SUMMARY
Interleukin-21 (IL-21) is a pleiotropic cytokine that induces expression of transcription factor BLIMP1 (encoded by Prdm1), which regulates plasma cell differentiation and T cell homeostasis. We identified an IL-21 response element downstream of Prdm1 that binds the transcription factors STAT3 and IRF4, which are required for optimal Prdm1 expression. Genome-wide ChIP-Seq mapping of STAT3- and IRF4-binding sites showed that most regions with IL-21-induced STAT3 binding also bound IRF4 in vivo and furthermore revealed that the noncanonical TTCnnnTAA GAS motif critical in Prdm1 was broadly used for STAT3 binding. Comparing genome-wide expression array data to binding sites revealed that most IL-21-regulated genes were associated with combined STAT3-IRF4 sites rather than pure STAT3 sites. Correspondingly, ChIP-Seq analysis of Irf4−/− T cells showed greatly diminished STAT3 binding after IL-21 treatment, and Irf4−/− mice showed impaired IL-21-induced Tfh cell differentiation in vivo. These results reveal broad cooperative gene regulation by STAT3 and IRF4.
INTRODUCTION
B lymphocyte-induced maturation protein-1 (BLIMP1) is a transcriptional repressor that was discovered as a zinc finger protein expressed upon plasmacytic differentiation of the Bcl-1 B cell lymphoma line (Turner et al., 1994). BLIMP1 is encoded by the PRDM1 gene in humans and the Prdm1 gene in mice. BLIMP1 regulates terminal differentiation of plasma cells in which it represses the expression of the Bcl6 and Pax5 genes (Martins and Calame, 2008); BCL6 in turn can repress Prdm1 expression (Shaffer et al., 2000). BLIMP1 has been most thoroughly studied in a B cell context, but it is also expressed in T cells, granulocytes, macrophages, epithelial cells, and germ cells and regulates T cell homeostasis and peripheral tolerance (Kallies et al., 2006; Martins et al., 2006; Ohinata et al., 2005).
Interleukin-21 (IL-21) is a type I cytokine produced after T cell activation that is most related to IL-2, IL-4, and IL-15 (Parrish-Novak et al., 2000; Spolski and Leonard, 2008). The IL-21 receptor was discovered as an orphan receptor (Ozaki et al., 2000; Parrish-Novak et al., 2000). Like IL-2, IL-4, IL-7, IL-9, and IL-15, IL-21 shares the common cytokine receptor γ chain, γc, as a critical receptor component (Spolski and Leonard, 2008). γc is encoded by the IL2RG gene, mutation of which results in X-linked severe combined immunodeficiency in humans, a disease in which T and NK cells are absent and B cells are present but nonfunctional (Leonard, 2001; Noguchi et al., 1993). IL-21 receptors are expressed by T cells, B cells, NK cells, myeloid cells, and keratinocytes, corresponding to pleiotropic actions of IL-21 on multiple lineages. IL-21 can act as a comitogen for T cells (Parrish-Novak et al., 2000), can potently drive CD8+ T cell expansion when combined with IL-7 or IL-15 (Zeng et al., 2005), and promotes the differentiation of T helper 17 (Th17) CD4+ T cells (Spolski and Leonard, 2008) and development of T follicular helper (Tfh) cells (Nurieva et al., 2008; Vogelzang et al., 2008). Within the B cell lineage, IL-21 critically regulates immunoglobulin production, particularly IgG1, and mice lacking expression of both IL-4 and IL-21R exhibit defective germinal center development and pan-hypogammaglobulinemia, which is likely to explain the B cell phenotype in humans with XSCID (Ozaki et al., 2002). Moreover, IL-21 potently induces BLIMP1 and plasma cell differentiation (Ozaki et al., 2004).
We have elucidated the molecular basis for IL-21-mediated BLIMP1 induction by finding an IL-21 response element 3′ of the Prdm1 gene whose activity requires both STAT3 and IRF4. STAT3 (signal transducer and activator of transcription 3) and IRF4 (interferon regulatory factor-4) mediate signaling in response to a range of cytokines, including IL-21 (Zeng et al., 2007; Huber et al., 2008). IRF4 is a lymphocyte-restricted transcription factor that is part of a family of DNA-binding proteins critical for the function and homeostasis of mature B and T cells, as well as for the development of subpopulations of dendritic cells (Gabriele and Ozato, 2007; Tamura et al., 2008). We identified a functional cooperation between STAT3 and IRF4 in IL-21-induced Prdm1 expression, extending the range of known actions for IRF4. Moreover, Solexa-based ChIP-Seq genome-wide analyses revealed that most genomic areas binding STAT3 after IL-21 treatment also constitutively bind IRF4 and that there was greatly diminished STAT3 binding in the absence of IRF4. We demonstrate that the expression of other IL-21-regulated genes also depends on both STAT3 and IRF4, revealing the broad functional cooperation of these transcription factors. Finally, we showed that the Prdm1 IL-21 response element uses a single-nucleotide variant (TTCnnnTAA) of the canonical TTCnnnGAA STAT3 motif, and this variant is a broadly used GAS motif in the genome.
