Abstract
Mutations of fibroblast growth factor receptor 3 (FGFR3) are responsible for achondroplasia (ACH) and related dwarfing conditions in humans. The pathogenesis involves constitutive activation of FGFR3, which inhibits proliferation and differentiation of growth plate chondrocytes. Here we report that activating mutations in FGFR3 increase the stability of the receptor. Our results suggest that the mutations disrupt c-Cbl-mediated ubiquitination that serves as a targeting signal for lysosomal degradation and termination of receptor signaling. The defect allows diversion of actively signaling receptors from lysosomes to a recycling pathway where their survival is prolonged, and, as a result, their signaling capacity is increased. The lysosomal targeting defect is additive to other mechanisms proposed to explain the pathogenesis of ACH.
Achondroplasia (ACH) is the prototype of human chondrodysplasias (1–3). It is one of an allelic series of disorders that results from heterozygous mutations of the gene encoding the fibroblast growth factor receptor 3 (FGFR3). This series includes the more severe thanatophoric dysplasia (TD) types I and II (TDI, TDII), severe achondroplasia with developmental delay and acanthosis nigricans, and the less clinically severe hypochondroplasia. From studies of mice null for FGFR3 and mice transgenic for human FGFR3 mutations and extensive in vitro studies, FGFR3 has been established as a major negative regulator of linear bone growth, acting to inhibit growth plate chondrocyte proliferation and terminal differentiation (4–11). These inhibitory signals are propagated through STAT (signal transducers and activators of transcription), mitogen-activated protein kinase, phosphatidylinositol 3-kinase, and other pathways (12–18). Most observations suggest that the bone growth disturbances in the human disorders result from augmentation of these normal inhibitory functions of FGFR3 rather than from the acquisition of new or different functions by the mutant receptors.
As one member of a family of four receptors (FGFR1–4), FGFR3 is normally activated by ligand-induced dimerization that activates the intrinsic tyrosine kinase activity of the receptor. This leads to transphosphorylation of key tyrosine residues in the cytoplasmic domain of the receptor that serve as docking sites for adaptor proteins and effectors that propagate FGFR3 signals (19–21).
Several mechanisms have been proposed to explain how activating mutations of FGFR3 enhance signals. For example, the typical achondroplasia mutation, G380R, which maps to the transmembrane domain, is thought to stabilize ligand-induced dimers (22). Monsonego-Ornan et al. (23) have suggested that it slows receptor internalization and prolongs signaling on the cell surface (23). Mutations associated with TDI, such as R248C, introduce free cysteine residues into the proximal extracellular domain of the receptor that are believed to form disulfide bonds, resulting in dimerization and receptor activation in the absence of ligand (24). In contrast, the K650E and K650M mutations found in TDII and severe achondroplasia with developmental delay and acanthosis nigricans, respectively, are thought to alter receptor conformation, leading to constitutive activation of kinase activity (21, 25).
It has become clear in recent years that many activated tyrosine kinase-coupled transmembrane receptors continue to propagate signals after internalization, and that lysosomal degradation may be required to terminate signaling (26, 27). Failure to deliver activated receptors to lysosomes may allow their diversion into a default pathway that recycles integral membrane proteins where they may continue to propagate signals. Lysosomal targeting of receptors is best illustrated for the epidermal growth factor receptor (EGFR) (28, 29). Although many details are still controversial, the EGFR targeting mechanism involves the attachment of ubiquitin to lysine residues on the cytoplasmic tail of activated EGFR by the adaptor protein c-Cbl. Ubiquitin serves as a targeting signal to direct transport of the activated EGFR to lysosomes; failure to attach the ubiquitin signal allows recycling of activated receptors.
Little is known about the fate of activated FGFR3, and most of what is known is inferred from limited knowledge about the fate of other FGFRs. For instance, Sorokin et al. (30) observed that PKC-mediated phosphorylation of Y766 is required for internalization of FGFR1. Citores et al. (31, 32) reported that FGFR4 may be recycled or degraded, depending on the presence of undefined targeting signals in the cytoplasmic tail of the receptor. Wong et al. (33) have detected a ligand-induced interaction between c-Cbl and FGFR that leads to ubiquitination of the receptor. Most recently, Monsonego-Ornan et al. (34) have demonstrated that FGFR3 is ubiquitinated in a kinase-dependent fashion.
These observations suggest that activated FGFR3 may behave in a fashion similar to EGFR and other receptors that target themselves for lysosomal degradation on activation. To explore this possibility and that lysosomal targeting may be disturbed in achondroplasia and related disorders, we studied the fate of activated WT and mutant FGFR3. We report that activated FGFR3 is normally targeted for lysosomal degradation through c-Cbl-mediated ubiquitination, and that FGFR3 harboring achondroplasia and TDII mutations escapes lysosomal targeting to recycle as signal-propagating receptor.
Methods
Cell Lines. Cos-7 cells (American Type Culture Collection) and Phoenix packaging cells were propagated in DMEM with 10% (vol/vol) FBS in a 5% CO2/95% air humidified incubator. RCJ 3.1c5.18 cells (a gift from Jane Aubin, University of Toronto, Toronto) were maintained as described (35). To generate stable cell lines, Cos-7 cells and RCJ cells were transfected with plasmids encoding WT, ACH, and TDII FGFR3-Myc fusion proteins and selected with 400 μg/ml G418 for 1 wk. Transfections were carried out by using Lipofectamine Plus (Invitrogen) according to the manufacturer's recommendations. For GFP fusion proteins, Phoenix packaging cells were transfected with plasmids encoding WT, ACH, or TDII FGFR3-GFP. Viral supernatant was collected between 48 and 72 h, during which time Cos-7 or RCJ cells were plated in 100-mm dishes (5 × 105 cells per plate). The collected supernatants were filtered through low protein-binding Acrodisc 0.45-μm syringe filters (Gelman) and Polybrene (Sigma) added to a final concentration of 4 μg/ml for Cos-7 cell infection and 8 μg/ml for RCJ cell infection. The filtered supernatant was applied to the Cos-7 and RCJ cells at a theoretical multiplicity of infection of 1:500, ensuring only one copy per infected cell. The cells were incubated with the viral supernatant at 32°C for 48 h. Transduced cells were split 1:5 and selected with hygromycin B at 350 μg/ml.
