Abstract
RAS mutations are common in myeloid malignancies; however, it is not known whether oncogenic RAS can initiate leukemia. We show that expressing mutant K-RasG12D protein from the endogenous murine locus rapidly induces a fatal myeloproliferative disorder with 100% penetrance characterized by tissue infiltration, hypersensitivity to growth factors, and hyperproliferation. Hematopoietic cells from diseased mice demonstrated increased levels of Ras-GTP, but effector kinases were not constitutively phosphorylated and responded normally to growth factors. Oncogenic RAS is sufficient to initiate myeloid leukemogenesis in mice, and this provides an in vivo system for biologic and preclinical studies.
Mutations in KRAS2, NRAS, and HRAS are the most common oncogenic alterations found in human cancer (1). RAS genes encode a family of 21-kDa proteins that regulate cell fates by cycling between inactive GDP-bound (Ras-GDP) and active GTP-bound (Ras-GTP) conformations (2, 3). Ras-GTP interacts with downstream effectors to elicit diverse cellular responses such as proliferation, differentiation, and survival (2–4). Cancer-associated RAS alleles encode proteins that accumulate in the GTP-bound conformation as a result of defects in both intrinsic GTPase activity and resistance to GTPase-activating proteins, which normally accelerate GTP hydrolysis on Ras. Oncogenic RAS cooperates with other oncogenes and tumor suppressors to transform cultured cells, which demonstrate elevated Ras-GTP levels and aberrant activation of downstream kinase effector cascades.
Overexpressing mutant Ras in wild-type mouse embryonic fibroblasts (MEFs) causes a growth arrest that is reminiscent of replicative senescence, which can be overcome by mutating Ink4a or Tp53 (5). These data suggest a requirement for cooperating mutations to circumvent Ras-induced senescence and imply that RAS mutations might occur late in tumorigenesis, when prior mutations have created a permissive environment. This notion has been challenged by studies in strains of mice engineered to express oncogenic Ras proteins from the endogenous locus. Johnson et al. (6) constructed a latent mutant Kras allele that was activated in vivo by spontaneous homologous recombination. These animals uniformly exhibited aggressive lung tumors, and some also developed lymphomas. This model was refined by inserting a transcriptional repression domain flanked by loxP sites (LSL cassette) into the mutant Kras locus (7). Activating this allele by tracheal instillation of an adenovirus encoding Cre recombinase efficiently caused lung cancer. Most recently, a cytomegalovirus (CMV)-Cre transgene was used to induce widespread tissue expression of a different conditional mutant Kras allele (8). MEFs from these KrasV12 mice failed to undergo replicative senescence; however, tumorigenesis was largely restricted to the lung. The authors hypothesized that the oncogenic potential of Kras is highly tissue-specific and that many somatic tissues tolerate mutant K-Ras protein expression without adverse phenotypic consequences (8). Although RAS mutations are found in advanced tumors from a diverse range of tissues, whether oncogenic RAS can initiate tumorigenesis outside the lung is unknown.
NRAS and KRAS2 mutations occur in ≈30% of acute myeloid leukemias (AML), myeloproliferative disorders (MPD), and myelodysplastic syndromes (MDS) (1). Expansion of myeloid blasts with suppression of normal hematopoiesis is the hallmark of AML, whereas MPD is associated with overproliferation of one or more lineages that retain the capacity to differentiate, and MDS is characterized by low blood cell counts and aberrant bone marrow morphology. Many leukemia-associated genetic lesions such as the PML-RARA and AML1-ETO fusions are associated with discrete disease phenotypes; by contrast, oncogenic point mutations in the NRAS and KRAS2 genes are found in AML, MPD, and MDS. The existence of RAS mutations in diverse myeloid malignancies raises the possibility that they invariably represent secondary events that cooperate with a spectrum of initiating genetic lesions. Consistent with this possibility, some AMLs contain subclones with independent RAS mutations, and RAS mutations that are detected at diagnosis may disappear over time in patients with persistent or relapsed disease (9–11). The failure of CMV-Cre, KrasV12 mice to develop myeloid leukemia (8) further suggests that Ras mutations do not initiate leukemogenesis.
Attempts to produce a tractable mouse model of myeloid leukemia initiated by oncogenic RAS have yielded inconsistent results, with disease arising inefficiently (12) or exclusively in the lymphoid lineage (13, 14). MacKenzie et al. (12) found that ≈60% of irradiated recipient mice injected with bone marrow that had been transduced with a retrovirus encoding an oncogenic Nras allele developed a spectrum of myeloid malignancies. However, the phenotypic variability, incomplete penetrance, and prolonged latency combined with both impaired in vitro proliferation and a high rate of apoptosis in Nras-infected cells suggested that cooperating mutations were required (12). The difficulties encountered in engineering a penetrant murine model might indicate that oncogenic RAS does not confer a durable proliferative advantage in leukemia-initiating cells or could reflect either absence of Ras expression in a relevant target cell population or failure to modulate transgene expression during hematopoietic differentiation.
To address this issue directly, we harnessed the IFN-inducible Mx1-Cre transgene to direct Cre recombinase expression to hematopoeitic cells. Here we show that activating the KrasG12D allele engineered by Jackson et al. (7) initiates an aggressive MPD with 100% penetrance that is associated with hyperproliferation, aberrant responses to growth factors, and rapid demise.
Materials and Methods
Mouse Strains and Polyinosinic–Polycytidilic Acid (pIpC) Treatment. The procedures were approved by the University of California, San Francisco, Committee on Animal Research. LSL-KrasG12D and Mx1-Cre (7, 15) pups were injected i.p. with 250 μg of pIpC at 21 days of age (Sigma). LSL-KrasG12D genotyping was performed as described (7). Primer sequences and amplification conditions for genotyping Mx1-Cre mice are available on request.
