Abstract
Genome packaging into preformed viral procapsids is driven by powerful molecular motors. The small terminase protein is essential for the initial recognition of viral DNA and regulates the motor’s ATPase and nuclease activities during DNA translocation. The crystal structure of a full-length small terminase protein from the Siphoviridae bacteriophage SF6, comprising the N-terminal DNA binding, the oligomerization core, and the C-terminal β-barrel domains, reveals a nine-subunit circular assembly in which the DNA-binding domains are arranged around the oligomerization core in a highly flexible manner. Mass spectrometry analysis and four further crystal structures show that, although the full-length protein exclusively forms nine-subunit assemblies, protein constructs missing the C-terminal β-barrel form both nine-subunit and ten-subunit assemblies, indicating the importance of the C terminus for defining the oligomeric state. The mechanism by which a ring-shaped small terminase oligomer binds viral DNA has not previously been elucidated. Here, we probed binding in vitro by using EPR and surface plasmon resonance experiments, which indicated that interaction with DNA is mediated exclusively by the DNA-binding domains and suggested a nucleosome-like model in which DNA binds around the outside of the protein oligomer.
Keywords: bacteriophage SPP1, DNA packaging, virus assembly, X-ray crystallography
The virus genome in tailed dsDNA bacteriophages and in the evolutionarily related herpes viruses is packaged into a preformed empty procapsid (1–3). A powerful ATP-fueled molecular machine drives the DNA with a speed of up to 1,800 bp/s through the portal protein embedded in a unique vertex of the icosahedral procapsid (2–4). The molecular motor usually consists of the small and large terminase proteins. The small terminase plays a dual role in virus particle assembly: It (i) recognizes viral DNA during the initiation of packaging and (ii) modulates the ATPase and nuclease activities of the large terminase during DNA translocation (5, 6). After filling a procapsid, the rest of the DNA is then docked to the portal entrance of another procapsid where the process of DNA translocation is repeated (7).
X-ray structures have been determined for portal proteins from bacteriophages φ29 (8), SPP1 (9), and P22 (10) and also for large terminases from bacteriophages T4 (6), RB49 (6), and SPP1 (11). Three-dimensional information on small terminases is limited to the cryo-EM structure of phage P22 small terminase (12), the NMR structure of the DNA-binding domain of phage λ gpNu1 (13), and the crystal structure of phage Sf6 small terminase (14). In the absence of accurate three-dimensional data for all three motor components of one particular phage, mapping of functional information to the structure and modeling molecular interactions between individual components is challenging. We have addressed this issue by extending the structural information on Bacillus subtilis bacteriophages SPP1 and SF6, two very closely related viruses of the Siphoviridae family. Here we present five X-ray structures for several different constructs of the SF6 small terminase. We also present mass spectrometry data on oligomeric states of the small terminase. Structural observations on the full-length protein containing the N-terminal DNA-binding domains, together with DNA-binding data and normal mode analysis calculations, suggest a model for packaging initiation in which DNA-binding domains are adjusted to form a periodical nucleoprotein assembly.
Results
Structure Determination.
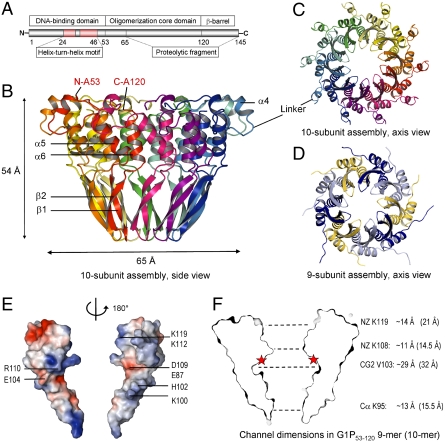
G1P, the small terminase of bacteriophage SF6, comprises 145 residues (Fig. 1A). We first determined the crystal structure of the oligomerization core domain of G1P, residues 53–120, in which the N-terminal DNA-binding and the C-terminal β-barrel domains were truncated (G1P53–120). This structure, determined at 1.85-Å resolution by using selenomethionine-substituted protein (SI Materials and Methods and Table S1), revealed a ten-subunit assembly (Fig. 1 B and C) and was used as a molecular replacement model to determine structures of a nine-subunit assembly (1.68-Å resolution, Fig. 1 D–F) and a second crystal form of the ten-subunit assembly (2.19-Å resolution) formed by the same truncated construct (G1P53–120). A further two structures, both revealing nine-subunit assemblies, were determined for (i) a proteolytic fragment G1P65–141 (3-Å resolution, Fig. 2A) containing the C-terminal β-barrel but missing the N-terminal DNA-binding domains and (ii) the full-length protein (approximately 4-Å resolution, Fig. 2 B and C and Fig. S1). An overview of all constructs used in various experiments is in SI Materials and Methods.
Fig. 1.
Structure of the oligomerization core domain (residues 53–120). (A) Domain organization showing location of the oligomerization core within the full-length protein. (B and C) Ribbon diagram of a ten-subunit assembly of G1P53–120 shown in two orthogonal views. The N and C termini as well as the secondary structure elements of a single subunit are labeled. “Linker” refers to residues 61–65 connecting α4 with the main body of the oligomerization core formed by helices α5 and α6. (D) Ribbon diagram of a nine-subunit assembly of the same construct, G1P53–120, shown in three alternating colors. (E) Electrostatic surface potential at subunit interface (ranging from -5, red, to +5 kT/e, blue) shown for two opposing subunits with the central axis vertical as in B. (F) Channel cross section indicating internal (van der Waals) diameters. The red stars indicate the position of the MTSSL in the S106C mutant used.
