Abstract
We determined the frequency of integrated viral DNA in the livers of three woodchucks chronically infected with the woodchuck hepatitis virus before and during 30 weeks of therapy with the nucleoside analog L-FMAU [1-(2-fluoro-5-methyl-beta, l-arabinofuranosyl)uracil, clevudine]. We found that although viral covalently closed circular DNA declined 20- to 100-fold, integrated viral DNA showed no discernable decrease over the course of treatment. Thus, chemotherapeutic clearance of covalently closed circular DNA did not involve the replacement of the infected hepatocyte population with uninfected progenitors, but rather, uninfected hepatocytes in the treated liver were derived from the infected hepatocyte population. The frequency of integrated DNA in chronically infected woodchucks was found to be 1 or 2 orders of magnitude higher than that in transiently infected woodchucks, implying that integration and other genomic damage accumulate over the duration of infection. Our results indicate that genetic changes from this damage remain in the liver even while virus infection is cleared and argue for early antiviral intervention in chronic hepatitis.
Chemotherapy of hepatitis B virus (HBV) infections has focused primarily on a strategy of inhibiting viral DNA synthesis through the use of nucleoside analogs that are highly specific for the viral DNA polymerase (1–9). Treatment of patients with such inhibitors can be remarkably effective in reducing viremias to very low levels (2, 3). The reduction of viremia has been shown to occur with biphasic kinetics, with the first phase of rapid reduction attributed to the inhibition of virus production by infected cells, combined with viral clearance from the blood by mechanisms probably unrelated to the action of the drug. The slower second phase has been attributed to the elimination of infected cells from the liver, by either normal or immune-enhanced mechanisms, and the accumulation of uninfected cells (10, 11). The gradual replacement of infected by uninfected hepatocytes has been observed during antiviral therapy of woodchucks chronically infected with the woodchuck hepatitis virus (WHV), an animal model for HBV (7). In 1-(2-f luoro-5-methyl-beta, l-arabinofuranosyl)uracil (L-FMAU)-treated woodchucks, the onset of the reduction in infected cells lags behind the decline of the covalently closed circular DNA (cccDNA), the viral transcriptional template in the nucleus. It has been proposed that the removal of cccDNA occurs by hepatocyte turnover, and that dilution of cccDNA copy numbers in dividing hepatocytes results in eventual segregation of uninfected progeny cells (12). Alternatively, uninfected hepatocytes may arise by differentiation of uninfected progenitors (13) or be generated through a slow loss of cccDNA from infected hepatocytes (14, 15).
During hepadnavirus infection, a small fraction of hepatocytes undergo recombination with viral DNA molecules in the nucleus and acquire an integrated viral DNA that may be stably retained in that hepatocyte lineage (16–20). The common precursor to integrated viral DNA is a full-length linear double-stranded DNA, with a left end just upstream of the viral core gene, that is formed as a by-product of viral genome synthesis (21, 22). Typically, these aberrant molecules are present at 5–25% the abundance of the normal relaxed circular genomes (23, 24). We have recently used integrated WHV DNA as a genetic marker to show that the uninfected hepatocytes populating the liver after immune clearance of a transient infection were derived from the infected hepatocyte population that was present before clearance (25). In the present study, we measured the frequency of integrated viral DNA during therapy of three woodchucks with L-FMAU (7) to determine whether integrated viral DNA persisted in the liver during the decline in cccDNA and the accumulation of uninfected cells.
