Abstract
Using a method of expression profiling called differential analysis of cDNA library expression (DAzLE), we report the expression profile of late response genes in a model of activity-dependent neuronal survival and neurite outgrowth. Using DAzLE, we isolated differentially expressed genes from cultured rat embryonic cortical neurons after KCl (50 mM)-mediated membrane depolarization. We identified 469 activity-dependent regulated genes, of which 174 are genes of unknown function. The regulation of 63 genes was found to be nitric oxide (NO)-dependent. Identifiable genes fell into several major categories, including signal transduction pathways, neuronal development, DNA replication, gene transcription, protein metabolism, energy regulatory proteins, and antiapoptotic proteins. These genes may be important in activity-dependent neuron survival and development. Furthermore, these late response genes provide the tools to begin to investigate downstream events in activity-dependent neuronal survival and development. The major advantage of DAzLE is that it provides a nearly complete and relatively comprehensive differential screening profile that has the potential to be a powerful and useful tool in other fields of study.
Activity-dependent neuronal survival requires increases in intracellular calcium and the induction of new gene transcription (1–4). Nitric oxide (NO) seems to play important roles in modulating activity-dependent neuronal survival (M. Gonzalez-Zulueta, V.L.D., and T.M.D., unpublished observations) (5). The molecular characterization of the immediate and early events accompanying activity-dependent neuronal survival has provided tremendous insight into the transcriptional regulators that control downstream expression of immediate early genes (3, 4, 6). Activation of the transcription factors cAMP-response element-binding protein (CREB) and MEF2 may play important roles as calcium-regulated transcription factors controlling the calcium-dependent survival of neurons (4, 7). Some early response genes such, as brain-derived neurotrophic factor, play a role, in part, as activity-induced neuronal survival proteins (2, 8). However, the secondary events and late response genes that control, and are responsible for, activity-dependent neuronal survival are not well characterized.
To begin to understand the secondary events that promote the calcium-dependent survival of neurons and the role of NO in this process, we conducted a screen for late response genes triggered by calcium influx into immature primary cerebral cortical neurons induced by membrane depolarization after serum withdrawal. Various strategies have been used to identify differentially expressed genes (9–16). The most sensitive and powerful methods rely on PCR and restriction endonuclease digestion. However, these methods only sample mRNA transcripts that contain the restriction enzyme site (11, 12). Microarray analysis can be used to retrospectively analyze gene expression (9), and a number of relatively complete genomic sequences are available for a variety of organisms. However, these databases still suffer from gene identification and annotation deficiencies that have hindered comprehensive identification of messenger RNA (17). Furthermore, many of these methods are not sensitive enough to identify very rare transcripts. Here, we describe the differential analysis of primary library expression (DAzLE) to identify calcium-and NO-induced genes that may play important roles in activity-dependent neuronal survival.
Materials and Methods
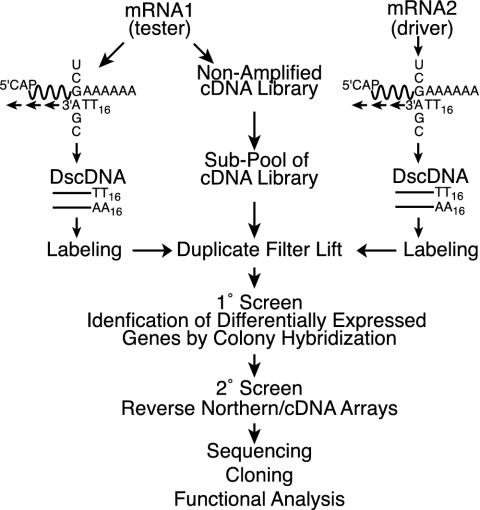
DAzLE. The DAzLE method (Fig. 1) is based on the screening of a primary nonamplified cDNA library with the probes containing poly(A/T) tailless cDNAs (Fig. 1). This method involves construction of a full-length cDNA library, followed by preparation of a pooled cDNA library, followed by library screening with poly(A/T) tailless [32P]cDNA probes reverse transcribed from mRNA samples of control or KCl-treated cortical neurons. After hybridization, differentially expressed clones were picked as primary positive genes.
Fig. 1.
Schematic outline of the DAzLE protocol. This scheme contains three steps: (i) construction and primary screening of a nonamplified cDNA library, (ii) differential hybridization analysis of cDNA clones, and (iii) clone analysis by sequencing and reverse Northern blot analysis. Differential hybridization analysis of the cDNA library was first performed with screening of random clones on duplicated nylon membranes. Positive clones that are isolated from primary screening were confirmed by reverse Northern blot analysis. Finally, the Northern blot analysis was used to confirm the differential expression of cDNA clones.