Pathologic Examination. Blood smears were stained with Wright–Giemsa (Sigma). Organs were collected in 10% formalin and processed by the Mouse Pathology Shared Resource at the University of California, San Francisco, Comprehensive Cancer Center. Tail vein blood (≈50 μl) was obtained in Microvet EDTA tubes (Becton Dickinson) for complete blood counts using a Hemavet 850FS (DREW Scientific). Criteria for killing mice included disheveled appearance, hunching, abnormal gait, and pallor.
Flow Cytometry. Staining with FITC- or phycoerythrinconjugated antibodies and flow cytometry were performed according to the manufacturer's instructions (Pharmingen). Data were collected by using a FACScan flow cytometer and cellquest software (Becton Dickinson), and analyzed with flowjo (TreeStar, Ashland, OR).
Colony Assays. Nucleated bone marrow cells (1 × 105) or splenocytes (2 × 105) were suspended in methylcellulose medium (M3231, Stem Cell Technologies) containing recombinant murine granulocyte/macrophage colony-stimulating factor (GM-CSF) or IL-3 (PeproTech, Boston) and plated in 1-ml duplicate cultures. The colonies were counted on day 8. Cellular content was evaluated by harvesting colonies into PBS followed by counting, cytospin preparations, and Wright–Giemsa staining. For self-renewal assays, quadruplicate cultures were established in 1 ml of methylcellulose medium supplemented with saturating doses of IL-3, IL-6, stem cell factor, and erythropoietin (M3434, Stem Cell Technologies) (16). The colonies were counted every 7 days, harvested, and replated in fresh medium.
Bone Marrow Mast Cell Cultures. Bone marrow mononuclear cells (5 × 106) were seeded in 10 ml of RPMI medium 1640 (Gibco) with 10% FCS (HyClone), 1 mM glutamine, 50 μM 2-mercaptoethanol, penicillin, streptomycin, and 2.5 ng/ml murine IL-3 (PeproTech). Nonadherent cells were replated in fresh medium at a density of 3–5 × 105 cells per ml every 2–4 days.
Spectral Karyotype Analysis. Short-term (24-h) cultures of Mx1-Cre KrasG12D splenocytes, metaphase cell preparation, and spectral karyotyping were performed as described (17). At least 10 metaphase cells were analyzed per case.
Adoptive Transfer. Recipients received a single fraction of 900 cGy. Sublethal irradiation was administered at a dose of 400 cGy. Eight-week-old recipient mice were injected with donor marrow mononuclear cells immediately after irradiation via the dorsal tail vein. Recipients received prophylactic antibiotics for 2 weeks.
Proliferation Assay. Mice received 150 mg/kg BrdUrd (Sigma) i.p. and, 6 h later, marrow cells and splenocytes were collected and resuspended in Iscove's modified Dulbecco's medium containing 20% FCS. Red cells were lysed, and remaining cells were washed with PBS. Cells were fixed overnight in 1% paraformaldehyde, 0.01% Tween 20 at 4°C, washed twice, and resuspended in DNase (Sigma) for 30 min at 37°C. After two washes, cells were stained with anti-BrdUrd–FITC (Pharmingen) for 20 min. Cells were washed twice, then analyzed by flow cytometry.
Biochemical Assays. Bone marrow mononuclear cells were harvested in Iscove's modified Dulbecco's medium containing 0.1% FCS, incubated at room temperature for 4 h, then stimulated with GM-CSF for 2–30 min. Cells were then washed with ice-cold PBS containing 1 mM sodium orthovanadate and lysed as described (18). Ras-GTP levels were measured as described elsewhere (18). Western blotting protocols were used to detect mitogen-activated protein (MAP)/extracellular signal-related kinase (ERK) kinase (MEK) (Cell Signaling 9122, 1:1,000), phospho-MEK (Cell Signaling 9121, 1:1,000), Akt (Cell Signaling 9272, 1:1,000) and phospho-Akt (1:5000; gift of D. Stokoe, University of California, San Francisco) by using ECL (Amersham Pharmacia).
Results
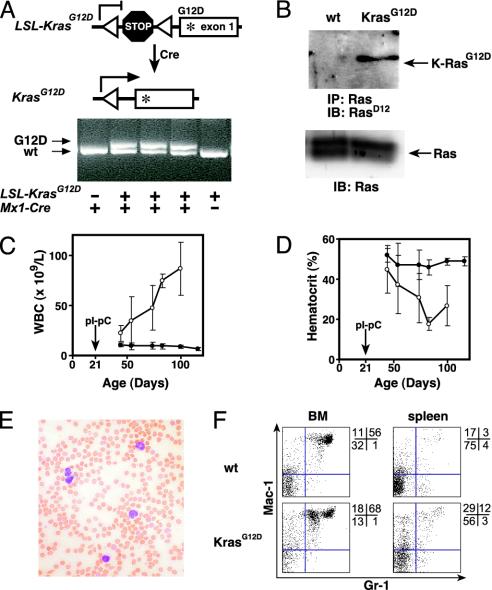
LSL-KrasG12D and Mx1-Cre mice were mated to generate compound heterozygous animals on a uniform F1 129Sv/Jae × C57BL/6 background. The Mx1 promoter can be activated by administering pIpC to stimulate endogenous IFN production (15). Mx1-Cre LSL-KrasG12D pups were treated with pIpC to induce Cre recombinase expression, and leukocyte DNA was analyzed 5 weeks later. PCR analysis using allele-specific primers showed that Mx1-Cre LSL-KrasG12D mice injected with pIpC uniformly demonstrated the recombined KrasG12D allele, which was absent in LSL-KrasG12D animals lacking the Mx1-Cre transgene (Fig. 1A). We refer to animals that were treated with pIpC to excise the LSL element as Mx1-Cre KrasG12D mice. Immunoblotting with an antibody specific for the aspartate substitution at codon 12 demonstrated expression of the mutant K-Ras protein in Mx1-Cre KrasG12D splenocytes (Fig. 1B).
