Abstract
Background
The role of n-3 fatty acids in prevention of breast cancer is well recognized, but the underlying molecular mechanisms are still unclear. In view of the growing need for early detection of breast cancer, Graham et al. (2010) studied the microarray gene expression in histologically normal epithelium of subjects with or without breast cancer. We conducted a secondary analysis of this dataset with a focus on the genes (n = 47) involved in fat and lipid metabolism. We used stepwise multivariate logistic regression analyses, volcano plots and false discovery rates for association analyses. We also conducted meta-analyses of other microarray studies using random effects models for three outcomes--risk of breast cancer (380 breast cancer patients and 240 normal subjects), risk of metastasis (430 metastatic compared to 1104 non-metastatic breast cancers) and risk of recurrence (484 recurring versus 890 non-recurring breast cancers).
Results
The HADHA gene [hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), alpha subunit] was significantly under-expressed in breast cancer; more so in those with estrogen receptor-negative status. Our meta-analysis showed an 18.4%-26% reduction in HADHA expression in breast cancer. Also, there was an inconclusive but consistent under-expression of HADHA in subjects with metastatic and recurring breast cancers.
Conclusions
Involvement of mitochondria and the mitochondrial trifunctional protein (encoded by HADHA gene) in breast carcinogenesis is known. Our results lend additional support to the possibility of this involvement. Further, our results suggest that targeted subset analysis of large genome-based datasets can provide interesting association signals.
Keywords: Breast cancer, HADHA gene, Gene expression profiling, Microarray, Meta-analysis
Background
Early detection of malignant breast neoplasms is critical to cancer prevention and treatment. Cancer chemoprevention (also called as treatment of carcinogenesis) is a primordial prevention step that is receiving considerable attention. In that context, the identification of an ideal biomarker for breast cancer has become increasingly important. In spite of the vast number of studies conducted in the past; a recent, comprehensive and elegant review argues that there is still a lack of clarity regarding the understanding of the process of breast carcinogenesis [1]. Interestingly, it has been demonstrated that the mammary gland basal cells have features consistent with the progenitor stem cells and that they can differentiate into benign or malignant lesions intraductally [2]. It has also been shown in murine models that differentiated intact mammary glands can exert a negative influence on the development of breast cancer [3]. However, the search for an ideal breast cancer biomarker is still on [4].
A logical undertaking in this direction is the use of microarrays to study the differential gene expressions in breast cancer. Consistent with the spirit of research that encourages very early detection of carcinogenesis, Graham et al. [5] recently studied histologically normal epithelium from subjects with and without breast cancer and identified an 86-gene signature that indicated a genomic change prior to carcinogenesis. They found that many of these genes belonged to the family of growth factors, cytokines, oxidative stress modifiers, p38 MAP kinase pathway members, transcription regulators or determinants of nucleic acid stability [5].
Interestingly they did not find genes associated with fatty acid or lipid metabolism to be differentially expressed in histologically normal epithelium. Derangements of fatty acid and lipid metabolism have been implicated in oncogenesis in many studies, especially in the cancer of breast [6]. It is generally believed that diets rich in n-6 polyunsaturated fatty acids (n-6 PUFA) and saturated fatty acids (SFA) increase the risk of tumorigenesis while diets rich in n-3 polyunsaturated fatty acids (n-3 PUFA) reduce the risk of cancer development [7-10]. Lipids have the ability to influence the process of neoplasia via their effects on hormone status, cell membrane integrity, signal transduction, immune modulation and regulation of gene expression [11,12]. In this study, we specifically examined whether the genes related to fatty acid and lipid metabolism are also differentially associated with breast cancer status. For this, we used a targeted subset analysis of the microarray data from Graham et al. [5] and also conducted meta-analyses of other microarray datasets.
Methods
The primary dataset
The microarray dataset used in the present study is available for public use on the Gene Expression Omnibus website http://www.ncbi.nlm.nih.gov/sites/GDSbrowser?acc=GDS3716 of the National Institutes of Health, USA. Details of the study subjects on whom these microarray studies were conducted have been described previously [5]. Briefly, the dataset comprises microarray data collected through Affymetrix Human Genome U133A platform that measures expression of 22,283 genes. The data were collected using histologically normal epithelium from four sets of subjects--those who underwent reduction mammoplasty (n = 18), those who underwent preventive mastectomy (n = 6), estrogen receptor positive (ER+) breast cancer patients (n = 9) and estrogen receptor negative (ER-) breast cancer patients (n = 9). The data were available in normalized format.
Targeted subset analysis
Our main aim was to assess if genes related with fatty acid and lipid metabolism were differentially expressed in the study dataset. For this we first culled a list of genes that have been implicated in the fatty acid and lipid metabolism. We used the DAVID http://david.abcc.ncifcrf.gov and KEGG Pathway http://www.genome.jp/kegg/metabolism.html websites and generated a list of 136 genes implicated in one or more of the following pathways: fatty acid metabolism, fatty acid biosynthesis, peroxisome proliferator-activated receptor (PPAR) signaling pathway, lipopolysaccharide biosynthesis, lipid metabolism and fat digestion and absorption. A full list with functional annotation of these 136 genes is provided as Additional file 1: Table S1. We then used the DAVID http://david.abcc.ncifcrf.gov and Clone/Gene ID Converter http://idconverter.bioinfo.cnio.es/IDconverter.php programs to find out which of these 136 genes were included in the Affymetrix Human Genome U133A platform. We found 47 probe sets related to genes (Table 1) that partake in lipid or fatty acid metabolism to be represented in the study datasets. We conducted our analyses on the potential differential expression of these 47 genes. Complete functional annotation for these 47 genes is provided in Additional file 2: Table S2.
Table 1.