Primary Cell Culture. Rat embryonic day (E) 17 cortical neurons were maintained in Eagle's minimal essential medium (MEM) plus 10% horse serum and 10% FCS on a six-well NUNC plate to 70–80% confluence at 37°C in 5% CO2 overnight before treatment. All inhibitors were added to plates 30 min before the KCl stimulation. All treatments were performed 1 day after plating. Membrane depolarization was attained with 50 mM KCl in MEM plus 5% horse serum and 5% FCS. Treatments included KCl (50 mM), KCl (50 mM) plus l-NAME (NG-nitro-l-arginine methyl ester; 100 μM)/nitro-Arg (N-Arg; 100 μM), KCl (50 mM) plus nifedipine (5 μM), NO donors (NOR3, 10 μM, Sigma; NOR3-NO–, 10 μM), and KCl (50 mM) plus EGTA (10 μM), EGTA (10 μM).
RNA Isolation. Total RNA was isolated by the TRIzol reagent according to the manufacturer's protocol from untreated, KCl (50 mM), KCl (50 mM) plus l-NAME (100 μM)/N-Arg (100 μM), KCl (50 mM) plus nifedipine (5 μM), NO donors (NOR3, 10 μM; NOR3-NO–, 10 μM), KCl (50 mM) plus EGTA (10 μM), or EGTA (10 μM)-treated neurons. Poly(A)-RNA was purified from total RNA by using an oligo(dT) cellulose column (GIBCO/BRL).
cDNA Library Construction. The SuperScript Plasmid System (Life Technologies, Grand Island, NY) was used for the construction of the cDNA library. Total RNA was first isolated from KCl-induced rat E17 primary cultured cortical neurons. Poly(A) RNA was then prepared from total RNA by subjecting it twice to oligo(dT) affinity chromatography. In the first strand synthesis reaction (i.e., reverse transcription), a modified oligo(dT) primer adapter containing a NotI site was used. Nick translational replacement of the mRNA was used to synthesize the second strand of cDNA. Second strand synthesis was catalyzed by Escherichia coli DNA polymerase I in combination with E. coli RNase H and E. coli DNA ligase. The product of the first strand and second strand reactions was blunt ended cDNA, so a SalI adapter was added to the 5′ end of the cDNA by T4 DNA ligase. These resulting cDNAs contained a NotI site downstream of the poly(A) tail, and a SalI site on the 5′ end. The cDNAs were size-fractionated by sephacryl column chromatography. Only the fractions containing cDNAs >1,000 bp were used for library construction. The cDNAs were digested with SalI and NotI and ligated into a SalI/NotI-digested pSPORT vector. Vector-ligated cDNAs were introduced into DH10B component cells by electroporation.
cDNA Library Screening. The plasmid cDNA library was screened according to the procedure described by Vogeli and Kaytes (18). The library was grown on Hybond-NX nylon transfer membranes (Amersham Biosciences). For the primary screening, the library was divided into 100 pools containing ≈2,000 colonies each. Duplicate replicas were made, and each set of membranes was probed by using appropriate cDNA probes that were specifically synthesized with A-, C-, and G-anchored poly(T)16–18 primer mix to fix the size of the poly(A/T) tail of the cDNAs. For one reaction, 5 μg of mRNA was used as template, with 0.5 μg of each of primers (i) 3′-A(T16)-5′, (ii) 3′-C(T16)-5′, and (iii) 3′-G(T16)-5′ (A-, C-, and G-anchored poly(T)16 primers) to synthesize the double-stranded cDNA with the SuperScript cDNA synthesis system (Life Technologies). The synthesized cDNA fragments from one reaction were divided into four parts as the templates for radiolabeling. In one reaction of radiolabeling, 50 μCi (1 Ci = 37 GBq) of [α-32P]dCTP was used in the labeling mix of the Ready-to-Go oligolabeling kit from Amersham Biosciences. The radioactive probes were purified with G50 Sephadex (Amersham Biosciences) columns before being added into the hybridization bags. Hybridization with the probe was carried out at 65°C in a solution containing 5× SSPE (1× SSPE = 0.15 M NaCl, 10 mM NaH2PO4, 1 mM EDTA), 5× Denhardt's solution [50× Denhardt's reagent contains 5 g of Ficoll (Type 400, Pharmacia), 5 g of polyvinylpyrrolidone, 5 g of BSA (Fraction V, Sigma)], and denatured salmon sperm DNA for 24 h. Posthybridization, washing was done with three changes of 3× SSC (1× SSC = 0.15 M NaCl, 15 mM sodium citrate)/0.5% SDS and three changes of 0.5× SSC/0.5% SDS at room temperature for 10 min each. The membranes were exposed to x-ray films. The positive clones were identified and further confirmed by reverse Northern hybridization.
Reverse Northern Blot Hybridization. A total of 980 individual primary positive clones were picked, and the plasmid DNA was extracted with the QIAprep 8 miniprep kit (Qiagen, Valencia, CA). After purification, 2 μg of plasmid DNA of each clone was digested with the NotI restriction enzyme to linearize the cDNA insert, and these plasmid DNAs were denatured and alkaline blotted onto nylon membranes. Spotted DNA was cross-linked to the filter with a UV crosslinker (Stratagene). The filters were hybridized with equivalent amounts of 32P-labeled double-strand cDNA of approximately equal specific activity derived from control and KCl-stimulated mRNAs, respectively. Filters were washed (see above) and exposed to a PhosphorImager (Molecular Dynamics) plate for 2–4 days before signal analysis.