RESULTS
IL-21 Rapidly Induces Prdm1 Gene Expression
We previously showed that IL-21 can induce BLIMP1 expression (Ozaki et al., 2004), and indeed, IL-21 can induce Prdm1 expression in multiple B lymphoma lines (Figure 1A). When primary splenic B cells were preactivated with anti-CD40, with or without anti-IgM, and rested, subsequent IL-21-induced Prdm1 expression was higher in cells preactivated without anti-IgM (Figure 1B). IL-21 also induced BLIMP1 expression in preactivated B cells (Figure 1C). Induction of Prdm1 mRNA was seen by 1 hr, with peak mRNA expression typically at 24 hr (Figure S1A and S1B available online). LPS also induced Prdm1 expression, whereas IL-4 did not (Figure S1B). IL-21 reproducibly induced Prdm1 gene expression slightly more quickly than did LPS, although LPS was more potent in its effect at 48 hr (Figure S1B).
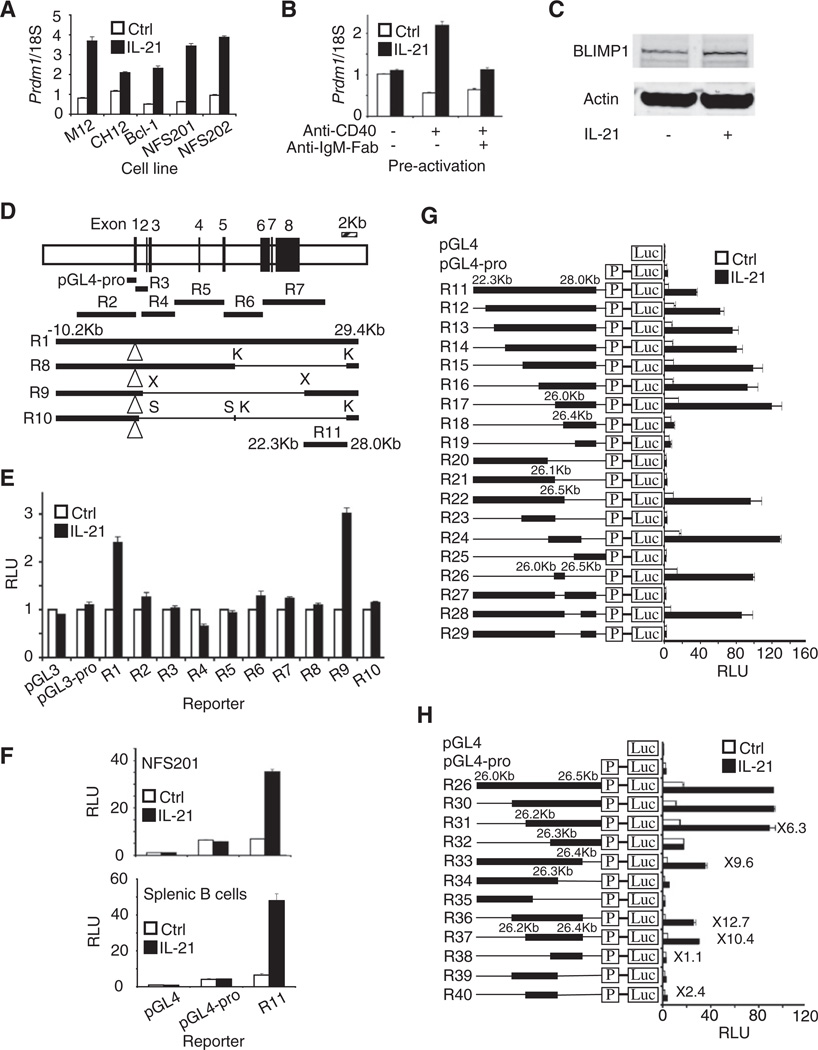
Figure 1. Characterization of an IL-21 Response Element 3′ of the Prdm1 Gene.
(A) IL-21 induces Prdm1 gene expression. Prdm1 mRNA (means ± SEM of ≥3 independent experiments, total combined samples ≥4) was determined by quantitative RT-PCR 6 hr after treatment with 50 ng/ml IL-21.
(B) Cells were activated with 1 µg/ml anti-CD40 and/or 3 µg/ml anti-IgM-Fab for 3 days, rested 24 hr, and treated with 100 ng/ml IL-21. Prdm1 mRNA (means ± SEM of two independent experiments, total combined samples = 4) was determined 24 hr later by quantitative RT-PCR.
(C) Cells were activated with 1 µg/ml anti-CD40 for 3 days, rested for 24 hr, and treated with 100 ng/ml IL-21 for 24 hr and immunoblotted with antibodies to BLIMP1 or actin. Representative results of three independent experiments.
(D) Schematic of the Prdm1 gene (defined by NM_007548 version 2; version 3 of May 2008 starts 61 bp downstream and lacks the 79 bp second exon) and locations of luciferase reporter constructs. pGL4-pro, minimal promoter (−972 to +183) reporter; K, KpnI; X, XhoI; S, SmaI; open triangle is the luciferase insertion site in exon 1 for R1, R8, R9, and R10.
(E) Luciferase assays in NFS201 cells were performed 18 hr after treatment with 50 ng/ml IL-21.
(F) Shown is luciferase activity 6 hr after treatment with 50 ng/ml IL-21 for NFS201 (means ± SEM of ≥4 independent experiments, total combined samples ≥6) and with 100 ng/ml IL-21 for splenic B cells (means ± SEM of ≥2 independent experiments, total combined samples ≥4).
(G and H) More detailed mapping of the Prdm1 IL-21 response element in preactivated splenic B cells. Shown is luciferase activity 6 hr after treatment with 100 ng/ml IL-21 (means ± SEM of ≥3 independent experiments, total combined samples ≥6).