Fig. 1.
K-RasG12D expression induces MPD in Mx1-Cre KrasG12D mice. (A) Excision of the stop cassette by Cre recombinase shown in schematic (Upper) and by PCR (Lower) using primers flanking the loxP sites (triangles). Under these conditions, the wild type (KrasWT) and mutant (KrasG12D) give the indicated products, and the unrearranged allele does not amplify. PCR was performed on DNA purified from peripheral blood from mice with the indicated genotypes 3 weeks after injection with pIpC. (B) Expression of K-RasG12D protein in Mx1-Cre KrasG12D splenocytes but not those from a littermate control after treatment with pIpC. Immunoprecipitation of total Ras was followed by immunoblotting with an antibody specific for the K-RasG12D protein (Upper). Immunoblot of total cell lysates (Lower) demonstrates equal loading. (C and D) Progressive leukocytosis (C) and anemia (D) in a cohort of Mx1-Cre KrasG12D mice (n = 11; open circles) and littermate controls (n = 19; filled circles) that had serial blood counts performed after pIpC injection at weaning (±SEM). (E) Blood smear showing leukocytosis with mature myeloid cells in mice with MPD. (F) Flow cytometry of bone marrow and spleen cells from wild-type and Mx1-Cre KrasG12D mice, 7 weeks after pIpC injection. Proportions of cells within each quadrant are shown to the right of each plot.
Mx1-Cre KrasG12D mice developed progressive leukocytosis that was already evident in blood specimens collected 3 weeks after plpC injection, which was associated with anemia and normal platelet counts (Fig. 1 C and D, and data not shown). Blood smears showed myeloid cells at various stages of differentiation (Fig. 1E). The predominant population had the morphology of mature monocytes, with some nuclear pleomorphism and cytoplasmic vacuoles. Extensive immunophenotyping of blood leukocytes revealed a major population that was CD45+, Mac-1+, CD16/32+, CD86+, CD31+, and negative for CD59, CD34, c-kit, Sca-1, Ter119, CD71, CD41, and a panel of B and T cell markers. These findings are consistent with monocytic maturation, although CD59 is usually expressed by murine monocytes. Histopathologic examination of bone marrow from Mx1-Cre KrasG12D mice revealed abundant myeloid cells at various stages of differentiation, but a paucity of erythroid precursors. Flow cytometry demonstrated an expanded population of Mac-1+, Gr-1lo cells; this phenotype is consistent with immature monocytic cells (Fig. 1F).
We injected 2 × 106 bone marrow cells from diseased Mx1-Cre KrasG12D mice into lethally and sublethally irradiated syngeneic hosts. Four of eight lethally irradiated recipients died 4–5 weeks later without overt signs of MPD. The remaining mice succumbed with MPD 5–7 weeks after adoptive transfer. All eight sublethally irradiated recipients survived for 12 weeks after transplantation with normal blood counts and absence of the recombined KrasG12D allele in blood leukocytes. Interestingly, four of these animals died over the next 6 weeks, including two that acquired the recombined allele in leukocytes. Whereas these data indicate that the MPD that arises in Mx1-Cre KrasG12D mice can be transplanted into lethally irradiated recipients, it is uncertain whether it can be transferred into sublethally irradiated mice.
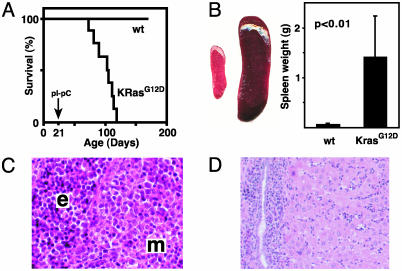
Mx1-Cre KrasG12D mice survived for a median of 84 days after pIpC injection (Fig. 2A). The spleens of moribund mice were massively enlarged and infiltrated with myeloid and erythroid cells (Fig. 2 B and C). Consistent with this observation, the Mac-1+ and Ter119+ populations were markedly expanded, and the Mac-1+, Gr-1lo population that was prominent in the marrow was also readily detected (Fig. 1F). A significant fraction of Mx1-Cre KrasG12D splenocytes expressed c-kit (data not shown), which is found on immature hematopoietic cells and mast cells. Spectral karyotype analysis of Mx1-Cre KrasG12D splenocytes was normal in all cases (n = 7). Mice with MPD also showed moderate hepatomegaly with myeloid infiltration, particularly in periportal areas (Fig. 2D). Under guidelines published recently (19), this disease is classified as MPD with excess monocytes. Importantly, Mx1-Cre KrasG12D mice do not develop AML; we did not observe abnormal expansion of immature blast forms in blood or marrow.
Fig. 2.
Survival and pathologic features of MPD in Mx1-Cre KrasG12D mice. (A) Kaplan–Meier survival curve in a cohort of Mx1-Cre KrasG12D (KrasG12D; n = 11) and littermate control wild-type (wt; n = 19) mice. Mx1-Cre KrasG12D mice have a median lifespan of 105 days (84 days after pIpC treatment), and all of the littermates were alive after 1 year of observation. The 100% penetrance of MPD was confirmed in 20 additional Mx1-Cre KrasG12D mice. (B) Massive splenomegaly, with a typical example of wild-type (Left) and Mx1-Cre KrasG12D (Right) spleens, and a graphical comparison of means and standard deviations (n = 19 Mx1-Cre KrasG12D vs. 17 control specimens) of spleen weights. (C and D) Hematoxylin and eosin-stained sections of Mx1-Cre KrasG12D spleen (C) and liver (D), showing extensive myeloid infiltration (m) and splenic erythroid hyperplasia (e).
