Abstract
Purpose
To identify non-housekeeping genes definitively expressed in the human trabecular meshwork (TM).
Methods
Microarray gene expression data on TM cultured cells from four studies were compared. Genes that were queried in at least three studies and assessed to be expressed in at least three studies were considered definitively expressed genes of the human TM. Housekeeping genes were removed from this set of genes. The non-housekeeping TM gene profile was analyzed for pathway enrichment and microRNA targeting, using bioinformatics tools. The results were compared with results of previous non-array based studies.
Results
Nine hundred and sixty-two genes were identified as non-housekeeping TM expressed genes. Analysis of these by Kyoto Encyclopedia of Genes and Genomes led to identification of two enriched biologic pathways that achieved a highly significant Bonferroni p-value (p≤0.01): focal adhesion and extracellular matrix (ECM)-receptor interaction. Many of the genes were previously implicated in TM-related functions and the TM-associated disease glaucoma; however, some are novel. MicroRNAs known to be expressed in the trabecular meshwork were predicted to target some of the genes. Ten genes identified here, ALDH1A1 (aldehyde dehydrogenase 1 family, member A1), CDH11 (cadherin 11, type 2, OB-cadherin), CXCR7 (chemokine (C-X-C motif) receptor 7), CHI3L1 (chitinase 3-like 1), FGF2 (fibroblast growth factor 2), GNG11 (guanine nucleotide binding protein [G protein], gamma 11), IGFBP5 (insulin-like growth factor binding protein 5), PTPRM (protein tyrosine phosphatase, receptor type, M), RGS5 (regulator of G-protein signaling 5), and TUSC3 (tumor suppressor candidate 3), were also reported as TM expressed genes in three earlier non-microarray based studies.
Conclusions
A transcriptome consisting of 962 non-housekeeping genes definitively expressed in the human TM was identified. Multiple genes and microRNAs are proposed for further study for a better understanding of TM physiology.
Introduction
The trabecular meshwork (TM) is located at the angle formed by the cornea and iris. The tissue is a major component of the conventional outflow pathway of aqueous humor and, thus, significantly modulates outflow of this fluid from the anterior chamber to venous blood via Schlemm’s canal [1]. Decreased outflow resulting from increased TM resistance causes increased intraocular pressure (IOP), which is a major risk factor for primary open angle glaucoma (POAG) [2-4]. The TM and aqueous humor also have roles in physiologic processes involving detoxification reactions and the immune system [5]. The precise mechanisms by which the TM affects these processes and regulates intraocular pressure are largely unknown. Analysis of the transcriptome of the TM and changes in the transcriptome due to conditions relevant to glaucoma may help illuminate these mechanisms. To this end, several studies have been performed within the past decade. Initially, human TM expressed genes were assessed by sequencing some clones of TM cDNA libraries [6,7]. Since the introduction of high-density chips for assessing gene expression profiles, four genome-wide TM transcriptome analyses that addressed glaucoma-relevant parameters and that included data on untreated TM cells have been published [8-11]. In one of the studies, gene expression profiles of cultured and native TM cells were compared, and it was reported that there was more than 90% similarity between expressed genes [10]. The remaining three studies used cultured TM cells. The data on the control cultures that were untreated in each of the four global studies allow assessment of the TM transcriptome. However, the profiles for the control cultures in these studies were not the same. The variations are due to a combination of factors, including individual variations, the complement of genes queried on the different chips used, and technical parameters of the hybridization reactions and analyses protocols. Here, we report results of a meta-analysis based on the four studies, with the objective of defining a TM transcriptome that is most likely to include only true positive non-housekeeping genes expressed in primary TM cultures of various sources. Housekeeping genes were designated by two recent studies on gene expression profiles of multiple human tissues using microarray and RNA deep sequencing data [12,13]. The derived non-housekeeping TM gene expression profile was analyzed using bioinformatics tools. The results were compared with those of previous studies on gene expression in normal human TM, including results of a recent study based on serial analysis of gene expression (SAGE) [6,7,14].
Methods
Microarray gene expression data on control TM cultured cells from four studies were downloaded [8-11] (Gene Expression Omnibus [GEO] accession numbers GSE492, GSE4316, GSE7144, and GSE27275). Features of the microarray studies, including source of TM RNAs, chips used, and number of genes queried on the chips, are presented in Table 1. For the three studies in which Affymetrix chips were used, a hybridization threshold signal of 200 was used for expressed genes [8-10]. The same cutoff value was applied in the microarray study used here to define housekeeping genes; the cutoff was based on results of hybridizations to negative control probes placed on these chips and derived with the objective of achieving an optimal false positive to false negative ratio [12]. Others have also used the same cutoff [15]. Earlier studies based on instrument parameters and statistical analysis have recommended that signals lower than 200 on Affymetrix chips should not be considered in gene expression studies [16,17]. For the Illumina chips, a hybridization cutoff signal was chosen so that approximately the same fraction (50%) of genes queried on Affymetrix and Illumina chips would be considered genes expressed in the human TM. Hybridization signal of 200 was surpassed for almost all genes queried on the Illumina chips. Genes were grouped based on the number of studies in which they were queried. Subsequently, expressed genes were assessed with consideration of the number of studies in which hybridization thresholds were met and the number of studies in which they were queried. Only genes that were queried in at least three studies, and whose hybridization signals met the threshold in at least three studies, were considered definitively expressed genes of the human TM. Housekeeping (HK) genes were removed from this set of genes. Results of two recent studies that aimed to identify ubiquitously expressed genes in human cells were used to define housekeeping genes. One study, based on publically available microarray gene expression data on 18 human tissues, identified 2,403 genes expressed in at least 16 of those tissues [12]. The second study was more recent, and it was based on RNA deep sequencing data derived from diverse tissues and cell culture samples; it identified 7,897 ubiquitously expressed genes. Genes included in one or both of these studies (8,416 genes) were removed from the aforementioned list of definitively expressed genes of the human TM to derive a list of definitively expressed non-housekeeping genes of the human TM (non-HK TM genes). This set of genes was used in subsequent in silico analyses. Enriched functional pathways and functional categories or gene ontology terms annotated by Kyoto Encyclopedia of Genes and Genomes (KEGG) [18] and GO [19] were identified using The Database for Annotation, Visualization and Integrated Discovery (DAVID) bioinformatics resource [20]. A cutoff Bonferroni-p value of 0.01 was used for enriched KEGG pathways or GO functions. Non-HK TM genes whose expressions were altered by glaucoma-relevant experimental manipulations were identified. MicroRNAs whose target genes were enriched in the non-HK TM genes were also identified, using BioProfiling. Finally, non-HK TM genes identified here were compared to TM transcriptomes assessed by non-microarray protocols. The significance of the findings is discussed.
Table 1. Features of microarray studies used to assess gene expression in human trabecular meshwork.
| Reference of study | Source of data | Microarray chip | No. genes queried* | No. genes expressed (%)** |
|---|---|---|---|---|
| [8] |
Pooled RNA of 5 pairs of eyes; average of duplicate chips |
Affymetrix Genome U95Av2Human |
8963 |
4419 (49.3%) |
| [9] |
Pooled RNA of 5 pairs of eyes; average of duplicate chips |
Affymetrix Human Genome U133A |
12962 |
7117 (54.9%) |
| [10] |
RNA of 3 pairs of eyes; one chip per individual; average of 3 chips |
Affymetrix Human Genome U133 plus 2.0 |
20203 |
10356 (51.2%) |
| [11] | RNA of 2 pairs of eyes; two chips per individual; average of 4 chips | Illumina HumanRf-8 V2 | 17133 | 8566 (50.0%) |
* http://www.affymetrix.com/support/technical/libraryfilesmain.affx and http://www.illumina.com/support/annotation_files.ilmn. ** As per criteria described in text.