Fig. 2.
Structure of N-terminal deletion construct (residues 65–141) and full-length small terminase. (A) Ribbon diagram of the proteolytic fragment G1P65–141 containing the C-terminal β-barrel in addition to the main body of the oligomerization core domain shown with the central axis vertical, with individual subunits in alternating colors. The indicated van der Waals diameter of the β-barrel (9.8 Å) corresponds to the most constricted part of the channel. (B) Ribbon diagram of the full-length G1P shown along the axis with DBDs in red. Two DBDs of the asymmetric part (DBD-1 and DBD-2), that were observed in the electron density maps, are shown as ribbons, and the putative position of the third DBD (DBD-3) is depicted by dashed circles. (C) Models of full-length G1P shown with all DBDs either in DBD-1 (left) or DBD-2 (right) orientation. For clarity, DBDs of only five subunits are shown, with a 10-bp dsDNA-DBD complex modeled only for one subunit. The rotational and translational DBD movements derived from the normal mode analysis are indicated on the left. (D) Stereo figure comparing superposed individual subunits of small terminases from Siphoviridae SF6 phage (two adjacent subunits, red and pink), Podoviridae Sf6 phage (blue), and the putative small terminase from prophage phBC6A51 (yellow). The structures were aligned to fit the position of the channel axis, the first of the oligomerization α-helices and the C-terminal β-strand. Although the position of the two helices is conserved in the oligomer, the fold of the monomer differs in the Podoviridae Sf6 small terminase, where helix α6 occupies the same position as α6 of an adjacent subunit in the small terminases from the other two phages.
Oligomerization Core Domain.
The conformation of individual subunits is essentially the same in the nine-subunit and ten-subunit oligomeric states of the G1P53–120 construct (Fig. 1 B–D). A segment of helix α4, present at the N terminus of this construct, and the following peptide (residues 65–69) that link the N-terminal DNA-binding domain with the oligomerization part are located at the outer surface of the oligomer and do not contribute to subunit interactions. The main body of the oligomerization core, residues 70–120, is formed by two short antiparallel α-helices (α5 and α6) connected by a β-hairpin (β1β2) (Figs. 1B and 2D). This segment mediates multiple subunit-subunit interactions comprising eleven hydrogen bonds, several hydrophobic contacts, and two salt bridges formed between residues His102/Glu104 and Glu115/Lys112 (Fig. 1E and Table S2). Interactions are very similar for both oligomeric states, resulting in similar intersubunit buried surface areas of approximately 1,000 Å2. However, the two oligomeric states differ in the dimensions of the central channel. The most constricted part of the oligomerization core domain has a van der Waals diameter of approximately 11 Å in the 9-mer and approximately 14.5 Å in the 10-mer (Fig. 1F). The channel reaches its maximum dimension in its middle part with a van der Waals diameter of approximately 29 Å in the 9-mer and approximately 32 Å in the 10-mer.
The C-Terminal Segment Forms an Intersubunit β-Barrel.
A proteolytic fragment, G1P65–141, comprising the C-terminal β-barrel and the main body of the oligomerization core domain, forms a nine-subunit assembly (Fig. 2A). In this structure, the N-terminal domains which were shown to be responsible for DNA binding (15, 16) and are further on designated as DBDs (DNA-binding domains), were missing because of degradation occurring during the period required for crystal growth. However, the C-terminal segment (residues 121–141), lacking in the previously crystallized construct, is clearly visible in the electron density maps. The C-terminal segments of all subunits assemble together in a parallel β-barrel with each subunit contributing one β-strand (β3, residues 125–139). The internal van der Waals diameter of the β-barrel varies from approximately 9.9 (measured for side-chain atoms of Gln134) to 14.7 Å (side-chain atoms of Thr132). The slope of the β-strands with respect to the barrel axis is approximately 55°, in good agreement with the theoretical value of 56.3° for this type of β-barrel (17).
The N-Terminal DNA-Binding Domains Are Connected to the Oligomer Via Flexible Linkers.
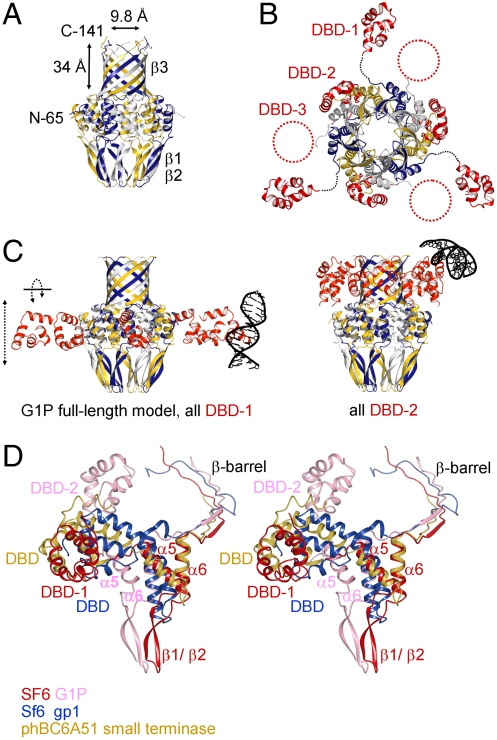
The X-ray data for the full-length protein are anisotropic and complete only to approximately 6-Å resolution, although in directions perpendicular to the c axis they extend to approximately 4 Å (Table S1). In this structure, shown in Fig. 2 B and C, the C-terminal β-barrel as well as the N-terminal DBDs of two out of three subunits in the asymmetric unit are defined in the electron density maps (Fig. S1). The positions and orientations of the two defined DBDs with respect to the oligomerization core domain differ markedly, with their centroids shifted along the oligomer axis by approximately 33 Å. The DBD of the third subunit of the asymmetric unit has no defined electron density indicating variability in its position, which is permitted by the lack of symmetry-related molecules in the vicinity of this domain. Taken together, the structural observations indicate significant variability in the positioning of DBDs.