Genes included in the analyses
| # | Symbol | Affymetrix Probe Set Id | Gene Name |
|---|---|---|---|
| 1 | ACAA1 | 202025_x_at | acetyl-Coenzyme A acyltransferase 1 |
| 2 | ACADL | 206068_s_at | acyl-Coenzyme A dehydrogenase, long chain |
| 3 | ACADM | 202502_at | acyl-Coenzyme A dehydrogenase, C-4 to C-12 straight chain |
| 4 | ACAT1 | 205412_at | acetyl-Coenzyme A acetyltransferase 1 |
| 5 | ACSBG2 | 221716_s_at | acyl-CoA synthetase bubblegum family member 2 |
| 6 | ACSL3 | 201660_at | acyl-CoA synthetase long-chain family member 3 |
| 7 | ACSL4 | 202422_s_at | acyl-CoA synthetase long-chain family member 4 |
| 8 | ACSL5 | 218322_s_at | acyl-CoA synthetase long-chain family member 5 |
| 9 | ADH1A | 207820_at | alcohol dehydrogenase 1B (class I), beta polypeptide; alcohol dehydrogenase 1A (class I), alpha polypeptide; alcohol dehydrogenase 1C (class I), gamma polypeptide |
| 10 | ADH6 | 207544_s_at | alcohol dehydrogenase 6 (class V) |
| 11 | ADIPOQ | 207175_at | adiponectin, C1Q and collagen domain containing |
| 12 | AGPAT2 | 210678_s_at | 1-acylglycerol-3-phosphate O-acyltransferase 2 (lysophosphatidic acid acyltransferase, beta) |
| 13 | ANGPTL4 | 221009_s_at | angiopoietin-like 4 |
| 14 | APOA4 | 206894_at | apolipoprotein A-IV |
| 15 | APOC3 | 205820_s_at | apolipoprotein C-III |
| 16 | AQP7 | 206955_at | aquaporin 7 |
| 17 | ARSA | 204443_at | arylsulfatase A |
| 18 | ASAH1 | 210980_s_at | N-acylsphingosine amidohydrolase (acid ceramidase) 1 |
| 19 | CERK | 218421_at | ceramide kinase |
| 20 | CETP | 206210_s_at | cholesteryl ester transfer protein, plasma |
| 21 | CYP7A1 | 207406_at | cytochrome P450, family 7, subfamily A, polypeptide 1 |
| 22 | DGAT1 | 202344_at | diacylglycerol O-acyltransferase homolog 1 (mouse) |
| 23 | EHHADH | 205222_at | enoyl-Coenzyme A, hydratase/3-hydroxyacyl Coenzyme A dehydrogenase |
| 24 | FABP2 | 207475_at | fatty acid binding protein 2, intestinal |
| 25 | FUT2 | 208505_s_at | fucosyltransferase 2 (secretor status included) |
| 26 | FUT4 | 209892_at | fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific) |
| 27 | FUT5 | 210398_x_at | fucosyltransferase 5 (alpha (1,3) fucosyltransferase) |
| 28 | FUT9 | 207696_at | fucosyltransferase 9 (alpha (1,3) fucosyltransferase) |
| 29 | GCDH | 203500_at | glutaryl-Coenzyme A dehydrogenase |
| 30 | GK2 | 215430_at | glycerol kinase 2 |
| 31 | GLA | 214430_at | galactosidase, alpha |
| 32 | HADHA | 208629_s_at | hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), alpha subunit |
| 33 | LTA | 206975_at | lymphotoxin alpha (TNF superfamily, member 1) |
| 34 | MTTP | 205675_at | microsomal triglyceride transfer protein |
| 35 | NPC1L1 | 220106_at | NPC1 (Niemann-Pick disease, type C1, gene)-like 1 |
| 36 | NR1H3 | 203920_at | nuclear receptor subfamily 1, group H, member 3 |
| 37 | OLR1 | 210004_at | oxidized low density lipoprotein (lectin-like) receptor 1 |
| 38 | PCK2 | 202847_at | phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| 39 | PLTP | 200661_at | phospholipid transfer protein |
| 40 | PPARG | 208510_s_at | peroxisome proliferator-activated receptor gamma |
| 41 | RXRB | 209148_at | retinoid × receptor, beta |
| 42 | RXRG | 205954_at | retinoid × receptor, gamma |
| 43 | SGMS1 | 212989_at | sphingomyelin synthase 1 |
| 44 | ST8SIA1 | 210073_at | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 |
| 45 | UCP1 | 221384_at | uncoupling protein 1 (mitochondrial, proton carrier) |
| 46 | UCP3 | 207349_s_at | uncoupling protein 3 (mitochondrial, proton carrier) |
| 47 | UGCG | 204881_s_at | UDP-glucose ceramide glucosyltransferase |
Replication of the results: meta-analyses
We also aimed to ensure that the results obtained from one microarray dataset were robust and could be replicated in other datasets. We queried the Oncomine database and retrieved microarray data from other relevant studies. We studied the association of gene expression with three outcomes--risk of breast cancer, risk of metastasis and risk of recurrence. We then combined these datasets meta-analytically using the random effects model of DerSimonian and Laird [13,14]. For these analyses the effect size was measured and expressed as the standardized mean difference (SMD) and its 95% confidence intervals. The Oncomine website reports the results as means, medians, quartiles and minimum and maximum values. Since the random-effects model assumes normal distribution of the effect measures, we first estimated the mean and standard error for each group (for example, for subjects with breast cancer; subjects with a metastatic event or subjects with recurring breast cancer) using the method described by Hozo et al. [15] We then estimated the SMDs. To depict the potential variability in the HADHA expression based on the probes used by individual studies, we conducted the meta-analyses separately for each combination of the study and the probe used. Each comparison represented a specific combination of the included study and the reporter used in the study. The between-study heterogeneity in this meta-analysis was examined using the I2 statistic. Since expression data on all individual subjects was available for the outcome of risk of breast cancer, we also conducted individual patient data (IPD) meta-analysis [16]. For this we used the clustered unconditional logistic regression analyses [16,17] with disease status as a dichotomous dependent variable, comparison-specific z-scores as the predictor variable and comparison indicator as the clustering variable. Comparison-specific z-scores were estimated as the relative deviates (mean expression/standard deviation of expression) within each comparison group.