An IL-21 Response Element Is 3′ of the Prdm1 Gene
To identify an IL-21-responsive region, we transfected NFS201 cells with a series of Prdm1 luciferase reporter constructs (Figure 1D). A “full-length” construct denoted R1 (reporter construct 1) extending from −10.2 to +29.4 kb relative to the main transcription start site (TSS) exhibited IL-21-induced activity, but a series of smaller constructs, R2–R7, did not (Figures 1D and 1E). We digested R1 with restriction enzymes to generate internal deletion constructs R8, R9, and R10 (Figure 1D). Of these, only R9 exhibited IL-21-induced luciferase activity (Figure 1E), indicating that the IL-21 response region is between 22.3 and 28.0 kb. Indeed, construct R11 containing this specific region 5′ to the Prdm1 minimal promoter (Figure 1D) responded to IL-21 in NFS201 cells and primary splenic B cells (Figure 1F).
We therefore generated deletion mutants of R11 (Figure 1G). 5′ deletion to 26.0 kb (constructs R12 to R17) exhibited IL-21-induced activity, but further deletion (R18 and R19) greatly diminished IL-21-induced activity. With 3′ deletion mutants, R22 retained activity, but R20 and R21 did not. Together, the activities of R17 and R22 indicated the critical role of the 26 to 26.5 kb region. Moreover, R23 and R25, which lack this region, were inactive, whereas R24 and R26 reporters, which span this region, potently responded to IL-21 (Figure 1G). Furthermore, deletion of the 26–26.5 kb region from R11 to yield R27 abrogated IL-21-induced activity (Figure 1G). To refine the mapping, we generated deletion mutants of R26 (Figure 1H). 5′ deletion to 26.2 kb (R31) retained activity but deletion to 26.3 kb (R32) eliminated activity. 3′ deletion to 26.4 kb (R33) lowered basal activity (open bar) but not inducibility (closed bar), whereas deletion to 26.3 kb (R34) or beyond (R35) eliminated IL-21-induced activity. Thus, both the 26.2–26.3 kb and 26.3–26.4 kb regions were essential, and R36 and R37, which span the 26.2 to 26.4 kb region, both exhibited substantial activity, whereas smaller fragments (R38, R39, and R40) had greatly decreased activity. These data were consistent with two factors binding within the 212 bp region (extending from +4678 to +4889 relative to the ATTAAA polyadenylation signal of the Prdm1 gene; see NM_007548.3), one in the R38 region and one in the R40 region, to mediate IL-21-induced Prdm1 expression.
STAT3 and IRF4 Bind to the Prdm1 IL-21 Response Element In Vivo
IL-21 can activate STAT1, STAT3, and STAT5, with STAT3 being the most important for IL-21 signaling (Diehl et al., 2008; Zeng et al., 2007). We used transcription-factor-binding prediction software (MatInspector) to identify potential γ-interferon-activated sequence (GAS) motifs that can bind these STATs (Levy and Darnell, 2002). Canonical GAS motifs have a TTCnnnGAA sequence, but the 212 bp Prdm1 IL-21 response element has a single-nucleotide variant TTCCAGTAA motif (underlined in Figure 2A). Electrophoretic mobility shift assays (EMSAs) with NFS201 nuclear extracts and a WT probe (probe #1 in Figure 2A) revealed an IL-21-induced complex (Figure 2B, lane 2, solid arrow), but not when the motif was mutated to agaCAGTAA (lane 4). The complex was super-shifted by anti-STAT3 but not by antibodies to STAT1, STAT5A, or STAT5B (lanes 5–8). IL-21 also induced formation of a faster mobility complex (lane 2, open arrow) whose intensity was enhanced with the mutant probe (lane 4). Examination of the DNA sequence revealed a putative IRF-binding motif (Tamura et al., 2008) (TTTC, boxed in Figure 2A) immediately 3′ of the STAT3-binding site. Indeed, anti-IRF4 super-shifted the faster complex, whereas anti-STAT3 did not (Figure 2B, lanes 9–11), indicative of IRF4 binding.
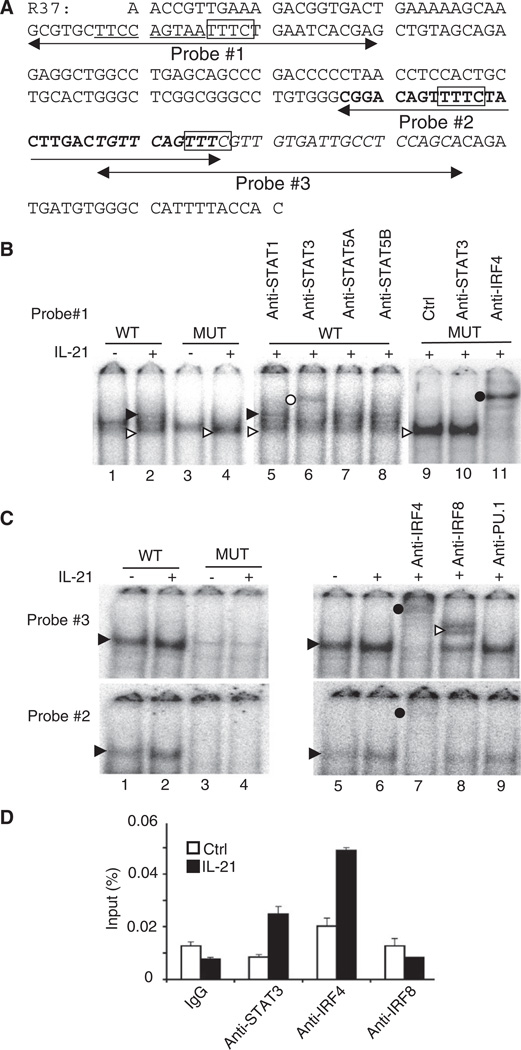
Figure 2. STAT3 and IRF4 Bind to the Prdm1 IL-21 Response Element.