To investigate further possible conformational changes in the small terminase and, in particular, the potential adjustments in the orientation of the DBDs, we performed normal mode analysis calculations. These showed low-energy modes corresponding to up-and-down movements of the DBDs (along the channel axis) as well as rotational adjustments (Movies S1 and S2). These data indicate a large degree of flexibility in the orientation of the DBDs, consistent with the structural observations described above.
Full-Length Small Terminase Exclusively Forms Nine-Subunit Assemblies Stabilized by the C-Terminal β-Barrel.
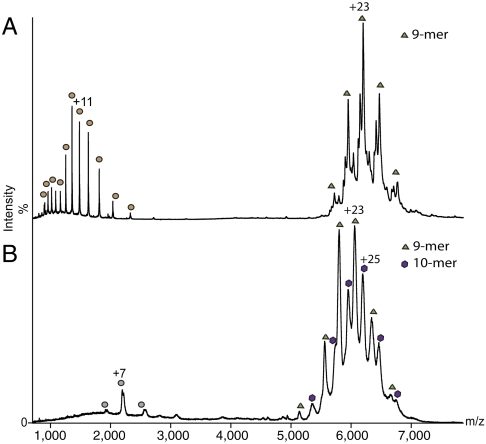
To understand whether the nine-subunit state, observed for the full-length terminase, predominates in solution or whether it was selected from a mixture of different oligomeric states during crystallization, we performed mass spectrometry analysis of the full-length protein under conditions where subunit interactions are maintained (18, 19). The only oligomer observed corresponded to the nine-subunit assembly (Fig. 3A). Partial microheterogeneity was observed at the C terminus of the protein, leading to peak splitting for the 9-mer species (Fig. S2). We also used mass spectrometry to probe the oligomeric state of the protein in which the C-terminal segment was removed (G1P1–120). Mass spectra for this truncated form indicated the presence of both nine-subunit and ten-subunit species in an approximately 1∶1 ratio (Fig. 3B and Fig. S2).
Fig. 3.
Mass spectrometry analysis. (A) Mass spectrum recorded for full-length G1P reveals a 9-mer centered at 6,000 m/z (green triangles). G1P has some minor C-terminal truncations (see Fig. S2), which arise from partial cleavage of residues 128–145. To resolve these peaks, increased acceleration voltage is applied leading to dissociation into monomers (low m/z of 1,000–2,000, brown circles). (B) Mass spectrum recorded for the construct G1P1–120 reveals the formation of both 9-mers (green triangles) and 10-mers (purple hexagon) in an approximately 1∶1 ratio. Gray circles correspond to monomers.
These observations indicate that, although the C-terminal segment is not essential for G1P oligomerization, it appears to stabilize the nine-subunit state, most probably through formation of the intersubunit β-barrel observed in the crystal structure. This hypothesis is supported by the electron paramagnetic resonance (EPR) data (Table S3) showing that the construct G1P53–145 containing the C-terminal β-barrel forms predominantly oligomers. Consistent with mass spectrometry analysis, variation of the subunit number in the crystal structures was observed only when the C-terminal β-barrel was truncated.
Comparison with Other Small Terminases.
Sequence alignment of 33 small terminases from phages closely related to SPP1 indicated several conserved residues that are exposed on the molecular surface (SI Materials and Methods and Figs. S3 and S4 B and C). One group of such residues (Lys5, Lys9, Arg12, and Gly32) is found in the helix-turn-helix (HTH) motif involved in DNA binding. Another group of conserved residues is present in the central channel (Glu87, Lys112, and Lys119). These residues are involved in electrostatic interactions that stabilize the oligomeric assembly and may, potentially, also mediate interactions with DNA. No conserved residues can be found at the outer surface of the oligomerization core.
Structural comparisons with the small terminase of phage Sf6 (14) belonging to the Podoviridae family and a putative small terminase from the Bacillus cereus prophage phBC6A51 [Protein Data Bank (PDB) ID code 2ao9] reveal that, despite a lack of sequence similarity, the folds are conserved (Fig. 2D). In all three proteins the central oligomerization motif formed by α-helices α5 and α6 in G1P and the C-terminal β-barrel are conserved features. The fold of the DBD is also very similar (pairwise Cα rmsd of 1.8–3.1 Å for approximately 45 DBD residues) although its position with respect to the oligomerization core varies. G1P differs from the other two terminases by the presence of an extended β-hairpin (β1/β2 in Fig. 2D) that connects the conserved α-helices α5 and α6. Interestingly, the structure of the T4-like bacteriophage 44RR2 small terminase reported in the accompanying paper (20) also reveals an oligomerization motif comprising two α-helices, as indicated by earlier mutational analysis (21).