Analysis of cDNA Insert. The cDNA inserts of the positive clones were sequenced on the 5′ end and 3′ end by the dideoxy chain termination method by using sequenase 2.0 (United States Biochemical) and by using T7 and Sp6 primers. Sequencing of the full-length clones was performed by using an automated DNA sequencer (ABI PRISM 377 DNA sequencer, Applied Biosystems) in some cases. The cDNA inserts from these positive clones were partially sequenced and analyzed for homology in National Center for Biotechnology Information (www.ncbi.nlm.nih.gov) databases. Protein structure analysis of the novel genes was performed with the program search for conserved domains (www.ncbi.nlm.nih.gov/blast).
Northern Blotting. Five micrograms of poly(A) RNA was sizefractionated in a 1.5% formaldehyde agarose gel and subsequently transferred onto a nylon membrane (Hybond N+ membrane, Amersham Pharmacia) with 20× SSC for 24 h. Filters were hybridized at 65°C with the probes and washed twice in 2× SSC/0.5% SDS at room temperature for 30 min, and twice in 0.5× SSC/0.55 SDS at 65°C for 30 min.
Neuronal NO Synthase (nNOS) Knockout Analysis. Cortical neuronal samples were collected from E19 nNOS knockout mice and E19 wild-type control C57B6. Total RNA was extracted and purified as described above. The mRNAs were used as the templates to synthesize double-stranded cDNAs. These cDNAs were used as probes with [α-32P]dCTP labeling to hybridize to dot-blots. Values were normalized to GAPDH, and the difference in expression between nNOS knockouts and wild-type cortex is presented in Table 1, which is published as supporting information on the PNAS web site.
Data Analysis. For all of the experiments, two to three replicates were performed. For standardization in the primary screening, the same amount of cDNA was used as the templates for the random labeling; the same amount of radioactivity was used to do the hybridization with the filters that contained sub-pools of the cDNA library. We used GAPDH as a standard control in the dot-blot/Northern blot experiments. The relative expression level of these genes was normalized by the average value of 14 clones including GAPDH, which exhibit stable expression under KCl treatment at different time points. The signals of the individual colonies on dot-blot images were scanned and quantitated with IPLab image quantitation software (Scanalytics, Fairfax, VA), and these data were processed with Microsoft excel. The criteria for each temporal expressions profile group were determined by the temporal profile of the level of expression of each gene. Individual genes were segregated into the appropriate temporal expression profile. The Z-score as a statistical method was used to determine differences between groups.
Results
DAzLE Is a Strategy for Differential Gene Analysis. Activity-dependent survival of developing rat cortical neurons was induced by using established protocols by membrane depolarization with high concentrations of potassium (50 mM KCl) after serum reduction (8, 19). We compared the expression profile of potassium-stimulated cultures at 6 h to identify the secondary response genes that follow the immediate and early responses of calcium influx mediated by potassium depolarization. The expression profile in response to potassium stimulation was determined by DAzLE (Fig. 1). The DAzLE method is based on screening nonamplified primary libraries by using poly(A/T) tailless cDNAs. A full-length cDNA library from potassium-stimulated cortical neuronal cultures was constructed. This cDNA library construction was followed by the preparation of a pooled cDNA library for library screening with poly(A/T) tailless 32P double-stranded cDNAs probes reverse transcribed from mRNA samples of unstimulated (driver) neurons and potassium-depolarized (tester) neurons. Probes were synthesized with A-, C-, G-anchored poly(T)16 primer mix to fix the size of the poly(A/T) tail of the cDNA. The use of poly(dA/T) tailless double-stranded DNA as probes limits cross hybridization among the 3′ end of the sequences so that rare transcripts are easily recovered as positive clones. Only clones that are dramatically up-regulated on visual inspection are picked. DAzLE dramatically increases the frequency of dynamically regulated genes and allows the direct isolation of differentially expressed clones for arraying, characterization, and storage (Fig. 1).