Reagents and Antibodies. Basic fibroblast growth factor (FGF2; Calbiochem) was used at a final concentration of 25 ng/ml in the presence of heparin (1 μg/ml). Cycloheximide (Sigma) was used at a concentration of 200 μg/ml. Anti-Myc, anti-hemagglutinin (HA), anti-FGFR3, and anti-c-Cbl (C15) antibodies were obtained from Santa Cruz Biotechnology. Anti-phosphotyrosine antibody 4G10 was a gift from Brian Druker (Oregon Health and Science University, Portland). Antibodies to extracellular signal-regulated kinase (Erk) 1/2 and phospho-Erk1/2 were purchased from Cell Signaling Technology (Beverly, MA) and anti-GFP was from Clontech. Fluorescence-tagged transferrin (transferrin-Alexa 594) was obtained from Molecular Probes.
Plasmids. Three sets of mouse FGFR3 expression vectors were used. FGFR3-WT and FGFR3-K644E, which corresponds to the K650E mutation in human TDII, were used for stability experiments (36). pCSFR3iiicmyc constructs (WT, ACH, and TDII, the latter two corresponding to human ACH-G380R and TDII-K650E mutations, respectively) were provided by Michael Naski and David Ornitz (Washington University, St. Louis) (7). They were designated WT, ACH, and TDII FGFR3-Myc. The FGFR3-GFP constructs were generated by subcloning the HindIII–ClaI fragment from pCSFR3iiicmyc into pEGFP-N2 (Clontech). The GFP-tagged FGFR3 receptors were comparable to Myc-tagged FGFR3 receptors in activating downstream signaling pathways, including mitogen-activated protein kinase, STAT1, and phospholipase C-γ (data not shown). pcDNA3.1c-Cbl and pcDNA3.170Zc-Cbl were generated by subcloning the XhoI–BamHI fragment from pJZenNeoc-Cbl and pJZenNeo70Zc-Cbl, respectively (gifts from Kate Kolibaba and Brian Druker, Oregon Health and Science University) into pcDNA3.1(–) (37). To generate the FGFR3 retroviral constructs, the Myc- and GFP-tagged FGFR3 were subcloned into a modified pRevTRE (Clontech), in which the tetracycline response element promoter was replaced with the cytomegalovirus (CMV) promoter from pEGFP-N1. pMT123(HA-Ub) was a gift from Dirk Bohmann (University of Rochester, Rochester, NY) (38).
Protein Stability Experiments. Cos-7 cells (2.5 × 105 cells per well) in 12-well plates were transfected with 0.25–1.0 μg of expression vector. At 19 h of transfection, cysteine and methionine were depleted by incubating cells in 0.5 ml of DMEM (–Met/Cys) with 10% FBS for 1 h. Cells were labeled with [35S]Met/[35S]Cys in 0.5 ml of DMEM (–Met/Cys) with 10% FBS, containing 80 μCi (1 Ci = 37 GBq) per well of ProMix (Amersham Biosciences) for 4 h. In pulse–chase experiments, the medium was replaced with DMEM with 10% FBS (time 0). At the time indicated, cells in each well were washed twice with PBS and lysed in 1 ml of the lysis buffer (50 mM Tris, pH 7.5/150 mM NaCl/2 mM EDTA/1% Triton X-100/1% deoxycholate/0.1 mM sodium orthovanadate/10 μM leupeptin/0.1 mM PMSF/2 μg/ml aprotinin). The lysate was passed through a 25-gauge needle three times and centrifuged at 15,000 × g at 4°C for 20 min. FGFR3 proteins were immunoprecipitated with 1 μg of FGFR3 antibody and protein A-Sepharose (Pharmacia) at 4°C overnight after clearance with 1 μg of rabbit IgG (Pierce). Beads were washed with 1 ml of the lysis buffer three times, and 50 μl of the SDS-loading buffer was added. Samples were treated at 100°C for 10 min before loading 20 μl of sample on SDS/7.5% polyacrylamide gels. Gels were stained with Coomassie brilliant blue, dried, analyzed by using the Cyclone Storage Phosphor System (Packard), and subjected to autoradiography. Control cells, which were mock-treated or transfected with vector without insert, showed no detectable expression of FGFR3.
Pulse–chase experiments were also done in the presence of cycloheximide (200 μg/ml) to inhibit protein synthesis during the chase. Cos-7 cells were transfected with 1 μg of FGFR3-Myc expression plasmid encoding WT, ACH, or TDII FGFR3 and pulsed with [35S]Met/[35S]Cys as before. The medium was replaced with medium containing 200 μg/ml cycloheximide 36 h after transfection (0-h time point). Cell lysates were harvested at 0, 2, 4, 6, and 8 h and analyzed by immunoprecipitation and Western blotting using anti-Myc antibodies. For luciferase assays, Cos-7 cells plated in six-well dishes (4 × 105 cells per well) were cotransfected with 0.25 μg of pCMV-luciferase (a gift from Paul Howard and Richard Maurer, Oregon Health and Science University) and 0.75 μg of WT, TDII, or ACH FGFR3-Myc construct. After 24 h, cells were lysed and assayed in triplicate for luciferase activity by using Luciferase Assay System (Promega).