Previously, structural data were also obtained for the isolated DNA-binding domain of the small terminase from bacteriophage λ (13). Structural comparisons show that, although SF6 (PDB ID code 2cmp) and Sf6 (14) small terminases share the same fold in their DBDs, it differs markedly from the fold observed in bacteriophage λ small terminase (Fig. S1). Although in all three cases, the polypeptide chain folds into four α-helices, in the case of λ small terminase the HTH motif is formed by the first and second α-helices, whereas in the other two terminases it is formed by the second and third α-helices. Unlike the SF6 and Sf6 terminases, where the recognition helix (α3) is almost immediately followed by another helix (α4), in bacteriophage λ small terminase the recognition helix (α2) is followed by an extended hairpin, necessitating classification of this fold as “winged HTH.” Fold differences result in a very different orientation of the HTH motif with respect to helix α4, which connects the DBD with the C-terminal oligomerization part of the protein (Fig. S1). So far, no structural data have been obtained for the oligomerization part of the bacteriophage λ small terminase, thereby preventing further structural comparison, although analytical ultracentrifugation data obtained for a complex of large and small λ terminases indicated the formation of macromolecular assemblies comprising eight subunits of the small terminase (22), thus suggesting a potential similarity with the multisubunit assemblies found in other small terminases.
Although all five small terminases (44RR2, P22, SF6, phBC6A51, and Sf6), for which oligomerization domains have been resolved by structural analysis, form ring-shaped oligomers, they differ in their oligomeric state with subunit numbers ranging from eight to eleven. Differences in oligomeric state result in different diameters of the internal channel.
DNA Binding by G1P is Exclusively Mediated by the DNA-Binding Domains.
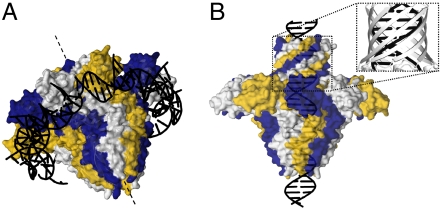
The central channel as well as the presence of multiple, peripheral DNA-binding domains per oligomer posed a question about potential modes of interaction with DNA. We hypothesized that, apart from interacting with the peripheral DBDs (Fig. 4A), DNA could be accommodated in the central channel during translocation (Fig. 4B). This assumption is supported by the presence of Lys/Arg rings in the central channel (Fig. S4A) and the intriguing finding that the geometry of the G1P β-barrel matches the geometry of B-form DNA. Indeed, the strand angle of the β-barrel matches the helical rise of double-stranded B-DNA (Fig. 4B, Inset). We probed interaction with the two potential DNA-binding surfaces (the internal channel and the surface around the outside of the oligomer formed by DBDs) by surface plasmon resonance (SPR) and EPR.
Fig. 4.
Potential models for the G1P-DNA complex. (A) DNA binding around the outside of the oligomer mediated by the DNA-binding domains. Each DBD in complex with a 10-bp oligonucleotide is reoriented so that DNA segments can form a continuous molecule. (B) DNA binding in the channel. The Inset demonstrates the match between the geometry of the C-terminal β-barrel and B-form DNA.
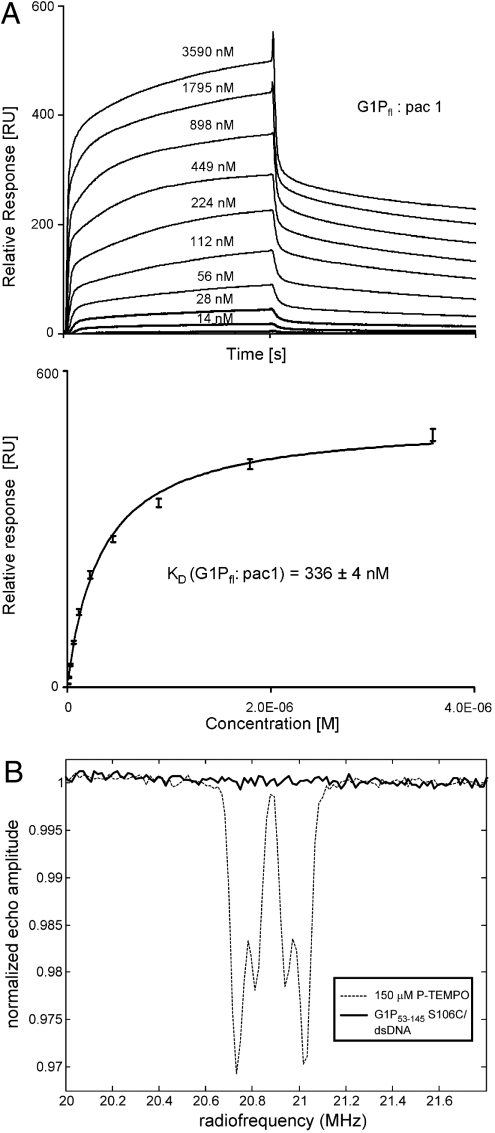
SPR experiments were performed with a diverse range of constructs for G1P and the SPP1 recognition pac site DNA, as specified in Materials and Methods. The strongest interaction was observed for the 428-bp pac DNA (pac1, comprising pacL, pacC, and pacR) (Fig. 5A). Full-length G1P bound to this DNA with a KD of 336 ± 4 nM and distinct association and dissociation phases. The binding affinity decreased when fragments of the pac site DNA were used (Table S4 and Fig. S5). Whereas the presence of the C-terminal β-barrel did not increase the affinity significantly, a significant reduction was observed for constructs lacking the DBDs. We hypothesized that the residual weak binding was due to nonspecific association of the DNA with basic residues located at the outer surface of the oligomerization core, in contrast to binding in the central channel. Indeed, the mutation of solvent-exposed positively charged Lys86 (Fig. S4) resulted in a significant further drop in affinity (Table S4 and Fig. S5) verifying our assumption of nonspecific binding. Therefore, the interaction with DNA as measured by SPR is predominantly dependent on the presence of the DBDs.