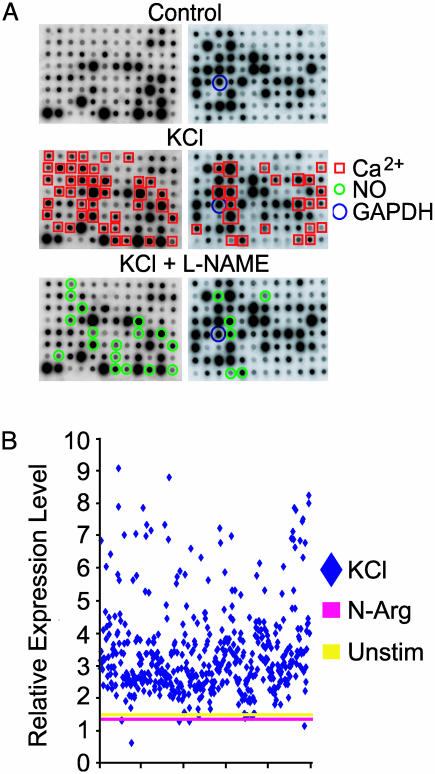
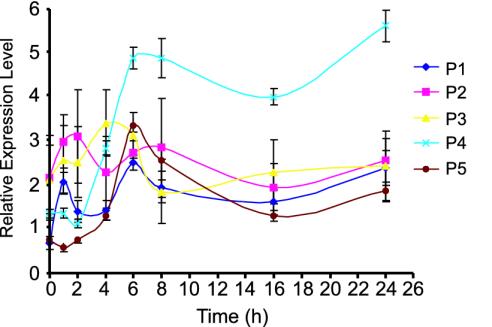
Primary Screening and Expression Pattern Analysis of Differentially Expressed Genes. Individual clones (200,000) were screened by DAzLE after membrane depolarization of primary cortical neurons. Nine hundred eighty (0.49%) colonies that showed higher intensity on x-ray film with the potassium-treated neuronal probe were picked, cultured, sequenced, and cataloged. These clones were arrayed and further screened for differential expression by reverse Northern analysis (Fig. 2A). Four hundred sixty-nine genes were found to be differentially expressed after membrane depolarization of rat cortical neurons. All 469 genes are calcium-dependent and are designated calcium-induced genes (CIGs). Sixty-three are NO-dependent (see below) and are designated NO-induced genes (NOIGs). Of the 469 genes, only a single clone was identified for 432 genes, and two or more clones were identified for 37 genes (data not shown). Quantitation of the differentially expressed gene (Fig. 2B) reveals that the majority of the genes are up-regulated 2- to 4-fold. The time course of the expression pattern of the differentially expressed genes was performed by comparing the expression levels after membrane depolarization at 1, 2, 4, 6, 8, 16, and 24 h. We are able to group the genes into five main expression patterns based on the temporal profile of gene expression (Fig. 3). Approximately 64% of the genes can be grouped into pattern four, in which expression begins to be detected 4 h after membrane depolarization, peaks at 6 h, and is sustained for the entire 24-h period. The second most abundant group, representing ≈25% of the genes, is pattern five, in which peak expression occurs at 6 h, followed by a rapid drop in signal intensity. A expected, no immediate response genes were identified, and <9% of the genes were induced at 1–2 h after potassium depolarization. Although different expression patterns were identified, all of the genes had increased expression levels at 24 h after membrane depolarization.
Fig. 2.
Signal level and pattern analysis of differentially expressed genes. (A) Reverse Northern blot analysis and classification of differentially expressed genes by DAzLE. Two micrograms of DNA of each clone was dot-blotted onto a nylon membrane and probed with [32P]cDNA synthesized from mRNA samples of KCl, KCl plus l-NAME, or l-NAME-treated rat cortical neurons. Clones (469 of 980) identified from primary screening are confirmed to show differential expression by reverse Northern blot analysis. These experiments were performed twice under the same conditions. (B) Quantitative changes in gene expression of 469 clones (KCl stimulation of rat cortical neurons). The relative expression level of these genes was normalized by the average value of 14 clones including GAPDH, which exhibit stable expression under KCl treatment at different time points. The average value of levels of unstimulated and N-Arg treatment at 6 h and 24 h were also used as controls.
Fig. 3.
Gene expression pattern analysis. Five gene expression patterns were identified after a time course analysis of the KCl-induced genes. Pattern 4 represents the majority of the gene group (64.2%). The expression level of this group peaks at 6 h after KCl induction and is sustained for 24 h. The percentages of genes in the other four expression patterns are as follows: pattern 1, 6.2%; pattern 2, 2.8%; pattern 3, 2.1%; and pattern 5, 24.7%.
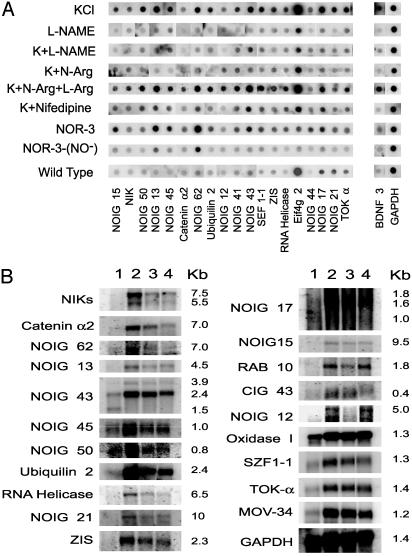
Analysis of Differentially Expressed Genes in Cortical Neurons. We subjected the calcium-induced genes to further analysis by identifying a subset of genes that are regulated by NO. Primary rat cortical neurons were stimulated with KCl in the presence or absence of NOS inhibitors. Sixty-three genes are identified as NO-dependent based on a 25% or higher reduction in signal intensity after membrane depolarization in the presence of the NOS inhibitors l-NAME or N-Arg. Membrane depolarization-induced expression of 20 genes is blocked >50% by NOS inhibitors, and the remainders are blocked between 25% and 50% (see Table 1). NOS inhibitors regulate the expression of 39 genes at 24 h, and 28 of the genes are down-regulated in neuronal cultures from mice lacking the gene for neuronal NO synthase (Table 1). We subjected a subset of the NO-dependent genes to further analysis (Fig. 4) and find that the expression of these genes is blocked or significantly attenuated by l-NAME and N-Arg. Consistent with NO dependence, this blockade is reversed by l-arginine (l-Arg). Furthermore, the NO donor NOR3 induces expression of all of the NO-dependent genes, and NOR3 depleted of NO has no effect. Supporting the notion that these are calcium–dependent genes is the observation that the voltage-sensitive calcium-channel blocker nifedipine prevents the upregulation of these genes by membrane depolarization (Fig. 4A).