Results
Approximately 50% of the genes queried on the chips of each study were expressed in the human TM (Table 1). A total of 21,813 genes were queried; the number of genes queried one, two, three, or four times are shown in Figure 1, and the genes are listed in Appendix 1. Of these, 5,379 genes met expression threshold criteria in at least three studies, and they are considered definitively expressed genes of the human TM. The majority of these (4,417 genes; 82%) were housekeeping genes; 962 (18%) were non-HK TM expressed genes (Appendix 1). Analysis of the 962 genes by KEGG led to identification of two enriched biologic pathways that achieved a highly significant Bonferroni p-value (p≤0.01): focal adhesion (Bonferroni p=0.00275) and extracellular matrix (ECM)-receptor interaction (Bonferroni p=0.00824). A third pathway, melanoma, achieved a significant Bonferroni p-value (p=0.01634). The non-HK TM genes involved in the focal adhesion and ECM-receptor interaction pathways are presented in Table 2. Analysis of the 962 genes by GO led to identification of many terms that achieved a highly significant Bonferroni p-value within the categories of biologic processes, molecular function, and cell component (Table 3). Related biologic processes were merged manually, on the basis of similarity between the processes and the fraction of non-HK TM genes associated with these processes, as shown in Figure 2. Genes whose expressions had been reported previously as changed due to glaucoma-relevant experimental manipulations were considered next [8-11]. The distribution of these genes among housekeeping and non-HK TM expressed genes is shown in Table 4, and the genes included among the non-HK genes are presented in supplementary Appendix 2. There was a trend toward preferential distribution of the genes within the non-HK TM class of genes in all four studies tested, although the difference reached statistical significance in only two of the studies. Notably, the preference is not observed in a study related to a non-TM ocular tissue [21] that was tested for comparison (Table 4). Subsequently, the 962 non-HK genes were submitted to BioProfiling to identify miRNAs that may preferentially target non-HK TM genes. No miRNA with a significant p-value was identified. However, enriched miRNA targets were identified for functionally clustered non-HK TM genes. Initially, the 962 non-HK TM genes were clustered into 26 groups by the Gene Functional Classification option within DAVID (Appendix 3). Eleven of the groups contained more than ten non-HK TM genes, and each of these groups was submitted separately to BioProfiling; ten is the default minimum number of genes accepted by this software. MicroRNAs with target gene enrichment (p≤0.05) were identified for four of the groups (Table 5).
Figure 1.
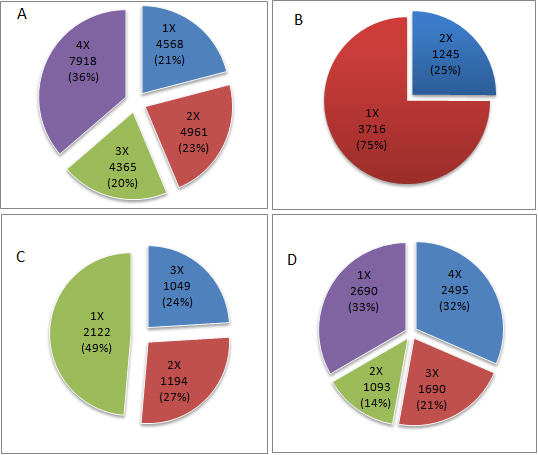
Number of genes expressed in human TM/number of times genes were queried on chips. A: Number of genes queried on chips of 1, 2, 3, or 4 of the microarray studies of this meta-analysis (% of all 21,813 genes queried at least once). B: Among genes queried 2×, number of genes assessed to be expressed in TM 1× or 2× (% genes queried 2×). C: Among genes queried 3×, number of genes assessed to be expressed in TM 1, 2, or 3× (% genes queried 3×). D: Among genes queried 4×, number of genes assessed to be expressed in TM 1, 2,3, or 4× (% genes queried 4×).
Table 2. Biologic pathways enriched by KEGG analysis of non-housekeeping human TM expressed genes (Bonferroni p≤0.01).
| Pathway | Number of genes | Gene |
|---|---|---|
| Focal adhesion (Bonferroni p=0.00275) |
30 |
CAV2, CAV1, TLN2, PIP5K1C, VTN, MYL9, COL6A3, COL6A2, PDGFC, COL11A1, THBS2, EGFR, COL4A2, COL4A1, PIK3CD, MET, ITGA2, HGF, FLNC, COL5A2, COL5A1, VEGFC, LAMA4, CCND2, ITGA5, ITGA7, PDGFRA, PDGFRB, COL1A1, MYLK |
| ECM-receptor interaction (Bonferroni p=0.00824) | 17 | COL4A2, COL4A1, HSPG2, ITGA2, VTN, COL5A2, COL5A1, LAMA4, SDC1, ITGA5, ITGA7, COL6A3, COL6A2, SV2A, COL1A1, THBS2, COL11A1 |
Table 3. Gene ontology terms enriched by analysis of non-housekeeping human TM expressed genes (Bonferroni p≤0.01).
| Category | Term | Gene annotation | No. genes | Bonferroni p-value |
|---|---|---|---|---|
| Biologic process |
GO:0048731 |
System development |
227 |
1.88E-18 |
| Biologic process |
GO:0032502 |
Developmental process |
280 |
2.45E-18 |
| Biologic process |
GO:0048856 |
Anatomic structure development |
239 |
4.89E-18 |
| Biologic process |
GO:0007275 |
Multicellular organismal development |
259 |
2.44E-17 |
| Biologic process |
GO:0048513 |
Organ development |
177 |
4.28E-15 |
| Biologic process |
GO:0009653 |
Anatomic structure morphogenesis |
134 |
1.13E-13 |
| Biologic process |
GO:0042127 |
Regulation of cell proliferation |
95 |
1.88E-10 |
| Biologic process |
GO:0032501 |
Multicellular organismal process |
320 |
7.73E-10 |
| Biologic process |
GO:0030198 |
Extracellular matrix organization |
28 |
1.08E-08 |
| Biologic process |
GO:0051270 |
Regulation of cell motion |
38 |
2.41E-08 |
| Biologic process |
GO:0007399 |
Nervous system development |
109 |
3.33E-07 |
| Biologic process |
GO:0042060 |
Wound healing |
36 |
3.35E-07 |
| Biologic process |
GO:0040012 |
Regulation of locomotion |
36 |
3.89E-07 |
| Biologic process |
GO:0042221 |
Response to chemical stimulus |
122 |
5.17E-07 |
| Biologic process |
GO:0022603 |
Regulation of anatomic structure morphogenesis |
38 |
1.10E-06 |
| Biologic process |
GO:0043062 |
Extracellular structure organization |
32 |
1.52E-06 |
| Biologic process |
GO:0006928 |
Cell motion |
60 |
2.88E-06 |
| Biologic process |
GO:0030334 |
Regulation of cell migration |
32 |
3.86E-06 |
| Biologic process |
GO:0001944 |
Vasculature development |
40 |
4.58E-06 |
| Biologic process |
GO:0032879 |
Regulation of localization |
70 |
5.99E-06 |
| Biologic process |
GO:0009888 |