(A) DNA sequence of the 212 bp (26,237 to 26,448 region relative to the TSS) R37 construct containing the IL-21 response element. The STAT3-binding motif is underlined, IRF4 and IRF8-binding motifs are boxed, and probes #1, #2, and #3 are indicated.
(B) EMSAs with nuclear extracts from NFS201 cells treated for 7 hr with 50 ng/ml IL-21. The solid and open arrows indicate IL-21-induced complexes, with the solid arrow corresponding to the STAT3-DNA complex. The open and closed circles indicate the supershifted STAT3 and IRF4 complexes, respectively. The WT probe is probe #1 in (A).
(C) EMSAs were performed with probes #2 and #3 (bold and italic sequences in A) with NFS201 nuclear extracts. The solid arrowhead indicates an IL-21-induced complex, the closed circle indicates supershifted IRF4-DNA complexes, and the open arrowhead indicates a supershifted IRF8-DNA complex.
(D) ChIP was performed with crosslinked splenic B cell lysates prepared 30 min after treatment with 100 ng/ml IL-21 (means ± SEM). Input was determined by quantitative PCR. Shown are representative results from two (B and C) or three (D) independent experiments.
To further investigate binding to the 212 bp IL-21 response element, we performed EMSAs with a series of 30 bp probes spanning this region. We found IL-21-induced complexes to two other probes, #2 and #3 (Figures 2C), which also contain putative TTTC IRF-binding motifs (boxed in Figure 2A). When we mutated the TTTC motifs to gagC, DNA-protein complexes were no longer observed (Figure 2C, lanes 3 and 4 versus 1 and 2). Moreover, complex formation was blocked and/or super-shifted by antibodies to IRF4 and IRF8. Signals were stronger with probe #3 than #2, perhaps because the GTTTC sequence is better than TTTTC for binding IRF4. We also tested antibodies to PU.1, a cofactor for IRF4 (Eisenbeis et al., 1995), but no supershift was observed (Figure 2C).
To evaluate STAT3, IRF4, and IRF8 binding in vivo, we used splenic B cells and chromatin immunoprecipitation (ChIP). IL-21 induced binding of STAT3 and IRF4 but not of IRF8 to the Prdm1 gene (Figure 2D), whereas none of these factors bound to a control intronic region of the mouse β-actin (Actb) gene (data not shown). Thus, STAT3 and IRF4 bind to the Prdm1 response element in primary B cells in vivo.
STAT3 and IRF4 Are Critical for IL-21-Induced Prdm1 Expression in B Cells
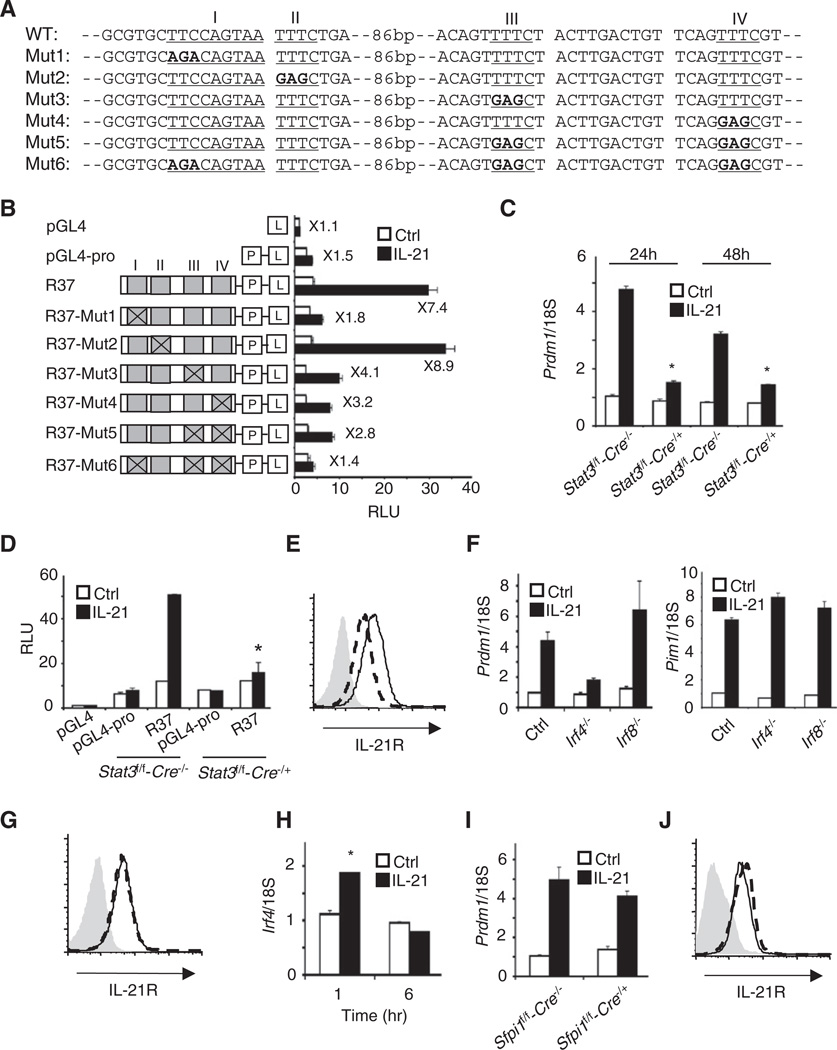
We next studied the role of STAT3 in IL-21-induced Prdm1 expression. When we mutated the TTCCAGTAA STAT3 site to agaCAGTAA (Mut1, Figure 3A), IL-21-induced luciferase activity was markedly reduced (R37-Mut1 in Figure 3B). Moreover, Prdm1 mRNA expression was lower in splenic B cells from Stat3f/f-CD19-Cre+/− mice, which lack STAT3 (Figure 3C), and correspondingly, IL-21-induced activity of the R37 construct was markedly reduced in these cells (Figure 3D). Thus, STAT3 was essential for activity of the Prdm1 IL-21 response element. IL-21 receptor expression was also slightly low in Stat3f/f-CD19-Cre+/− B cells (Figure 3E).