Fig. 4.
Analysis of differentially expressed genes in cortical neurons. (A) Reverse Northern blot analysis of differentially expressed genes isolated by DAzLE. Differentially expressed genes were dot blotted onto nylon membranes and hybridized with [32P]cDNA samples of rat E17 cortical neurons. The neurons were treated with KCl (50 mM), l-NAME (100 μM), KCl (50 mM) plus l-NAME (100 μM), KCl (50 μM) plus N-Arg (100 μM), KCl (50 mM) plus N-Arg (100 μM) plus l-Arg (10 mM), KCl (50 mM) plus nifedipine (5 μM), NOR3 (10 μM), or NOR3-NO– (10 μM), for 6 h. (B) Northern blot analysis of genes differentially expressed in KCl-induced cortical neurons. Northern blot analysis was performed on mRNA (5 μg) isolated from primary culture of rat E17 cortical neurons, which received the following treatments: (i) unstimulated, (ii) KCl (50 mM), (iii) KCl (50 mM) plus N-Arg (100 μM), and (iv) KCl (50 mM) plus nifedipine (5 μM). Control hybridizations were performed by using rat GAPDH cDNA.
Northern blot analysis was used to further characterize the differentially expressed transcripts. Shown in Fig. 4B are representative Northern blots of several calcium-induced genes (CIGs) and NO-induced genes (NOIGs). More than 90% of the genes that are identified as membrane depolarization-induced genes by DAzLE are also differentially expressed by Northern blot analysis (data not shown). We failed to detect a signal on routine Northern blot analysis on a number of genes, suggesting that they might be very rare transcripts (data not shown). Consistent with the reverse Northern analysis, expression of some NO-dependent genes is dramatically blocked by the NOS inhibitor N-Arg whereas other genes are modestly regulated by NOS inhibitors. Thus, there seems to be genes whose expression is modulated but not controlled entirely by NO formation. Furthermore, a subset of genes are identified whose expression is not blocked by nifedipine, suggesting that these genes are regulated by non-l-type calcium channels as their induction in response to membrane depolarization was completely blocked by the calcium chelator EGTA (data not shown).
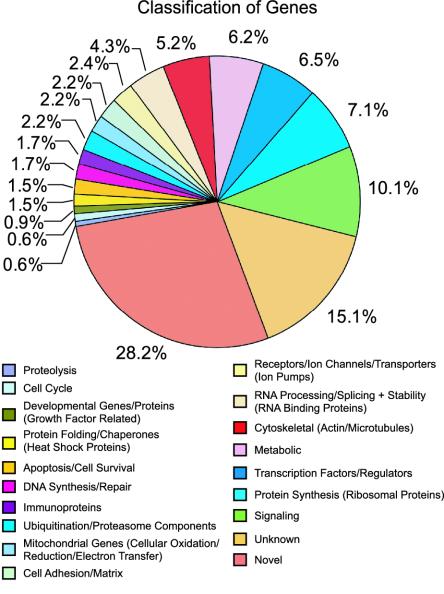
Functional Classification of KCl-Induced Genes. The sequences of the calcium-induced genes and NOIGs provide important clues to their possible functions (see Table 2, which is published as supporting information on the PNAS web site) and provide potential insight into the mechanisms by which membrane depolarization induces calcium-dependent neuronal survival. In particular, several genes were identified that play important roles in neuronal development, neurite outgrowth, cell survival, and calcium-regulated plasticity. The functional breakdown of the genes selectively expressed after membrane depolarization and cortical neurons is shown in Fig. 5. We see a broad spectrum of functional categories of activity-dependent genes (see Table 2 for a full list of genes). A number of genes implicated in cell survival are identified, including JNK-interacting protein 1, neuronal apoptosis inhibitory protein, brain-derived neurotrophic factor, 14-3-3, and calmodulin. We observed many protein kinases and phosphatases. Many proteins associated with the ubiquitin proteasomal and protein chaperone pathways are observed. We observe a number of transcription factors, small nuclear-associated proteins, proteins implicated in translation of RNA processing that are probably involved with the regulation of processes accompanying activity-dependent neuronal survival. There are also genes involved in cytoskeletal function and cell structure, which are probably required for the assembly and function of dendrites, growth cones, and axons. Finally, we observe 174 genes with no known function.
Fig. 5.
Functional classification of KCl-induced genes. Four hundred sixty-six unique genes were assigned into 19 categories based on the biological functions of known genes in the database of the National Center for Biotechnology Information. The percentage of up-regulated genes in each category is indicated. The full list of genes is provided in Table 2.