Tissue development |
74 |
7.37E-06 |
| Biologic process |
GO:0001568 |
Blood vessel development |
39 |
7.86E-06 |
| Biologic process |
GO:0050793 |
Regulation of developmental process |
74 |
1.33E-05 |
| Biologic process |
GO:0030154 |
Cell differentiation |
141 |
1.38E-05 |
| Biologic process |
GO:0048522 |
Positive regulation of cellular process |
154 |
2.02E-05 |
| Biologic process |
GO:0009887 |
Organ morphogenesis |
65 |
2.12E-05 |
| Biologic process |
GO:0040017 |
Positive regulation of locomotion |
23 |
2.16E-05 |
| Biologic process |
GO:0051272 |
Positive regulation of cell motion |
23 |
2.16E-05 |
| Biologic process |
GO:0051239 |
Regulation of multicellular organismal process |
92 |
3.57E-05 |
| Biologic process |
GO:0008284 |
Positive regulation of cell proliferation |
52 |
5.50E-05 |
| Biologic process |
GO:0048869 |
Cellular developmental process |
143 |
6.15E-05 |
| Biologic process |
GO:0009611 |
Response to wounding |
61 |
7.10E-05 |
| Biologic process |
GO:0006950 |
Response to stress |
141 |
8.91E-05 |
| Biologic process |
GO:0022008 |
Neurogenesis |
66 |
1.06E-04 |
| Biologic process |
GO:0048518 |
Positive regulation of biologic process |
162 |
1.65E-04 |
| Biologic process |
GO:0009605 |
Response to external stimulus |
88 |
1.99E-04 |
| Biologic process |
GO:0048514 |
Blood vessel morphogenesis |
33 |
2.49E-04 |
| Biologic process |
GO:0007167 |
Enzyme linked receptor protein signaling pathway |
44 |
4.24E-04 |
| Biologic process |
GO:0001501 |
Skeletal system development |
42 |
4.53E-04 |
| Biologic process |
GO:0048699 |
Generation of neurons |
61 |
4.77E-04 |
| Biologic process |
GO:0048519 |
Negative regulation of biologic process |
146 |
4.87E-04 |
| Biologic process |
GO:0030335 |
Positive regulation of cell migration |
20 |
5.44E-04 |
| Biologic process |
GO:0048523 |
Negative regulation of cellular process |
136 |
5.95E-04 |
| Biologic process |
GO:0007155 |
Cell adhesion |
71 |
6.80E-04 |
| Biologic process |
GO:0022610 |
Biologic adhesion |
71 |
7.21E-04 |
| Biologic process |
GO:0007169 |
Transmembrane receptor protein tyrosine kinase signaling pathway |
33 |
0.001 |
| Biologic process |
GO:0065008 |
Regulation of biologic quality |
122 |
0.0015 |
| Biologic process |
GO:0016477 |
Cell migration |
37 |
0.0018 |
| Biologic process |
GO:0008285 |
Negative regulation of cell proliferation |
44 |
0.0019 |
| Biologic process |
GO:0032101 |
Regulation of response to external stimulus |
26 |
0.0033 |
| Biologic process |
GO:0043067 |
Regulation of programmed cell death |
76 |
0.0053 |
| Biologic process |
GO:0035295 |
Tube development |
31 |
0.0063 |
| Biologic process |
GO:0010941 |
Regulation of cell death |
76 |
0.0064 |
| Biologic process |
GO:0042981 |
Regulation of apoptosis |
75 |
0.0074 |
| Biologic process |
GO:0051674 |
Localization of cell |
38 |
0.0088 |
| Biologic process |
GO:0048870 |
Cell motility |
38 |
0.0088 |
| Cellular component |
GO:0044421 |
Extracellular region part |
109 |
7.78E-13 |
| Cellular component |
GO:0005576 |
Extracellular region |
172 |
7.52E-10 |
| Cellular component |
GO:0031012 |
Extracellular matrix |
51 |
7.34E-09 |
| Cellular component |
GO:0044459 |
Plasma membrane part |
179 |
1.96E-08 |
| Cellular component |
GO:0031226 |
Intrinsic to plasma membrane |
114 |
5.43E-08 |
| Cellular component |
GO:0005578 |
Proteinaceous extracellular matrix |
47 |
6.44E-08 |
| Cellular component |
GO:0005887 |
Integral to plasma membrane |
110 |
2.62E-07 |
| Cellular component |
GO:0005615 |
Extracellular space |
70 |
1.51E-05 |
| Cellular component |
GO:0005886 |
Plasma membrane |
257 |
2.57E-05 |
| Cellular component |
GO:0005581 |
Collagen |
13 |
3.45E-05 |
| Cellular component |
GO:0044420 |
Extracellular matrix part |
23 |
3.87E-05 |
| Cellular component |
GO:0005624 |
Membrane fraction |
73 |
9.01E-04 |
| Cellular component |
GO:0000267 |
Cell fraction |
89 |
0.0027 |
| Cellular component |
GO:0005626 |
Insoluble fraction |
73 |
0.0032 |
| Molecular function |
GO:0005515 |
Protein binding |
509 |
4.31E-06 |
| Molecular function |
GO:0005102 |
Receptor binding |
86 |
4.26E-05 |
| Molecular function |
GO:0005201 |
Extracellular matrix structural constituent |
19 |
4.31E-04 |
| Molecular function |
GO:0008201 |
Heparin binding |
20 |
0.0015 |
| Molecular function |
GO:0008083 |
Growth factor activity |
25 |
0.0038 |
| Molecular function |
GO:0005539 |
Glycosaminoglycan binding |
23 |
0.004 |
| Molecular function |
GO:0030247 |
Polysaccharide binding |
24 |
0.0059 |
| Molecular function |
GO:0001871 |
Pattern binding |
24 |
0.0059 |
| Molecular function | GO:0004714 | Transmembrane receptor protein tyrosine kinase activity | 15 | 0.0084 |
Figure 2.
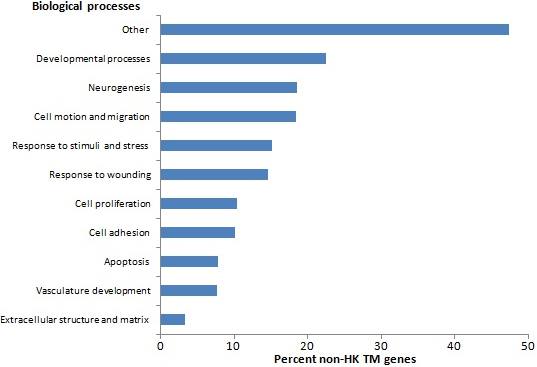
Percent of non-HK TM expressed genes enriched in related gene ontology biologic processes.
Table 4. Distribution of genes with altered expression due to experimental manipulations among housekeeping and non-HK TM expressed genes*.
| Experimental manipulation | Reference | (No.) % HK TM genes affected by experimental manipulation* | (No.) % non- HK TM genes affected by experimental manipulation* | Statistical significance | % non-HK/% HK TM genes affected by experimental manipulation |
|---|---|---|---|---|---|
| Treatment with prostoglandin analogs |
8 |
(25) 0.57 |
(8) 0.83 |
NS (p<0.50) |
1.47 |
| TGFβ¥ treatment |
9 |
(35) 0.79 |
(49) 5.09 |
p<0.001 |
6.43 |
| Glaucoma versus non-glaucoma TM cells |
10 |
(6) 0.14 |
(5) 0.51 |
NS (p~0.10) |
3.82 |
|
PITX2 knockdown |
11 |
(11) 0.25 |
(9) 0.94 |
p~0.001 |
3.76 |
| Effect of aging on retina& | 21 | (8) 0.18 | (1) 0.10 | NS (p<0.75) | 0.56 |
*Affected genes are as reported in respective studies, and HK and non-HK genes refer to genes identified in present study; ¥ transforming growth factor β; & represents a control treatment relating to a non-TM ocular tissue; NS=not significant.