Figure 3. STAT3 and IRF4 Mediate IL-21-Induced Prdm1 Expression in B Cells.
(A) Sequences for STAT3 and IRF4-binding motif mutants in the R37 reporter.
(B) Luciferase assays were performed with WT or mutated R37 constructs shown in (A). Shown is luciferase activity 6 hr after treatment with 100 ng/ml IL-21 (means ± SEM of ≥3 independent experiments, total combined samples ≥ 6).
(C) Prdm1 mRNA expression (means ± SEM of two independent experiments, total combined samples = 4) was determined in preactivated splenic B cells by quantitative RT-PCR 24 and 48 hr after treatment with 100 ng/ml IL-21.
(D) Shown is luciferase activity of the R37 construct 6 hr after treatment with 100 ng/ml IL-21 (means ± SEM of two independent experiments, total combined samples = 4).
(E) IL-21 receptor expression in preactivated and rested Stat3-deficient B cells (dashed line). The solid line refers to WT mice. The isotype antibody control is shown in gray.
(F) Prdm1 and Pim1 mRNA expression (means ± SEM of ≥3 independent experiments, total combined samples ≥4) was determined by quantitative RT-PCR 24 hr after treatment with 100 ng/ml IL-21.
(G) IL-21 receptor expression of Irf4−/− B cells (dashed line) and WT control cells (solid line) was determined. The isotype antibody control is shown in gray.
(H) Splenic B cells were treated with 100 ng/ml IL-21 (means ± SEM of two independent experiments, total combined samples = 4) and Irf4 expression was determined.
(I) Prdm1 mRNA expression of IL-21 stimulated cells expressing PU.1 (Sfpi1f/f-Cre−/−) or lacking PU.1 (Sfpi1f/f-Cre−/+) was determined (means ± SEM of two independent experiments, total combined samples ≥ 4).
(J) IL-21 receptor expression of PU.1-deficient B cells (dashed line) and WT control (solid line). The isotype antibody control is shown in gray. In (C) and (D), *p < 0.05 (compared with IL-21-treated WT control mice). In (H), *p < 0.05 for IL-21-treated versus Ctrl.
We next separately mutated the IRF motifs from TTTC to gagC in the context of R37 (Figure 3A). The Mut2 mutation did not affect IL-21-induced luciferase activity, whereas Mut3, Mut4, and Mut5 (combination of Mut3 and Mut4) lowered activity, and simultaneous mutation of Mut3, Mut4, and the GAS motif (Mut6) dramatically decreased inducible activity (Figure 3B). We found greatly diminished IL-21-induced Prdm1 mRNA expression in Irf4−/− but not Irf8−/− B cells (Figure 3F, left panel), whereas IL-21-induced Pim1 expression was not diminished in either Irf4−/− or Irf8−/− B cells (right panel). There was no difference in IL-21R expression in Irf4−/− versus WT B cells (Figure 3G), and IL-21 slightly induced Irf4 expression (Figure 3H). Because IRF4 and PU.1 can interact (Eisenbeis et al., 1995; Tamura et al., 2008), we also examined the IL-21-induced Prdm1 expression in Sfpi1f/f-CD19-Cre+/− B cells and (Sfpi1 is the gene that encodes PU.1) found similar Prdm1 expression to wild-type (WT) cells (Figure 3I). IL-21R expression in Sfpi1f/f/CD19-Cre+/− B cells was similar to that in the WT control (Figure 3J). Thus, IRF4, but not PU.1 or IRF8, is required for Prdm1 expression.
