Abstract
We describe a mechanism of gene regulation involving formation of a complex between PSF protein and mouse VL30 (mVL30) retrotransposon RNA. PSF represses transcription of the insulin-like growth factor 1 (IGF1)-inducible gene P450scc by binding to an insulin-like growth factor response element (IGFRE) motif in the gene. The complex with mVL30 RNA releases PSF, allowing transcription to proceed. Retrovirally mediated transmission of mVL30 RNA to human tumor cells induced several genes, including oncogenes, which also are induced by IGF1, and promoted metastasis. In mice, steroid synthesis is activated in steroidogenic cells by pituitary hormones, which concomitantly induce transcription of mVL30 RNA in the cells. We showed that steroid synthesis could also be activated in mouse steroidogenic adrenal cells by transfection with cDNA encoding either mVL30 RNA tracts that form a complex with PSF or a small interfering RNA (siRNA) that degrades PSF transcripts. These results suggest that mVL30 RNA regulates steroidogenesis, and possibly other physiological processes of mice, by complex formation with PSF. Retrotransposons such as mVL30 apparently evolved not only as “junk” DNA but also as transcriptionally active noncoding DNA that acquired physiological and pathological functions.
The complete genome of a mouse VL30 (mVL30) retrotransposon is structurally similar to a retroviral genome, with 5′ and 3′ LTRs flanking an internal region containing ≈3.7 kb (1). In contrast to the internal region of an infectious retroviral genome that encodes gag, pol, and env proteins, the corresponding region of the mVL30 genome has numerous stop codons in all reading frames that probably block formation of a functional protein. The mouse genome contains multiple copies of transcriptionally active mVL30 DNA, and virtually all mouse cells contain mVL30 RNA although the level varies among different tissues and at different developmental stages (1). Because retroviral vectors for gene transfer and gene therapy usually are produced in packaging cells derived from mouse cells, these cells also contain mVL30 RNA. A remarkable property of retroviral vectors is the capacity to transmit mVL30 RNA from a packaging cell to cells infected by the retrovirus, which synthesize, integrate, and transcribe mVL30 cDNA (1). In an earlier report (2), we showed that retroviral-mediated transfection of tissue factor (TF) cDNA into a nonmetastatic human melanoma cell line increased the metastatic potential of the cells to which mVL30-1 RNA also had been transmitted. The increase in metastatic potential depended on expression of TF protein on the cell surface and also on transcription of mVL30-1 cDNA. Dependence on mVL30-1 RNA was surprising because the RNA seems to lack significant coding potential. Here, we report that mVL30-1 RNA forms a complex with PSF, a multifunctional regulatory protein that binds to RNA in spliceosomes (3, 4) and to an IGFRE motif in the insulin-like growth factor 1 (IGF1)-inducible gene P450ssc that represses transcription of the gene (5, 6). The complex with mVL30-1 RNA releases PSF, allowing transcription to proceed. We discuss the significance of this mechanism for the regulation of physiological and pathological cellular functions.
Materials and Methods
Human Tumor Cell Lines. The human melanoma lines Th, T4, Te, L1, L3, L4, and L8 were cloned from the parental line YU-SIT1 by infection with a retroviral vector (2). mVL30-1 RNA was retrovirally transmitted to the Th, L3, and L8 lines but not to the T4, Te, L1, and L4 lines. DU145 is a human prostate carcinoma line that lacks mVL30-1 RNA, and DU145T was cloned from DU145 by infection with a retroviral vector and contains mVL30-1 RNA. The cells were maintained in DMEM containing 10% FBS, penicillin, and streptomycin in a CO2 incubator at 37°C.
Microarray Analyses of Differential Gene Expression Induced by mVL30-1 RNA in Human Tumor Cells. Microarray slides OHU16K containing 16,000 oligos derived from different human genes were purchased from the Yale University Keck Facility. cDNA probes for the Th and Te clones were synthesized by reverse transcription by using oligo-deoxythymidine [oligo(dT)] as the primer and incorporating allyl amine-deoxyuridine triphosphate (aa-dUTP) into the cDNA. The reaction solution contained 125 μg/ml oligo(dT) primer, 0.5 mM each dATP, dCTP, and dGTP, 0.2 mM aa-dUTP, 0.3 mM dTTP, 10 mM DTT, and 1,280 units/ml Moloney murine leukemia virus (MMLV) reverse transcriptase (SuperScript II). The total volume was 40 μl. The dyes cyanine-3 (cy-3) or cyanine-5 (cy-5) were coupled to aa-dUTP in the cDNAs by using N-hydroxysuccinimide (NHS)-ester cy-3 or cy-5 (Pharmacia) and incubating at 25°C for 1 h in subdued light. The cDNA probes were hybridized to the microarray slides in hybridization buffer at 67°C for 18 h. After hybridization, the slides were washed once with 1× SSC plus 0.03% SDS at 25°C for 10 min, twice with 0.2× SSC, and once with 0.2× SSC. The hybridizations were done in triplicate, and the slides were scanned with a GSI Lumonics (Packard) or an Axon laser scanner (Axon Instruments, Foster City, CA). The fluorescent hybridization signals were measured with genepix software, and the data were analyzed with Microsoft excel. The genes showing a fluorescent intensity >200 with either the Th or Te probes were chosen for RT-PCR analysis.