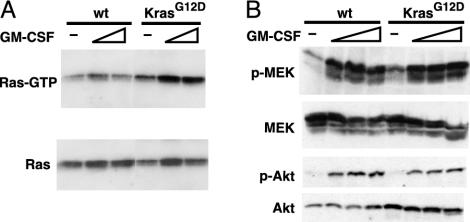
We measured the percentage of Ras-GTP and assayed activation of the Ras effectors Akt and MEK in bone marrow mononuclear cells isolated from Mx1-Cre KrasG12D and control mice. As expected, the mutant cells demonstrated elevated levels of Ras-GTP (Fig. 3A); however, this was not associated with constitutive phosphorylation of Akt or MEK. Mutant and control cells showed marked increases in Ras-GTP levels that were associated with equivalent activation of effector kinases in response to growth factor stimulation (Fig. 3B). We confirmed these surprising results under multiple experimental conditions in which growth factor concentration and time after stimulation varied widely (data not shown).
Fig. 3.
Ras activation and signaling in Mx1-Cre KrasG12D bone marrow. (A) Bone marrow cells from wild-type or Mx1-Cre KrasG12D mice were starved for 4 h in Iscove's modified Dulbecco's medium with 0.1% FCS, then stimulated for 3minwith1or3ng/ml GM-CSF. Ras-GTP was affinity-purified with Raf-RBD-agarose and detected by immunoblotting for total Ras (Upper). Equal input was demonstrated by immunoblotting (Lower). (B) Bone marrow was harvested and starved as above, then stimulated with GM-CSF at concentrations of 0, 1, 3, and 10 ng/ml for 5 min and analyzed for the indicated proteins by immunoblotting.
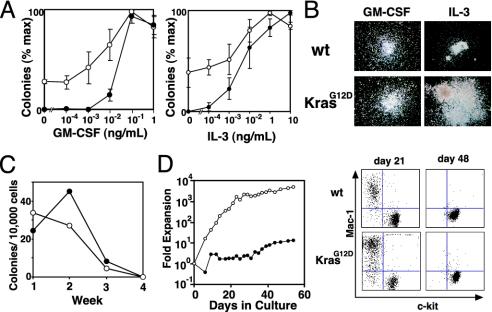
The growth of KrasG12D progenitors was assayed in methylcellulose medium. Bone marrow mononuclear cells from mice with MPD, but not controls, formed significant numbers of colony-forming unit granulocyte-macrophage (CFU-GM) colonies in the absence of exogenous cytokines. Low concentrations of GM-CSF that did not induce colony formation from wild-type cells dramatically increased the size and number of Mx1-Cre KrasG12D colonies (Fig. 4A). CFU-GM colonies grown from Mx1-Cre KrasG12D bone marrow and spleens in the presence of saturating concentrations of GM-CSF were very large and displayed an abnormal, spreading morphology (Fig. 4B). Mx1-Cre KrasG12D CFU-GM also showed a dramatic proliferative response to IL-3. We also counted the total number of cells present in methylcellulose cultures containing saturating levels of GM-CSF or IL-3. In wild-type cultures, the total number of nucleated cells increased 4-fold in response to GM-CSF and 20-fold with IL-3. Mx1-Cre KrasG12D cultures showed greatly enhanced proliferation, with a 9-fold increase in cell numbers without added cytokines, a 19-fold increase with GM-CSF, and a 37-fold increase with IL-3.
Fig. 4.
In vitro growth of Mx1-Cre KrasG12D (open circles) and control hematopoietic cells (filled circles). (A) Methylcellulose colony assays using GM-CSF or IL-3 at the indicated concentrations. Experiments were performed by using three mice of each genotype (±SEM). (B) Photomicrographs (original magnification ×40) of wild-type (wt) and mutant (KrasG12D) myeloid progenitor colonies grown in saturating concentrations of growth factors. (C) Colony formation with weekly serial replating of 10,000 unsorted bone marrow cells in methylcellulose. (D) Growth kinetics and flow cytometry of bone marrow cells cultured in the presence of IL-3.
To compare the self-renewal of Mx1-Cre KrasG12D and wild-type progenitors, we seeded 1 × 104 marrow cells in methylcellulose medium containing saturating concentrations of growth factors and counted colonies after 7 days. The cells were then collected and washed, and 1 × 104 cells were transferred into new methylcellulose each week. Mx1-Cre KrasG12D bone marrow cells showed normal replating efficiency in this assay (Fig. 4C). Bone marrow mast cell cultures established from Mx1-Cre KrasG12D mice demonstrated enormous proliferation of Mac1+ cells for the first few weeks, but reverted to wild-type growth rates over time with the expected switch to cells that were Mac1– and c-kit+ (Fig. 4D). Cytospin preparations during this proliferative stage confirmed a predominance of mature myeloid cells with some immature forms.
Diseased animals were injected with BrdUrd to measure proliferation in vivo. Mx1-Cre KrasG12D bone marrows contained a similar percentage of BrdUrd-positive cells as wild-type marrow, with but a marked increase within the Mac1+ fraction (Table 1). The percentage of Mx1-Cre KrasG12D splenocytes that incorporated BrdUrd was also elevated. Most proliferating cells in Mx1-Cre KrasG12D spleens were in the Mac1+/Gr1lo fraction or expressed the erythroid marker Ter119 (Table 1 and data not shown). In contrast, the few cycling cells identified in normal spleens were mostly lymphocytes (Table 1 and data not shown). These data show that the spleen contains a large number of proliferating cells in Mx1-Cre KrasG12D spleens and suggest that these populations contribute to the MPD.