Other statistical analysis
To quantify and test differential gene expression, we used two-tailed Student's t tests for unpaired samples. The clinical and statistical significance of the findings were presented as volcano plots. To account for multiple testing, we estimated the false discovery rates (q values) using the QVALITY software program [18]. Discriminant utility of each gene was assessed using non-parametric receiver operating characteristic (ROC) curve analysis. To group subjects based on their HADHA expression, we used a k-means clustering approach. All statistical analyses were conducted using Stata 10.0 software package (Stata Corp, College Station, Texas). We aimed for a type I error rate of 0.05 and a false discovery rate of 0.15.
Results
Differential expression analyses
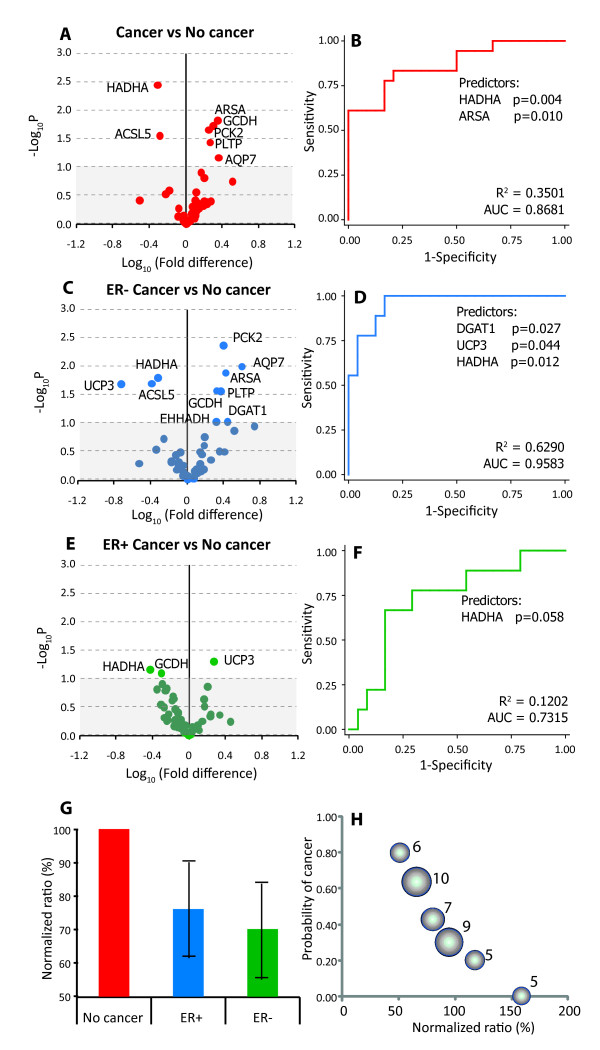
Using the shortlisted set of 47 genes shown in Table 1, we first determined if these genes were differentially expressed in subjects with cancer (n = 18) and those without (n = 24). The volcano plot (Figure 1A) showed that seven of the 47 genes were significantly differentially expressed between these study groups. These genes included five over-expressed genes (AQP7, PLTP, PCK2, GCDH and ARSA) and two under-expressed genes (ACSL5 and HADHA). Of these, HADHA was the most significant statistically. To account for the possible covariance among these gene expression values we conducted stepwise multivariate analyses using unconditional logistic regression and observed that only two genes--HADHA and ARSA--were retained in the final model (Figure 1B). This model explained 35% of inter-individual variability in breast cancer susceptibility with a predictive accuracy of 86.8%. Interestingly, when HADHA expression was removed from this model the ARSA lost its statistical significance but removal of ARSA did not affect the statistical significance of HADHA. This indicates that HADHA gene expression was the most important statistical predictor of altered risk of breast cancer.
Figure 1.
Association of fatty acid and lipid metabolism related genes with the risk of breast cancer. (A-F) Association analyses. Panels A, C and E show the volcano plots for cancer with no cancer, ER-versus no cancer and ER + versus no cancer comparisons, respectively. These plots depict the biological significance (log-fold change) on the X-axis and the statistical significance (-log P) on the Y-axis. Significance values above 0.1 are indicated by the grey shaded area in the volcano plots. Panels, B, D and F show the corresponding receiver-operating characteristic (ROC) curves for the final models from stepwise logistic regression analyses. The genes retained in the final model and their statistical significance is shown under the ROC curves, the variance explained by the model is shown as R2 and the predictive accuracy is indicated by the area under the ROC curve (AUC). (G) Comparative expression of the HADHA gene in the indicated study groups. Error bars indicate 95% confidence intervals. (H) Bubble plot showing the dose-response relationship between HADHA expression and the risk of breast cancer. Each bubble represents one of the six clusters generated using k-means clustering algorithm based on the HADHA expression. The radius of the bubble is proportional to the number of subjects in that cluster (indicated by numbers next to the bubbles).
Does ER status influence the expression of HADHA?
To examine if this association could be influenced by the ER status, we conducted three sets of analyses. First, we studied whether HADHA expression was different based on the ER status. We found that the mean HADHA was not significantly differentially expressed by ER status (mean HADHA expression in subjects with ER + breast cancer = 6.00; in subjects with ER-breast cancer = 5.90; p = 0.462). Second, we adjusted the standard error estimates for the ER status using clustered logistic regression and observed that the statistical significance for the HADHA gene expression further increased (p = 0.0001) while that of the ARSA gene decreased (p = 0.082) indicating that the association of HADHA was unlikely to have been influenced by the ER status. Third, we constructed volcano plots and conducted stepwise logistic regression analyses by comparing the ER + and ER-subjects separately with subjects without cancer as the reference group. We observed (Figure 1C-F) that HADHA gene expression was the only consistent predictor across ER status but more so in the ER-subjects. Indeed, the q value for the HADHA gene was 0.15 for the cancer versus no cancer comparison, 0.13 for the ER-versus no cancer comparison but 0.88 for the ER + versus no cancer comparison. Two other genes (UCP3 and DGAT1) were retained in the final model of stepwise regression analyses when ER-subjects were compared to the no cancer group however this association was not observed when ER + subjects were compared to the same reference group.