RT-PCR Analyses of Differential Gene Expression Induced by mVL30-1 RNA in Human Tumor Cells. The Th and Te lines were grown as a monolayer to ≈70% confluence, and total RNA was isolated with TRIzol. RT-PCR was performed as described (2). The beta-actin mRNA was used to normalize the amounts of RNA in the two cell samples. To select the optimal PCR conditions for each gene analyzed, the number of PCR cycles was varied, and the intensities of the resulting PCR bands were used to calculate the ratio of Th/Te intensities for each cycle. A PCR cycle in the range that yielded a constant Th/Te ratio for each gene was used for the experiments reported in Fig. 1. Each PCR was done in triplicate, and the Th/Te ratios were consistent for all genes. The PCR primers were as follows: GAGE6, 5′-GCCTCCTGAAGTGATTGGGCCTA-3′ and 5′-CAGGCGTTTTCACCTCCTCTGGA-3′; D4S234, 5′-GCTGAGTTCACCGTCAGCATCAC-3′ and 5′-GAACTGACATCCAGGGCGATACG-3′; APOD, 5′-GCCACCCCAGTTAACCTCACAGA-3′ and 5′-TTGGGGCAGTTCACCTGGTCTGT-3′; LYL1, 5′-CTACATTGGGCCAGCAGGACCTT-3′ and 5′-GCACCTCGTTCTTGCTCAGCTTC-3′; HMGCS1, 5′-CTCTAGGTGTGCTCCTGAATCAG-3′ and 5′-CCTACTTCAGACCTTGAAGTGGA-3′; COL18A1, 5′-TCCACCTGGTTGCGCTCAACAGC-3′ and 5′-CCTCAGGACGTCCTTGCCGTCAA-3′; LCN2, 5′-TATGTGGTAGGCCTGGCAGGGAA-3′ and 5′-TGGGACAGGGAAGACGATGTGGT-3′; P450SCC, 5′-ACCCAACCCGATGGCTGAGCAAA-3′ and 5′-TCACTGCTGGGTTGCTTCCTGGT-3′; PTK7, 5′-AAGTCACAGCCCCTCAGCACCAA-3′ and 5′-CACCATGGGGCATCTCTCCATGT-3′; IGFBP3, 5′-ACCCAGAACTTCTCCTCCGAGTC-3′ and 5′-CTGGGAGAGGCTGCCCATACTTA-3′; LDLR, 5′-AGCAATGGCGGCTGCCAGTATCT-3′ and 5′-CTGTGGTCTTCTGATAGACGGGG-3′; VGF, 5′-GACGACGTGGTCAGCATCATCGA-3′ and 5′-GGAGCAGCACGTGCTCGATGTAA-3′; and RPL26, 5′-CTTCCGACCGAAGCAAGAATCGC-3′ and 5′-GGCGAGATTTGGCTTTCCGTTCG-3′.
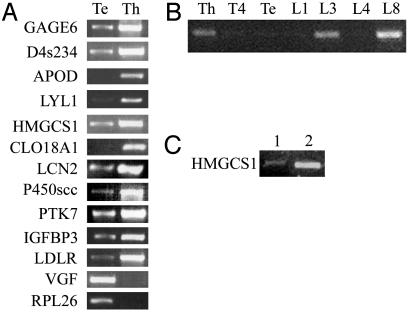
Fig. 1.
RT-PCR tests for an effect of mVL30-1 RNA on transcription of the genes identified by microarray analysis in Table 1. (A) Transcription of the genes in human melanoma clones Te and Th; mVL30-1 RNA is present in Th and absent in Te. (B) Transcription of gene HMGCS1 in a panel of human melanoma clones; mVL30-1 RNA is present in clones Th, L3, and L8 and absent in clones T4, Te, L1, and L4. The human melanoma clones were isolated from a human melanoma line after infection with a retroviral vector, which transmitted mVL30-1 RNA to ≈50% of the clones (2). (C) Transcription of gene HMGCS1 in the human prostatic tumor line DU145 (lane 1) and in clone DU145T, which was transfected with mVL30-1 cDNA (lane 2); mVL30-1 RNA is present in DU145T and absent in DU145. The RT-PCR assays were semiquantitative.
Plasmid-Mediated Transfection of mVL30-1 cDNA into Human Melanoma Cells. The complete mVL30-1 cDNA was ligated into the SpeI and NotI cloning sites of the plasmid pcDNA3.1 (Invitrogen). The transfection procedure for Te cells involved growing the cells in DMEM plus 10% FCS and transfecting 1 μg of the cDNA with Lipofectamine 2000 (Invitrogen). Colonies were selected in DMEM with zeocin and G418, and the cells were tested for mVL30-1 RNA by RT-PCR by using the primers 5′-CTTCATGACCAAACCCTTCA-3′ and 5′-GTATGAGTTTCTTCTGCCA-3′ as described above.
Deletion Mapping of mVL30-1 cDNA. The internal region of the mVL30-1 cDNA was synthesized by PCR by using the following primers: 5′-ATCGACCT TA AGT T TGGTGCAT TGGCCGGGAA-3′ containing an AflII site, and 5′-TACAGTGCGGCCGCCCACTTCT TCTTGTAAGTA-3′ containing a NotI site. The PCR product was cloned into the pcDNA 3.1 plasmid by using the AflII and NotI cloning sites. Various deletions of the internal region of the cDNA were generated from the 5′ end and the 3′ end by using Erase-a-Base kit (Promega), and the size of each fragment was analyzed by agarose gel electrophoresis.
Nuclear Lysates. Cultures of the human melanoma clones Th and Te were grown to about 70% confluence, washed twice with PBS, removed from the plastic substrate by scraping in PBS, and pelleted by centrifugation. The cell pellets were resuspended in a hypotonic buffer (10 mM Hepes, pH 7.9/1.5 mM MgCl2/10 mM KCl/0.2 mM PMSF/0.5 mM DTT) and disrupted with 20 strokes of a Dounce homogenizer. The nuclei were pelleted by centrifugation for 5 min at 3,300 × g. The nuclear pellet was equilibrated 30 min in extraction buffer (20 mM Hepes, pH 7.9/25% glycerol/1.5 mM MgCl2/0.6 M KCl/0.2 mM EDTA/0.2 mM PMSF/0.5 mM DTT) with continuous gentle mixing, and then was centrifuged at 25,000 × g for 30 min. The supernatant was dialyzed against 50 vol of dialysis buffer (20 mM Hepes, pH 7.9/20% glycerol/100 mM KCl/0.2 mM EDTA/0.2 mM PMSF/0.5 mM DTT). The dialysate was centrifuged for 20 min, and the supernatant fluid that serves as the nuclear lysate was stored at –80°C.