Phosphorylation and Ubiquitination Assays. To examine activation status of FGFR3, lysates (in lysate buffer as above) were prepared from RCJ cells stably expressing WT or mutant FGFR3-GFP under baseline conditions and after FGF2 treatment. After immunoprecipitation of receptor with anti-GFP antibodies and transfer, Western blots were probed with antiphosphotyrosine antibodies. Western blots of the cell lysates were also probed with anti-Erk1/2 and anti-phospho-Erk1/2. To assess c-Cbl phosphorylation, RCJ cells stably expressing WT or TDII FGFR3-myc under baseline conditions and after FGF2 treatment were immunoprecipitated for c-Cbl and blotted for phosphotyrosine. To examine ubiquitination of FGFR3, Cos-7 cells stably expressing WT or mutant FGFR3-Myc were transfected with pMT123(HA-Ub). Some cultures were also cotransfected with pcDNA3.1c-Cbl or pcDNA3.170Zc-Cbl and/or treated with FGF2 (25 ng/ml). After 48 h, cells were harvested. Cell lysates were immunoprecipitated with anti-Myc antibody and blotted with anti-HA antibody. Immunoblot results were scanned, and images were analyzed by using scion image (Scion, Frederick, MD). Band intensity was quantified by using the Gel Plot 2 macro. Results expressed as fold change in phosphorylation or ubiquitination relative to WT FGFR3 under baseline conditions were combined from independent experiments to determine mean values and standard deviations, and in some cases differences were validated by Student's t test.
Microscopy. RCJ and Cos-7 cells stably expressing FGFR3-GFP fusion proteins were grown on Labtek four-well chamber slides (Apogent, Hudson, NH) and imaged as live cells on a Nikon Eclipse E800 epifluorescence microscope and/or on a Leica TCS SP2 laser scanning confocal system mounted on a DM IRE2 inverted microscope. Live cell imaging was performed at ambient temperature (24°C). Lysosomes were identified by incubating cells in 50 μM LysoTracker-Red DND-99 (Molecular Probes) for 30 min at 37°C before epifluorescence imaging. For transferrin colocalization, Cos-7 cells were transfected with WT or mutant FGFR3-GFP constructs. After 48 h they were incubated for 30 min at 37°C with transferrin-Alexa594, rinsed thoroughly in PBS, fixed in 2% paraformaldehyde for 30 min, and imaged by epifluorescence. The image files were exported as PSD or TIFF files and assembled into figures in photoshop 7.0 (Adobe Systems, Mountain View, CA).
Results
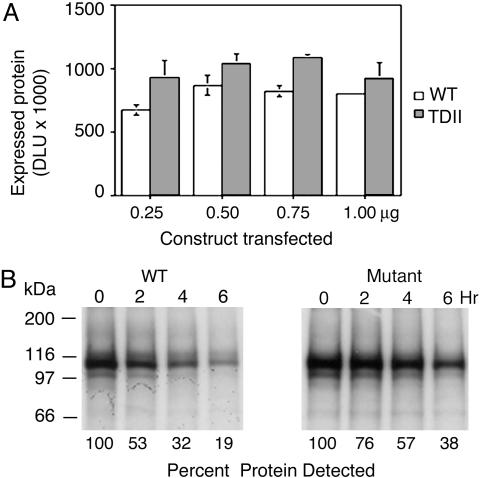
The effect of the activating mutations on FGFR3 protein turnover was examined by metabolically labeling transiently expressed FGFR3 corresponding to WT and TDII (FGFR3-K644E) mutant proteins with [35S]Met/[35S]Cys in Cos-7 cells. About 30% more protein was observed in cells transfected with FGFR3-K644E compared with those with FGFR3-WT (Fig. 1A). The level of mutant receptor was increased regardless of the amount of construct transfected. In addition, analysis of receptor turnover by pulse–chase showed that mutant protein was more stable than the WT receptor (Fig. 1B). The half-life of FGFR3-WT was estimated to be slightly >2 h, whereas FGFR3-K644E had a half-life of >4 h. Repeating the pulse–chase experiment with the addition of cycloheximide to block protein synthesis during the chase showed similar results (data not shown). To control for positive regulatory effects that FGFR3 signaling might have on the promoter of the FGFR3 constructs, cells expressing WT and mutant FGFR3 were cotransfected with pcDNA3.1-luciferase. Subsequent assays showed similar levels of luciferase activity in cells expressing WT or TDII FGFR3, indicating that the increase in levels of FGFR3 was mediated posttranscriptionally (data not shown).
Fig. 1.
Activating mutations increase stability and prolong the half-life of FGFR3. (A) Cos-7 cells transfected with WT or TDII FGFR3 and metabolically labeled with [35S]Met/[35S]Cys exhibit higher levels of mutant protein regardless of amount of construct transfected as indicated. DLU, digital light units. (B) Pulse–chase analysis of labeled receptors shows longer survival for mutant receptors with a half-life estimated at >4 h for TDII FGFR3 compared to slightly more than 2 h for WT FGFR3.