Table 5. MicroRNAs with target enrichment in functionally grouped non-HK TM genes*.
| Group no. | miRNA with enriched targets | No. target genes (No. genes in group) | Target genes | p value |
|---|---|---|---|---|
| 3 |
HSA-MIR-29A.5 |
8 (10) |
COL1A1, COL4A1, COL4A2, COL4A5, COL5A1, COL5A2, COL7A1, COL11A1 |
2.85E-09 |
| 3 |
HSA-MIR-29C.5 |
8 (10) |
COL1A1, COL4A1, COL4A2, COL4A5, COL5A1, COL5A2, COL7A1, COL11A1 |
5.12E-09 |
| 3 |
HSA-MIR-29B.5 |
7 (10) |
COL4A1, COL4A2, COL4A5, COL5A1, COL5A2, COL7A1, COL11A1 |
3.91E-07 |
| 3 |
HSA-MIR-29A.4 |
8 (10) |
COL1A1, COL4A1, COL4A2, COL4A5, COL5A1, COL5A2, COL7A1, COL11A1 |
8.04E-07 |
| 3 |
HSA-MIR-29B.4 |
8 (10) |
COL1A1, COL4A1, COL4A2, COL4A5, COL5A1, COL5A2, COL7A1, COL11A1 |
9.36E-07 |
| 3 |
HSA-MIR-29C.4 |
8 (10) |
COL1A1, COL4A1, COL4A2, COL4A5, COL5A1, COL5A2, COL7A1, COL11A1 |
1.05E-06 |
| 3 |
HSA-LET-7A.4 |
6 (10) |
COL1A1, COL4A1, COL4A2, COL4A5, COL5A1, COL5A2 |
0.0014 |
| 3 |
HSA-LET-7C.4 |
6 (10) |
COL1A1, COL4A1, COL4A2, COL4A5, COL5A1, COL5A2 |
0.0015 |
| 3 |
HSA-LET-7B.4 |
6 (10) |
COL1A1, COL4A1, COL4A2, COL4A5, COL5A1, COL5A2 |
0.0017 |
| 3 |
HSA-LET-7G.4 |
5 (10) |
COL1A1, COL4A1, COL4A2, COL4A5, COL5A2 |
0.01 |
| 3 |
HSA-LET-7F.4 |
5 (10) |
COL1A1, COL4A1, COL4A2, COL4A5, COL5A2 |
0.01 |
| 3 |
HSA-LET-7E.4 |
5 (10) |
COL1A1, COL4A1, COL4A2, COL4A5, COL5A2 |
0.01 |
| 3 |
HSA-LET-7I.4 |
5 (10) |
COL1A1, COL4A1, COL4A2, COL4A5, COL5A2 |
0.01 |
| 3 |
HSA-MIR-98.4 |
5 (10) |
COL1A1, COL4A1, COL4A2, COL4A5, COL5A2 |
0.01 |
| 11 |
HSA-MIR-657.4 |
5 (13) |
FKTN, UST, GALNT10, XYLT1, GLT8D2 |
0.01 |
| 13 |
HSA-MIR-517B.4 |
5 (49) |
SLC2A3, SCARA3, ORAI2, MBOAT2, C9ORF91 |
0.01 |
| 13 |
HSA-MIR-188–3P.4 |
5 (49) |
SLC7A11, FAM119B, D4S234E, FAM171A1, FAM155A |
0.03 |
| 14 |
HSA-MIR-26A.5 |
6 (33) |
CDK6, DAPK1, PDGFRA, MAP3K2, STK39, TRIB2 |
0.01 |
| 14 |
HSA-MIR-34A.5 |
5 (33) |
AXL, CDK6, EPHA4, MET, PDGFRA |
0.01 |
| 14 |
HSA-MIR-34C-5P.4 |
7 (33) |
AXL, CDK6, EPHA4, MET, PDGFRA, IKBKE, STK39 |
0.01 |
| 14 |
HSA-MIR-34A.4 |
6 (33) |
AXL, CDK6, EPHA4, MET, PDGFRA, PRKD1 |
0.01 |
| 14 |
HSA-MIR-26B.4 |
7 (33) |
CDK6, DAPK1, PDGFRA, WEE1, MAP3K2, STK39, TRIB2 |
0.02 |
| 14 |
HSA-MIR-381.4 |
7 (33) |
EPHA4, GRK5, MET, RPS6KA3, TEK, WEE1, ZAK |
0.02 |
| 14 |
HSA-MIR-449B.4 |
5 (33) |
AXL, CDK6, EPHA4, PDGFRA, IKBKE |
0.02 |
| 14 |
HSA-MIR-26A.4 |
7 (33) |
CDK6, DAPK1, PDGFRA, WEE1, MAP3K2, STK39, TRIB2 |
0.02 |
| 14 |
HSA-MIR-519C-5P.4 |
8 (33) |
AXL, CDK6, EPHA4, MYLK, PTK7, RPS6KA3, MAP3K2, ZAK |
0.02 |
| 14 |
HSA-MIR-26B.5 |
5 (33) |
CDK6, DAPK1, MAP3K2, STK39, TRIB2 |
0.02 |
| 14 |
HSA-MIR-485–3P.4 |
5 (33) |
CDK6, DAPK1, MET, MELK, TRIB2 |
0.03 |
| 14 |
HSA-MIR-323–3P.4 |
6 (33) |
DAPK1, MET, RPS6KA3, SGK1, MELK, MAP3K2 |
0.03 |
| 14 |
HSA-MIR-32.4 |
6 (33) |
AXL, CDK6, SGK1, AATK, STK39, ZAK |
0.04 |
| 18 |
HSA-MIR-124.4 |
7 (19) |
AHR, E2F3, ELF4, ETS1, MITF, PLAGL2, SOX9 |
0.01 |
| 18 |
HSA-MIR-145.4 |
5 (19) |
KLF5, ETS1, MAF, PLAGL2 SOX9 |
0.02 |
| 18 | HSA-MIR-32.4 | 5 (19) | E2F3, GATA2, MITF, SOX4, HAND1 | 0.02 |
* miRNAs identified using BioProfiling under stringent setting; gene function groups identified by classification tool of DAVID.
Finally, the TM expressed genes identified here were compared to TM expressed genes reported earlier on the basis of sequencing in two TM cDNA libraries and one SAGE experiment [6,7,14]. As expected, most housekeeping genes commonly used as reference genes in expression studies, and expected to be expressed at high levels, were identified in all four studies (Table 6A). HPRT1 (hypoxanthine phosphoribosyltransferase 1) was identified only in the microarray analysis presented here. Among the five known glaucoma-causing genes– MYOC (myocilin), CYP1B1 (cytochrome P450, family 1, subfamily B, polypeptide 1), OPTN (optineurin), WDR36 (WDR36), and LTBP2 (latent transforming growth factor beta binding protein 2)–all except WDR36 were identified in three of the studies (Table 6B) [22,23]. WDR36 was not identified in any of the studies. FOXC1 (forkhead box C1) and PITX2 (paired-like homeodomain 2), genes associated with glaucoma-related Axenfeld-Rieger syndrome, were also identified in three of the studies [24]. The study in which the glaucoma-related genes were reported identified the lowest (386) number of TM expressed genes and, probably, only those highly expressed [6]. Only ten non-HK TM genes identified in the present analysis were identified in all four studies, and these are most likely to be highly expressed and consistently expressed non-housekeeping genes of the human TM (Table 7). The ten genes were ALDH1A1 (aldehyde dehydrogenase 1 family, member A1), CDH11 (cadherin 11, type 2, OB-cadherin), CXCR7 (chemokine (C-X-C motif) receptor 7), CHI3L1 (chitinase 3-like 1), FGF2 (fibroblast growth factor 2), GNG11 (guanine nucleotide binding protein (G protein), gamma 11), IGFBP5 (insulin-like growth factor binding protein 5), PTPRM (protein tyrosine phosphatase, receptor type, M), RGS5 (regulator of G-protein signaling 5), and TUSC3 (tumor suppressor candidate 3). Among the interesting features of the ten genes is that five (ALDH1A1, CDH11, CH13L1, FGF2, and IGFBP5) were predicted by TargetScanHuman to be targeted by microRNA-23a and −23b cluster microRNAs (mir-23ab, mir-24, mir-27ab) and mir-204/211 (Table 7). Mir-24 and mir-204/211 are microRNAs experimentally confirmed to be expressed in the TM and to affect expression of some glaucoma-relevant genes [25,26]. Furthermore, the expression of mir-27, which was predicted to target CH13L1 and CXCR7, may be regulated by the glaucoma-relevant transcription factor PITX2 [27]. The ten genes identified in all the studies are further discussed in the discussion section below.