Binding of RNA or DNA to Proteins in a Nuclear Lysate. For RNA samples, the nuclear lysate containing 2 μg of total protein was mixed with 5 ng of 32P-labeled RNA, 800 ng of poly(I-C) (Sigma), and 6% glycerol. Binding buffer (10 mM Tris, pH 7.6/5 mM MgCl2/1 mM DTT/100 mM KCl) was added to a final volume of 10 μl, and the samples were incubated at 25°C for 20 min. For DNA samples, 5 ng of 32P-labeled oligonucleotides were added to the nuclear extract mixed with 500 ng of poly(dI-dC) (Sigma) and 6% glycerol; the rest of the procedure was as described for RNA. After incubation, the samples were placed on ice and irradiated at 254 nm for 30 min. After irradiation of RNA samples, RNase T1 was added, and the samples were incubated at 25°C for 20 min. The irradiated samples were fractionated by 7.5% PAGE, and the gel was autoradiographed.
Affinity Chromatography of a Nuclear Protein. Preparative amounts of polyadenylated mVL30-1 RNA were synthesized by transcription with T7 RNA polymerase of a mVL30-1 cDNA insert in the plasmid pSP64 (Promega). The mVL30-1 RNA was immobilized on oligo(dT) Dynabeads (Dynal, Great Neck, NY) as follows. The Dynabeads were washed first with 0.5× SSC followed with binding buffer (10 mM Tris, pH7.5/100 mM KCl/2 mM MgCl/0.2 ml) and suspended in 50 μl of binding buffer, and 5 μg of the polyadenylated mVL30-1 RNA was added. The mixture was incubated at 25°C for 30 min on a rotating wheel, unbound RNA was removed, and the Dynabeads were washed twice with binding buffer. The Dynabeads containing immobilized mVL30-1 RNA were incubated with a nuclear lysate from human melanoma cells at 4°C for 60 min on a rotating wheel, and the Dynabeads were collected with a magnet and washed in binding buffer. The bound proteins were eluted first with 0.5 M followed with 1.0 M KCl, and an aliquot of each eluate was tested for binding to a nuclear protein as described above. The remainder of each eluate was fractionated by SDS/PAGE, and the protein band was excised from the gel and digested with 1% trypsin. The tryptic peptides were analyzed by the Yale Keck Facility by using HPLC and matrix-assisted laser desorption-ionization mass spectrometry. Partial sequencing was done for three major peptides, and protein databases were screened for a matching protein based on the size and sequence of a tryptic peptide.
Western Immunoassay for PSF. The nuclear proteins purified by affinity chromatography on mVL30-1 RNA were fractionated by 7.5% PAGE and transferred to a nitrocellulose membrane. The membrane was blocked for 2 h in PBST containing 5% powdered dried milk and incubated for 1 h with an anti-PSF antibody (provided by J. Patton, Vanderbilt University, Nashville, TN). The membrane was washed in PBST and incubated for 1 h with anti-rabbit IgG conjugated to horseradish peroxidase. The antibody bands were visualized with an ECL detection kit (Amersham Pharmacia Biotech).
Construction of a DNA Encoding a Small Interfering RNA (siRNA) That Degrades PSF Transcripts. The DNA encoding the siRNA contained a U6 promoter followed by a 19-nt sense strand of siRNA, a 9-nt loop (5′-TTCAAGAGA-3′), a 19-nt antisense strand of siRNA, and a stretch of five thymine residues. The U6 promoter was derived from the plasmid pmU6(-315/1)pBSKAmp provided by C. Guerrier-Takada (7). The DNA was synthesized by PCR as follows (8). In the first round of PCR, the reverse primer (5′-TCTCT TGA ACT TA AT TCT TCCTGGCGTCCCA AACAAGGCTTTTC-3′) consisted of the 9-nt loop, a 19-nt antisense sequence of PSF, and a 16-nt sequence complementary to 3′ end of the U6 promoter. The forward primer (5′-TTGGAGCTCCACCGCGGTGG-3′) was complementary to the upstream sequence of the U6 promoter in the plasmid. The PCR conditions were 94°C for 2 min, 30 cycles of 94°C for 10 s, 60°C for 30 s, and 72°C for 30 s, followed by 72°C for 5 min. The PCR products were separated by agarose gel electrophoresis and purified with a QIAquick gel purification kit (Qiagen, Valencia, CA). The PCR products were used for a second round of PCR, as follows. The forward primer was the same as used for the first PCR. The reverse primer (5′-AAAAAGACGCCAGGAAGAATTAAGTCTCTTGAA-3′) consisted of five adenine residues, a 19-nt sense strand of PSF, and a complementary sequence to the 9-nt loop. The PCR conditions were 94°C for 2 min, 30 cycles of 94°C for 10 s, 70°C for 30 s, and 72°C for 30 s, followed by 72°C for 5 min. The PCR products were purified with a QIAquick PCR purification kit (Qiagen). The effect of the siRNA on the expression of PSF was tested by RT-PCR by using the mouse PSF-specific primers (5′-CCAGTCATTGTGGAACCACTTGA-3′and 5′-CTTCTCTCCCTCTACCATATCCT-3′).
Progesterone Synthesis in Y1 Cells. A plasmid encoding the pbt-A RNA segment was constructed as follows. The U6 promoter from plasmid pmU6(-315/1)pBSKAmp was inserted into plasmid pcDNA 3.1 by using the BglII and ApaI sites, followed by pbt-A cDNA and a stretch of five thymine residues. The control plasmid contained mVL30-1 cDNA from nucleotide 1523 to nucleotide 1841 without the pbt-A and pbt-B sequences, which inserted into the AflII and XbaI sites of pcDNA 3.1. The Y1 cells were transfected with 1 μg of the control plasmid or the plasmid encoding the pbt-A RNA, or with 0.3 μg of the siRNA expression fragment. Lipofectamine 2000 was used for all transfections. After 48 h, the cells were seeded in a 24-well plate at a density of 0.5 × 105 cells per well and incubated at 37°C in a CO2 incubator for 24 h, and the medium was then replaced with fresh DMEM. For the sample treated with a pituitary hormone, 10 nM adrenocorticotropic hormone (Sigma) was added to the medium of cells transfected with the control plasmid. All samples were kept at 37°C in a CO2 incubator for 3 h, and the concentration of progesterone in the medium was determined by ELISA with a Progesterone EIA kit (Bio-Analysis, Santa Monica, CA); 20α-hydroxyprogesterone was used for standard curve.