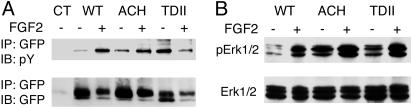
To determine the activation status of the accumulated receptors, cell lysates of RCJ cells stably transfected with WT or mutant FGFR3-GFP were immunoprecipitated with antibodies to GFP and immunoblotted with antibodies to phosphotyrosine. Fig. 2A shows that phosphorylation of WT FGFR3 and also ACH FGFR3 is substantially enhanced by ligand (FGF2), whereas TDII FGFR3 is constitutively phosphorylated. Erk1/2, which is known to be phosphorylated downstream of FGFR3 activation, is also phosphorylated in the absence of ligand in cells expressing mutant but not WT FGFR3 (Fig. 2B). These findings suggest that the FGFR3 that accumulates in ACH and TDII is both activated and propagating downstream signals.
Fig. 2.
Mutant FGFR3 accumulates as an activated, signal-propagating receptor. (A) Western analysis of receptors from stably transfected RCJ cells indicates that tyrosine phosphorylation of WT FGFR3 and ACH FGFR3 is enhanced substantially in response to FGF2, whereas TDII receptors are phosphorylated in the absence of FGF2. IP, immunoprecipitate; IB, immunoblot; pY, phosphotyrosine. (B) Maximal phosphorylation of Erk1/2, a downstream signaling target of FGFR3, requires ligand in the presence of WT FGFR3 but is constitutive in the presence of mutant FGFR3.
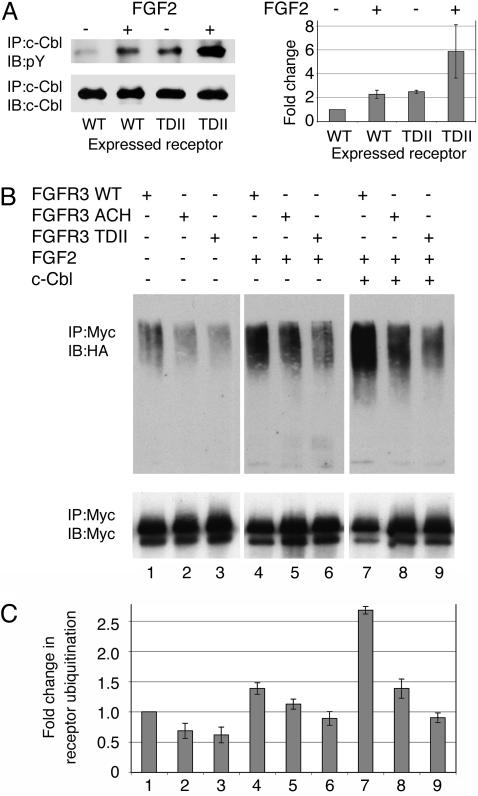
Given the recent implication of c-Cbl-mediated ubiquitination in the turnover of activated FGFR, phosphorylation of c-Cbl and ubiquitination of WT and mutant FGFR3 were assessed. First, immunoblotting showed that c-Cbl is phosphorylated under baseline conditions, and that phosphorylation is increased substantially in response to FGF2 in RCJ cells stably expressing WT FGFR3 (Fig. 3A). Baseline c-Cbl phosphorylation in cells expressing TDII FGFR3 is comparable to that of cells expressing WT FGFR3 after ligand stimulation. It increases further with ligand stimulation.
Fig. 3.
FGFR3 activation leads to phosphorylation of c-Cbl and ubiquitination of FGFR3. (A) Western analysis of c-Cbl in RCJ cells stably transfected with WT or TDII FGFR3-Myc and studied under baseline conditions or after FGF2 treatment. In the presence WT FGFR3, c-Cbl is phosphorylated under baseline conditions, and this is increased by the addition of FGF2. Baseline phosphorylation of c-Cbl in the presence of TDII FGFR3 is comparable to that of stimulated WT FGFR3, and it increases further with ligand. A representative blot (Left) and the results of three independent experiments (Right) are shown. The results are expressed as fold change in c-Cbl phosphorylation relative to c-Cbl phosphorylation in WT FGFR3-expressing cells not treated with ligand. (B and C) Western analysis of FGFR3 ubiquitination in Cos-7 cells stably expressing WT, ACH, or TDII FGFR3 and transiently transfected with ubiquitin-HA. (B) A representative blot. (C) Results of four independent experiments in which ubiquitination is expressed as fold change relative to receptor ubiquitination in cells expressing WT FGFR3 under baseline conditions. The numbered lanes, which refer to both B and C, were loaded for equivalent amounts of FGFR3-Myc. The relative ubiquitination of WT FGFR3 was increased by ligand stimulation compared to baseline (lane 4 vs. lane 1) and further increased by cotransfection with c-Cbl (lane 7 vs. lane 4). Mutant receptors were less ubiquitinated than WT receptors under all three conditions (lanes 2 or 3 vs. lane 1; lanes 5 or 6 vs. lane 4; and lanes 8 or 9 vs. lane 7). All of these differences were statistically significant with P < 0.05 (Student's t test).
Next, Cos-7 cells stably expressing WT or mutant FGFR3-Myc were transiently transfected with HA-tagged ubiquitin. After 48 h, Myc-immunoprecipitated FGFR3 was immunoblotted for HA (Fig. 3 B and C). Under baseline conditions, FGFR3 ubiquitination of both WT and mutant FGFR3-Myc was detected, but it was less for mutant FGFR3-Myc (lanes 1–3). Ubiquitination of both WT and mutant FGFR3-Myc was increased by FGF2, but WT receptors continued to be ubiquitinated more than mutant receptors, and TDII FGFR3-Myc was ubiquitinated less than ACH FGFR3-Myc (lanes 4–6). To determine whether this differential ubiquitination pattern reflected saturation of endogenous c-Cbl, the experiment was repeated in cells cotransfected with c-Cbl. The presence of additional c-Cbl increased the extent of ubiquitination but did not change the pattern (lanes 7–9). When the experiment was repeated in cells cotransfected with 70Z-Cbl, a c-Cbl mutant lacking ubiquitin ligase activity, ubiquitination levels similar to those observed in cells treated with ligand were detected (lanes 4–6) (data not shown). Attempts to immunoprecipitate c-Cbl with antibodies to FGFR3 and vice versa were unsuccessful. These observations suggest that both WT and mutant FGFR3 are ubiquitinated by c-Cbl in response to ligand, and that activating mutations alter this process.