Discussion
These results demonstrate that DAzLE is a simple and efficient method for generating cDNAs highly enriched for differentially expressed genes. Due to the nature of our screen, we believe that we have a fairly comprehensive picture of the secondary events that occur during activity-dependent neuronal survival. DAzLE is a large-scale gene expression analysis system that can be used to comprehensively assess all of the mRNA transcripts of expressed genes in cells or tissue. In this study, DAzLE was engaged to screen 2 × 105 colonies of an unamplified cDNA library, which represents a 2.5-fold screen of the number of potential gene transcripts (≈80,000 gene transcripts in a single cell) (11). Additional screening would be required to confirm a comprehensive picture because the majority of the transcripts identified were unique in our screen. Multiple rounds of DAzLE at different time points after membrane depolarization would provide a comprehensive temporal profile of gene expression. Cherry picking genes that are differentially expressed by DAzLE coupled with custom microarray analysis will provide a powerful strategy to study the differential expression of genes in any given cell type, tissue, or experimental paradigm. Adaptation of this technique to include custom cDNA microarrays allows the analysis and initial identification of differentially expressed transcripts within 3 months and was successfully used to identify N-methyl-d-aspartate (NMDA)–glutamate receptor-induced late response genes (43).
A variety of strategies has been used to identify differentially expressed genes, including differential RNA display (20, 21), serial analysis of gene expression (SAGE) (11, 22), subtraction hybridization (23–25), reciprocal subtraction differential RNA display (26), representational difference analysis (RDA) (27), RNA fingerprinting by arbitrarily primed PCR (28), electronic subtraction (29), combinatorial matrix gene analysis (30), and rapid subtraction hybridization (31). All these methods have advantages and disadvantages. SAGE allows one to comprehensively and quantitatively genotype and compare gene expression between different populations of cells or different experimental paradigms. However, it has become clear with the cloning of the human genome that the 9- to 12-bp ditags isolated by SAGE are not sufficient to identify a single gene in the computer database. A search of short cDNA sequences can pull up hundreds of “hits” on the database. Longer ditags are required to increase the sensitivity of SAGE. Additionally, the method relies on a complete or complex computer database for the species genome being investigated. Lastly, SAGE works well for the identification of and description of relative changes in known genes. Functional analysis can then be performed. However, if the gene is novel, 9–12 bp is not sufficient to clone a gene. Conventional library screening, differential analysis, and subtractive hybridization have been successfully used to identify differentially expressed genes on a limited scale. Usually “10s” of genes are reported by using this methodology (23–25). Although RDA is a relatively powerful differential screening methodology, it is able to detect only 75–80% of the mRNA transcript population in a sample because the procedure depends on restriction endonucleases that may not recognize some of the cDNAs in the sample. RDA also typically identifies 10s of genes on routine screening (27, 32–34). DAzLE, is a technique designed to gain a direct and quantitative measure of gene expression, and it has many advantages. DAzLE is based mainly on representation of mRNA (cDNA) by long sequence fragments from a cDNA library, screening of these fragments with poly(A/T)-tailless–cDNA probes, and collection of differentially expressed colonies to elucidate the gene expression profile. Coupled with custom cDNA microarray, it is simple, elegant, and powerful. DAzLE dramatically increases the frequency of dynamically regulated genes by eliminating the characterization of constitutively and abundantly expressed genes and allows the direct isolation of differentially expressed clones for arraying, characterization, and storage. DAzLE is performed on nonamplified libraries. A nonamplified library ensures that a smaller cDNA population needs to be screened to obtain positive clones. Thus, DAzLE not only is able to detect medium- to high-abundance transcripts, but also reliably detects low-abundance transcripts, which is a significant advance over previous gene screening strategies. This capability is based on the fact that, by screening nonamplified libraries, rarely expressed genes have the same possibility to be surveyed with higher expressed genes. Screening is accomplished with poly(dA/T) tailless double-stranded DNA as the probes. Thus, DAzLE limits cross hybridization among the 3′ end of the sequences, further ensuring that rare transcripts are easily recovered as positive clones. The screening can ultimately cover 100% of the transcripts of the cell until positive clones have been duplicated. As an illustration of the power of the technique, <10% of the confirmed activity-dependent genes identified here and NMDA-dependent genes (43) were duplicated on one round of screening. Furthermore, a large percentage of the transcripts identified in both screens are genes of unknown function.
Our gene set represents a heretofore uncharacterized picture of the response of immature cortical neurons to membrane depolarization. It has minimal overlap with genes that were previously identified to be up-regulated by membrane depolarization because most prior screens focused on the identification of immediate and early response genes (35–38) and our screen was designed to identify late response genes that may be important for the activity-dependent neuronal survival that follows membrane depolarization. Accordingly, we failed to identify many previously characterized immediate and early response genes whose expression returns to baseline before 6 h. Some previously characterized activity-dependent induced genes were identified in our screen and include Hsc70, Sydecan, Ryndocan, PC3, and brain-derived neurotrophic factor (36, 39–41). Furthermore, most prior screens were performed in mature neurons, and this screen focused on the identification of activity-dependent genes in immature neurons and thus represents a previously uncharacterized gene set. Thus, the set of genes reported here might play potentially important roles in neurodevelopment and activity-dependent processes during maturation of the nervous system.