Results
Microarray and RT-PCR Analyses of Differential Gene Transcription. Slides containing 16,000 human ESTs from the Yale University Keck Facility were hybridized with cDNA probes derived from the human melanoma clones Th and Te; mVL30-1 RNA is present in Th and absent from Te (2). Each probe was labeled with a different fluorescent dye and hybridized together, and the Th/Te hybridization ratio for each EST was calculated. The genes corresponding to ESTs with a ratio >3.5 or >0.40 are listed in Table 1. The differential transcription of these genes in Th and Te cells was confirmed by semiquantitative RT-PCR assays (Fig. 1A).
Table 1. Effect of mVL30-1 RNA on gene transcription in human cell lines.
| Gene ID | Th/Te ratio | Gene symbol | Gene description |
|---|---|---|---|
| U19147 | 10.4 | GAGE6 | G antigen 6 |
| M98528 | 5.5 | D4S234E | Neuron-specific protein |
| J02611 | 5.1 | APOD | Apolipoprotein D |
| M22637 | 4.8 | LYLI | Lymphoblastic leukemia-derived sequence 1 |
| X66435 | 4.5 | HMGCSI | 3-Hydroxy-3-methylglutaryl-coenzyme A synthase 1 (soluble) |
| AF018081 | 4.5 | COLI8A1 | Collagen, type XVIII, alpha 1 |
| X99133 | 4.4 | LCN2 | Lipocalin 2 (oncogene 24p3) |
| M14565 | 4.1 | P450scc | P-450 cholesterol side-chain cleavage enzyme |
| U33635 | 4.0 | PTK7 | PTK7 protein tyrosine kinase 7 |
| M35878 | 3.9 | IGFBP3 | Insulin-like growth factor binding protein 3 |
| L00352 | 3.8 | LDLR | Low-density lipoprotein receptor |
| Y12661 | 0.39 | VGF | VGF nerve growth factor inducible |
| X69392 | 0.38 | RPL26 | Ribosomal protein L26 |
Microarray slides containing 16,000 human ESTs were hybridized with cDNA probes prepared from the cloned human melanoma lines Th and Te. Both lines were retrovirally transfected with TF cDNA and express high levels of TF protein, but only the Th line contains mVL30-1 RNA transmitted by the retroviral vector (2). The genes corresponding to the microarray spots showing a >3.8-fold increase or 2.5-fold decrease in the Th/Te ratio were further tested by RT-PCR assays for Th and Te cells (see Fig. 1A). The genes satisfyng the criteria for both differential hybridization and differential RT-PCR are listed in Table 1.
The HMGCS1 gene was chosen for further testing of induced gene transcription by mVL30-1 RNA. One test involved additional human melanoma clones that contain or lack VL30-1 RNA and express high or low levels of TF (2). The results (Fig. 1B) show that induced transcription of the HMGCS1 gene depends only on the presence of mVL30-1 RNA in the melanoma clones, in contrast to metastasis, which depends also on a high level of TF expression. Another test involved two clones derived from the human prostatic tumor line DU145; one clone was transfected with mVL30-1 cDNA and contained mVL30-1 RNA, and the other clone lacked mVL30-1 RNA. Transcription of HMGCS1 was induced in the transfected clone (Fig. 1C), indicating that the effect of mVL30-1 RNA on gene transcription is not limited to one type of human tumor.
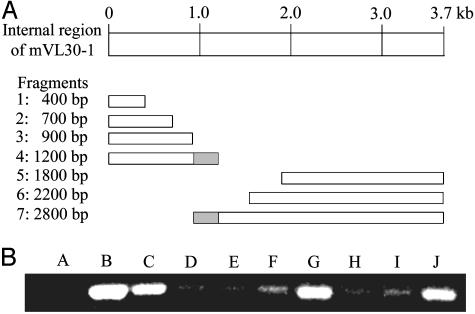
Mapping the Region in mVL30-1 RNA That Regulates Gene Transcription. Fragments of the internal region of mVL30-1 cDNA were generated by nuclease digestion starting from the 5′ and 3′ ends of the molecule, and the sizes of the fragments were determined by electrophoresis in agarose gel (Fig. 2A). To test for induced gene transcription, each cDNA fragment was transfected into the melanoma clone Te, which does not contain mVL30-1 RNA, and the transfected clones were tested by semiquantitative RT-PCR assays for transcription of the HMGCS1 gene. Fragments 4 and 7 showed strong transcription of HMGCS1 (Fig. 2B). The two clones overlap by a 318-bp region spanning nucleotides 1523 to 1841, which should contain the sequences in mVL30-1 RNA that regulate gene transcription.
Fig. 2.
Mapping the region of mVL30-1 that induces transcription of gene HMGCS1 in human melanoma cells. (A) Deletion map of the internal region of mVL30-1. Deletion fragments were generated from a 3.7-kb cDNA clone of the internal region of mVL30-1 by using an Erase-a-Base kit (Promega). The fragments were transfected into the human melanoma clone Te, which does not contain mVL30-1 RNA, and clones containing individual fragments were isolated. The size of the fragment in each clone was determined by agarose gel electrophoresis. (B) Effect of the mVL30-1 deletion fragments on transcription of gene HMGCS1 in clone Te. Transcription was assayed by RT-PCR by using primers that generate a 0.4-kb PCR fragment of HMGCS1. Lane A, clone Te; lane B, clone Th (contains retrovirally transmitted mVL30-1 RNA); lane C, Te clone transfected with full-length mVL30-1 cDNA. Lanes D–J, Te clones transfected with a mVL30-1 cDNA fragment, as follows: lane D, fragment 1; lane E, fragment 2; lane F, fragment 3; lane G, fragment 4; lane H, fragment 5; lane I, fragment 6; lane J, fragment 7. The RT-PCR assays were semiquantitative.