Our assay for left-end viral-cell junctions of integrated DNA, based on selective PCR, has been described in detail elsewhere (25). Briefly, cellular DNA extracted from infected liver was cut with PvuII and the PvuII ends were ligated to a double-stranded linker designed to regenerate the PvuII site. WHV DNA is not cut by PvuII, consequently linkers were ligated specifically to woodchuck cellular sequences. Linker-ligated DNA was diluted and distributed into the wells of a PCR microplate. Individual viral-cell sequences were amplified by PCR using a virus-specific primer, oriented leftward in the minus strand direction, and a linker-specific primer oriented rightward toward the ligated cellular sequences. The products of left end viral-cell junction templates were then specifically amplified by inverse PCR. For inverse PCR, the linker sequence was removed by PvuII digestion to generate a 5′ phosphate, which allowed circularization of the amplified viral-cell junction with T4 DNA ligase. Circular molecules were cut with HindIII in the viral sequences and amplified with nested viral-specific primers. Individual viral-cell junction products arising in the wells were confirmed by sequencing. The frequency of viral-cell junctions was calculated from the number of products detected in a known amount of cellular DNA.
Three chronically WHV-infected adult woodchucks were orally administered 10 mg/kg L-FMAU daily for 30 weeks. Biopsies obtained before treatment and after 6, 15, and 30 weeks of treatment were analyzed for the frequency of viral-cell junctions. The amounts of intrahepatic viral DNA and fractions of infected cells in this study have been published (7), and the results of these analyses are summarized in Table 1 along with the frequency of integrated DNA determined in this study. Whereas cccDNA declined 20- to 100-fold over the course of treatment, integrated DNA, detected as viral-cell junctions, remained at about the same level. This result implies that clearance of virus from the liver by L-FMAU did not involve wholesale replacement of infected hepatocytes by an uninfected progenitor hepatocyte population (13).
Table 1. Frequency of integrated viral DNA during L-FMAU therapy.
| Animal | Weeks of therapy | cccDNA per hepatocyte* | Cells assayed† | Viral-cell joints detected | Viral-cell joints per cell |
|---|---|---|---|---|---|
| WC343 | 0 | 63 | 440 | 6 | 0.014 |
| 6 | 8 | 380 | 5 | 0.013 | |
| 15 | 8 | 500 | 8 | 0.016 | |
| 30 | 0.8 | 420 | 12 | 0.029 | |
| WC345 | 0 | 41 | 206 | 10 | 0.049 |
| 6 | 18 | 75 | 4 | 0.053 | |
| 15 | 19 | 270 | 8 | 0.030 | |
| 30 | 2.2 | 400 | 14 | 0.035 | |
| WC346 | 0 | 19 | 280 | 6 | 0.021 |
| 6 | 6 | 190 | 8 | 0.042 | |
| 15 | 1.7 | 300 | 9 | 0.030 | |
| 30 | 0.2 | 100 | 5 | 0.050 |
From ref. 7.
Based on the amount of DNA analyzed, assuming 10 pg of woodchuck DNA per cell.
The frequency of integrated viral DNA in these chronically infected woodchucks was 10- to 100-fold higher than that we observed in transiently infected woodchucks, determined by the same method in our previous study (25). This difference suggests that integrated viral DNA increases with the duration of infection (≈3 months vs. 1.5–2 years). By our assay, integrated DNA was found at frequencies of up to 1 per 20 liver cells in chronically infected woodchucks. Because many integrated viral DNAs would not be detected by the specific assay we used, this number is surely a minimum measure of the frequency of integration.
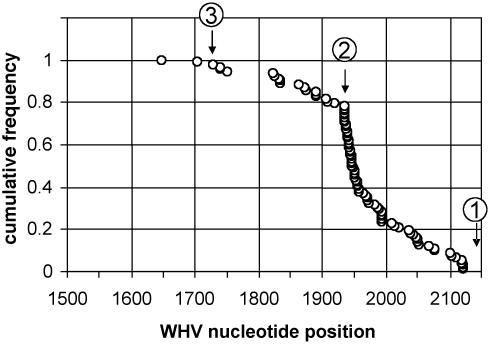
We determined the positions on the viral genome of the sites of recombination in the 95 viral-cell junctions amplified in this study. The cumulative distribution of these sites is shown in Fig. 1. Approximately 98% of all recombination events analyzed occurred downstream of nucleotide 1729, the 3′ end of DR2, and 78% were at or downstream of nucleotide 1935, the left end of in situ primed linear DNA. This distribution is consistent with our previous proposal that two distinct linear viral DNAs with left ends located at positions 1730 and 1935 give rise to integrated viral DNA by nonhomologous end joining with cellular DNA. Analysis of the flanking cellular DNA sequences did not reveal preferential sites of integration on the woodchuck genome.