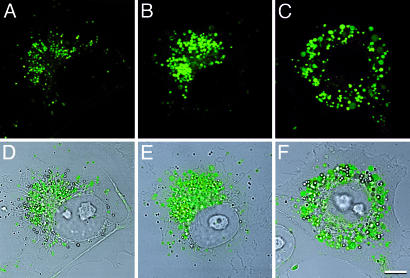
Epifluorescence and confocal microscopy consistently showed more intense fluorescence in RCJ and Cos-7 cells stably expressing mutant compared to WT FGFR3-GFP fusion proteins. Confocal imaging of live cells localized the fusion proteins to vesicular structures that were more abundant and larger in cells expressing the mutant receptors, especially cells expressing TDII FGFR3-GFP (Fig. 4). Most of the GFP-positive vesicles were juxtanuclear in location (Fig. 4C), although some vesicles could be detected at the cell surface.
Fig. 4.
FGFR3 resides in vesicles that are increased in number and size for mutant receptors. Live cell confocal microscopy of Cos-7 cells expressing WT FGFR3-GFP (A and D), ACH FGFR3-GFP (B and E), and TDII FGFR3-GFP (C and F), showing GFP fluorescence in A–C and GFP fluorescence superimposed on transmitted light images in D–F. Note the predominant juxtanuclear distribution of receptors. (Bar = 8 μm.)
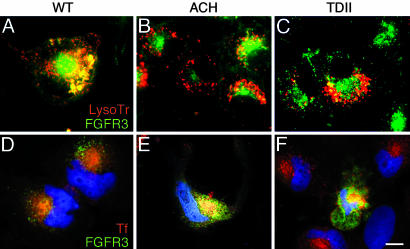
LysoTracker-Red was used to visualize lysosomal targeting of the FGFR3-GFP fusion proteins in live RCJ cells. Fig. 5 A–C shows that WT FGFR3-GFP colocalizes with the lysosomal marker to a greater extent than either ACH or TDII FGFR3-GFP, suggesting defective targeting of mutant receptors. To determine whether they are diverted into the recycling pathway, the FGFR3-GFP fusion proteins were colocalized with transferrin receptor, a marker of this pathway. Fig. 5 D–F shows substantially greater colocalization for mutant vs. WT FGFR3-GFP in Cos-7 cells transfected with the fusion protein constructs and incubated with conjugated transferrin.
Fig. 5.
Mutant FGFR3 is diverted from lysosomes to recycling compartment. (A–C) Localization of receptors to lysosomes in RCJ cells stably expressing WT FGFR3-GFP (A), ACH FGFR3-GFP (B), or TDII FGFR3-GFP (C) and treated with LysoTracker-Red. Colocalization is greater for WT than for mutant FGFR3-GFP. (D–F) Colocalization of receptors with transferrin-Alexa594 as marker of recycling endosomes in Cos-7 cells transfected with WT FGFR3-GFP (D), ACH FGFR3-GFP (E), or TDII FGFR3-GFP (F). Nuclei are stained with Hoechst 33342 in D–F. Localization to recycling endosomes is greater for mutant (E and F) than for WT (D) receptors. (Bar = 12 μm.)
Discussion
Our observations indicate that activated FGFR3 is normally targeted for lysosomal degradation through a mechanism that involves c-Cbl-mediated ubiquitination, much as activated EGFR is targeted for lysosomal degradation and signal termination. They suggest that activating FGFR3 mutations associated with ACH and TDII disturbs normal targeting, allowing the mutant receptors to escape into a recycling pathway where they accumulate as actively signaling receptors with a half-life of about twice that of WT receptors. The consequence is amplification of the normal inhibitory FGFR3 signals in the growth plate.
Our results show that FGF ligand stimulation increases both baseline phosphorylation of c-Cbl and ubiquitination of WT FGFR3. The increase in receptor ubiquitination observed when WT c-Cbl, but not when mutant c-Cbl, was coexpressed with the receptor provides further evidence for c-Cbl mediation of FGFR3 ubiquitination. Wong et al. (ref. 33; J. Schlessinger, personal communication) recently reported similar findings from experiments that are probably more relevant to FGFR1 than to FGFR3, because they did not distinguish between FGFRs, and they used FGFR1 in experiments in which FGFR was expressed. Nevertheless, they showed that c-Cbl mediates ubiquitination of FGFR in a ligand-dependent manner through an indirect interaction that requires the adaptor proteins FRS2α and Grb2. Their model proposes that ligand activation of FGFR leads to recruitment and activation of FRS2α, which in turn recruits c-Cbl-bound Grb2 to form a ternary complex that directs ubiquitination of both FGFR and FRS2α (33).