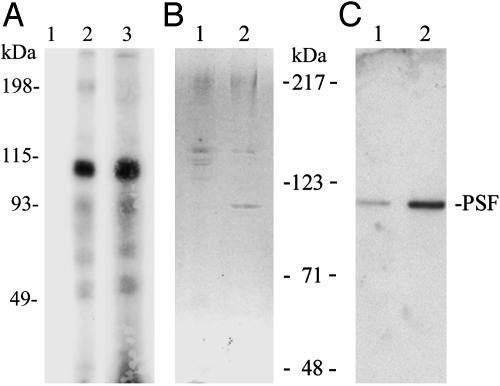
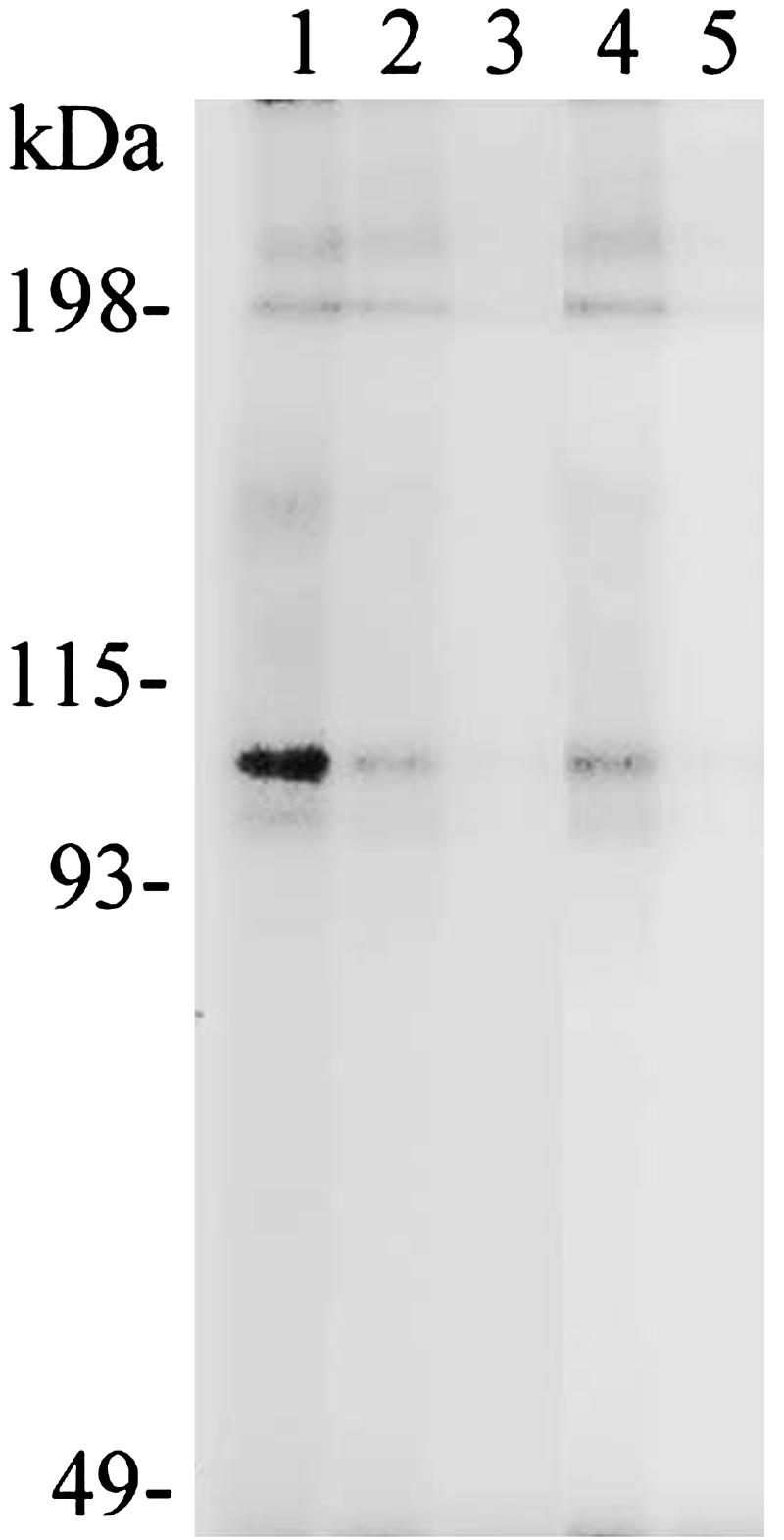
Complex Formation of mVL30-1 RNA with the Nuclear Protein PSF. A nuclear extract from melanoma clone Te was mixed with 32P-labeled mVL30-1 RNA from the entire internal region and also from fragment 4 (see Fig. 2A). Each sample was irradiated by UV and fractionated by SDS/PAGE, and the gel was autoradiographed. A strong band corresponding to a protein of ≈100 kDa was present in both samples (Fig. 3A). To identify the 100-kDa protein, the nuclear extract was first fractionated by affinity chromatography with mVL30-1 RNA attached to oligo(dT) beads, and the bound proteins were eluted first with 0.5M KCl and then with 1.0M KCl and analyzed by SDS/PAGE (Fig. 3B). The major protein band at the 100-kDa position was recovered from the gel and digested with 1% trypsin, and the tryptic peptides were purified by HPLC and analyzed by matrix-assisted laser desorption-ionization mass spectrometry and Edman sequencing., The analysis identified the protein as PSF, and the identification was confirmed by a Western assay using an anti-PSF antibody (Fig. 3C).
Fig. 3.