Table 6. TM expressed genes identified in microarray, cDNA library, and SAGE studies.
|
|
Study |
||||
|---|---|---|---|---|---|
|
|
Microarray studiesα |
|
|
|
|
| Gene | All genes | Non-HK genes | cDNA library 1β | cDNA library 2γ | SAGEδ |
|
A. Commonly used housekeeping genes | |||||
|
GAPDH |
+ |
- |
+ |
+ |
+ |
|
ACTB |
+ |
- |
+ |
+ |
+ |
|
B2M |
+ |
- |
+ |
+ |
+ |
|
HPRT1 |
+ |
- |
- |
- |
- |
|
B. Known glaucoma causing genes | |||||
|
CYP1B1 |
+ |
+ |
- |
+ |
+ |
|
MYOC |
+ |
+ |
- |
+ |
+ |
|
OPTN |
+ |
- |
- |
+ |
+ |
|
WDR36 |
- |
- |
- |
- |
- |
|
LTBP2 |
+ |
- |
- |
+ |
+ |
|
FOXC1 |
+ |
+ |
- |
- |
+ |
| PITX2 | + | + | - | + | + |
Table 7. Ten genes identified in all studies and miRNAs predicted to target these genes.
| Gene | miRNAs* |
|---|---|
|
ALDH1A1 |
mir-23ab |
|
CDH11 |
mir-27ab, mir-144, mir-128, mir-26ab/1297, mir-214/761, mir-200bc/429, mir-101 |
|
CXCR7 |
mir-27ab, mir-142, mir-607, mir-524–5p, mir-520d-5p, mir-let7 |
|
CHI3L1 |
mir-24 |
|
FGF2 |
mir-23ab |
|
GNG11 |
mir-587 |
|
IGFBP5 |
mir-24, mir-204/211, mir-130/301, mir-137, mir-139, mir-140, mir-193ab |
|
PTPRM |
mir-130/301, mir-34abc, mir-205, mir-218, mir-148, mir-152 |
|
RGS5 |
mir-182 |
| TUSC3 | mir-30abc |
* Predicted by TargetScan.
Discussion
The objective of this study was to identify non-housekeeping genes definitively expressed in human TM cultured cells. We aimed to minimize the consequences of individual variability and experimental techniques. To this end, we performed meta-analysis on four available microarray-based global gene expression studies on human TM cultured cells. The data was based on gene expression in 30 eyes of 15 individuals, 16–86 years of age. Cultured cells were used as surrogates for in vivo conditions, due to obvious limitations in procuring native tissue samples. Furthermore, as cultured cells are more likely to be used by others because of the same limitations, this analysis on cultured cells may be prove to be of greater use to other investigators. Greater than 90% similarity between transcriptomes of TM cultured and native cells has been reported, albeit the differences may be important [10]. Classification of housekeeping genes is somewhat arbitrary; we accepted all genes considered housekeeping in the most recent microarray and/or RNA deep sequencing based studies to be housekeeping genes. This low stringency was used to minimize the possibility of inclusion of housekeeping genes in our list of non-HK TM expressed genes. Ultimately, 962 TM expressed genes are proposed to be part of the non-housekeeping transcriptome of human TM cultured cells. Because stringent criteria were set for selection of these genes, they are highly likely to be TM expressed genes. Nevertheless, as only approximately half (4,417/8,416) of the genes considered here to be housekeeping genes met TM expression criteria, it is expected that the 962 non-HK genes are not inclusive of all non-HK genes expressed in the human TM. Indeed, a few genes whose expressions in the TM are strongly supported by experimental evidence were not among the 962 genes reported here, such as MEIS2 (Meis homeobox 2) [11]. True non-HK genes that are not included among the 962 genes are likely biased toward those with lower levels of expression; they are unlikely to be biased with respect to function. The identified genes, therefore, are expected to reflect the morphological and functional properties of the human TM. They identify molecular markers to be addressed in studies on TM-related biology, including the TM-related disease glaucoma.
Analysis of the 962 genes by the DAVID bioinformatics tool identified focal adhesion and ECM-receptor KEGG-pathways to be enriched in the human TM at a highly significant Bonferroni p value level. A recently published gene expression profile of the normal human TM based on SAGE data also included many genes related to extracellular matrix, cell signaling, and cell structure/adhesion functions [14]. The extracellular matrix of the TM is important with respect to current understanding of TM function [28-31]. The importance of the ECM with respect to glaucoma is emphasized by the observation that two known glaucoma-causing genes are associated with this structure. The protein product of MYOC, the first identified POAG gene, interacts with components of the ECM [32,33]. Furthermore, the protein product of ADAMTS10 (ADAM metallopeptidase with thrombospondin type 1 motif, 10), which has been identified as causative of glaucoma in a canine model of the disease, is involved in ECM formation [34,35]. Focal adhesions are specialized structures formed at contact points between cells and the ECM. In addition to structural links, some constituents of focal points are signaling molecules whose activities culminate in reorganization of the actin cytoskeleton that, in turn, affects cell shape, cell motility, and gene expression. Experimental evidence for involvement of focal adhesion proteins such as paxillin in signaling pathways in the human TM have been reported [36-38]. Among the 34 non-HK TM genes identified in the focal adhesion and ECM-receptor pathways, 17 were common to both; the overlap is to be expected, given the nature of the pathways (Table 2). Eight of the 17 common genes were collagen-coding genes. Common sequence variants near CAV1 (caveolin 1) and CAV2, two of the genes only associated with focal adhesions, were recently reported to be associated with primary open-angle glaucoma [39]. The protein products of these genes affect IOP levels and are involved in nitric oxide and TGF-β (transforming growth factor beta) signaling and in the formation of caveolae [39]. Caveolae have been strongly implicated in signal transduction. HGF (hepatocyte growth factor), another of the focal adhesion associated genes, encodes hepatocyte growth factor. The concentration of this protein has been shown to be significantly higher in the aqueous fluid of glaucomatous eyes [40]. Furthermore, association between sequence variations in HSP and glaucoma has recently been reported in the Nepalese population [41]. Analysis of the 962 genes with DAVID also identified multiple enriched GO terms. ECM-related terms were enriched in all three term categories—biologic process, cellular component, and cellular function. Among the 14 cellular component-enriched terms, seven and five, respectively, were related to the ECM and plasma membrane. The genes associated with these terms constituted 400 of the 962 non-HK TM expressed genes. The high representation of ECM and plasma membrane-related terms is consistent with enriched focal adhesion and ECM-receptor KEGG pathways, and emphasizes that biologic processes related to these terms are integral to the biologic roles of the TM.
The glaucoma-relevant parameters that were tested in the genome-wide TM transcriptome analyses on which the present study is based, were treatments with prostaglandin analogs, transforming growth factor β, glaucoma status, and PITX2 knockdown [8-11]. It was observed that these treatments preferentially affected the expression of genes classified here as non-HK TM expressed genes (Table 4). Although discussion of relevance of the genes with respect to the treatments is beyond the scope of this presentation, their enrichment in the non-HK class of genes suggests that the non-HK genes identified will be of value in studies aimed at achieving a better understanding of TM physiology.