Our results are generally consistent with this model, although we did not assess FRS2α in our experiments. It should be noted, however, that FRS2α may interact differently with FGFR1 than with FGFR3. For instance, comparison of binding of the juxtamembrane region implicated in FRS2α–FGFR interaction has shown much stronger binding of FRS2α to FGFR1 than to FGFR3 (39–41). Of additional interest, we found c-Cbl phosphorylation to depend substantially on receptor activation by ligand for WT FGFR3 or by mutation, in contrast to Wong et al. (33), who observed that c-Cbl was constitutively phosphorylated. This difference undoubtedly reflects technical differences in how the experiments were carried out; however, it might also indicate differences between FGFR1 and FGFR3 activation of c-Cbl.
When ubiquitination of mutant FGFR3 was compared to that of WT FGFR3, we detected less ubiquitination of the mutant receptors both under baseline conditions and after ligand stimulation. The relative deficiency was greater for TDII than for ACH in both cases, and this difference was exaggerated when WT c-Cbl was coexpressed with the receptors to alleviate the possible saturation of endogenous c-Cbl. In fact, although coexpression of c-Cbl more than doubled ubiquitination of WT FGFR3, it had little if any effect on mutant FGFR3.
Our results point to a defect in c-Cbl-mediated ubiquitination of mutant FGFR3 that is proportionate to the severity of the dwarfism associated with the ACH and TD mutations. They differ from those of Monsonego-Ornan et al. (34), who observed increased ubiquitination rather than decreased ubiquitination of the mutant receptors. There are several possible explanations for the disparity, none of which are mutually exclusive. Our experiments were performed in Cos-7 cells, whereas theirs were done mainly in HEK293 cells. To determine ligand effect, we assayed cells under steady-state exposure to FGF2, whereas they assayed cells after 30 min to 6 h of FGF9 stimulation. Monsonego-Ornan et al. (34) used transient transfection to express receptors, whereas we used retroviral transfer, which in theory should have produced lower levels of receptor and a lower likelihood of overloading saturable degradation pathways (42). Although we strongly favor the notion that the ACH and TDII mutations lead to defective ubiquitination of FGFR3, because of the differences in experimental approach, we cannot exclude the alternate possibility.
It is not clear how the mutations interfere with ubiquitination of activated receptors, although there are several possibilities. Based on our observation that c-Cbl is phosphorylated more by mutant than by WT FGFR3, one possibility is that the excessive kinase activity associated with activating mutations phosphorylates inhibitory tyrosine residues on c-Cbl. Such a mechanism has been proposed to explain how Smad pathways propagating transforming growth factor β signals can be inhibited (43). Another possibility is excessive activation of one or more of the recently described negative feedback loops involving FRS2α and Grb2 that have been implicated in the propagation of mitogen-activated protein kinase signals and potentially in c-Cbl-mediated ubiquitination of FGFRs (33, 44, 45). A third possibility involves sequestration of c-Cbl by Sprouty 2 (Spry2). Spry2 is phosphorylated at a conserved tyrosine residue (Tyr-55) in response to FGF stimulation, creating a binding site for c-Cbl that could lead to its sequestration and inability to ubiquitinate targets, such as FGFRs (45–47). Given recent evidence that mono- and polyubiquitinated proteins are routed differently within cells, it is also conceivable that the mutations alter the stoichiometry of FGFR3 ubiquitination (48, 49). It remains to be determined whether the WT and mutant receptors are mono- or polyubiquitinated in our experimental system.
Regardless of how lysosomal targeting of activated FGFR3 is disturbed, our results suggest that mutant FGFR3 is diverted to the pathway that recycles integral membrane proteins, including receptors, such as the transferrin receptor. This notion is consistent with previous reports of recycling of FGFR4 (31). Our findings are supported by observations from tissues from patients with activating FGFR3 mutations. For example, Hwang and Ghadially (50) reported “globular smooth-tubule aggregates” in the cytoplasm of chondrocytes from four infants with TD. Perinuclear accumulation of FGFR3 was observed in primary chondrocytes from an infant with TDI by Legeai-Mallet et al. (14). Delezoide et al. (51) reported increased immunostaining for FGFR3 protein in hypertrophic and proliferative chondrocytes in growth plate tissues from fetuses with TDI compared with controls. Of note, the staining tended to be perinuclear in distribution. Although not reported here, we have observed accumulation of TDI FGFR3-GFP similar to that of ACH and TDII FGFR-GFP in RCJ and Cos-7 cells, which is consistent with the notion that FGFR3 mutations associated with dwarfism involve faulty lysosomal targeting of activated FGFR3.
Our model of decreased lysosomal degradation of mutant FGFR3 is compatible with other mechanisms proposed to explain the molecular pathogenesis of ACH, i.e., stabilized dimers, dimerization induced by free cysteine residues, and conformational changes that constitutively activate kinase activity. It is additive to these mechanisms and, in fact, raises some interesting possibilities for synergy with respect to intensifying FGFR3 signals. For example, current models for receptor recycling suggest that dimerized receptors revert to their monomeric, nonsignaling form during endosomal recycling. The dimer-stabilizing effects of the ACH mutation could delay or even prevent this reversion of activated FGFR3, which would prolong the duration of signal propagation and augment the signal intensity.
In summary, we offer substantial evidence that activated FGFR3 is targeted for lysosomal degradation by c-Cbl-mediated ubiquitination, and that activating mutations found in patients with ACH and related chondrodysplasias disturb this process, leading to recycling of activated receptors and amplification of FGFR3 signals. We suggest this mechanism contributes to the molecular pathogenesis of ACH and represents a potential target for therapeutic intervention.
Acknowledgments
We thank Jane Aubin, Michael Naski, David Ornitz, Kate Kolibaba, Brian Druker, Paul Howard, Richard Maurer, and Dirk Bohmann for their gifts of cells and reagents; Joseph Schlessinger for sharing unpublished data; and Phillip Stork, Matthew Thayer, Caroline Enns, and Brian Druker for helpful suggestions. This work was supported by research grants from the Shriners Research Program (no. 8540) and the D. Miller Foundation.