Binding of mVL30-1 RNA to PSF in a nuclear extract from the human melanoma clone Te. (A) 32P-labeled mVL30-1 RNA was added to the nuclear extract, and the mixture was irradiated with UV and fractionated by SDS/PAGE. The gel was autoradiographed. Lanes 1 and 2, internal region of mVL30-1 RNA without (lane 1) or with (lane 2) the nuclear extract; lane 3, fragment 4 of mVL30-1 RNA (see Fig. 2) with the nuclear extract. (B)Affinity chromatography of the nuclear extract on oligo(dT) beads containing immobilized mVL30-1. Lanes 1 and 2, the bound proteins were eluted first with 0.5M KCl (lane 1) and then with 1.0M KCl (lane 2), and the eluates were fractionated by SDS/PAGE (7.5%) and stained with Coomassie blue. The band at 100 kDa was eluted and analyzed by matrix-assisted laser desorption-ionization mass spectroscopy. (C) Western immunoassay of the gel from B, using an anti-human PSF antibody provided by J. Patton. The antibody was visualized with an ECL detection kit.
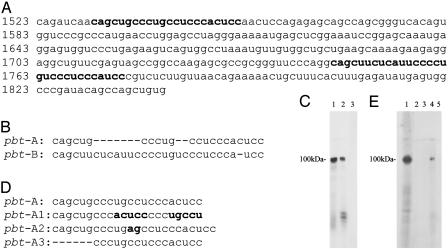
Mapping the Tracts in mVL30-1 RNA That Form a Complex with PSF. The 318-bp region overlapping fragments 4 and 7 in mVL30-1 RNA (Fig. 2) was digested to generate 6 fragments. Each fragment was 32P-labeled and tested for binding to proteins in a nuclear extract from the human melanoma clone Te. Only fragments from nucleotide 1523 to nucleotide 1626 and from nucleotide 1741 to nucleotide 1841 showed a major band at the 100-kDa position of the PSF protein. A sequence comparison of the two PSF-binding fragments identified a tract in each fragment, called pbt-A and pbt-B, that has significant sequence homology (Fig. 4 A and B). Each pbt was 32P-labeled and tested for binding to proteins in a nuclear extract from the human melanoma clone Te. Both pbt showed a major band at the position of the PSF protein (Fig. 4C), indicating formation of a complex with PSF. To test the importance of the sequence as well as composition of pbt-A for binding PSF, the sequence but not the composition was changed in pbt-A1 (Fig. 4D), which did not form a complex with PSF (Fig. 4E), demonstrating the importance of the pbt-A sequence for binding PSF. Other sequence changes that strongly inhibited complex formation with PSF were addition of two purines in pbt-A2 (Fig. 4 D and E) and deletion of the purine-rich sequence in pbt-A3 (Fig. 4 D and E).
Fig. 4.
Binding of pbt from VL30-1 RNA to PSF. (A) Sequence of the 318-bp region of mVL30-1 RNA (nucleotides 1523–1841) that binds to PSF. The pbt-A and pbt-B are shown in bold. (B) Sequence comparison of pbt-A and pbt-B. (C) Binding of pbt-A and pbt-B to PSF. 32P-labeled RNA from each tract was added to a nuclear extract from the human melanoma clone Te, and the mixture was irradiated with UV. The irradiated samples were fractionated by SDS/PAGE (7.5%), and the gel was autoradiographed. Lane 1, pbt-A RNA; lane 2, pbt-B RNA; lane 3, RNA fragment from nucleotides 1561–1681 that does not contain pbt-A or pbt-B sequences. (D) Mutations in pbt-A. The inversion in pbt-A1 and the insertion in pbt-A2 are shown in bold, and the deletions in pbt-A3 are shown as dashes. (E) Effect of mutations in the pbt-A on binding to PSF. The procedure is as described for C. Lane 1, pbt-A; lane 2, pbt-A1; lane 3, pbt-A2; lane 4, pbt-A3; lane 5, pbt-A without the nuclear extract.
Inhibition and Dissociation of PSF Binding to an IGFRE Motif. PSF represses the porcine gene P450scc, by binding to the IGFRE motif in the gene (5, 6). Our finding that mVL30-1 RNA induces transcription of the homologous human P450scc gene (Table 1) and forms a complex with PSF (Fig. 3A) suggests that mVL30-1 RNA might inhibit the binding of PSF to IGFRE. To test for such an inhibitory effect, 32P-labeled IGFRE DNA was mixed with a nuclear extract from the human melanoma clone Te to form a labeled complex with PSF, and pbt-A RNA was added to the mixture either before or after formation of the complex. The samples were irradiated by UV and fractionated by SDS/PAGE, and the gel was autoradiographed (Fig. 5). The 100-kDa band is missing from the samples in which pbt-A was added either before or after formation of the complex. The first result indicates that pbt-A competes with IGFRE for binding PSF, and that the complex formed with pbt-A excludes formation of a complex with IGFRE. The second result indicates that pbt-A RNA alone, or as part of mVL30-1 RNA, can release PSF bound to IGFRE. Most of the genes induced in human melanoma cells by mVL30-1 RNA (Table 1) also are induced by addition of IGF1 to the culture medium (data not shown), consistent with the proposed regulatory mechanism involving release of PSF from an IGFRE motif.
Fig. 5.
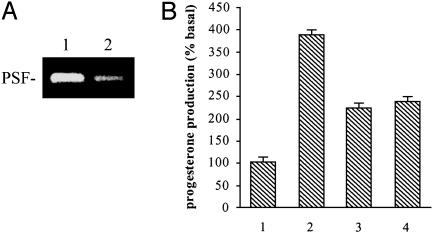
Inhibitory effect of pbt-A RNA on the binding of PSF to the IGFRE motif in gene P450scc. 32P-labeled IGFRE DNA was mixed with a nuclear extract from the human melanoma clone Te, the samples were irradiated with UV and fractionated by SDS/PAGE (7.5%), and the gel was autoradiographed. Lane 1, without added pbt-A RNA; lanes 2 and 3, with pbt-A RNA added at the same time as IGFRE DNA; lanes 4 and 5, with pbt-A RNA added 20 min after IGFRE DNA. The molar ratio of pbt-A RNA to IGFRE DNA was 5 in lanes 2 and 4 and 50 in lanes 3 and 5.