IL-21-Induced Prdm1 Gene Expression in T Cells Is Dependent on STAT3 and IRF4
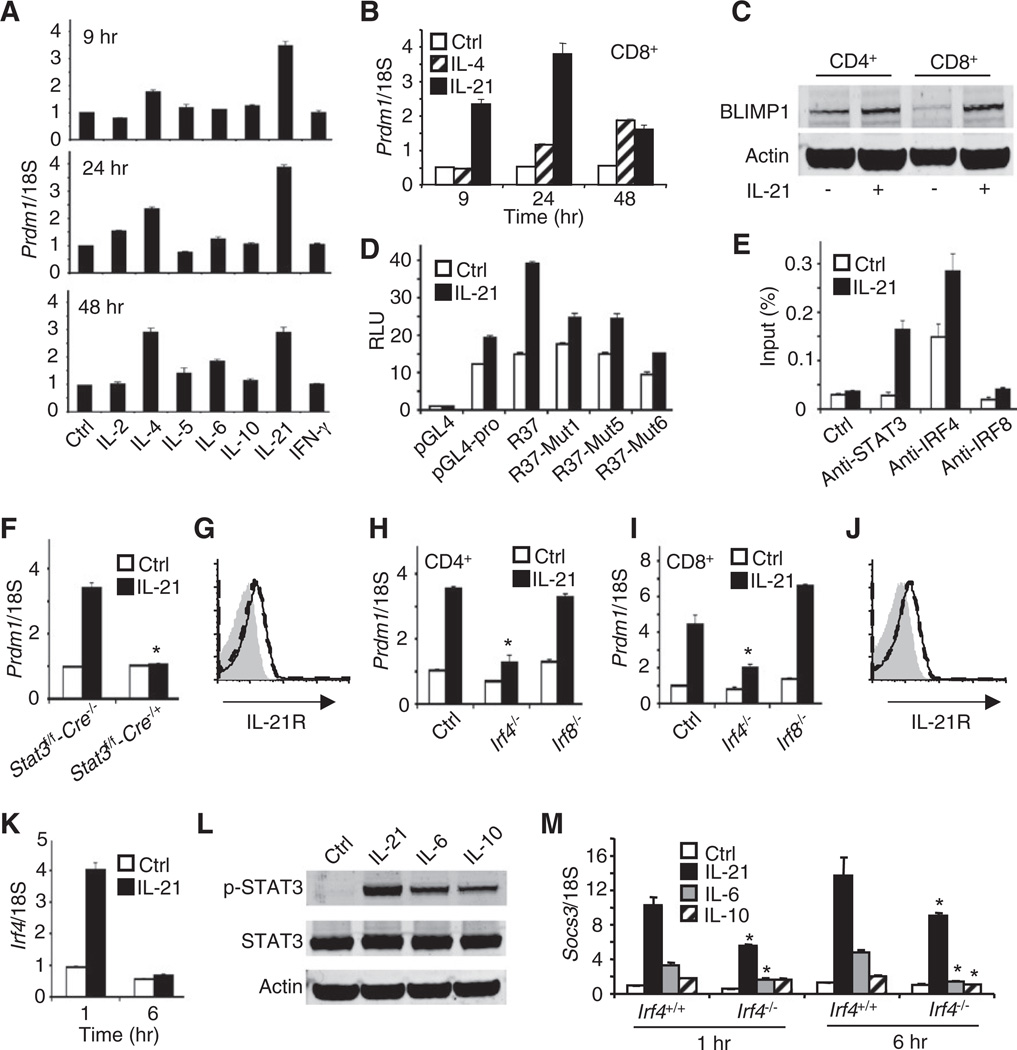
We next examined Prdm1 induction in preactivated splenic T cells. Whereas IL-2, IL-5, IL-6, IL-10, and IFN-γ had little if any effect, IL-4 induced Prdm1 expression, and IL-21 was the most potent inducer in both CD4+ (Figure 4A) and CD8+ T cells (Figure 4B), with more rapid Prdm1 induction than was seen with IL-4. IL-21 also induced BLIMP1 protein (Figure 4C). As in B cells, IL-21 markedly increased activity of the 212 bp R37 reporter in T cells, and mutation of the GAS motif (Mut1) or IRF motifs (Mut5) or both together (Mut6) decreased activity (Figure 4D). ChIP experiments confirmed STAT3 and IRF4 binding to the IL-21 response element in splenic CD4+ T cells (Figure 4E), with no binding of these factors to an intronic region of the Actb gene (data not shown). IL-21-induced Prdm1 expression was abrogated in splenic Stat3-deficient CD4+ T cells (Figure 4F) and markedly diminished in CD4+ and CD8+ T cells from Irf4−/− but not Irf8−/− mice (Figures 4H and 4I). IL-21R expression was not altered in Stat3-deficient and Irf4−/− T cells (Figure 4G and 4J). In contrast to B cells, IL-21 markedly induced Irf4 mRNA expression in CD4+ T cells at 1 hr (Figure 4K). Although IL-21, IL-6, and IL-10 each activated STAT3, IL-21 was most potent (Figure 4L), corresponding to its more potent induction of Prdm1 (Figure 4A) and Socs3 (Figure 4M) expression. At 6 hr, IL-6-, IL-10-, and IL-21-induced Socs3 expression was reduced in Irf4−/− CD4+ T cells (Figure 4M).
Figure 4. STAT3 and IRF4 Control IL-21-Induced Prdm1 Expression in CD4+ T Cells.
(A) T cells were preactivated with anti-CD3 plus soluble anti-CD28, and IL-2 for 3 days and rested for 24 hr in fresh RPMI medium, and Prdm1 mRNA in CD4+ T cells was determined by quantitative RT-PCR 9, 24, and 48 hr after treatment with IL-2, IL-4, IL-5, IL-6, IL-10, IL-21, or IFN-γ (see Experimental Procedures for details). Shown are means ± SEM of ≥5 independent experiments (total combined samples ≥10).
(B) Prdm1 mRNA expression was determined in preactivated CD8+ T cells by quantitative RT-PCR at the indicated times after treatment with 100 ng/ml IL-21.