A number of genes involved in cell survival were identified, including brain-derived neurotrophic factor, neuronal apoptosis inhibitory protein, JNK-interacting protein-1, calmodulin, and 14-3-3, and these genes, as well as some of the genes of unknown function, may play important roles in activity-dependent neuronal survival. It is likely that after membrane depolarization there are a number of genes that contribute to neuronal survival. In addition, we identified a number of genes with diverse function that likely play important roles in the synaptic modeling/plasticity that follows membrane depolarization. A unique aspect of this screen was the identification of a subset of genes that are regulated by NO. We isolated 63 NO-dependent genes, including 29 genes of unknown function. Although NO has been clearly implicated as a mediator of NMDA-induced neurotoxicity (42), it is also emerging as a regulator of cell survival, and it plays important roles in activity-dependent neuronal survival of PC-12 cells (5) and immature cortical neurons (M. Gonzalez-Zulueta, V.L.D., and T.M.D., unpublished observations). Characterization of these transcripts may provide important insight into the role of the NO-induced transcriptional response that leads to neuronal survival after membrane depolarization.
It is not unexpected that membrane depolarization would induce such a complicated and diverse transcriptional response. It is clear from the expression data that several gene products contribute to the neuronal survival induced by activity and calcium influx. Characterization of many of these cell survival genes and their potential role in activity-dependent neuronal survival will lead to better understanding of the response of neurons to increases in intracellular calcium and the role of NO in these processes. Furthermore, identification and characterization of the genes of unknown function await further study but will provide the framework for a more comprehensive understanding of the molecular mechanisms underlying activity-dependent neuronal survival.
Supplementary Material
Acknowledgments
We thank Weza Cotman for manuscript preparation. This work was supported by U.S. Public Health Service Grants NS 37090 and DA 00266 and the National Alliance for Research on Schizophrenia and Depression (NARSAD) Foundation.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: DAzLE, differential analysis of primary cDNA library expression; NMDA, N-methyl-d-aspartate; En, embryonic day n; l-NAME, NG-nitro-l-arginine methyl ester; NOIG, NO-induced gene; SAGE, serial analysis of gene expression.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. CD567765–CD568226).
References
- 1.West, A. E., Chen, W. G., Dalva, M. B., Dolmetsch, R. E., Kornhauser, J. M., Shaywitz, A. J., Takasu, M. A., Tao, X. & Greenberg, M. E. (2001) Proc. Natl. Acad. Sci. USA 98, 11024–11031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Du, J., Feng, L., Yang, F. & Lu, B. (2000) J. Cell Biol. 150, 1423–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finkbeiner, S. & Greenberg, M. E. (1998) J. Neurobiol. 37, 171–189. [PubMed] [Google Scholar]
- 4.Finkbeiner, S., Tavazoie, S. F., Maloratsky, A., Jacobs, K. M., Harris, K. M. & Greenberg, M. E. (1997) Neuron 19, 1031–1047. [DOI] [PubMed] [Google Scholar]
- 5.Kim, T. W., Lee, C. H., Choi, C. Y., Kwon, N. S., Baek, K. J., Kim, Y. G. & Yun, H. Y. (2003) Neurosci. Lett. 344, 209–211. [DOI] [PubMed] [Google Scholar]
- 6.Grewal, S. S., York, R. D. & Stork, P. J. (1999) Curr. Opin. Neurobiol. 9, 544–553. [DOI] [PubMed] [Google Scholar]
- 7.Mao, Z., Bonni, A., Xia, F., Nadal-Vicens, M. & Greenberg, M. E. (1999) Science 286, 785–790. [DOI] [PubMed] [Google Scholar]
- 8.Ghosh, A., Carnahan, J. & Greenberg, M. E. (1994) Science 263, 1618–1623. [DOI] [PubMed] [Google Scholar]
- 9.Crawford, D. R., Kochheiser, J. C., Schools, G. P., Salmon, S. L. & Davies, K. J. (2002) Gene Expression 10, 101–107. [PMC free article] [PubMed] [Google Scholar]
- 10.Chilingaryan, A., Gevorgyan, N., Vardanyan, A., Jones, D. & Szabo, A. (2002) Math. Biosci. 176, 59–69. [DOI] [PubMed] [Google Scholar]
- 11.Velculescu, V. E., Zhang, L., Vogelstein, B. & Kinzler, K. W. (1995) Science 270, 484–487. [DOI] [PubMed] [Google Scholar]