Regulation of Steroid Synthesis in Mice. Pituitary hormones induce steroid synthesis in steroidogenic cells of the ovary, testis, adrenal cortex, and placenta of the mouse (9, 10), and also in the mouse adrenal tumor line Y1 (11). A seminal finding was the concomitant induction of mVL30 RNA by pituitary hormones in mouse steroidogenic cells and Y1 cells (12, 13), suggesting a role for mVL30 RNA in regulating steroid synthesis. According to the proposed regulatory mechanism, mVL30 RNA can induce one or more genes in the steroidogenic pathway by forming a complex that releases PSF from the genes. To test this mechanism, a transient transfection procedure was used to deliver to Y1 cells a cDNA encoding pbt-A RNA to form a complex with PSF, or a siRNA to degrade PSF transcripts. Both transfections induced synthesis of progesterone, indicating activation of the steroidogenic pathway (Fig. 6).
Fig. 6.
Effect of pbt-A RNA and siRNA that degrades PSF transcripts on progesterone synthesis in Y1 cells. (A) Effect of an anti-PSF siRNA on PSF synthesis in Y1 cells. PSF transcription was determined by RT-PCR. Lane 1, Y1 cells without siRNA; lane 2, Y1 cells transiently transfected with a DNA encoding the siRNA. (B) Effect of pbt-A RNA and siRNA on progesterone synthesis in Y1 cells. Lane 1, Y1 cells transiently transfected with a control DNA encoding nucleotides spanning 1523–1841 in mVL30-1 from which the pbt-A and pbt-B sequences were deleted; lane 2, Y1 cells induced with 10 nM adrenocorticotropic hormone; lane 3, Y1 cells transiently transfected with a pbt-A region expression plasmid; lane 4, Y1 cells were transfected with a siRNA expression DNA fragment. The error bars show the mean value ± SE (n = 3).
Discussion
This study expands on an earlier finding that a retroviral vector used to transfect TF cDNA into a human melanoma line also transmitted mVL30-1 RNA from the retrovirual packaging cell line (2). The transfected melanoma cells that expressed high levels of TF protein and mVL30-1 RNA together, but not separately, showed a strong increase in metastatic potential. Here, we described a mechanism of gene regulation involving formation of a complex between mVL30 RNA and the regulatory protein PSF that could account for the effect of mVL30 RNA on metastasis, and also on the regulation of steroid synthesis.
PSF is a multifunctional protein, containing an RNA-binding domain (RBD) and a DNA-binding domain (DBD). The protein was originally identified as a component of spliceosomes (3) and later shown to function as a repressor of the IGF1-inducible gene P450ssc, which controls a rate-limiting step in steroid synthesis (5). Repression of P450scc involves binding of PSF to an IGFRE motif of the gene (5), and the complex formed with mVL30-1 RNA inhibits and dissociates the binding of PSF to the IGFRE (see Fig. 5), allowing transcription to proceed. Complex formation with PSF and the resulting regulation of gene transcription were mapped to two short pyrimidine-rich PSF-binding tracts in mVL30 RNAs, called pbt-A and pbt-B (see Figs. 4 A and B). Binding of the tracts to the RBD of PSF could generate an allosteric alteration that reduces the affinity of the DBD for the cognate DNA motif. The inhibitory effect will depend on the concentration of mVL30 RNA and its affinity for the DNA motif to which PSF binds. In mice, mVL30 RNA is transcribed from endogenous genomic retrotransposons and virtually all mouse cells contain mVL30 RNA, but the level varies among different cell types and different stages of development (1). Thus, the key parameters that determine whether a gene is repressed by PSF or induced by mVL30 RNA are the type of gene, the type of cell, and the stage of development.
The mouse genome contains a large ancient family of mVL30 retrotransposons that have undergone repeated cycles of transposition and mutation. The mutations introduced into mVL30 during its evolution from a progenitor retrovirus not only eliminated coding functions but also generated noncoding functions for mVL30 RNA. The pbt sequences are tightly conserved within the mVL30 family, particularly for pbt-A, and changing the sequence but not the composition of pbt-A inhibits binding to PSF (see Fig. 4 D and E), suggesting that the existing pbt sequences are optimal for binding to PSF.
There is compelling evidence that PSF and mVL30 RNA are involved in the regulation of steroid synthesis in mice. The steroid biosynthetic pathway is activated in steroidogenic tissues of the ovary, testis, adrenal cortex, placenta, and corpus luteum by pituitary hormones (9, 10), which concomitantly induce synthesis of mVL30 RNA in these tissues (12, 13). A key step in steroid synthesis is expression of P450scc, which is repressed by the binding of PSF to the IGFRE motif in the gene (5) and induced by IGF1 and mVL30 RNA. Other steroidogenic genes induced by IGF1 and mVL30 RNA include LDLR and HMGCS1 (see Table 1 and Fig. 1). We showed that steroid synthesis is activated in the mouse adrenal tumor line Y1 by transfection of a cDNA encoding pbt-A from mVL30-1 RNA or a siRNA that degrades the PSF transcript. These results suggest that the role of pituitary hormones in steroid synthesis in mice is indirect, involving induced synthesis of mVL30 RNA that forms a complex with PSF. The complex would mediate transcription of P450scc and other genes in the steroid biosynthetic pathway repressed by PSF. In addition to steroidogenic genes, mVL30 RNA induces the gene D4s234, which encodes a neuron-specific protein (14), and the gene COL18AI, which encodes type XVIII collagen required for normal eye development (15), suggesting that mVL30 RNA is involved in the regulation of other developmental pathways besides steroidogenesis. A seminal study of neurogenesis in the Drosophila eye showed that transcription of one or more members of the 17.6 family of retroviral-like retrotransposons was essential for the development of a normal pattern of neuronal connections between the eye and the brain (16). It seems that that transcriptionally active retrotransposons acquired essential developmental functions as early as the invertebrate stage of evolution.