(C) Preactivated splenic CD4+ and CD8+ T cells were treated with 100 ng/ml IL-21 for 24 hr and BLIMP1 or Actin protein levels were determined by immunnoblotting.
(D) Luciferase assays were performed with WT or mutated forms of R37 for STAT3 and/or IRF4-binding motifs 6 hr after treatment of CD4+ T cells with 100 ng/ml IL-21.
(E) ChIP of STAT3 and IRF4 was performed with splenic CD4+ T cell lysates prepared 20 min after treatment with 100 ng/ml IL-21. Input was determined by quantitative PCR. Results are representative of three independent experiments.
(F) Prdm1 mRNA expression was measured in splenic CD4+ T cells from Stat3f/f-Cre−/− or Stat3f/f-Cre−/+ mice; cells were not treated or were stimulated with IL-21. The asterisk indicates no induction as compared to IL-21-treated Stat3f/f-Cre−/− cells.
(G) IL-21R expression was measured in CD4+ T cells from Stat3-deficient (dashed line) or WT control (solid line) mice. The isotype antibody control is shown in gray.
(H and I) Prdm1 mRNA expression was measured in WT (control), Irf4−/−, and Irf8−/− CD4+ T cells (H) or CD8+ T cells (I) by quantitative RT-PCR 24 hr after treatment with 0 or 100 ng/ml IL-21mRNA levels (means ± SEM of two independent experiments, total combined samples = 4). The asterisk indicates decreased induction as compared to Ctrl.
(J) IL-21R expression was measured in CD4+ T cells from Irf4−/− (dashed line) or WT (solid line) mice. The isotype antibody control is shown in gray.
(K) IL-21 significantly induces Irf4 expression in splenic CD4+ T cells. Cells were treated with 100 ng/ml IL-21 (means ± SEM of two independent experiments, total combined samples = 4).
(L) CD4+ T cells were preactivated and then not stimulated or stimulated with IL-21, IL-6, or IL-10, and subsequently, lysates were immunoblotted with antibodies to phospho-STAT3, STAT3, or Actin. A representative result from three independent experiments is shown.
(M) CD4+ T cells from Irf4+/+ or Irf4−/− mice were preactivated and then not stimulated or stimulated with IL-21, IL-6, or IL-10 for 1 or 6 hr. Socs3 mRNA expression (means ± SEM of two independent experiments, total combined samples = 4) was determined by quantitative RT-PCR. *p < 0.05 (comparing similar samples to cells from Irf4+/+ and Irf4−/− mice).
Because STAT3 and IRF4 both bind to the IL-21 response element and functionally cooperate for Prdm1 expression, we hypothesized that these proteins might associate. Although we saw weak coprecipitation in some experiments, this was not reproducible (data not shown), consistent with the lack of interaction reported in another study that used STAT3 as a negative control for coprecipitation with IRF4 (Gupta et al., 1999). Thus, if STAT3 and IRF4 interact, the association is either transient or unstable under the experimental conditions used.
STAT3 and IRF4 Often Bind to Overlapping Sites on the Genome
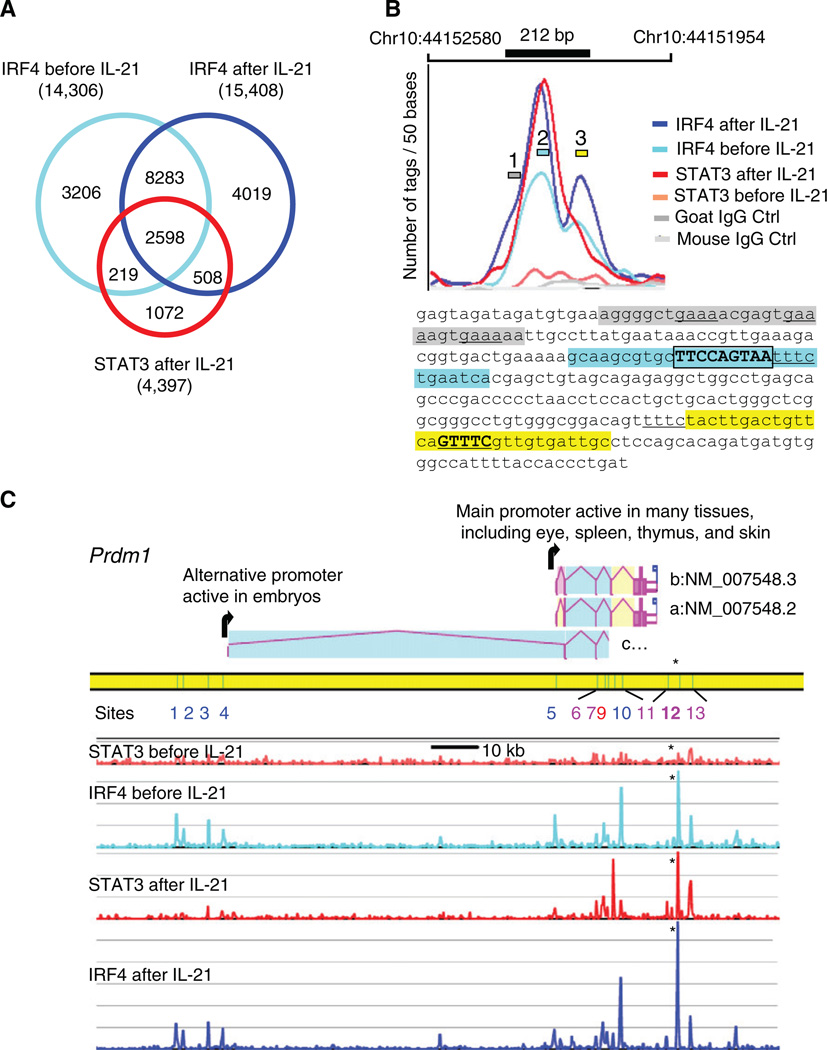
Because STAT3 and IRF4 cooperatively activated the Prdm1 IL-21 response element, we investigated the extent to which binding sites for these factors colocalized. Using ChIP followed by Solexa-based sequencing (Chip-Seq), we mapped (see Experimental Procedures and Supplemental Experimental Procedures and Figures S4 and S5 for details) the STAT3- and IRF4-binding sites on the whole genome in CD4+ T cells that were preactivated, rested, and not treated or treated with IL-21. A total of 4.7 to 6.3 million uniquely mapped tags were obtained from two independent experiments that were merged (Table S1). We identified (see Experimental Procedures) 4,422 STAT3 and 18,883 IRF4 sites, covering 1.74 Mb and 7.36 Mb (0.07% and 0.3% of the genome), respectively. Strikingly, the binding profiles of STAT3 and IRF4 strongly overlapped: 76% of the STAT3-binding regions also bound IRF4 (Figure 5A). The a priori probability of this level of colocalization is very low (p < 10−5000). Table S2 shows for all STAT3-binding sites the characteristics of the peaks, their coordinates on the genome, the number of STAT3 and IRF4 tags at each site before and after IL-21 treatment, and the list of neighboring genes, which are candidates for regulation by STAT3 and IRF4.