Fig. 5.
DNA binding. (A) SPR experiments for full-length G1P and the pac DNA recognition site comprising 428 bp (pac1). Equilibrium dissociation constant KD was estimated from a steady state analysis of three individual experiments. (B) EPR experiments. Mims ENDOR spectra for G1P53–145 S106C labeled with MTSSL at the inner channel surface as shown in Fig. 1F. The control experiment with 4-phosphonoxy-2,2,6,6-tetramethyl-piperidine-1-oxyl (dashed line) clearly shows phosphorous coupling, but no coupling was observed for the oligomer incubated with a 22-bp dsDNA (solid line).
To distinguish whether the weak residual binding after removing the DBDs was due to binding in the channel or whether it was due to nonspecific interaction with the positively charged rings present at the outer surface of the oligomerization domain, we performed EPR experiments with the spin label attached in the internal groove of the channel, where there is sufficient space to accommodate the label without affecting the diameter of the channel (Fig. 1F). Pulsed EPR measurements were performed by using the G1P53–145 S106C mutant that lacks the DNA-binding domains, labeled with the MTSSL (Materials and Methods). Our modeling indicated that if DNA is present in the channel, the distance between the spin label and the nearest 31P nucleus of the DNA would be < 1 nm. This distance is sufficiently short for the hyperfine interactions between the electron spin of the spin label in the protein and the nuclear spin of the 31P nucleus in the DNA to be observed by electron nuclear double resonance (ENDOR). We used the Mims ENDOR pulse sequence at the Q band, which was shown to be most sensitive to longer electron-31P distances (23). Substoichiometrically labeled oligomers (containing approximately one spin label per oligomer) were used to prevent spin-spin interactions between spin labels on adjacent subunits in the oligomer. The control experiment using a phosphorylated spin label (with ca. 0.6 nm nitroxide-31P distance) clearly showed phosphorous coupling in the Mims ENDOR spectra with an excellent signal-to-noise ratio (Fig. 5B). However, ENDOR spectra of substoichiometrically labeled G1P oligomers incubated with a 22-bp SPP1 dsDNA oligonucleotide showed no detectable phosphorous coupling.
The combination of SPR and EPR results demonstrates that a complex of G1P with DNA in the inner channel could not be assembled in vitro, at least under the conditions tested.
Discussion
Small terminases play two crucial roles during DNA packaging. Initially, the small terminase binds to a specific recognition site within the viral chromosome and recruits the large terminase. The assembled terminase-DNA complex then docks onto the procapsid and the small terminase performs its second role, regulating the enzymatic activities of the large terminase.
Previous models describing the interaction between the small terminase and the pac site DNA have suggested that the DNA wraps around the circular oligomer. However, the presence of the central channel suggested that it could accommodate DNA (24). In this model of packaging initiation, the DNA would likely interact first with the DNA-binding domains and then be translocated through the central channel of the small terminase. This mode of DNA binding has been increasingly favored (2, 14, 24). However, the observed inner diameters of the eight-subunit assembly (14) and nine-subunit assembly (Figs. 1F and 2A) are too narrow to accommodate the DNA double helix. Moreover, small terminases from different phages differ in their oligomeric states, containing either eight [Sf6 (14)], nine [SF6, P22 (24), phBC6A51], or 11/12 [44RR2 (20)] subunits, indicating that the number of subunits and consequently the diameter of the inner channel are not critical for the function of the small terminase. Indeed, in the bacteriophage P22 small terminase, both nine- and ten-subunit assemblies are functional for packaging in vivo (24). In agreement with these observations, the combination of our SPR and EPR data presented here strongly indicate that the G1P-DNA interaction is mediated exclusively by DNA-binding domains and not by the inner channel.