Analysis that led to identification of miRNAs with enriched targets within one of the functionally related non-HK TM group of genes (group 3) proved interesting. All the genes placed within this group coded collagen proteins. Much evidence on the role of collagens in TM physiology and glaucoma pathogenesis is available in the literature [42-44]. Based on an association study, it was recently reported that the collagen-coding gene COL5A1 (collagen, type V, alpha 1) affects central corneal thickness, which is a glaucoma risk factor [42]. Expressional elevation of COL8A2 (collagen, type VIII, alpha 2) in TM cells in response to dexamethasone treatment has also been observed, and it has been suggested that this is part of the pathological process leading to steroid-induced glaucoma [45,46]. Notably, rare COL8A2 mutations have been found in some glaucoma patients [47]. Results of experiments on cultured bovine TM cells have suggested that increased expression of collagen type 4 causes resistance to aqueous outflow [48]. Enriched target genes for all three members (miR-29a, b, and c) of the miR-29 family were found within group 3 genes. It is already known that miR-29 family members regulate expression of ECM proteins, including collagen types 1, 4, and 5, in human TM cultured cells [49] and in non-ocular tissues [50-55]. Experimental evidence for their effect on collagen type 7, which is predicted here, has not been reported. Effects of TGF-β and oxidative stress on ECM synthesis in the human TM may be partially mediated by miRNAs of this family [49]. Targets of six HSA-LET-7 family members were also significantly enriched in group 3 genes. To the best of our knowledge, experimental evidence for effects of these miRNAs on collagen expression is not available. HSA-LET-7F was also predicted to target ELF4 (E74-like factor 4), ETS1 (v-ets erythroblastosis virus E26 oncogene homolog 1), PLAGL2 (pleiomorphic adenoma gene-like 2), TEAD3 (TEA domain family member 3), and HAND1 (heart and neural crest derivatives expressed 1) in group 18 genes, although the p value (p=0.06) in this case did not reach statistical significance (data not shown). Finally, numerous target genes for miR-26 and miR-34 family members were identified among group14 non-HK TM expressed genes.
In addition to the genes discussed above, some non-HK TM expressed genes identified in this study, as well as in previous TM transcriptome studies, are of particular interest. CH13L1 has already been proposed as a candidate TM marker, due to its high expression in the TM and restricted expression in other tissues [10]. CH13L1 codes a carbohydrate binding lectin. The protein directly interacts with type 1 collagen, and this interaction may affect tissue remodeling [56]. Involvement of CH13L1 in the pathogenesis of glaucoma has been considered [57-59]. ALDH1A1 codes an aldehyde dehydrogenase that may be involved in minimizing the deleterious effects of oxidative damage caused largely by exposure of the anterior chamber to ultraviolet radiation [11,60]. In addition, its expression in human TM cells is affected by TGF-β treatment [9]. TGF-β signaling and oxidative stress are both important components of glaucoma etiology [61]. FGF-2 encodes basic fibroblast growth factor 2 and has roles in angiogenesis. Neovascular glaucoma is a rare form of glaucoma [62]. It has been shown that the hormone ghrelin inhibits FGF-2 mediated angiogenesis [63]. Notably, it has been reported recently that ghrelin levels are significantly lower in the aqueous humor of open-angle glaucoma patients, than in control individuals [64]. RGS5 is of interest because an alternatively spliced form of its mRNA was identified in human ocular tissues, with highest expression of this form in the TM [65]. In addition, an IGFBP-5 mRNA isoform with a unique 3′-untranslated region only expressed in the TM has been identified [66]. It would be of interest to determine if mir-24 and mir-204/211, predicted to target IGFBP-5 mRNA, may selectively affect specific isoforms. It has already been shown that these miRNAs regulate multiple functions in TM [25,26]. To the best of our knowledge, TM-specific properties for CDH11, CXCR7, GNG11, PTPRM, and TUSC3 have not been reported. CDH11 is an adhesion molecule, and CXCR7, GNG11, and PTPRM are involved in cell signaling processes.
In conclusion, multiple protein coding genes and miRNAs are introduced here as candidates for study for better understanding of TM physiology. The observation that experimental data have already implicated many of the identified genes and miRNAs in ocular functions supports the premise that the remaining are also good candidates for further study.
Acknowledgments
We acknowledge the Ophthalmic Research Center, Shahid Beheshti University of Medical Sciences and the Iran National Science Foundation for funding this research.
Appendix 1
Appendix 1. Four microarray/human TM gene expression studies: housekeeping and non-housekeeping genes expressed.To access the data, click or select the words “Appendix 1.” This will initiate the download of a compressed (pdf) archive that contains the file.
Appendix 2
Appendix 2. Non-HK TM genes with altered expression due to glaucoma-relevant exprimental manipulations. To access the data, click or select the words “Appendix 2.” This will initiate the download of a compressed (pdf) archive that contains the file.
Appendix 3
Appendix 3. Non-HK human TM expressed genes grouped based on function. To access the data, click or select the words “Appendix 3.” This will initiate the download of a compressed (pdf) archive that contains the file.
References
- 1.Gasiorowski JZ, Russell P. Biological properties of trabecular meshwork cells. Exp Eye Res. 2009;88:671–5. doi: 10.1016/j.exer.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sommer A. Intraocular pressure and glaucoma. Am J Ophthalmol. 1989;107:186–8. doi: 10.1016/0002-9394(89)90221-3. [DOI] [PubMed] [Google Scholar]
- 3.Bill A, Phillips CI. Uveoscleral drainage of aqueous humour in human eyes. Exp Eye Res. 1971;12:275–81. doi: 10.1016/0014-4835(71)90149-7. [DOI] [PubMed] [Google Scholar]
- 4.Quigley HA. Open-angle glaucoma. N Engl J Med. 1993;328:1097–106. doi: 10.1056/NEJM199304153281507. [DOI] [PubMed] [Google Scholar]
- 5.Tezel G, Wax MB. The immune system and glaucoma. Curr Opin Ophthalmol. 2004;15:80–4. doi: 10.1097/00055735-200404000-00003. [DOI] [PubMed] [Google Scholar]
- 6.Gonzalez P, Epstein DL, Borras T. Characterization of gene expression in human trabecular meshwork using single-pass sequencing of 1060 clones. Invest Ophthalmol Vis Sci. 2000;41:3678–93. [PubMed] [Google Scholar]
- 7.Tomarev SI, Wistow G, Raymond V, Dubois S, Malyukova I. Gene expression profile of the human trabecular meshwork: NEIBank sequence tag analysis. Invest Ophthalmol Vis Sci. 2003;44:2588–96. doi: 10.1167/iovs.02-1099. [DOI] [PubMed] [Google Scholar]
- 8.Zhao X, Ramsey KE, Stephan DA, Russell P. Effects of Prostaglandin Analogues on Human Ciliary Muscle and Trabecular Meshwork Cells. Invest Ophthalmol Vis Sci. 2003;44:1945–52. doi: 10.1167/iovs.02-0920. [DOI] [PubMed] [Google Scholar]
- 9.Zhao X, Ramsey KE, Stephan DA, Russell P. Gene and protein expression changes in human trabecular meshwork cells treated with transforming growth factor β. Invest Ophthalmol Vis Sci. 2004;45:4023–34. doi: 10.1167/iovs.04-0535. [DOI] [PubMed] [Google Scholar]