Graded risk of breast cancer based on HADHA expression
We next considered whether the association of HADHA gene expression with risk of breast cancer exhibited a threshold effect or whether it was a graded dose-response. For this, we used two approaches. First, we normalized the gene expression in the no cancer group to 100%. We found (Figure 1G) that the HADHA expression had fallen to 73% (95% CI 64%-83%) in subjects with cancer; with a higher expression in ER + subjects (76% of the no cancer group, 95% CI 61%-91%) than in ER-subjects (70% of the no cancer group, 95% CI 55%-85%). Second, the k-means clusters (which explained 95.9% of the variability in HADHA expression) clearly demonstrated a dose-response association (Figure 1H) such that more severe down-regulation of HADHA was associated with a greater risk of being in the breast cancer group.
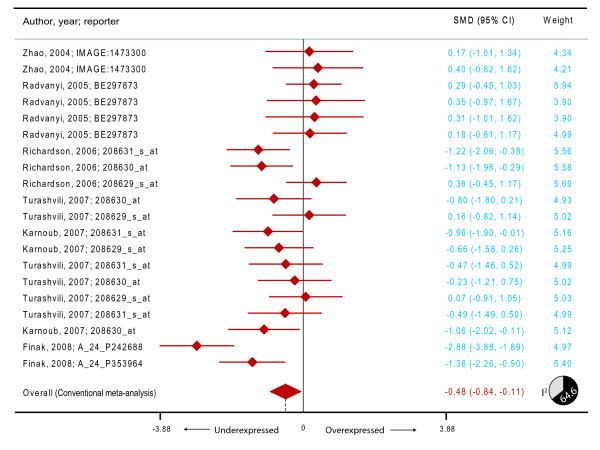
Meta-analyses of the differential expression of HADHA
Lastly, we examined the robustness of the differential expression of HADHA by conducting meta-analysis of published microarray studies comparing cases of breast cancer with subjects without breast cancer. Querying the Oncomine database, we found six studies [19-24] that represented 20 different comparisons of breast cancer patients with normal subjects (Table 2). The reasons for this larger number of comparisons were the different reporters used in the microarray experiments as well as the different subtypes of breast cancer reported by the studies.
Table 2.
Comparisons included in the meta-analysis of differential HADHA expression
| No | Author, Year | Ref | Controls | Cases | Breast cancer histology | Reporter |
|---|---|---|---|---|---|---|
| 1 | Zhao, 2004 | [24] | 3 | 37 | Invasive ductal carcinoma | IMAGE:1473300 |
| 2 | Zhao, 2004 | [24] | 3 | 21 | Lobular carcinoma | IMAGE:1473300 |
| 3 | Radvanyi, 2005 | [21] | 9 | 7 | Invasive lobular carcinoma | BE297873 |
| 4 | Radvanyi, 2005 | [21] | 9 | 32 | Invasive ductal carcinoma | BE297873 |
| 5 | Radvanyi, 2005 | [21] | 9 | 3 | Invasive mixed carcinoma | BE297873 |
| 6 | Radvanyi, 2005 | [21] | 9 | 3 | Ductal carcinoma in situ | BE297873 |
| 7 | Richardson, 2006 | [22] | 7 | 40 | Ductal carcinoma | 208629_s_at |
| 8 | Richardson, 2006 | [22] | 7 | 40 | Ductal carcinoma | 208630_at |
| 9 | Richardson, 2006 | [22] | 7 | 40 | Ductal carcinoma | 208631_s_at |
| 10 | Karnoub, 2007 | [20] | 15 | 7 | Invasive ductal carcinoma | 208629_s_at |
| 11 | Karnoub, 2007 | [20] | 15 | 7 | Invasive ductal carcinoma | 208630_at |
| 12 | Karnoub, 2007 | [20] | 15 | 7 | Invasive ductal carcinoma | 208631_s_at |
| 13 | Turashvili, 2007 | [23] | 20 | 5 | Invasive ductal carcinoma | 208629_s_at |
| 14 | Turashvili, 2007 | [23] | 20 | 5 | Invasive ductal carcinoma | 208629_s_at |
| 15 | Turashvili, 2007 | [23] | 20 | 5 | Invasive ductal carcinoma | 208630_at |
| 16 | Turashvili, 2007 | [23] | 20 | 5 | Invasive lobular carcinoma | 208630_at |
| 17 | Turashvili, 2007 | [23] | 20 | 5 | Invasive lobular carcinoma | 208631_s_at |
| 18 | Turashvili, 2007 | [23] | 20 | 5 | Invasive lobular carcinoma | 208631_s_at |
| 19 | Finak, 2008 | [19] | 6 | 53 | Invasive breast carcinoma | A_24_P242688 |
| 20 | Finak, 2008 | [19] | 6 | 53 | Invasive breast carcinoma | A_24_P353964 |
We first observed that the mean expression levels for HADHA probes (expressed as log transformed values) were widely different across the six studies (Zhao et al. [24]:-0.33, Radvanyi et al. [21]: 3.08, Richardson et al. [22]: 5.29, Karnoub et al. [20]: 2.97, Turashvili et al. [23]: 3.92 and Finak et al. [19]:-2.65). We therefore transformed these values into comparison-specific z-scores (mean expression for a comparison/standard deviation of expression for that comparison). Upon this z-transformation, all the studies had a mean z-score of 0 and a standard deviation of 1. We conducted meta-analyses on the HADHA expression z-scores. Using the DerSimonian and Laird model, we observed (Figure 2) that the summary SMD (filled diamond in Figure 2) was-0.48 (95% CI-0.84--0.11). Considering the statistical properties of SMD it is possible to transform this into probability [25]. This transformation indicated that there was an average 18.4% reduction in expression of HADHA (95% CI 4.5%-30.0%) in cases of breast cancer as compared to normal subjects. Interestingly, this significant reduction in the expression of HADHA was observed in spite of the high degree of heterogeneity (I2 64.6, p < 0.001, pie-chart in Figure 2) between the comparisons due to different cancer subtypes, reporters used in various studies and other study characteristics.
Figure 2.