We then sought to model the interaction between DNA and the small terminase oligomer comprising multiple DNA-binding domains, as observed in SF6, Sf6, and phBC6A51. We did not use DNA-interaction data available for the bacteriophage λ small terminase (13, 25) because of significant differences in DBD folds and oligomeric states (dimeric species reported for λ are not observed in the SF6, Sf6, and phBC6A51 small terminases) and because of DNA bending in λ involving an additional host factor (26). Interestingly, when modeling the DNA, guided by HTH-DNA complexes of closest structural homologues (27), the DNA orientation is roughly parallel to the oligomer axis. This mode of interaction suggests that one DNA molecule could connect the DBDs of several small terminase oligomers. However, from the mass difference accumulating to the chip surface in our SPR experiments, we calculated that there was only about one G1P oligomer per approximately 100 bp DNA. Moreover, previous DNase footprinting experiments with G1P bound to pac DNA revealed nine protected sites within an approximately 100-nucleotide segment, one site every 10 ± 1 bp (every turn of the DNA helix); these sites were separated by DNase-sensitive regions (16). Taken together, these data indicate that in the complex an approximately 100-bp DNA fragment wraps around the protein oligomer interacting with multiple HTH motifs separated by approximately 34 Å (16). The approximately 34-Å separations would be in good agreement with the DBD separation observed in crystal structures and with their flexibility. This model suggests that small terminases with varying numbers of subunits would still be functional as long as the DBDs were appropriately spaced.
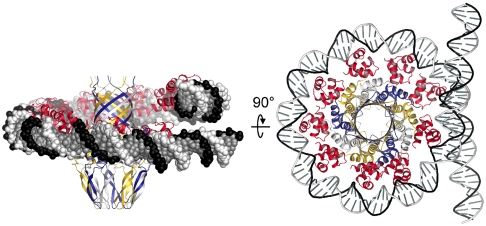
Our structure of full-length G1P has two out of three DNA-binding domains in the asymmetric unit defined, because no electron density was observed for the third domain. The two DBDs for which electron density was observed are in different positions relative to the oligomerization domain, which indicates significant flexibility in their position. The observed flexibility is further supported by normal mode analysis calculations. This flexibility may be essential for binding DNA because to bind DNA in the expected circular/helicoidal orientation, each DBD must reorient in order to match its HTH motif with the corresponding segment of DNA. On the basis of the observed flexibility of the DBD positions, we generated a model starting with the two extreme DBD positions defined in our crystal structure. We interpolated the positions of the other DBDs around the oligomer, adjusted them to be equidistant from the oligomer and each other, and aligned the HTH motifs. Intriguingly, the two starting positions provide enough spatial separation to allow a helicoidal assembly where the DNA wraps completely around the oligomer (Fig. 6). In contrast, in a model with all DBDs adopting the same constant position relative to the oligomerization domain, the DNA molecule cannot encircle the G1P oligomer without unrealistic distortion.
Fig. 6.
Model of the small terminase-DNA complex during DNA recognition. Positions of DBDs were remodeled so that the vertical (channel) coordinate of the two extreme positions corresponds to coordinates observed in the crystal structure with all other DBDs occupying intermediate positions; orientation of each DBD was adjusted so that helix-turn-helix (HTH) motifs fit the helicoidal DNA, with approximately 34-Å spacing between adjacent HTH motifs.
The small terminase performs at least two distinct tasks during DNA packaging: First, it assures the specific recognition of the pac DNA, and second, it regulates the enzymatic activities of the large terminase during DNA translocation, which requires that the small terminase is engaged in the molecular motor. We show that the DNA recognition task involves the small terminase DBDs and that the DBDs are the dominant interaction site for the formation of a nucleoprotein complex. Our structure of full-length G1P, in which the DBD exists in two very different positions and orientations, together with normal mode analysis, DNA-binding data, and the DNase footprinting results (16) allowed us to model the interaction with DNA prior to recruiting the large terminase to perform the initial cut. We argue that the interaction must involve remodeling at the level of the tertiary structure and aligning the DBDs with the DNA to form a helicoidal, nucleosome-like assembly. After directing the DNA cleavage, the small terminase up-regulates ATP hydrolysis and thereby DNA translocation while inhibiting nuclease activity (5, 28, 29). Despite the conserved overall structure of the small terminase, no conserved residues were located on the exterior surface of G1P, indicating that interaction with the large terminase or portal protein is mediated by the overall architecture of the molecular motor and by the distinctive shape of the small terminase.
Materials and Methods
Protein Production, Crystallization, and X-Ray Structure Determination.
The gene encoding full-length small terminase (G1Pfl, residues 1–145) of bacteriophage SF6 was cloned, overexpressed, and purified as described in SI Materials and Methods. Crystals of G1Pfl grew within 1 wk by vapor diffusion at 293 K from solution containing 30% Jeffamine 600 M and 0.1 M sodium cacodylate, pH 6.0. The 4-Å–resolution data were collected at ID-14 beamline at European Synchrotron Radiation Facility (ESRF). A single better-diffracting crystal from the G1Pfl sample grew after 6 mo from solution containing 0.2 M MgCl2, 8% (wt/vol) PEG 20,000, 8% (wt/vol) PEG 550 monomethyl ether, buffered with Tris·HCl, pH 8.0. The 3-Å–resolution data from this crystal were collected in house by using a Rigaku RU200 X-ray generator with a rotating anode. Crystals of G1P53–120-SeMet were obtained under several crystallization conditions: 0.1 M malonic acid, imidazole, and boric acid buffer, pH 7.0, 25% (wt/vol), PEG 1500 (crystal form #1); 0.2 M KCl, 50 mM Hepes, pH 7.5, 35% (vol/vol) pentaerythritol propoxylate (5/4 PO/OH) (crystal form #2) and 2.7 M (NH4)2SO4, 0.1 M Tris·HCl, pH 8.5 (crystal form #3). Single-wavelength anomalous dispersion X-ray diffraction data at the selenium absorption edge (λ = 0.9716 Å) for crystal forms #1 and #2 were collected at the I04 beam line (Diamond). Data for crystal form #3 were collected at BM14 (ESRF). Most crystallographic calculations were performed by using the CCP4 program package (30). G1Pfl and G1P65–141 datasets were processed with MOSFLM/SCALA (Table S1). All other data were processed by using HKL2000 (31). A detailed description of structure determination is given in SI Materials and Methods. Coordinates have been deposited with the Protein Data Bank under accession ID codes 3ZQM (G1P53–120, #1), 3ZQN (G1P53–120, #2), 3ZQO (G1P53–120, #3), 3ZQP (G1P65–141), and 3ZQQ (G1Pfl).