- 10.Liton PB, Luna C, Challa P, Epstein DL, Gonzalez P. Genome wide expression profile of human trabecular meshwork cultured cells, nonglaucomatous and primary open angle glaucoma tissue. Mol Vis. 2006;12:774–90. [PMC free article] [PubMed] [Google Scholar]
- 11.Paylakhi SH, Fan JB, Mehrabian M, Sadeghizadeh M, Yazdani S, Katanforoush A, Rezaei Kanavi M, Ronaghi M, Elahi E. Effect of PITX2 knockdown on transcriptome of primary human trabecular meshwork cell cultures. Mol Vis. 2011;17:1209–21. [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu J, He F, Song S, Wang J, Yu J. How many human genes can be defined as housekeeping with current expression data? BMC Genomics. 2008;9:172–82. doi: 10.1186/1471-2164-9-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramsköld D, Wang ET, Burge CB, Sandberg R. An Abundance of Ubiquitously Expressed Genes Revealed by Tissue Transcriptome Sequence Data. PLOS Comput Biol. 2009;5:e1000598. doi: 10.1371/journal.pcbi.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y, Munro D, Layfield D, Dellinger A, Walter J, Peterson K, Rickman CB, Allingham RR, Hauser MA. Serial analysis of gene expression (SAGE) in normal human trabecular meshwork. Mol Vis. 2011;17:885–93. [PMC free article] [PubMed] [Google Scholar]
- 15.Huminiecki L, Wolfe KH. Divergence of spatial gene expression profiles following species-specific gene duplications in human and mouse. Genome Res. 2004;14(10A):1870–9. doi: 10.1101/gr.2705204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lyng H, Badiee A, Svendsrud DH, Hovig E, Myklebost O, Stokke T. Profound influence of microarray scanner characteristics on gene expression ratios: analysis and procedure for correction. BMC Genomics. 2004;5:10. doi: 10.1186/1471-2164-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X, Kim J, Zhou J, Gu W, Quigg R. Use of signal thresholds to determine significant changes in microarray data analyses. Genet Mol Biol. 2005;28:191–200. [Google Scholar]
- 18.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sherman BT. Huang da W, Tan Q, Guo Y, Guo Y, Bour S, Liu D, Stephens R, Baseler MW, Lane HC, Lempicki RA. DAVID Knowledgebase: a gene-centered database integrating heterogeneous gene annotation resources to facilitate high-throughput gene functional analysis. BMC Bioinformatics. 2007;8:426. doi: 10.1186/1471-2105-8-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoshida S, Yashar BM, Hiriyanna S, Swaroop A. Microarray analysis of gene expression in the aging human retina. Invest Ophthalmol Vis Sci. 2002;43:2554–60. [PubMed] [Google Scholar]
- 22.Wiggs JL. Genetic etiologies of glaucoma. Arch Ophthalmol. 2007;125:30–7. doi: 10.1001/archopht.125.1.30. [DOI] [PubMed] [Google Scholar]
- 23.Narooie-Nejad M, Paylakhi SH, Shojaee S, Fazlali Z, Rezaei Kanavi M, Nilforushan N, Yazdani S, Babrzadeh F, Suri F, Ronaghi M, Elahi E, Paisán-Ruiz C. Loss of function mutations in the gene encoding latent transforming growth factor beta binding protein 2, LTBP2, cause primary congenital glaucoma. Hum Mol Genet. 2009;18:3969–77. doi: 10.1093/hmg/ddp338. [DOI] [PubMed] [Google Scholar]
- 24.Strungaru MH, Dinu I, Walter MA. Genotype-phenotype correlations in Axenfeld-Rieger malformation and glaucoma patients with FOXC1 and PITX2 mutations. Invest Ophthalmol Vis Sci. 2007;48:228–37. doi: 10.1167/iovs.06-0472. [DOI] [PubMed] [Google Scholar]
- 25.Luna C, Li G, Qiu J, Epstein L, Gonzalez P. MicroRNA-24 Regulates the processing of latent TGFβ1 during cyclic mechanical stress in human trabecular meshwork cells through direct targeting of FURIN. J Cell Physiol. 2011;226:1407–14. doi: 10.1002/jcp.22476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li G, Luna C, Qiu J, Epstein DL, Gonzalez P. Role of miR-204 in the regulation of apoptosis, endoplasmic reticulum stress response, and inflammation in human trabecular meshwork cells. Invest Ophthalmol Vis Sci. 2011;52:2999–3007. doi: 10.1167/iovs.10-6708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lozano-Velasco E, Contreras A, Crist C, Hernández-Torres F, Franco DE, Aránega A. Pitx2c modulates Pax3+/Pax7+ cell populations and regulates Pax3 expression by repressing miR27 expression during myogenesis. Dev Biol. 2011;357:165–78. doi: 10.1016/j.ydbio.2011.06.039. [DOI] [PubMed] [Google Scholar]
- 28.Johnson M. What controls aqueous humour outflow resistance? Exp Eye Res. 2006;82:545–57. doi: 10.1016/j.exer.2005.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.González-Avila G, Ginebra M, Hayakawa T, Vadillo-Ortega F, Teran L, Selman M. Collagen metabolism in human aqueous humor from primary open-angle glaucoma: decreased degradation and increased biosynthesis play a role in its pathogenesis. Arch Ophthalmol. 1995;113:1319–23. doi: 10.1001/archopht.1995.01100100107039. [DOI] [PubMed] [Google Scholar]
- 30.Samples JR, Alexander JP, Acott TS. Regulation of the levels of human trabecular matrix metalloproteinases and inhibitor by interleukin-1 and dexamethasone. Invest Ophthalmol Vis Sci. 1993;34:3386–95. [PubMed] [Google Scholar]
- 31.Yue BY. The extracellular matrix and its modulation in the trabecular meshwork. Surv Ophthalmol. 1996;40:379–90. doi: 10.1016/s0039-6257(96)80066-x. [DOI] [PubMed] [Google Scholar]
- 32.Filla MS, Liu X, Nguyen TD, Polansky JR, Brandt CR, Kaufman PL, Peters DM. In vitro localization of TIGR/MYOC in trabecular meshwork extracellular matrix and binding to fibronectin. Invest Ophthalmol Vis Sci. 2002;43:151–61. [PubMed] [Google Scholar]
- 33.Ueda J, Wentz-Hunter K, Yue BY. Distribution of myocilin and extracellular matrix components in the juxtacanalicular tissue of human eyes. Invest Ophthalmol Vis Sci. 2002;43:1068–76. [PubMed] [Google Scholar]
- 34.Kuchtey J, Olson LM, Rinkoski T, Mackay EO, Iverson TM, Gelatt KN, Haines JL, Kuchtey RW. Mapping of the Disease Locus and Identification of ADAMTS10 As a Candidate Gene in a Canine Model of Primary Open Angle Glaucoma. PLoS Genet. 2011;7:e1001306. doi: 10.1371/journal.pgen.1001306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Apte SS. A disintegrin-like and metalloprotease (reprolysin-type) with thrombospondin type 1 motif (ADAMTS) superfamily: functions and mechanisms. J Biol Chem. 2009;284:31493–7. doi: 10.1074/jbc.R109.052340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turner CE. Paxillin: a cytoskeletal target for tyrosine kinases. Bioessays. 1994;16:47–52. doi: 10.1002/bies.950160107. [DOI] [PubMed] [Google Scholar]
- 37.Burridge K, Turner CE, Romer LH. Tyrosine phosphorylation of paxillin and ppl25 FAK accompanies cell adhesion to extracellular matrix: a role in cytoskeletal assembly. J Cell Biol. 1992;119:893–903. doi: 10.1083/jcb.119.4.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turner CE, Petra KM, Taylor DS, Molloy CJ. Angiotensin II stimulation of rapid paxillin tyrosine phosphorylation correlates with formation of focal adhesions in rat aortic smooth muscle cells. J Cell Sci. 1995;108:333–42. doi: 10.1242/jcs.108.1.333. [DOI] [PubMed] [Google Scholar]