Fig. 1.
Cumulative distribution of viral recombination sites of 95 left-end viral-cell junctions. Symbols indicate the cumulative frequency of recombination sites as a function of their positions on the WHV genome, from right to left. The numbered arrows mark the positions on the genome of the following: 1, the sequencing primer (3′ end at nucleotide 2145); 2, the left end of in situ primed linear DNA (nucleotide 1935); 3, the 3′ end of DR2 (nucleotide 1729). Ninety-eight percent of all viral recombination sites are to the right (downstream) of nucleotide 1729 (arrow 3).
Integration of hepadnaviral DNA in a chicken cell line has been shown to be stimulated by cell DNA damage (26), and we have recently shown that double-stranded breaks are targets for hepadnaviral DNA integration (C. Bill and J.S., unpublished work). Taken together, these two observations raise the possibility that integrations normally occur at sites of cell DNA damage. Thus, integration of viral DNA not only can cause genomic damage and mutation but also may indicate the existence of more widespread genetic changes at other sites that have not been marked by integration of viral sequences. Genomic damage is a by-product of reactive oxygen species produced during chronic inflammation, and chronic inflammation has been demonstrated to be associated with the development of liver cancer in transgenic mice expressing hepatitis B viral surface antigens (27, 28). The present study indicates that, in the short term, antiviral chemotherapy does not clear the liver of the genetic changes incurred by infected cells during the period of chronic hepatitis because the uninfected hepatocytes that are generated during therapy are derived from infected cells. Therefore, such antiviral treatment, in theory, would be more effective in preventing genetic consequences of chronic hepatitis, i.e., hepatocellular carcinoma, the earlier it can be administered in the course of infection.
Acknowledgments
We thank Yuao Zhu for providing the liver samples used in this study, Bai Hua Zhang for excellent technical assistance, and Jianming Hu for critical reading of the manuscript. This work was supported by Department of Health and Human Services Grants CA-84017 and CA-18641.
Abbreviations: cccDNA, covalently closed circular DNA; L-FMAU, 1-(2-fluoro-5-methyl-beta, l-arabinofuranosyl)uracil; WHV, woodchuck hepatitis virus.
References
- 1.Marion, P. L., Salazar, F. H., Winters, M. A. & Colonno, R. J. (2002) Antimicrob. Agents Chemother. 46, 82–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leung, N. W., Lai, C. L., Chang, T. T., Guan, R., Lee, C. M., Ng, K. Y., Lim, S. G., Wu, P. C., Dent, J. C., Edmundson, S., et al. (2001) Hepatology 33, 1527–1532. [DOI] [PubMed] [Google Scholar]
- 3.Lau, D. T., Khokhar, M. F., Doo, E., Ghany, M. G., Herion, D., Park, Y., Kleiner, D. E., Schmid, P., Condreay, L. D., Gauthier, J., et al. (2000) Hepatology 32, 828–834. [DOI] [PubMed] [Google Scholar]
- 4.Foster, W. K., Miller, D. S., Marion, P. L., Colonno, R. J., Kotlarski, I. & Jilbert, A. R. (2003) Antimicrob. Agents Chemother. 47, 2624–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Addison, W. R., Walters, K. A., Wong, W. W., Wilson, J. S., Madej, D., Jewell, L. D. & Tyrrell, D. L. (2002) J. Virol. 76, 6356–6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peek, S. F., Cote, P. J., Jacob, J. R., Toshkov, I. A., Hornbuckle, W. E., Baldwin, B. H., Wells, F. V., Chu, C. K., Gerin, J. L., Tennant, B. C. & Korba, B. E. (2001) Hepatology 33, 254–266. [DOI] [PubMed] [Google Scholar]