Table 1. Percentages of nucleated spleen and bone marrow cells staining with Mac-1, TER119, or BrdUrd.
| % positive cells
|
||||||
|---|---|---|---|---|---|---|
| Source | Genotype | BrdUrd | Mac-1 | TER119 | BrdUrd and Mac-1 | BrdUrd and TER119 |
| Bone marrow | Kraswt | 18 | 45 | 10 | 5 | 5 |
| Bone marrow | KrasG12D | 17 | 65 | 5 | 10 | 2 |
| Spleen | Kraswt | 6 | 16 | 4 | 2 | 1 |
| Spleen | KrasG12D | 26 | 40 | 12 | 7 | 8 |
Discussion
We find that somatic activation of a latent KrasG12D allele rapidly induces a fatal MPD with complete penetrance. By contrast, mice in which oncogenic Kras was activated by spontaneous recombination (6) or by a CMV–Cre transgene (8) did not develop myeloid malignancies after months of observation. It is possible that the latent Kras allele was not activated in a susceptible leukemia-initiating cell in either strain. The Mx1-Cre transgene directs efficient recombination in bone marrow (15) and in hematopoietic cells with high repopulating potential (16). Furthermore, the KrasV12 allele designed by Guerra et al. (8) was modified with a 3′ internal ribosome entry site (IRES)–βgeo cassette, which may have altered the expression of Kras transcripts. Consistent with this idea, we find that activating KrasG12D expression in pancreatic and colonic epithelia induces proliferation and initiates tumor formation in vivo (D.A.T. and T.E.J., unpublished data). Thus, the proposal that oncogenic Ras can only initiate tumorigenesis in a narrow range of cell types (8) must be reevaluated.
Ras-GTP levels are consistently elevated in Mx1-Cre KrasG12D bone marrow cells. However, our analysis of MEK and Akt phosphorylation surprisingly uncovered no differences between mutant and control cells, despite profound effects of KrasG12D expression on myeloid proliferation. Similarly, mouse embryonic fibroblasts expressing KrasV12 from the endogenous locus paradoxically demonstrated reduced levels of phosphorylated ERK at steady state, and showed only modest prolongation of ERK and Akt activation after starvation and stimulation (8). Our data show that the biochemical consequences of expressing oncogenic Ras in primary cells are remarkably different from what has been observed in transformed cell lines. Furthermore, although mutant Ras proteins are frequently described as “constitutively active,” neither Ras-GTP nor downstream effector cascades are saturated in KrasG12D marrow, which respond to growth factors with enhanced myeloid progenitor colony formation and by activating MEK, ERK, and Akt. Because hematopoietic cells at various stages of differentiation are present in KrasG12D mice with MPD, aberrant activation of effector cascades within a minor subpopulation would not be detected in lysates of whole bone marrow. For example, the highly proliferative colonyforming unit granulocyte-macrophage fraction comprises ≈0.1% of the nucleated cells present in Mx1-Cre KrasG12D marrow. Assessing the activation status of signaling cascades in rare subsets of primary cells is a challenging problem that will likely require new methodologies such as the flow cytometry-based approach recently described by Perez and Nolan (20).
Primary cells might also respond to chronic Ras activation by modifying signaling networks. Recent studies have uncovered extensive biochemical crosstalk between linear pathways, feedback from downstream components, and effects of previous exposure to a ligand on subsequent responses (21). Bhalla and colleagues (22) found that ERK activation of NIH 3T3 cells by platelet-derived growth factor (PDGF) induced MAP kinase phosphatase (MKP) expression, which extinguished signaling by dephosphorylating ERK. MKP levels remained elevated for some time after initial exposure to PDGF, and this dampened the response to subsequent stimulation. It is possible that hematopoietic cells adapt to chronically elevated Ras-GTP levels by attenuating effector cascades. This idea is consistent with recent data showing that acute and chronic loss of Rb1 function had differential effects on quiescence and senescence phenotypes in mouse embryonic fibroblasts, which were explained, in part, by modulating levels of p19ARF and p107 (23).
Somatic activation of KrasG12D expression results in pronounced proliferation of myeloid lineage cells that are hyperresponsive to hematopoietic growth factors. Nf1 encodes neurofibromin, a GTPase-activating protein for Ras (4, 24). Interestingly, although murine Nf1-deficient myeloid progenitors are hypersensitive to GM-CSF in vitro, this defect is less severe than in Mx1-Cre KrasG12D marrow (25, 26). Furthermore, Nf1 inactivation initiates an MPD that is less aggressive than the corresponding disorder in Mx1-Cre KrasG12D mice (ref. 25 and our unpublished data). Whereas oncogenic RAS mutations both markedly impair the intrinsic Ras GTPase activity and confer resistance to GTPase-activating proteins, inactivation of Nf1 is a less severe biochemical lesion that does not alter intrinsic GTP hydrolysis. The correlation between the predicted biochemical consequences of expressing KrasG12D or ablating Nf1 and phenotypic severity argues in favor of a quantitative relationship between the activation status of Ras and responsiveness to growth factors in myeloid cells. Mx1-Cre KrasG12D progenitors demonstrate normal self-renewal in colony replating assays, which is in contrast to cells that are immortalized by AML1-ETO or MLL fusion proteins (27, 28). However, the effects of KrasG12D on the self-renewal of hematopoietic stem cells can only be definitively addressed in competitive repopulation and serial transplantation experiments.
Ineffective erythropoiesis is prominent in MDS and is observed in some patients with MPD. Similarly, Mx1-Cre KrasG12D mice show large numbers of proliferating splenic Ter119+ cells in the face of a progressive anemia. Although we have not observed morphologic features of dysplasia, the uniform finding of marked splenic erythropoiesis with a increased proportion of immature elements in anemic Mx1-Cre KrasG12D mice suggests that erythropoiesis is ineffective. Interestingly, transduction of human or murine erythroid progenitors with oncogenic RAS results in hyperproliferation (29–31) but with defective differentiation resulting in reduced production of mature erythrocytes. Our in vivo data are consistent with these findings and support the notion that mutant RAS directly impairs erythropoiesis in patients with MPD and MDS. Harnessing the culture system recently described by Zhang et al. (29) to analyze erythroid cells from Mx1-Cre KrasG12D mice will help to elucidate how expressing K-RasG12D from its normal locus perturbs erythroid development.