Meta-analysis of the differential expression of the HADHA gene in breast cancer compared to subjects without breast cancer. The figure shows a forest plot with filled diamonds indicating the point estimates and error bars indicating the 95% confidence interval around the standardized mean difference. The summary effect measure is shown as a filled diamond whose center (dashed vertical line) indicates the point estimate and the width indicates the 95% confidence interval.
We observed that the invasive ductal carcinoma (p = 0.046) and unspecified invasive breast carcinoma (p = 0.005) showed a significant under-expression of HADHA gene but lobular carcinoma (p = 0.781), invasive lobular carcinoma (p = 0.780) or invasive mixed carcinoma (p = 0.717) did not show a significant alteration of HADHA gene expression. Alternatively, we conducted the IPD meta-analysis using logistic regression analyses. We found that the odds ratio for breast cancer was 0.74 (95% CI 0.60-0.92) after clustered analyses. Thus, there was a 26% reduction in the risk of breast cancer per unit increase in z-scores. These values show a striking resemblance with the findings observed in the Graham et al. dataset and demonstrate the replicability of our findings.
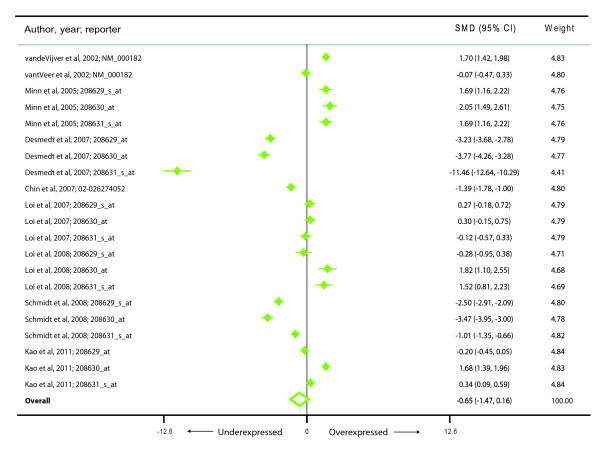
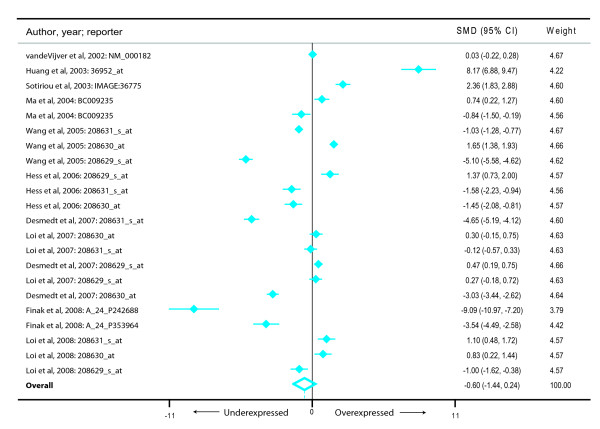
We also investigated if HADHA expression was associated with an altered risk of metastasis and recurrence. For risk of a metastatic event we found nine studies [26-33] representing 430 metastatic events and 1104 metastasis-free cancers (Figure 3). Subjects who developed a metastatic event during follow-up had a reduced HADHA expression (summary effect size-0.65, 95% CI-1.47-0.16) but this was not statistically significant (p = 0.117). Also, there was a very high degree of between-study heterogeneity (I2 98.8%). Similarly, for the outcome of the risk of recurrence (Figure 4), we found that there were 10 studies [19,27,29,30,32,34-38] representing 484 recurring and 890 non-recurring breast cancers. Meta-analysis demonstrated that although there was a consistent decrease in average HADHA expression in patients with a recurring form of breast cancer (summary effect size-0.60, 95% CI-1.44-0.24), the finding was neither statistically significant (p = 0.160) nor homogeneous (I2 = 98.7%) across studies.
Figure 3.
Meta-analysis of the differential expression of the HADHA gene in breast cancer patients with and without metastatic events. The figure shows a forest plot with filled diamonds indicating the point estimates and error bars indicating the 95% confidence interval around the standardized mean difference. The summary effect measure is shown as a hollow diamond whose center (dashed vertical line) indicates the point estimate and the width indicates the 95% confidence interval.
Figure 4.
Meta-analysis of the differential expression of the HADHA gene in breast cancer patients with and without recurrence events. The figure shows a forest plot with filled diamonds indicating the point estimates and error bars indicating the 95% confidence interval around the standardized mean difference. The summary effect measure is shown as a hollow diamond whose center (dashed vertical line) indicates the point estimate and the width indicates the 95% confidence interval.
Discussion
Our analyses of the microarray dataset based on the Graham et al. [5] study demonstrated a consistent, strong and significant association of the HADHA gene expression in histologically normal epithelium with the likelihood of breast cancer. Moreover, this observation was further substantiated by the meta-analysis of other published studies. Only one study has previously reported differential association of this gene with regard to BRCA1 positive, BRCA2 positive and sporadic malignant tumors of the breast [39]. Our results further support the putative involvement of HADHA in breast cancer susceptibility.
Biological plausibility
Biological significance of our novel observations should be considered in the light of the following facts. First, the HADHA gene (chromosomal location 2p23) codes for the four alpha chains in the 8-meric mitochondrial trifunctional protein (TFP) [40]. This enzyme performs three cardinal functions in the β-oxidation of long chain fatty acids by catalyzing the activities of the 2-enoyl-CoA hydratase (ECH), L-3-hydroxyacyl-CoA dehydrogenase (HACD) and 3-ketoacyl-CoA thiolase (KACT). Of these three, the first two enzymes (ECH and HACD) are specifically catalyzed by the alpha chains of TFP. Severe deficiency (< 50% of normal activity) of TFP is known to be associated with life-threatening manifestation of the long chain 3-hydroxyacyl-CoA dehydrogenase deficiency [41]. However, the effects of a milder deficiency of TFP (for example, activity between 50%-80% of the normal) are currently unknown. Our results indicate that breast cancer patients had 18-30% decreased expression of HADHA gene. We therefore hypothesize that there may be a compromised metabolism of long chain fatty acids in breast cancer due to a relative deficiency of the alpha chains of TFP. In this context, it is noteworthy that a recent large genome-wide association study [42] found a strong association of breast cancer with a polymorphism in the gene encoding enoyl CoA hydratase domain containing 1 (ECHDC1), which also partakes in the integrity of the TFP.