- 7.Zhu, Y., Yamamoto, T., Cullen, J., Saputelli, J., Aldrich, C. E., Miller, D. S., Litwin, S., Furman, P. A., Jilbert, A. R. & Mason, W. S. (2001) J. Virol. 75, 311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Le Guerhier, F., Pichoud, C., Jamard, C., Guerret, S., Chevallier, M., Peyrol, S., Hantz, O., King, I., Trepo, C., Cheng, Y. C. & Zoulim, F. (2001) Antimicrob. Agents Chemother. 45, 1065–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luscombe, C., Pedersen, J., Uren, E. & Locarnini, S. (1996) Hepatology 24, 766–773. [DOI] [PubMed] [Google Scholar]
- 10.Nowak, M. A., Bonhoeffer, S., Hill, A. M., Boehme, R., Thomas, H. C. & McDade, H. (1996) Proc. Natl. Acad. Sci. USA 93, 4398–4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perelson, A. S. (2002) Nat. Rev. Immunol. 2, 28–36. [DOI] [PubMed] [Google Scholar]
- 12.Mason, W. & Litwin, S. (2002) in HBV Human Virus Guide, eds. Locarnini, S. & Lai, C. L. (Int. Med. Press, London), pp. 99–113.
- 13.Thorgeirsson, S. S. (1996) FASEB J. 10, 1249–1256. [PubMed] [Google Scholar]
- 14.Guo, J. T., Pryce, M., Wang, X., Barrasa, M. I., Hu, J. & Seeger, C. (2003) J. Virol. 77, 1885–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Civitico, G. M. & Locarnini, S. A. (1994) Virology 203, 81–89. [DOI] [PubMed] [Google Scholar]
- 16.Brechot, C., Scotto, J., Charnay, P., Hadchouel, M., Degos, F., Trepo, C. & Tiollais, P. (1981) Lancet 318, 765–768. [DOI] [PubMed] [Google Scholar]
- 17.Shafritz, D. A., Shouval, D., Sherman, H. I., Hadziyannis, S. J. & Kew, M. C. (1981) N. Engl. J. Med. 305, 1067–1073. [DOI] [PubMed] [Google Scholar]
- 18.Koshy, R., Maupas, P., Muller, R. & Hofschneider, P. H. (1981) J. Gen. Virol. 57, 95–102. [DOI] [PubMed] [Google Scholar]
- 19.Dejean, A., Vitvitski, L., Brechot, C., Trepo, C., Tiollais, P. & Charnay, P. (1982) Virology 121, 195–199. [DOI] [PubMed] [Google Scholar]
- 20.Kam, W., Rall, L. B., Smuckler, E. A., Schmid, R. & Rutter, W. J. (1982) Proc. Natl. Acad. Sci. USA 79, 7522–7526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang, W. & Summers, J. (1999) J. Virol. 73, 9710–9717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gong, S. S., Jensen, A. D., Chang, C. J. & Rogler, C. E. (1999) J. Virol. 73, 1492–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Staprans, S., Loeb, D. D. & Ganem, D. (1991) J. Virol. 65, 1255–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang, W., Mason, W. S. & Summers, J. (1996) J. Virol. 70, 4567–4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Summers, J., Jilbert, A. R., Yang, W., Aldrich, C. E., Saputelli, J., Litwin, S., Toll, E. & Mason, W. S. (2003) Proc. Natl. Acad. Sci. USA 100, 11652–11659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petersen, J., Dandri, M., Burkle, A., Zhang, L. & Rogler, C. E. (1997) J. Virol. 71, 5455–5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakamoto, Y., Guidotti, L. G., Kuhlen, C. V., Fowler, P. & Chisari, F. V. (1998) J. Exp. Med. 188, 341–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hagen, T. M., Huang, S., Curnutte, J., Fowler, P., Martinez, V., Wehr, C. M., Ames, B. N. & Chisari, F. V. (1994) Proc. Natl. Acad. Sci. USA 91, 12808–12812. [DOI] [PMC free article] [PubMed] [Google Scholar]