The MPD that arises in Mx1-Cre KrasG12D mice closely resembles chronic myelomonocytic leukemia (CMML) and juvenile myelomonocytic leukemia (JMML). RAS mutations are common in CMML (32) and genetic analysis of JMML supports the idea that hyperactive Ras can initiate myeloid leukemogenesis. Children with neurofibromatosis type 1 and Noonan syndrome (NS) are predisposed to JMML (33, 34). The NF1 tumor suppressor undergoes homozygous inactivation in JMML bone marrow, which results in hyperactive Ras (26, 35). Germ-line mutations in PTPN11 cause ≈50% of NS. PTPN11 encodes SHP-2, a tyrosine phosphatase that relays signals from activated growth factor receptors to Ras (36, 37). Germ-line and somatic PTPN11 mutations that activate SHP-2 phosphatase activity have recently been detected in JMML (38). Importantly, RAS mutations are also found in ≈25% of JMML samples, and are largely restricted to cases that do not show NF1 or PTPN11 mutations (38, 39). Together, these data support the idea that JMML is initiated by hyperactive Ras, which results from a mutation in RAS, NF1, or PTPN11. Mx1-Cre KrasG12D mice provide a tractable model system for investigating the pathogenesis of CMML and JMML.
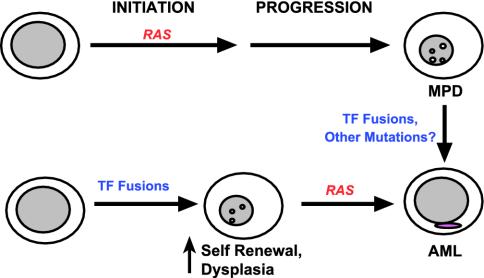
Leukemia-associated mutations that result in hyperactive Ras such as BCR-ABL, mutant FLT3, and loss of NF1 all induce MPD in mice (25, 40, 41). Together with these observations, our data argue that deregulated Ras signaling in a susceptible hematopoietic precursor initiates MPD in vivo (Fig. 5). If this is true, why are RAS mutations also found in patients with AML and MDS? Genetic data from studies of AML specimens support a model in which RAS point mutations and other genetic lesions that deregulate signal transduction pathways cooperate with transcription factor fusion proteins in leukemogenesis (42). In most children who develop AML associated with AML1-ETO, the fusion gene can be detected in neonatal blood specimens, which implies the need for secondary mutations (43). Furthermore, transgenic or knock-in mice engineered to express AML1-ETO, PML-RARA, or MLL-AF9 either do not spontaneously develop AML or show incomplete penetrance and prolonged latency (16, 44, 45). Based on these data, it is likely that AML is initiated by a transcription factor fusion and that oncogenic RAS serves as a cooperating event (Fig. 5). Although less is known about MDS, we envision a similar role of oncogenic RAS as in AML (i.e., a mutation that is acquired during disease evolution). Thus, we propose that oncogenic RAS can either initiate leukemogenesis or occur as a secondary event, and that the disease phenotype will be dictated by both the constellation of mutations and by the order in which they are acquired. This idea is consistent with observations in chronic myeloid leukemia (CML), which is caused by the BCR-ABL fusion. Patients with CML present with MPD but subsequently develop a phenotype of acute leukemia during the “blast crisis” phase of the disease, which may be accompanied by translocations involving transcription factors such as AML1 or HOXA9 (46, 47). We speculate that intercrossing mice that express AML-associated fusion proteins with the Mx1-Cre LSL-Kras strain will generate accurate and highly penetrant models that recapitulate the clinical and biologic features of human AML.
Fig. 5.
Proposed role of RAS mutations in MPD and AML. Our results and other human and murine data support the idea that initiating events that result in hyperactive Ras induce MPD (upper pathway). Transcription factor (TF) fusions are detected in some cases of CML at the time of evolution to blast crisis. Other acquired mutations are likely to exist as well. By contrast, leukemia-associated TF fusions may initiate AML by perturbing cell fates but appear to require a cooperating mutation that drives proliferation to cause overt disease. Here, oncogenic RAS (or activated FLT3) represent secondary mutations that contribute to leukemogenesis (lower pathway). The ability of oncogenic RAS to either initiate disease or cooperate with preexisting genetic lesions is likely to account for the finding of RAS mutations in myeloid malignancies with distinct phenotypes.
The proteins encoded by mutant alleles of FLT3 and CKIT and the BCR-ABL fusion not only result in hyperactive Ras, but interact biochemically with other signaling molecules cascades (48, 49). Despite this, a broad implication of this study is that hyperactive Ras is a pivotal biochemical alteration that promotes the growth of malignant myeloid lineage cells in the context of each of these genetic lesions. This idea is consistent with studies showing that RAS mutations are rare in CML (50), in JMML samples with mutations in either PTPN11 or NF1 (38, 39), and in AML specimens with FLT3 mutations (51). Hyperactive Ras therefore represents a common molecular target in myeloid malignancies. The acquisition of resistance to imatinib (52) suggests that combining this drug with a second agent directed against Ras or one of its effectors is a rational therapeutic strategy for patients with de novo or resistant CML (53). Hematopoietic cells from Mx1-Cre KrasG12D mice provide a tractable system for exploring therapeutic approaches directed against hyperactive Ras in myeloid malignancies that may be applicable to many human cancers.
Acknowledgments
We are indebted to David Stokoe for providing the Akt antibodies and to Leisa Johnson for the antibody against the K-RasG12D protein. We are also grateful to Drs. Gary Gilliland and Iris Chan for helpful discussions. This work was supported in part by National Institutes of Health Grants CA72614, CA84221, and CA84306 and by the Jeffrey and Karen Peterson Family Foundation. B.S.B. was supported by the Frank A. Campini Foundation, National Institutes of Health Training Grant T32 DK07636, and the Leukemia and Lymphoma Society of America. T.E.J. is an Investigator of the Howard Hughes Medical Institute.