- 39.Thorleifsson G, Walters GB, Hewitt AW, Masson G, Helgason A, DeWan A, Sigurdsson A, Jonasdottir A, Gudjonsson SA, Magnusson KP, Stefansson H, Lam DS, Tam PO, Gudmundsdottir GJ, Southgate L, Burdon KP, Gottfredsdottir MS, Aldred MA, Mitchell P, St Clair D, Collier DA, Tang N, Sveinsson O, Macgregor S, Martin NG, Cree AJ, Gibson J, Macleod A, Jacob A, Ennis S, Young TL, Chan JC, Karwatowski WS, Hammond CJ, Thordarson K, Zhang M, Wadelius C, Lotery AJ, Trembath RC, Pang CP, Hoh J, Craig JE, Kong A, Mackey DA, Jonasson F, Thorsteinsdottir U, Stefansson K. Common variants near CAV1 and CAV2 are associated with primary open-angle glaucoma. Nat Genet. 2010;42:906–9. doi: 10.1038/ng.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu DN, Ritch R. Hepatocyte growth factor is increased in the aqueous humor of glaucomatous eyes. J Glaucoma. 2001;10:152–7. doi: 10.1097/00061198-200106000-00002. [DOI] [PubMed] [Google Scholar]
- 41.Awadalla MS, Thapa SS, Burdon KP, Hewitt AW, Craig JE. The association of hepatocyte growth factor (HGF) gene with primary angle closure glaucoma in the Nepalese population. Mol Vis. 2011;17:2248–54. [PMC free article] [PubMed] [Google Scholar]
- 42.Vithana EN, Aung T, Khor CC, Cornes BK, Tay WT, Sim X, Lavanya R, Wu R, Zheng Y, Hibberd ML, Chia KS, Seielstad M, Goh LK, Saw SM, Tai ES, Wong TY. Collagen-related genes influence the glaucoma risk factor, central corneal thickness. Hum Mol Genet. 2011;15:649–58. doi: 10.1093/hmg/ddq511. [DOI] [PubMed] [Google Scholar]
- 43.Tane N, Dhar S, Roy S, Pinheiro A, Ohira A, Roy S. Effect of excess synthesis of extracellular matrix components by trabecular meshwork cells: Possible consequence on aqueous outflow. Exp Eye Res. 2007;84:832–42. doi: 10.1016/j.exer.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 44.Villarreal G, Jr, Oh DJ, Kang MH, Rhee DJ. Coordinated Regulation of Extracellular Matrix Synthesis by the MicroRNA-29 Family in the Trabecular Meshwork. Invest Ophthalmol Vis Sci. 2011;52:3391–7. doi: 10.1167/iovs.10-6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rozsa FW, Reed DM, Scott KM, Pawar H, Moroi SE, Kijek TG, Krafchak CM, Othman MI, Vollrath D, Elner VM. Gene expression profile of human trabecular meshwork cells in response to long-term dexamethasone exposure. Mol Vis. 2006;12:125–41. [PubMed] [Google Scholar]
- 46.Wilson K, McCartney MD, Miggans ST, Clark AF. Dexamethasone induced ultrastructural changes in cultured human trabecular meshwork cells. Curr Eye Res. 1993;12:783–93. doi: 10.3109/02713689309020383. [DOI] [PubMed] [Google Scholar]
- 47.Wyatt D, Taheri S, Triana M, Abdrabou W, Desronvil T, DelBono E, Olivier M, Wigg J. Distribution of DNA sequence variants in COL8A1 and COL8A2 in glaucoma patients with thin CCT. ARVO Annual Meeting; 2009 May 3-7; Fort Lauderdale (FL). [Google Scholar]
- 48.Tane N, Dhar S, Roy S, Pinheiro A, Ohira A, Roy S. Effect of excess synthesis of extracellular matrix components by trabecular meshwork cells: possible consequence on aqueous outflow. Exp Eye Res. 2007;84:832–42. doi: 10.1016/j.exer.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 49.Luna C, Li G, Qiu J, Epstein DL, Gonzalez P. Role of miR-29b on the regulation of the extracellular matrix in human trabecular meshwork cells under chronic oxidative stress. Mol Vis. 2009;15:2488–97. [PMC free article] [PubMed] [Google Scholar]
- 50.Li Z, Hassan MQ, Jafferji M, Aqeilan RI, Garzon R, Croce CM, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J Biol Chem. 2009;284:15676–84. doi: 10.1074/jbc.M809787200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, Hill JA, Olson EN. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci USA. 2008;105:13027–32. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ogawa T, Iizuka M, Sekiya Y, Yoshizato K, Ikeda K, Kawada N. Suppression of type I collagen production by microRNA-29b in cultured human stellate cells. Biochem Biophys Res Commun. 2010;391:316–21. doi: 10.1016/j.bbrc.2009.11.056. [DOI] [PubMed] [Google Scholar]
- 53.Du B, Ma LM, Huang MB, Zhou H, Huang HL, Shao P, Chen YQ, Qu LH. High glucose down-regulates miR-29a to increase collagen IV production in HK-2 cells. FEBS Lett. 2010;584:811–6. doi: 10.1016/j.febslet.2009.12.053. [DOI] [PubMed] [Google Scholar]
- 54.Kapinas K, Kessler CB, Delany AM. MiR-29 suppression of osteonectin in osteoblasts: regulation during differentiation and by canonical Wnt signaling. J Cell Biochem. 2009;108:216–24. doi: 10.1002/jcb.22243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sengupta S, den Boon JA, Chen IH, Newton MA, Stanhope SA, Cheng YJ, Chen CJ, Hildesheim A, Sugden B, Ahlquist P. MicroRNA 29c is down-regulated in nasopharyngeal carcinomas, up-regulating mRNAs encoding extracellular matrix proteins. Proc Natl Acad Sci USA. 2008;105:5874–8. doi: 10.1073/pnas.0801130105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bigg HF, Wait R, Rowan AD, Cawston TE. The mammalian chitinase-like lectin, YKL-40, binds specifically to type I collagen and modulates the rate of type I collagen fibril formation. J Biol Chem. 2006;281:21082–95. doi: 10.1074/jbc.M601153200. [DOI] [PubMed] [Google Scholar]
- 57.Lo WR, Rowlette LL, Caballero M, Yang P, Hernandez MR, Borrás T. Tissue differential microarray analysis of dexamethasone induction reveals potential mechanisms of steroid glaucoma. Invest Ophthalmol Vis Sci. 2003;44:473–85. doi: 10.1167/iovs.02-0444. [DOI] [PubMed] [Google Scholar]
- 58.Miyahara T, Kikuchi T, Akimoto M, Kurokawa T, Shibuki H, Yoshimura N. Gene microarray analysis of experimental glaucomatous retina from cynomolgus monkey. Invest Ophthalmol Vis Sci. 2003;44:4347–56. doi: 10.1167/iovs.02-1032. [DOI] [PubMed] [Google Scholar]
- 59.Dozin B, Malpeli M, Camardella L, Cancedda R, Pietrangelo A. Response of young, aged and osteoarthritic human articular chondrocytes to inflammatory cytokines: molecular and cellular aspects. Matrix Biol. 2002;21:449–59. doi: 10.1016/s0945-053x(02)00028-8. [DOI] [PubMed] [Google Scholar]
- 60.Piatigorsky J. Review: A case for corneal crystallins. J Ocul Pharmacol Ther. 2000;16:173–80. doi: 10.1089/jop.2000.16.173. [DOI] [PubMed] [Google Scholar]
- 61.Tezel G, Yang X, Luo C, Peng Y, Sun SL, Sun D. Mechanisms of immune system activation in glaucoma: oxidative stressstimulated antigen presentation by the retina and optic nerve head glia. Invest Ophthalmol Vis Sci. 2007;48:705–14. doi: 10.1167/iovs.06-0810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kuang TM, Liu CJ, Chou CK, Hsu WM. Clinical experience in the management of neovascular glaucoma. J Chin Med Assoc. 2004;67:131–5. [PubMed] [Google Scholar]
- 63.Conconi MT, Nico B, Guidolin D, Baiguera S, Spinazzi R, Rebuffat P, Malendowicz LK, Vacca A, Carraro G, Parnigotto PP, Nussdorfer GG, Ribatti D. Ghrelin inhibits FGF-2-mediated angiogenesis in vitro and in vivo. Peptides. 2004;25:2179–85. doi: 10.1016/j.peptides.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 64.Katsanos A, Dastiridou A, Georgoulias P, Cholevas P, Kotoula M.Plasma and aqueous humour levels of ghrelin in open-angle glaucoma patients. Clin Experiment Ophthalmol 201139324–9.Tsironi EE [DOI] [PubMed] [Google Scholar]
- 65.Liang Y, Li C, Guzman VM, Chang WW, Evinger AJ, 3rd, Sao D, Woodward DF. Identification of a novel alternative splicing variant of RGS5 mRNA in human ocular tissues. FEBS J. 2005;272:791–9. doi: 10.1111/j.1742-4658.2004.04516.x. [DOI] [PubMed] [Google Scholar]
- 66.Wirtz MK, Xu H, Rust K, Alexander JP, Acott TS. Insulin-Like Growth Factor Binding Protein-5 Expression by Human Trabecular Meshwork. Invest Ophthalmol Vis Sci. 1998;39:45–53. [PubMed] [Google Scholar]