Abbreviations: Erk, extracellular signal-regulated kinase; ACH, achondroplasia; EGFR, epidermal growth factor receptor; FGF2, basic fibroblast growth factor 2; FGFR3, fibroblast growth factor receptor 3; HA, hemagglutinin; TD, thanatophoric dysplasia.
References
- 1.Horton, W. A. (1997) Curr. Opin. Pediatr. 9, 437–442. [DOI] [PubMed] [Google Scholar]
- 2.Horton, W. A. & Hecht, J. T. (2002) in Connective Tissue and its Heritable Disorders, eds. Royce, P. M. & Steinmann, B. (Wiley, New York), pp. 901–908.
- 3.Francomano, C. & Muenke, M. (2002) in Connective Tissue and Its Heritable Disorders, eds. Royce, P. M. & Steinmann, B. (Wiley, New York), pp. 961–991.
- 4.Colvin, J. S., Bohne, B. A., Harding, G. W., McEwen, D. G. & Ornitz, D. M. (1996) Nat. Genet. 12, 390–397. [DOI] [PubMed] [Google Scholar]
- 5.Deng, C., Wynshaw-Boris, A., Zhou, F., Kuo, A. & Leder, P. (1996) Cell 84, 911–921. [DOI] [PubMed] [Google Scholar]
- 6.Naski, M. C., Colvin, J. S., Coffin, J. D. & Ornitz, D. M. (1998) Development (Cambridge, U.K.) 125, 4977–4988. [DOI] [PubMed] [Google Scholar]
- 7.Naski, M. C., Wang, Q., Xu, J. & Ornitz, D. M. (1996) Nat. Genet. 13, 233–237. [DOI] [PubMed] [Google Scholar]
- 8.Ornitz, D. M., Xu, J., Colvin, J. S., McEwen, D. G., MacArthur, C. A., Coulier, F., Gao, G. & Goldfarb, M. (1996) J. Biol. Chem. 271, 15292–15297. [DOI] [PubMed] [Google Scholar]
- 9.Chen, L., Adar, R., Yang, X., Monsonego, E. O., Li, C., Hauschka, P. V., Yayon, A. & Deng, C. X. (1999) J. Clin. Invest. 104, 1517–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garofalo, S., Kliger-Spatz, M., Cooke, J. L., Wolstin, O., Lunstrum, G. P., Moshkovitz, S. M., Horton, W. A. & Yayon, A. (1999) J. Bone Miner. Res. 14, 1909–1915. [DOI] [PubMed] [Google Scholar]
- 11.Wang, Y., Spatz, M. K., Kannan, K., Hayk, H., Avivi, A., Gorivodsky, M., Pines, M., Yayon, A., Lonai, P. & Givol, D. (1999) Proc. Natl. Acad. Sci. USA 96, 4455–4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanai, M., Göke, M., Tsunekawa, S. & Podolsky, D. K. (1997) J. Biol. Chem. 272, 6621–6628. [DOI] [PubMed] [Google Scholar]
- 13.Su, W.-C. S., Kitagawa, M., Xue, N., Xie, B., Garofalo, S., Cho, J., Deng, C., Horton, W. A. & Fu, X.-Y. (1997) Nature 386, 288–291. [DOI] [PubMed] [Google Scholar]
- 14.Legeai-Mallet, L., Benoist-Lasselin, C., Delezoide, A. L., Munnich, A. & Bonaventure, J. (1998) J. Biol. Chem. 273, 13007–13014. [DOI] [PubMed] [Google Scholar]
- 15.Sahni, M., Ambrosetti, D. C., Mansukhani, A., Gertner, R., Levy, D. & Basilico, C. (1999) Genes Dev. 13, 1361–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weksler, N. B., Lunstrum, G. P., Reid, E. S. & Horton, W. A. (1999) Biochem. J. 342, 677–682. [PMC free article] [PubMed] [Google Scholar]
- 17.Choi, D. Y., Toledo-Aral, J. J., Lin, H. Y., Ischenko, I., Medina, L., Safo, P., Mandel, G., Levinson, S. R., Halegoua, S. & Hayman, M. J. (2001) J. Biol. Chem. 276, 5116–5122. [DOI] [PubMed] [Google Scholar]
- 18.Hart, K. C., Robertson, S. C., Kanemitsu, M. Y., Meyer, A. N., Tynan, J. A. & Donoghue, D. J. (2000) Oncogene 19, 3309–3320. [DOI] [PubMed] [Google Scholar]
- 19.Schlessinger, J. (2000) Cell 103, 211–225. [DOI] [PubMed] [Google Scholar]
- 20.Schlessinger, J., Plotnikov, A. N., Ibrahimi, O. A., Eliseenkova, A. V., Yeh, B. K., Yayon, A., Linhardt, R. J. & Mohammadi, M. (2000) Mol. Cell 6, 743–750. [DOI] [PubMed] [Google Scholar]
- 21.Hart, K. C., Robertson, S. C. & Donoghue, D. J. (2001) Mol. Biol. Cell 12, 931–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Webster, M. K. & Donoghue, D. J. (1996) EMBO J. 15, 520–527. [PMC free article] [PubMed] [Google Scholar]
- 23.Monsonego-Ornan, E., Adar, R., Feferman, T., Segev, O. & Yayon, A. (2000) Mol. Cell. Biol. 20, 516–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sorokin, A., Lemmon, M. A., Ullrich, A. & Schlessinger, J. (1994) J. Biol. Chem. 269, 9752–9759. [PubMed] [Google Scholar]