Abbreviations: MPD, myeloproliferative disorder; AML, acute myeloid leukemia; MDS, myelodysplastic syndrome; JMML, juvenile myelomonocytic leukemia; pIpC, polyinosinic–polycytidilic acid; GM-CSF, granulocyte/macrophage colony-stimulating factor; MAP, mitogen-activated protein; ERK, extracellular signal-related kinase; MEK, MAP/ERK kinase; CML, chronic myeloid leukemia.
References
- 1.Bos, J. L. (1989) Cancer Res. 49, 4682–4689. [PubMed] [Google Scholar]
- 2.Bourne, H. R., Sanders, D. A. & McCormick, F. (1990) Nature 348, 125–132. [DOI] [PubMed] [Google Scholar]
- 3.Bourne, H. R., Sanders, D. A. & McCormick, F. (1991) Nature 349, 117–127. [DOI] [PubMed] [Google Scholar]
- 4.Boguski, M. & McCormick, F. (1993) Nature 366, 643–653. [DOI] [PubMed] [Google Scholar]
- 5.Serrano, M., Lin, A. W., McCurrach, M. E., Beach, D. & Lowe, S. W. (1997) Cell 88, 593–602. [DOI] [PubMed] [Google Scholar]
- 6.Johnson, L., Mercer, K., Breenbaum, D., Bronson, R., Crowley, D., Tuveson, D. & Jacks, T. (2001) Nature 410, 1111–1116. [DOI] [PubMed] [Google Scholar]
- 7.Jackson, E. L., Willis, N., Mercer, K., Bronson, R. T., Crowley, D., Montoya, R., Jacks, T. & Tuveson, D. A. (2001) Genes Dev. 15, 3243–3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guerra, C., Mijimolle, N., Dhawahir, A., Dubus, P., Barradas, M., Serrano, M., Campuzano, V. & Barbacid, M. (2003) Cancer Cell 4, 111–120. [DOI] [PubMed] [Google Scholar]
- 9.Farr, C. J., Saiki, R. K., Erlich, H. A., McCormick, F. & Marshall, C. J. (1988) Proc. Natl. Acad. Sci. USA 85, 1629–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Padua, R. A., Carter, G., Hughes, D., Gow, J., Farr, C., Oscier, D., McCormick, F. & Jacobs, A. (1988) Leukemia 2, 503–510. [PubMed] [Google Scholar]
- 11.Bashey, A., Gill, R., Levi, S., Farr, C. J., Clutterbuck, R., Millar, J. L., Pragnell, I. B. & Marshall, C. J. (1992) Blood 79, 981–989. [PubMed] [Google Scholar]
- 12.MacKenzie, K. L., Dolnikov, A., Millington, M., Shounan, Y. & Symonds, G. (1999) Blood 93, 2043–2056. [PubMed] [Google Scholar]
- 13.Dunbar, C. E., Crosier, P. S. & Nienhuis, A. W. (1991) Oncogene Res. 6, 39–51. [PubMed] [Google Scholar]
- 14.Hawley, R. G., Lieu, F. H., Fong, A. Z. & Hawley, T. S. (1994) Gene Ther. 1, 136–138. [PubMed] [Google Scholar]
- 15.Kuhn, R., Schwenk, F., Aguet, M. & Rajewsky, K. (1995) Science 269, 1427–1429. [DOI] [PubMed] [Google Scholar]
- 16.Higuchi, M., O'Brien, D., Kumaravelu, P., Lenney, N., Yeoh, E. & Downing, J. R. (2002) Cancer Cell 1, 63–74. [DOI] [PubMed] [Google Scholar]
- 17.Le Beau, M. M., Bitts, S., Davis, E. M. & Kogan, S. C. (2002) Blood 99, 2985–2991. [DOI] [PubMed] [Google Scholar]
- 18.Donovan, S., See, W., Bonifas, J., Stokoe, D. & Shannon, K. M. (2002) Cancer Cell 2, 507–514. [DOI] [PubMed] [Google Scholar]
- 19.Kogan, S. C., Ward, J. M., Anver, M. R., Berman, J. J., Brayton, C., Cardiff, R. D., Carter, J. S., de Coronado, S., Downing, J. R., Fredrickson, T. N., et al. (2002) Blood 100, 238–245. [DOI] [PubMed] [Google Scholar]
- 20.Perez, O. D. & Nolan, G. P. (2002) Nat. Biotechnol. 20, 155–162. [DOI] [PubMed] [Google Scholar]
- 21.Jordan, J. D., Landau, E. M. & Iyengar, R. (2000) Cell 103, 193–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhalla, U. S., Ram, P. T. & Iyengar, R. (2002) Science 297, 1018–1023. [DOI] [PubMed] [Google Scholar]
- 23.Sage, J., Miller, A. L., Perez-Mancera, P. A., Wysocki, J. M. & Jacks, T. (2003) Nature 424, 223–228. [DOI] [PubMed] [Google Scholar]
- 24.Donovan, S., Shannon, K. M. & Bollag, G. (2002) BBA Rev. Cancer. 1602, 23–45. [DOI] [PubMed] [Google Scholar]
- 25.Largaespada, D. A., Brannan, C. I., Jenkins, N. A. & Copeland, N. G. (1996) Nat. Genet. 12, 137–143. [DOI] [PubMed] [Google Scholar]
- 26.Bollag, G., Clapp, D. W., Shih, S., Adler, F., Zhang, Y., Thompson, P., Lange, B. J., Freedman, M. H., McCormick, F., Jacks, T. & Shannon, K. (1996) Nat. Genet. 12, 144–148. [DOI] [PubMed] [Google Scholar]
- 27.Okuda, T., Cai, Z., Yang, S., Lenny, N., Lyu, C. J., van Deursen, J. M., Harada, H. & Downing, J. R. (1998) Blood 91, 3134–3143. [PubMed] [Google Scholar]