Second, the efficacy of β-oxidation of n-3 and n-6 long chain fatty acids can be tissue- and location- specific. For example, in rat livers it has been shown that the n-3/n-6 ratio influences peroxisomal but not mitochondrial β-oxidation [43]. In contrast, mitochondrial β-oxidation of long chain fatty acid has been implicated in breast cancer pathogenesis [42]. We also could not demonstrate a significant association of the genes involved in the PPAR-γ pathway reinforcing the possibility that mitochondrial rather than peroxisomal β-oxidation of long chain fatty acids may be more critical in breast carcinogenesis. Third, HADHA occupies an important position in the network of genes that have been implicated in autophagy and apoptosis [44]. Finally, triangulation of the following facts lends additional credence to our observations: i) intact epithelium of mammary glands has the ability to act as stem cells for carcinogenesis [2]; ii) n-3 long chain fatty acids have the ability to target such stem cells [45]; and iii) HADHA is involved in the mitochondrial β-oxidation of long chain fatty acids. Together these observations from published literature strongly support the biological plausibility of our finding that HADHA is differentially expressed in subjects with and without breast cancer.
Limitations
Our study has all the limitations implicit in any microarray association study and meta-analyses. In addition, there are three more limitations. First, although there is a strong circumstantial evidence that favors an inference of HADHA expression-breast cancer association, it must be realized that robust functional studies are required before this association can be conclusively claimed. Our study does not have a component of functional assays that can help put these results in a biological perspective. Second, due to limitations imposed by the microarray platform used in the primary study, we could not evaluate the potential association of a large number of additional lipid and fat metabolism related genes with the risk of breast cancer. Inclusion of those genes may not only affect the q values associated with HADHA but also may provide a more comprehensive understanding of the role of fatty acids in breast cancer. Thirdly, although consistent, the observed differential expression of HADHA with cancer progression (as reflected by risk of metastasis and recurrence) is statistically inconclusive.
Conclusions
Our study has three important implications--biological, methodological and epidemiologic. Biologically, our study has identified a novel target gene that corroborates the existing knowledge about the role of long chain fatty acids in breast cancer and provides interesting directions for further research in this area. Also, our findings put the focus on the putative functional aspects of mitochondria and TFP in breast carcinogenesis.
From a methodological standpoint, our study shows that high dimensionality of omics-type datasets is fraught with the vexing problem of finding strong associations at the cost of potentially missing weaker but biologically meaningful associations. Literature addressing the issue of multiple comparisons in large volume datasets focuses primarily on the possibility of finding false positive associations [46]. However, there exists a demonstrable probability that such high-volume datasets may also falsely mask true associations. It is likely that the Graham et al. study did not report a significant association of HADHA with the risk of breast cancer due to a large number of multiple comparisons. The fact that we discovered an association of HADHA with breast cancer shows that microarray dataset analysis (as well as analyses of other large datasets like genome-wide association studies, proteomics data or metabolomics datasets) may benefit by using targeted subset analyses based on functional annotation and conceptual understanding of the molecular mechanisms in disease. Finally, in an epidemiological context, our study shows that error in long chain fatty acid metabolism in the breast tissue might herald the onset of carcinogenesis and thus can be helpful for the primordial prevention of breast cancer.
Abbreviations
ER: Estrogen receptor; n-3 PUFA: Omega--3 polyunsaturated fatty acids; n-6 PUFA: Omega--6 polyunsaturated fatty acids; SFA: Saturated fatty acids; SMD: Standardized mean difference; CI: Confidence interval.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
MM and HK conceptualized the study, collected data, conducted analyses and wrote the manuscript. Both authors have read and approve the manuscript.
Supplementary Material
Table S1. Excel table containing detailed annotation of the 136 genes related to fat and lipid metabolism that were primarily selected for analyses.
Table S2. Excel table containing detailed annotation of the 47 genes included in this study related to fat and lipid metabolism that were primarily selected for analyses.
Contributor Information
Manju Mamtani, Email: manjuhemant@gmail.com.
Hemant Kulkarni, Email: hemant_kulkarnius@yahoo.com.
Acknowledgements
We sincerely acknowledge and deeply appreciate the Editorial assistance provided by Cindy Tumiel, Department of Genetics, Texas Biomedical Research Institute, San Antonio, Texas, USA.
