P. ‘Knobs-into-holes’ engineering of antibody CH3 domains for heavy chain heterodimerization. Protein Eng. 1996;9:617–621. doi: 10.1093/protein/9.7.617. [DOI] [PubMed] [Google Scholar]
- 63.Pietzsch J, et al. Anti-gp41 antibodies cloned from HIV-infected patients with broadly neutralizing serologic activity. J Virol. 2010;84:5032–5042. doi: 10.1128/JVI.00154-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li Y, et al. Broad HIV-1 neutralization mediated by CD4-binding site antibodies. Nat Med. 2007;13:1032–1034. doi: 10.1038/nm1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thali M, et al. Characterization of a discontinuous human immunodeficiency virus type 1 gp120 epitope recognized by a broadly reactive neutralizing human monoclonal antibody. J Virol. 1991;65:6188–6193. doi: 10.1128/jvi.65.11.6188-6193.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wolbank S, Kunert R, Stiegler G, Katinger H. Characterization of human class-switched polymeric (immunoglobulin M [IgM] and IgA) anti-human immunodeficiency virus type 1 antibodies 2F5 and 2G12. J Virol. 2003;77:4095–4103. doi: 10.1128/JVI.77.7.4095-4103.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.West AP, Jr., et al. Design and expression of a dimeric form of human immunodeficiency virus type 1 antibody 2G12 with increased neutralization potency. J Virol. 2009;83:98–104. doi: 10.1128/JVI.01564-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.West AP, Jr., et al. Evaluation of CD4-CD4i antibody architectures yields potent, broadly cross-reactive anti-human immunodeficiency virus reagents. J Virol. 2010;84:261–269. doi: 10.1128/JVI.01528-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dey B, Del Castillo CS, Berger EA. Neutralization of human immunodeficiency virus type 1 by sCD4-17b, a single-chain chimeric protein, based on sequential interaction of gp120 with CD4 and coreceptor. J Virol. 2003;77:2859–2865. doi: 10.1128/JVI.77.5.2859-2865.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang P, Yang X. Neutralization efficiency is greatly enhanced by bivalent binding of an antibody to epitopes in the V4 region and the membrane-proximal external region within one trimer of human immunodeficiency virus type 1 glycoproteins. J Virol. 2010;84:7114–7123. doi: 10.1128/JVI.00545-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Alam SM, et al. The role of antibody polyspecificity and lipid reactivity in binding of broadly neutralizing anti-HIV-1 envelope human monoclonal antibodies 2F5 and 4E10 to glycoprotein 41 membrane proximal envelope epitopes. J Immunol. 2007;178:4424–4435. doi: 10.4049/jimmunol.178.7.4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Alam SM, et al. Role of HIV membrane in neutralization by two broadly neutralizing antibodies. Proc Natl Acad Sci USA. 2009;106:20234–20239. doi: 10.1073/pnas.0908713106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Veiga AS, Pattenden LK, Fletcher JM, Castanho MA, Aguilar MI. Interactions of HIV-1 antibodies 2F5 and 4E10 with a gp41 epitope prebound to host and viral membrane model systems. ChemBioChem. 2009;10:1032–1044. doi: 10.1002/cbic.200800609. [DOI] [PubMed] [Google Scholar]
- 74.Scherer EM, Leaman DP, Zwick MB, McMichael AJ, Burton DR. Aromatic residues at the edge of the antibody combining site facilitate viral glycoprotein recognition through membrane interactions. Proc Natl Acad Sci USA. 2010;107:1529–1534. doi: 10.1073/pnas.0909680107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Julien JP, et al. Ablation of the complementarity-determining region H3 apex of the anti-HIV-1 broadly neutralizing antibody 2F5 abrogates neutralizing capacity without affecting core epitope binding. J Virol. 2010;84:4136–4147. doi: 10.1128/JVI.02357-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ofek G, et al. Relationship between antibody 2F5 neutralization of HIV-1 and hydrophobicity of its heavy chain third complementarity-determining region. J Virol. 2010;84:2955–2962. doi: 10.1128/JVI.02257-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Diskin R, Marcovecchio PM, Bjorkman PJ. Structure of a clade C HIV-1 gp120 bound to CD4 and CD4-induced antibody reveals anti-CD4 polyreactivity. Nat Struct Mol Biol. 2010;17:608–613. doi: 10.1038/nsmb.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mouquet H, et al. Polyreactivity increases the apparent affinity of anti-HIV antibodies by heteroligation. Nature. 2010;467:591–595. doi: 10.1038/nature09385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tiller T, et al. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J Immunol Methods. 2008;329:112–124. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li M, et al. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol. 2005;79:10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.