Mass Spectrometry.
Thirty microliters of a solution containing G1Pfl or G1P1–120 were buffer-exchanged into 1 M ammonium acetate (pH 7.5) by using Bio-Spin columns (Bio-Rad). MS analysis was conducted on a Q-ToF2 mass spectrometer (Waters) modified for high mass detection and conservation of noncovalent interactions between protein subunits (18, 19). Then 2 μL of this solution was introduced into the mass spectrometer from a gold-plated capillary needle. Instrument parameters used during experiments were capillary voltage, 1.7 kV; cone voltages, up to 110 V; extractor cone voltage, 5 V; and collision energy, ranging from 30 to 100 V. The mass spectra recorded were calibrated by using cesium iodide. All data were acquired and processed by using MassLynx v4.1 (Waters).
Surface Plasmon Resonance.
On the basis of DNA-interaction regions identified by DNAse footprinting of the G1P-pac DNA complex (16) with G1P found to bind to pacL and pacR but not to pacC, three different SPP1 pac site variants were generated by PCR using the SPP1 genome as template (pac1: complete pac site—i.e., including all three functional sites pacL-pacC-pacR—and spans nucleotides -326 to +108 relative to the cleavage site in pacC, length 428 bp; pac2: ends after pacL, spans nucleotides -326 to -82, length 253 bp; pac3: starts after pacL and covers pacC and pacR, nucleotides -82 to +108, length 190 bp). Further details are given in SI Materials and Methods.
EPR Measurements.
The protein was spin labeled with 1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl)-methanethiosulfonate (MTSSL) using a standard protocol. Continuous wave-EPR measurements were carried out at room temperature by using an X-band spectrometer, and Mims ENDOR was carried out by using a pulsed Q-band spectrometer at 50 K. Full details can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank Bernie Strongitharm and Andrew Leech (University of York) and also John Butler and Tim Fagge (Biacore) for help with SPR measurements. Ralf Flaig and Martin Walsh (Diamond, Oxford) are thanked for help during data collection. We are grateful to Andrey Lebedev, Eleonor Dodson, Johan Turkenburg, and Guy Dodson (University of York) for useful suggestions during data analysis. Ma Yun (University of York), Emma Carter, and Damien Murphy (University of Cardiff) are thanked for help with EPR spectroscopy. This work was supported by the Welcome Trust (fellowship 081916 to A.A.A.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. R.W.H. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3ZQM, 3ZQN, 3ZQO, 3ZQP, and 3ZQQ).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1110270109/-/DCSupplemental.
References
- 1.Rao VB, Feiss M. The bacteriophage DNA packaging motor. Annu Rev Genet. 2008;42:647–681. doi: 10.1146/annurev.genet.42.110807.091545. [DOI] [PubMed] [Google Scholar]
- 2.Casjens SR. The DNA-packaging nanomotor of tailed bacteriophages. Nat Rev Microbiol. 2011;9:647–657. doi: 10.1038/nrmicro2632. [DOI] [PubMed] [Google Scholar]
- 3.Earnshaw WC, Casjens SR. DNA packaging by the double-stranded DNA bacteriophages. Cell. 1980;21:319–331. doi: 10.1016/0092-8674(80)90468-7. [DOI] [PubMed] [Google Scholar]
- 4.Smith DE, et al. The bacteriophage ϕ29 portal motor can package DNA against a large internal force. Nature. 2001;413:748–752. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- 5.Camacho AG, Gual A, Lurz R, Tavares P, Alonso JC. Bacillus subtilis bacteriophage SPP1 DNA packaging motor requires terminase and portal proteins. J Biol Chem. 2003;278:23251–23259. doi: 10.1074/jbc.M301805200. [DOI] [PubMed] [Google Scholar]
- 6.Sun S, et al. The structure of the phage T4 DNA packaging motor suggests a mechanism dependent on electrostatic forces. Cell. 2008;135:1251–1262. doi: 10.1016/j.cell.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 7.Black LW. DNA packaging in dsDNA bacteriophages. Annu Rev Microbiol. 1989;43:267–292. doi: 10.1146/annurev.mi.43.100189.001411. [DOI] [PubMed] [Google Scholar]
- 8.Simpson AA, et al. Structure of the bacteriophage ϕ29 DNA packaging motor. Nature. 2000;408:745–750. doi: 10.1038/35047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lebedev AA, et al. Structural framework for DNA translocation via the viral portal protein. EMBO J. 2007;26:1984–1994. doi: 10.1038/sj.emboj.7601643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olia AS, Prevelige PE, Johnson JE, Cingolani G. Three-dimensional structure of a viral genome-delivery portal vertex. Nat Struct Mol Biol. 2011;18:597–603. doi: 10.1038/nsmb.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smits C, et al. Structural basis for the nuclease activity of a bacteriophage large terminase. EMBO Rep. 2009;10:592–598. doi: 10.1038/embor.2009.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nĕmeček D, Lander GC, Johnson JE, Casjens SR, Thomas GJ., Jr Assembly architecture and DNA binding of the bacteriophage P22 terminase small subunit. J Mol Biol. 2008;383:494–501. doi: 10.1016/j.jmb.2008.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Beer T, et al. Insights into specific DNA recognition during the assembly of a viral genome packaging machine. Mol Cell. 2002;9:981–991. doi: 10.1016/s1097-2765(02)00537-3. [DOI] [PubMed] [Google Scholar]