The proposed regulatory role for endogenous mVL30 RNA in mouse steroidogenesis could also account for the effect of exogenous mVL30 RNA in promoting metastasis of human tumor cells (2). Most of the 11 genes induced by mVL30-1 RNA in human tumor cells (Table 1) also are induced by exposing the cells to IGF1, and a high serum level of IGF1 is associated with an increased risk for developing several types of human cancer (17). At least 3 of the 11 genes (GAGE6, LCN2, and PTK7) are oncogenic (18–20). Both mVL30 RNA and IGF1 induce genes that are repressed by PSF, suggesting that transmission of mVL30 RNA to human cells could have similar oncogenic effects as a high serum level of IGF1. Retroviral vectors used for gene transfer and gene therapy can transmit VL30 RNA to human cells, which synthesize, integrate, and transcribe mVL30 cDNA. These cells could be transformed by integration of mVL30 cDNA, resulting in activation of an oncogene or inactivation of a tumor-suppressor gene depending on the integration site, or by formation of a complex of mVL30 RNA with PSF, resulting in induction of one or more oncogenes repressed by PSF.
The PSF protein sequence is tightly conserved between mice and humans (21), suggesting that a regulatory mechanism involving human PSF and a human homolog of mVL30 also could be conserved. We have identified complete retrotransposon sequences in the human genome that are >90% identical to the mVL30 sequence (unpublished results), suggesting that human VL30 RNA could have regulatory roles analogous to those of mVL30 RNA in mice. An exciting future challenge is to elucidate the regulatory interactions involving human VL30 RNA and PSF.
Acknowledgments
We thank Dr. James Patton for providing the anti-PSF antibody, Dr. Randall Urban for providing an expression plasmid encoding the PSF protein, and Dr. Cecilia Guerrier-Takada for providing the plasmid pmU6(-315/1)pBSKAmp. The research reported here was supported in part by Program Project Grant HL29019 from the National Institutes of Health and by generous gifts from private donors.
Abbreviations: mVL30, mouse VL30; TF, tissue factor; IGF1, insulin-like growth factor 1; IGFRE, insulin-like growth factor response element; oligo(dT), oligo-deoxythymidine; siRNA, small interfering RNA.
References
- 1.French, N. S. & Norton, J. D. (1997) Biochim. Biophys. Acta 1352, 33–47. [DOI] [PubMed] [Google Scholar]
- 2.Song, X., Wang, B., Bromberg, M., Hu, Z., Konigsberg, W. & Garen A. (2002) Proc. Natl. Acad. Sci. USA 99, 6269–6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patton, J. G., Porro, E. B., Galceran, J., Tempst, P. & Nadal-Ginard, B. (1993) Genes Dev. 7, 393–406. [DOI] [PubMed] [Google Scholar]
- 4.Peng, R., Dye, B. T., Perez, I., Barnard, D. C., Thompson, A. B. & Patton, J. G. (2002) RNA 8, 1334–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Urban, R. J., Bodenburg, Y., Kurosky, A., Wood, T. G. & Gasic, S. (2000) Mol. Endocrinol. 14, 774–782. [DOI] [PubMed] [Google Scholar]
- 6.Shav-Tal, Y. & Zipori, D. (2002) FEBS Lett. 531, 109–114. [DOI] [PubMed] [Google Scholar]
- 7.Das, G., Henning, D., Wright, D. & Reddy, R. (1988) EMBO J. 7, 503–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gou, D., Jin, N. & Liu, L. (2003) FEBS Lett. 548, 113–118. [DOI] [PubMed] [Google Scholar]
- 9.Waterman, M. R., Mason, J. I. & Simpson, E. R. (1988) Prog. Clin. Biol. Res. 274, 543–555. [PubMed] [Google Scholar]
- 10.Stocco, D. M. & Clark, B. J. (1996) Endocr. Rev. 17, 221–244. [DOI] [PubMed] [Google Scholar]
- 11.Yasumura, Y., Buonassisi, V. & Sato, G. (1966) Cancer Res. 26, 529–535. [PubMed] [Google Scholar]
- 12.Schiff, R., Itin, A. & Keshet, E. (1991) Genes Dev. 5, 521–532. [DOI] [PubMed] [Google Scholar]
- 13.Bohm, S., Bakke, M., Nilsson, M., Zanger, U. M., Spyrou, G. & Lund, J. (1993) J. Biol. Chem. 268, 3952–3963. [PubMed] [Google Scholar]
- 14.Carlock, L., Vo, T., Lorincz, M., Walker, P. D., Bessert, D., Wisniewski, D. & Dunbar, J. C. (1996) Brain Res. Mol. Brain 42, 202–212. [DOI] [PubMed] [Google Scholar]
- 15.Sertie, A. L., Sossi, V., Camargo, A. A., Zatz, M., Brahe, C. & Passos-Bueno, M. R. (2000) Hum. Mol. Genet. 9, 2051–2058. [DOI] [PubMed] [Google Scholar]
- 16.Mozer, B. A. & Benzer, S. (1994) Development 120, 1049–1058. [DOI] [PubMed] [Google Scholar]
- 17.Furstenberger, G. & Senn, H. J. (2002) Lancet Oncol. 3, 298–302. [DOI] [PubMed] [Google Scholar]
- 18.Van den Eynde, B., Peeters, O., De Backer, O., Gaugler, B., Lucas, S. & Boon, T. (1995) J. Exp. Med. 182, 689–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stoesz, S. P., Friedl, A., Haag, J. D., Lindstrom, M. J., Clark, G. M. & Gould, M. N. (1998) Int. J. Cancer 79, 565–572. [DOI] [PubMed] [Google Scholar]
- 20.Easty, D. J., Mitchell, P. J., Patel, K., Florenes, V. A., Spritz, R. A. & Bennett, D. C. (1997) Int. J. Cancer 71, 1061–1065. [DOI] [PubMed] [Google Scholar]
- 21.Shav-Tal, Y., Cohen, M., Lapter, S., Dye, B., Patton, J. G., Vandekerckhove, J. & Zipori, D. (2001) Mol. Biol. Cell 12, 2328–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]