- 28.Ayton, P. M. & Cleary, M. L. (2001) Oncogene 20, 5695–5707. [DOI] [PubMed] [Google Scholar]
- 29.Zhang, J., Socolovsky, M., Gross, A. W. & Lodish, H. F. (2003) Blood 102, 3938–3946. [DOI] [PubMed] [Google Scholar]
- 30.Darley, R. L., Hoy, T. G., Baines, P., Padua, R. A. & Burnett, A. K. (1997) J. Exp. Med. 185, 1337–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Darley, R. L., Pearn, L., Omidvar, N., Sweeney, M., Fisher, J., Phillips, S., Hoy, T. & Burnett, A. K. (2002) Blood 100, 4185–4192. [DOI] [PubMed] [Google Scholar]
- 32.Onida, F., Kantarjian, H. M., Smith, T. L., Ball, G., Keating, M. J., Estey, E. H., Glassman, A. B., Albitar, M., Kwari, M. I. & Beran, M. (2002) Blood 99, 840–849. [DOI] [PubMed] [Google Scholar]
- 33.Bader-Meunier, B., Tchernia, G., Miélot, F., Fontaine, J. L., Thomas, C., Lyonnet, S., Lavergne, J. M. & Dommergues, J. P. (1997) J. Pediatr. 130, 885–889. [DOI] [PubMed] [Google Scholar]
- 34.Stiller, C. A., Chessells, J. M. & Fitchett, M. (1994) Br. J. Cancer 70, 969–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Side, L., Taylor, B., Cayouette, M., Connor, E., Thompson, P., Luce, M. & Shannon, K. (1997) N. Engl. J. Med. 336, 1713–1720. [DOI] [PubMed] [Google Scholar]
- 36.Tartaglia, M., Mehler, E. L., Goldberg, R., Zampino, G., Brunner, H. G., Kremer, H., van der Burgt, I., Crosby, A. H., Ion, A., Jeffery, S., et al. (2001) Nat. Genet. 29, 465–468. [DOI] [PubMed] [Google Scholar]
- 37.Neel, B. G., Gu, H. & Pao, L. (2003) Trends Biochem. Sci. 28, 284–293. [DOI] [PubMed] [Google Scholar]
- 38.Tartaglia, M., Niemeyer, C. M., Fragale, A., Song, X., Buechner, J., Jung, A., Hahlen, K., Hasle, H., Licht, J. D. & Gelb, B. D. (2003) Nat. Genet. 34, 148–150. [DOI] [PubMed] [Google Scholar]
- 39.Kalra, R., Paderanga, D., Olson, K. & Shannon, K. M. (1994) Blood 84, 3435–3439. [PubMed] [Google Scholar]
- 40.Van Etten, R. A. (2002) Oncogene 21, 8643–8651. [DOI] [PubMed] [Google Scholar]
- 41.Kelly, L. M., Liu, Q., Kutok, J. L., Williams, I. R., Boulton, C. L. & Gilliland, D. G. (2002) Blood 99, 310–318. [DOI] [PubMed] [Google Scholar]
- 42.Kelly, L., Clark, J. & Gilliland, D. G. (2002) Curr. Opin. Oncol. 14, 10–18. [DOI] [PubMed] [Google Scholar]
- 43.Wiemels, J. L., Xiao, Z., Buffler, P. A., Maia, A. T., Ma, X., Dicks, B. M., Smith, M. T., Zhang, L., Feusner, J., Wiencke, J., et al. (2002) Blood 99, 3801–3805. [DOI] [PubMed] [Google Scholar]
- 44.Corral, J., Lavenir, I., Impey, H., Warren, A. J., Forster, A., Larson, T. A., Bell, S., McKenzie, A. N., King, G. & Rabbitts, T. H. (1996) Cell 85, 853–861. [DOI] [PubMed] [Google Scholar]
- 45.Brown, D., Kogan, S., Lagasse, E., Weissman, I., Alcalay, M., Pelicci, P. G., Atwater, S. & Bishop, J. M. (1997) Proc. Natl. Acad. Sci. USA 94, 2551–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamamoto, K., Nakamura, Y., Saito, K. & Furusawa, S. (2000) Br. J. Haematol. 109, 423–426. [DOI] [PubMed] [Google Scholar]
- 47.Nucifora, G., Begy, C. R., Kobayashi, H., Roulston, D., Claxton, D., Pedersen-Bjergaard, J., Parganas, E., Ihle, J. N. & Rowley, J. D. (1994) Proc. Natl. Acad. Sci. USA 91, 4004–4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gilliland, D. G. & Griffin, J. D. (2002) Blood 100, 1532–1542. [DOI] [PubMed] [Google Scholar]
- 49.Sawyers, C. L. (1999) N. Engl. J. Med. 340, 1330–1340. [DOI] [PubMed] [Google Scholar]
- 50.Collins, S. J., Howard, M., Andrews, D. F., Agura, E. & Radich, J. (1989) Blood 73, 1028–1032. [PubMed] [Google Scholar]
- 51.Thiede, C., Steudel, C., Mohr, B., Schaich, M., Schakel, U., Platzbecker, U., Wermke, M., Bornhauser, M., Ritter, M., Neubauer, A., et al. (2002) Blood 99, 4326–4335. [DOI] [PubMed] [Google Scholar]
- 52.Gorre, M. E., Mohammed, M., Ellwood, K., Hsu, N., Paquette, R., Rao, P. N. & Sawyers, C. L. (2001) Science 293, 876–880. [DOI] [PubMed] [Google Scholar]
- 53.Peters, D. G., Hoover, R. R., Gerlach, M. J., Koh, E. Y., Zhang, H., Choe, K., Kirschmeier, P., Bishop, W. R. & Daley, G. Q. (2001) Blood 97, 1404–1412. [DOI] [PubMed] [Google Scholar]