- 12.Kohroki, J., Tsuchiya, M., Fujita, S., Nakanishi, T., Itoh, N. & Tanaka, K. (1999) Biochem. Biophys. Res. Commun. 262, 365–367. [DOI] [PubMed] [Google Scholar]
- 13.Burgess, J. K. (2001) Clin. Exp. Pharmacol. Physiol. 28, 321–328.11251648 [Google Scholar]
- 14.Stein, J. & Liang, P. (2002) Cell Mol. Life Sci. 59, 1235–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tavares, A. B., Esplin, M. S. & Adashi, E. Y. (2001) Semin. Reprod. Med. 19, 167–173. [DOI] [PubMed] [Google Scholar]
- 16.von Stein, O. D. (2001) Methods Mol. Biol. 175, 263–278. [DOI] [PubMed] [Google Scholar]
- 17.Jenssen, T. K., Laegreid, A., Komorowski, J. & Hovig, E. (2001) Nat. Genet. 28, 21–28. [DOI] [PubMed] [Google Scholar]
- 18.Vogeli, G. & Kaytes, P. S. (1987) Methods Enzymol. 152, 407–415. [DOI] [PubMed] [Google Scholar]
- 19.Toescu, E. C. (1999) Neuroscience 94, 561–570. [DOI] [PubMed] [Google Scholar]
- 20.Liang, P. & Pardee, A. B. (1992) Science 257, 967–971. [DOI] [PubMed] [Google Scholar]
- 21.Shen, R., Su, Z. Z., Olsson, C. A. & Fisher, P. B. (1995) Proc. Natl. Acad. Sci. USA 92, 6778–6782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang, L., Zhou, W., Velculescu, V. E., Kern, S. E., Hruban, R. H., Hamilton, S. R., Vogelstein, B. & Kinzler, K. W. (1997) Science 276, 1268–1272. [DOI] [PubMed] [Google Scholar]
- 23.Jiang, H., Lin, J. J., Su, Z. Z., Goldstein, N. I. & Fisher, P. B. (1995) Oncogene 11, 2477–2486. [PubMed] [Google Scholar]
- 24.Su, Z. Z., Shi, Y. & Fisher, P. B. (1997) Proc. Natl. Acad. Sci. USA 94, 9125–9130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sagerstrom, C. G., Sun, B. I. & Sive, H. L. (1997) Annu. Rev. Biochem. 66, 751–783. [DOI] [PubMed] [Google Scholar]
- 26.Kang, D. C., LaFrance, R., Su, Z. Z. & Fisher, P. B. (1998) Proc. Natl. Acad. Sci. USA 95, 13788–13793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hubank, M. & Schatz, D. G. (1994) Nucleic Acids Res. 22, 5640–5648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McClelland, M., Mathieu-Daude, F. & Welsh, J. (1995) Trends Genet. 11, 242–246. [DOI] [PubMed] [Google Scholar]
- 29.Adams, M. D., Kerlavage, A. R., Fields, C. & Venter, J. C. (1993) Nat. Genet. 4, 256–267. [DOI] [PubMed] [Google Scholar]
- 30.Schena, M., Shalon, D., Davis, R. W. & Brown, P. O. (1995) Science 270, 467–470. [DOI] [PubMed] [Google Scholar]
- 31.Jiang, H., Kang, D. C., Alexandre, D. & Fisher, P. B. (2000) Proc. Natl. Acad. Sci. USA 97, 12684–12689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cooper, P., Mueck, B., Yousefi, S., Potter, S. & Jarai, G. (2000) Am. J. Physiol. 278, L284–L293. [DOI] [PubMed] [Google Scholar]
- 33.Iwama, A., Zhang, P., Darlington, G. J., McKercher, S. R., Maki, R. & Tenen, D. G. (1998) Nucleic Acids Res. 26, 3034–3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frazer, J. K., Pascual, V. & Capra, J. D. (1997) J. Immunol. Methods 207, 1–12. [DOI] [PubMed] [Google Scholar]
- 35.Qian, Z., Gilbert, M. E., Colicos, M. A., Kandel, E. R. & Kuhl, D. (1993) Nature 361, 453–457. [DOI] [PubMed] [Google Scholar]
- 36.Nedivi, E., Hevroni, D., Naot, D., Israeli, D. & Citri, Y. (1993) Nature 363, 718–722. [DOI] [PubMed] [Google Scholar]
- 37.Lyford, G. L., Yamagata, K., Kaufmann, W. E., Barnes, C. A., Sanders, L. K., Copeland, N. G., Gilbert, D. J., Jenkins, N. A., Lanahan, A. A. & Worley, P. F. (1995) Neuron 14, 433–445. [DOI] [PubMed] [Google Scholar]
- 38.Brakeman, P. R., Lanahan, A. A., O'Brien, R., Roche, K., Barnes, C. A., Huganir, R. L. & Worley, P. F. (1997) Nature 386, 284–288. [DOI] [PubMed] [Google Scholar]
- 39.Nedivi, E. (1999) J. Neurobiol. 41, 135–147. [PMC free article] [PubMed] [Google Scholar]
- 40.Hevroni, D., Rattner, A., Bundman, M., Lederfein, D., Gabarah, A., Mangelus, M., Silverman, M. A., Kedar, H., Naor, C., Kornuc, M., et al. (1998) J. Mol. Neurosci. 10, 75–98. [DOI] [PubMed] [Google Scholar]
- 41.Robertson, H. A. (1992) Biochem. Cell Biol. 70, 729–737. [DOI] [PubMed] [Google Scholar]
- 42.Samdani, A. F., Dawson, T. M. & Dawson, V. L. (1997) Stroke 28, 1283–1288. [DOI] [PubMed] [Google Scholar]
- 43.Hong, S.-J., Li, H., Becker, K. G., Dawson, V. L. & Dawson, T. M. (2003) Proc. Natl. Acad. Sci. USA, in press.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.