References
- Bonovas S, Tsantes A, Drosos T, Sitaras NM. Cancer chemoprevention: a summary of the current evidence. Anticancer Res. 2008;28(3B):1857–1866. [PubMed] [Google Scholar]
- Moriya T, Kanomata N, Kozuka Y, Hirakawa H, Kimijima I, Kimura M, Watanabe M, Sasano H, Ishida T, Ohuchi N. et al. Molecular morphological approach to the pathological study of development and advancement of human breast cancer. Med Mol Morphol. 2010;43(2):67–73. doi: 10.1007/s00795-010-0504-5. [DOI] [PubMed] [Google Scholar]
- Martins FC, Botelho MF, Cabrita AM, de Oliveira CF. Influence of normal mammary epithelium on breast cancer progression: the protective role of early pregnancy. Tumori. 2010;96(6):999–1003. [PubMed] [Google Scholar]
- Cazzaniga M, Decensi A, Bonanni B, Luini A, Gentilini O. Biomarkers for risk assessment and prevention of breast cancer. Curr Cancer Drug Targets. 2009;9(4):482–499. doi: 10.2174/156800909788486768. [DOI] [PubMed] [Google Scholar]
- Graham K, Ge X, de Las Morenas A, Tripathi A, Rosenberg CL. Gene expression profiles of estrogen receptor-positive and estrogen receptor-negative breast cancers are detectable in histologically normal breast epithelium. Clin Cancer Res. 2010;17(2):236–246. doi: 10.1158/1078-0432.CCR-10-1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels EK, Kairemo K. Fatty acid facts, part II: role in the prevention of carcinogenesis, or, more fish on the dish? Drug News Perspect. 2008;21(9):504–510. doi: 10.1358/dnp.2008.21.9.1290819. [DOI] [PubMed] [Google Scholar]
- Gillet L, Roger S, Bougnoux P, Le Guennec JY, Besson P. Beneficial effects of omega-3 long-chain fatty acids in breast cancer and cardiovascular diseases: voltage-gated sodium channels as a common feature? Biochimie. 2010;93(1):4–6. doi: 10.1016/j.biochi.2010.02.005. [DOI] [PubMed] [Google Scholar]
- Terry PD, Terry JB, Rohan TE. Long-chain (n-3) fatty acid intake and risk of cancers of the breast and the prostate: recent epidemiological studies, biological mechanisms, and directions for future research. J Nutr. 2004;134(12 Suppl):3412S–3420S. doi: 10.1093/jn/134.12.3412S. [DOI] [PubMed] [Google Scholar]
- Wall R, Ross RP, Fitzgerald GF, Stanton C. Fatty acids from fish: the anti-inflammatory potential of long-chain omega-3 fatty acids. Nutr Rev. 2010;68(5):280–289. doi: 10.1111/j.1753-4887.2010.00287.x. [DOI] [PubMed] [Google Scholar]
- Wendel M, Heller AR. Anticancer actions of omega-3 fatty acids-current state and future perspectives. Anticancer Agents Med Chem. 2009;9(4):457–470. doi: 10.2174/1871520610909040457. [DOI] [PubMed] [Google Scholar]
- Escrich E, Moral R, Grau L, Costa I, Solanas M. Molecular mechanisms of the effects of olive oil and other dietary lipids on cancer. Mol Nutr Food Res. 2007;51(10):1279–1292. doi: 10.1002/mnfr.200700213. [DOI] [PubMed] [Google Scholar]
- Escrich E, Solanas M, Moral R, Escrich R. Modulatory effects and molecular mechanisms of olive oil and other dietary lipids in breast cancer. Curr Pharm Des. 2010;17(8):813–830. doi: 10.2174/138161211795428902. [DOI] [PubMed] [Google Scholar]
- Chalmers TC, Berrier J, Hewitt P, Berlin J, Reitman D, Nagalingam R, Sacks H. Meta-analysis of randomized controlled trials as a method of estimating rare complications of non-steroidal anti-inflammatory drug therapy. Aliment Pharmacol Ther. 1988;2(Suppl 1):9–26. doi: 10.1111/j.1365-2036.1988.tb00761.x. [DOI] [PubMed] [Google Scholar]
- DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- Hozo SP, Djulbegovic B, Hozo I. Estimating the mean and variance from the median, range, and the size of a sample. BMC Med Res Methodol. 2005;5:13. doi: 10.1186/1471-2288-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopman L, van der Heijden GJ, Grobbee DE, Rovers MM. Comparison of methods of handling missing data in individual patient data meta-analyses: an empirical example on antibiotics in children with acute otitis media. Am J Epidemiol. 2008;167(5):540–545. doi: 10.1093/aje/kwm341. [DOI] [PubMed] [Google Scholar]
- Hedlund C, Rapoport AM, Dodick DW, Goadsby PJ. Zolmitriptan nasal spray in the acute treatment of cluster headache: a meta-analysis of two studies. Headache. 2009;49(9):1315–1323. doi: 10.1111/j.1526-4610.2009.01518.x. [DOI] [PubMed] [Google Scholar]
- Kall L, Storey JD, Noble WS. QVALITY: non-parametric estimation of q-values and posterior error probabilities. Bioinformatics. 2009;25(7):964–966. doi: 10.1093/bioinformatics/btp021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H, Chen H, Omeroglu G, Meterissian S, Omeroglu A. et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14(5):518–527. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- Karnoub AE, Dash AB, Vo AP, Sullivan A, Brooks MW, Bell GW, Richardson AL, Polyak K, Tubo R, Weinberg RA. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449(7162):557–563. doi: 10.1038/nature06188. [DOI] [PubMed] [Google Scholar]
- Radvanyi L, Singh-Sandhu D, Gallichan S, Lovitt C, Pedyczak A, Mallo G, Gish K, Kwok K, Hanna W, Zubovits J. et al. The gene associated with trichorhinophalangeal syndrome in humans is overexpressed in breast cancer. Proc Natl Acad Sci USA. 2005;102(31):11005–11010. doi: 10.1073/pnas.0500904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson AL, Wang ZC, De Nicolo A, Lu X, Brown M, Miron A, Liao X, Iglehart JD, Livingston DM, Ganesan S. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell. 2006;9(2):121–132. doi: 10.1016/j.ccr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- Turashvili G, Bouchal J, Baumforth K, Wei W, Dziechciarkova M, Ehrmann J, Klein J, Fridman E, Skarda J, Srovnal J. et al. Novel markers for differentiation of lobular and ductal invasive breast carcinomas by laser microdissection and microarray analysis. BMC Cancer. 2007;7:55. doi: 10.1186/1471-2407-7-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Langerod A, Ji Y, Nowels KW, Nesland JM, Tibshirani R, Bukholm IK, Karesen R, Botstein D, Borresen-Dale AL. et al. Different gene expression patterns in invasive lobular and ductal carcinomas of the breast. Mol Biol Cell. 2004;15(6):2523–2536. doi: 10.1091/mbc.E03-11-0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Li L, Korantzopoulos P, Goudevenos JA, Li G. Meta-analysis of association between C-reactive protein and immediate success of electrical cardioversion in persistent atrial fibrillation. Am J Cardiol. 2008;101(12):1749–1752. doi: 10.1016/j.amjcard.2008.02.066. [DOI] [PubMed] [Google Scholar]