- 25.Mohammadi, M., Schlessinger, J. & Hubbard, S. R. (1996) Cell 86, 577–587. [DOI] [PubMed] [Google Scholar]
- 26.Waterman, H. & Yarden, Y. (2001) FEBS Lett. 490, 142–152. [DOI] [PubMed] [Google Scholar]
- 27.Clague, M. J. & Urbe, S. (2001) J. Cell Sci. 114, 3075–3081. [DOI] [PubMed] [Google Scholar]
- 28.Levkowitz, G., Waterman, H., Zamir, E., Kam, Z., Oved, S., Langdon, W. Y., Beguinot, L., Geiger, B. & Yarden, Y. (1998) Genes Dev. 12, 3663–3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levkowitz, G., Waterman, H., Ettenberg, S. A., Katz, M., Tsygankov, A. Y., Alroy, I., Lavi, S., Iwai, K., Reiss, Y., Ciechanover, A., et al. (1999) Mol. Cell 4, 1029–1040. [DOI] [PubMed] [Google Scholar]
- 30.Sorokin, A., Mohammadi, M., Huang, J. & Schlessinger, J. (1994) J. Biol. Chem. 269, 17056–17061. [PubMed] [Google Scholar]
- 31.Citores, L., Wesche, J., Kolpakova, E. & Olsnes, S. (1999) Mol. Biol. Cell 10, 3835–3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Citores, L., Khnykin, D., Sorensen, V., Wesche, J., Klinenberg, O., Wiedtocha, A. & Olsnes, S. (2001) J. Cell Sci. 114, 1677–1689. [DOI] [PubMed] [Google Scholar]
- 33.Wong, A., Lamothe, B., Li, A., Schlessinger, J. & Lax, I. (2002) Proc. Natl. Acad. Sci. USA 99, 6684–6689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Monsonego-Ornan, E., Adar, R., Rom, E. & Yayon, A. (2002) FEBS Lett. 528, 83–89. [DOI] [PubMed] [Google Scholar]
- 35.Grigoriadis, A. E., Heersche, J. N. & Aubin, J. E. (1996) Differentiation 60, 299–307. [DOI] [PubMed] [Google Scholar]
- 36.Iwata, T., Li, C. L., Deng, C. X. & Francomano, C. A. (2001) Hum. Mol. Genet. 10, 1255–1264. [DOI] [PubMed] [Google Scholar]
- 37.Rao, N., Ghosh, A. K., Ota, S., Zhou, P., Reddi, A. L., Hakezi, K., Druker, B. K., Wu, J. & Band, H. (2001) EMBO J. 20, 7085–7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Treier, M., Staszewski, L. M. & Bohmann, D. (1994) Cell 78, 787–798. [DOI] [PubMed] [Google Scholar]
- 39.Xu, H., Lee, K. W. & Goldfarb, M. (1998) J. Biol. Chem. 273, 17987–17990. [DOI] [PubMed] [Google Scholar]
- 40.Lin, H. Y., Xu, J., Ischenko, I., Ornitz, D. M., Halegoua, S. & Hayman, M. J. (1998) Mol. Cell. Biol. 18, 3762–3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hadari, Y. R., Gotoh, N., Kouhara, H., Lax, I. & Schlessinger, J. (2001) Proc. Natl. Acad. Sci. USA 98, 8578–8583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.French, A. R., Sudlow, G. P., Wiley, H. S. & Lauffenburger, D. A. (1994) J. Biol. Chem. 269, 15749–15755. [PubMed] [Google Scholar]
- 43.Kretzschmar, M., Doody, J. & Massague, J. (1997) Nature 389, 618–622. [DOI] [PubMed] [Google Scholar]
- 44.Lax, I., Wong, A., Lamothe, B., Lee, A., Frost, A., Hawes, J. & Schlessinger, J. (2002) Mol. Cell 10, 709–719. [DOI] [PubMed] [Google Scholar]
- 45.Hanafusa, H., Torii, S., Yasunaga, T. & Nishida, E. (2002) Nat. Cell Biol. 4, 850–858. [DOI] [PubMed] [Google Scholar]
- 46.Egan, J. E., Hall, A. B., Yatsula, B. A. & Bar-Sagi, D. (2002) Proc. Natl. Acad. Sci. USA 99, 6041–6046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rubin, C., Litvak, V., Medvedovsky, H., Zwang, Y., Lev, S. & Yarden, Y. (2003) Curr. Biol. 13, 297–307. [DOI] [PubMed] [Google Scholar]
- 48.Haglund, K., Sigismund, S., Polo, S., Szymkiewicz, I., Di Fiore, P. P. & Dikic, I. (2003) Nat. Cell Biol. 5, 461–466. [DOI] [PubMed] [Google Scholar]
- 49.Mosesson, Y., Shtiegman, K., Katz, M., Zwang, Y., Vereb, G., Szollosi, J. & Yarden, Y. (2003) J. Biol. Chem. 278, 21323–21326. [DOI] [PubMed] [Google Scholar]
- 50.Hwang, W. S. & Ghadially, F. N. (1996) Ultrastruct. Pathol. 20, 219–222. [DOI] [PubMed] [Google Scholar]
- 51.Delezoide, A. L., Lasselin-Benoist, C., Legeai-Mallet, L., Brice, P., Senee, V., Yayon, A., Munnich, A., Vekemans, M. & Bonaventure, J. (1997) Hum. Mol. Genet. 6, 1899–1906. [DOI] [PubMed] [Google Scholar]