- 14.Zhao H, et al. Crystal structure of the DNA-recognition component of the bacterial virus Sf6 genome-packaging machine. Proc Natl Acad Sci USA. 2010;107:1971–1976. doi: 10.1073/pnas.0908569107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chai S, Kruft V, Alonso JC. Analysis of the Bacillus subtilis bacteriophages SPP1 and SF6 gene 1 product: A protein involved in the initiation of headful packaging. Virology. 1994;202:930–939. doi: 10.1006/viro.1994.1415. [DOI] [PubMed] [Google Scholar]
- 16.Chai S, Lurz R, Alonso JC. The small subunit of the terminase enzyme of Bacillus subtilis bacteriophage SPP1 forms a specialized nucleoprotein complex with the packaging initiation region. J Mol Biol. 1995;252:386–398. doi: 10.1006/jmbi.1995.0505. [DOI] [PubMed] [Google Scholar]
- 17.Murzin AG, Lesk AM, Chothia C. Principles determining the structure of beta-sheet barrels in proteins. I. A theoretical analysis. J Mol Biol. 1994;236:1369–1381. doi: 10.1016/0022-2836(94)90064-7. [DOI] [PubMed] [Google Scholar]
- 18.Sobott F, Hernandez H, McCammon MG, Tito MA, Robinson CV. A tandem mass spectrometer for improved transmission and analysis of large macromolecular assemblies. Anal Chem. 2002;74:1402–1407. doi: 10.1021/ac0110552. [DOI] [PubMed] [Google Scholar]
- 19.Hernandez H, Robinson CV. Determining the stoichiometry and interactions of macromolecular assemblies from mass spectrometry. Nat Protoc. 2007;2:715–726. doi: 10.1038/nprot.2007.73. [DOI] [PubMed] [Google Scholar]
- 20.Sun S, et al. The structure and function of the small terminase component of the DNA packaging machine in T4 like bacteriophages. Proc Natl Acad Sci USA. 2011 doi: 10.1073/pnas.1110224109. doi: 10.1073/pnas.1110224109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kondabagil KR, Rao VB. A critical coiled coil motif in the small terminase, gp16, from bacteriophage T4: Insights into DNA packaging initiation and assembly of packaging motor. J Mol Biol. 2006;358:67–82. doi: 10.1016/j.jmb.2006.01.078. [DOI] [PubMed] [Google Scholar]
- 22.Maluf NK, Gaussier H, Bogner E, Feiss M, Catalano CE. Assembly of bacteriophage Lambda terminase into a viral DNA maturation and packaging machine. Biochemistry. 2006;45:15259–15268. doi: 10.1021/bi0615036. [DOI] [PubMed] [Google Scholar]
- 23.Zänker P-P, Jeschke G, Goldfarb D. Distance measurements between paramagnetic centers and a planar object by matrix Mims electron nuclear double resonance. J Chem Phys. 2005;122:024515. doi: 10.1063/1.1828435. [DOI] [PubMed] [Google Scholar]
- 24.Nĕmeček D, et al. Subunit conformations and assembly states of a DNA-translocating motor: The terminase of bacteriophage P22. J Mol Biol. 2007;374:817–836. doi: 10.1016/j.jmb.2007.08.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ortega ME, Catalano CE. Bacteriophage lambda gpNu1 and Escherichia coli IHF proteins cooperatively bind and bend viral DNA: Implications for the assembly of a genome-packaging motor. Biochemistry. 2006;45:5180–5189. doi: 10.1021/bi052284b. [DOI] [PubMed] [Google Scholar]
- 26.Maluf NK, Yang Q, Catalano CE. Self-association properties of the bacteriophage lambda terminase holoenzyme: Implications for the DNA packaging motor. J Mol Biol. 2005;347:523–542. doi: 10.1016/j.jmb.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 27.Tahirov TH, et al. Mechanism of c-Myb-C/EBP beta cooperation from separated sites on a promoter. Cell. 2002;108:57–70. doi: 10.1016/s0092-8674(01)00636-5. [DOI] [PubMed] [Google Scholar]
- 28.Gual A, Camacho AG, Alonso JC. Functional analysis of the terminase large subunit, G2P, of Bacillus subtilis bacteriophage SPP1. J Biol Chem. 2000;275:35311–35319. doi: 10.1074/jbc.M004309200. [DOI] [PubMed] [Google Scholar]
- 29.Al-Zahrani AS, et al. The small terminase, gp16, of bacteriophage T4 Is a regulator of the DNA packaging motor. J Biol Chem. 2009;284:24490–24500. doi: 10.1074/jbc.M109.025007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Collaborative Computational Project N. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 31.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.