- Chin SF, Wang Y, Thorne NP, Teschendorff AE, Pinder SE, Vias M, Naderi A, Roberts I, Barbosa-Morais NL, Garcia MJ. et al. Using array-comparative genomic hybridization to define molecular portraits of primary breast cancers. Oncogene. 2007;26(13):1959–1970. doi: 10.1038/sj.onc.1209985. [DOI] [PubMed] [Google Scholar]
- Desmedt C, Piette F, Loi S, Wang Y, Lallemand F, Haibe-Kains B, Viale G, Delorenzi M, Zhang Y, d'Assignies MS. et al. Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin Cancer Res. 2007;13(11):3207–3214. doi: 10.1158/1078-0432.CCR-06-2765. [DOI] [PubMed] [Google Scholar]
- Kao KJ, Chang KM, Hsu HC, Huang AT. Correlation of microarray-based breast cancer molecular subtypes and clinical outcomes: implications for treatment optimization. BMC Cancer. 2011;11:143. doi: 10.1186/1471-2407-11-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loi S, Haibe-Kains B, Desmedt C, Lallemand F, Tutt AM, Gillet C, Ellis P, Harris A, Bergh J, Foekens JA. et al. Definition of clinically distinct molecular subtypes in estrogen receptor-positive breast carcinomas through genomic grade. J Clin Oncol: J Am Soc Clin Oncol. 2007;25(10):1239–1246. doi: 10.1200/JCO.2006.07.1522. [DOI] [PubMed] [Google Scholar]
- Loi S, Haibe-Kains B, Desmedt C, Wirapati P, Lallemand F, Tutt AM, Gillet C, Ellis P, Ryder K, Reid JF. et al. Predicting prognosis using molecular profiling in estrogen receptor-positive breast cancer treated with tamoxifen. BMC Genomics. 2008;9:239. doi: 10.1186/1471-2164-9-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, Viale A, Olshen AB, Gerald WL, Massague J. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436(7050):518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ. et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347(25):1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- van't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT. et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;41(6871):530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- Hess KR, Anderson K, Symmans WF, Valero V, Ibrahim N, Mejia JA, Booser D, Theriault RL, Buzdar AU, Dempsey PJ. et al. Pharmacogenomic predictor of sensitivity to preoperative chemotherapy with paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide in breast cancer. J Clin Oncol: J Am Soc Clin Oncol. 2006;24(26):4236–4244. doi: 10.1200/JCO.2006.05.6861. [DOI] [PubMed] [Google Scholar]
- Huang E, Cheng SH, Dressman H, Pittman J, Tsou MH, Horng CF, Bild A, Iversen ES, Liao M, Chen CM. et al. Gene expression predictors of breast cancer outcomes. Lancet. 2003;361(9369):1590–1596. doi: 10.1016/S0140-6736(03)13308-9. [DOI] [PubMed] [Google Scholar]
- Ma XJ, Wang Z, Ryan PD, Isakoff SJ, Barmettler A, Fuller A, Muir B, Mohapatra G, Salunga R, Tuggle JT. et al. A two-gene expression ratio predicts clinical outcome in breast cancer patients treated with tamoxifen. Cancer Cell. 2004;5(6):607–616. doi: 10.1016/j.ccr.2004.05.015. [DOI] [PubMed] [Google Scholar]
- Sotiriou C, Neo SY, McShane LM, Korn EL, Long PM, Jazaeri A, Martiat P, Fox SB, Harris AL, Liu ET. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc Natl Acad Sci USA. 2003;100(18):10393–10398. doi: 10.1073/pnas.1732912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, Talantov D, Timmermans M, Meijer-van Gelder ME, Yu J. et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365(9460):671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- Hedenfalk I, Duggan D, Chen Y, Radmacher M, Bittner M, Simon R, Meltzer P, Gusterson B, Esteller M, Kallioniemi OP. et al. Gene-expression profiles in hereditary breast cancer. N Engl J Med. 2001;344(8):539–548. doi: 10.1056/NEJM200102223440801. [DOI] [PubMed] [Google Scholar]
- Fould B, Garlatti V, Neumann E, Fenel D, Gaboriaud C, Arlaud GJ. Structural and functional characterization of the recombinant human mitochondrial trifunctional protein. Biochemistry. 2010;49(39):8608–8617. doi: 10.1021/bi100742w. [DOI] [PubMed] [Google Scholar]
- Spiekerkoetter U. Mitochondrial fatty acid oxidation disorders: clinical presentation of long-chain fatty acid oxidation defects before and after newborn screening. J Inherit Metab Dis. 2010;33(5):527–532. doi: 10.1007/s10545-010-9090-x. [DOI] [PubMed] [Google Scholar]
- Gold B, Kirchhoff T, Stefanov S, Lautenberger J, Viale A, Garber J, Friedman E, Narod S, Olshen AB, Gregersen P. et al. Genome-wide association study provides evidence for a breast cancer risk locus at 6q22.33. Proc Natl Acad Sci USA. 2008;105(11):4340–4345. doi: 10.1073/pnas.0800441105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi H, Kojima K, Sekine S, Murano Y, Aoyama T. Effect of dietary n-6/n-3 ratio on liver n-6/n-3 ratio and peroxisomal beta-oxidation activity in rats. J Oleo Sci. 2008;57(12):649–657. doi: 10.5650/jos.57.649. [DOI] [PubMed] [Google Scholar]
- Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466(7302):68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson KL, Hubbard NE. Fatty acids and breast cancer: the role of stem cells. Prostaglandins Leukot Essent Fatty Acids. 2010;82(4-6):237–241. doi: 10.1016/j.plefa.2010.02.019. [DOI] [PubMed] [Google Scholar]
- Delongchamp RR, Bowyer JF, Chen JJ, Kodell RL. Multiple-testing strategy for analyzing cDNA array data on gene expression. Biometrics. 2004;60(3):774–782. doi: 10.1111/j.0006-341X.2004.00228.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Excel table containing detailed annotation of the 136 genes related to fat and lipid metabolism that were primarily selected for analyses.
Table S2. Excel table containing detailed annotation of the 47 genes included in this study related to fat and lipid metabolism that were primarily selected for analyses.