Figure 5. STAT3- and IRF4-Binding Sites Colocalize.
(A) Venn diagram of IRF4-binding sites before and after IL-21 stimulation. Shown are STAT3 sites after stimulation; the rare STAT3 sites before IL-21 treatment were omitted.
(B) High-resolution mapping of STAT3 and IRF4 ChIP-Seq tags at the Prdm1 IL-21 response element. The number of tags/50 bp is plotted. The 212 bp IL-21 response element is drawn to scale and exactly overlaps the STAT3 and two IRF4-binding sites. The maxima of the curves coincide with the essential motifs shown in Figures 3 and 4; the sequences at these maxima are high-lighted in blue and yellow, with DNA-binding motifs for STAT3 and IRF4 boxed and underlined, respectively. The shoulder of the largest IRF4 peak suggests a secondary binding site (gray region).
(C) The mouse Prdm1 gene includes three alternative transcript variants a, b, and c (AceView at NCBI). The STAT3- and IRF4-binding sites before and after IL-21 treatment are aligned (scale bar is 10 kb and 20 tags/200 bp window vertically), with binding sites corresponding to peaks 1–13 (Table S11). Although only one STAT3-IRF4 site (site 12, see asterisk, shown in detail in B) was found to regulate IL-21-mediated Prdm1 expression, several other major STAT3 peaks were induced by IL-21, and all overlapped IRF4 peaks. In intron 5 of variant a, site 9 strongly binds STAT3 but weakly binds IRF4, whereas site 10, which is 1.59 kb 3′ of site 9, strongly binds IRF4 but weakly binds STAT3. Additionally, IRF4 associated to the two promoter areas at sites 1 to 5.
As expected, IL-21 induced STAT3 binding (4397 sites after IL-21 versus 335 before). In contrast, similar numbers of IRF4-binding sites were seen before (14,306) and after (15,408) IL-21 treatment, and their genome-wide pattern was partly reorganized (Figures 5A). The fact that 64% of the sites where STAT3 bound after IL-21 treatment were occupied by IRF4 prior to IL-21 suggested that IRF4 might promote recruitment of STAT3, and the strong overlap of STAT3- and IRF4-binding sites suggested that these factors might broadly cooperate to regulate gene expression.
High-Resolution Mapping of STAT3- and IRF4-Binding Sites in the Prdm1 Gene
We found that the genome-wide ChIP-Seq data corroborated and extended our characterization of the Prdm1 IL-21 response element. STAT3 and IRF4 both bound to the 212 bp IL-21 response element (Figure 5B), and the maxima of the ChIP-Seq tag densities lie within a few base pairs of the motifs we had defined. The STAT3 peak coincides with the TTCCAGTAA motif (#2 blue region in Figure 5B and sequence below) and the two IRF4 main peaks coincide with the TTTC motif (underlined in the blue region) and the GTTTC motif (#3 yellow region). A secondary IRF4-binding site (#1 gray region) was also observed. The pattern suggests that IL-21 induces STAT3 binding and a rearrangement of IRF4 binding nearby. Interestingly, additional STAT3 and IRF4 sites were found in the Prdm1 gene (Figure 5C and Table S2; see Table S10 for all IRF4-binding sites in mouse CD4+ T cells), but their functional significance was not indicated by our Prdm1 reporter assays (Figures 1E and 1F).
Correlation of STAT3- and IRF4-Binding Sites with IL-21-Regulated Genes
Given the functional cooperation of STAT3 and IRF4 for IL-21-induced Prdm1 expression, we investigated whether this extended to other genes. We performed an Affymetrix gene expression analysis (Figures S2 and S3 and Table S3) using preactivated T cells induced by IL-21 for 0, 1, 6, and 24 hr. Five biological replicas at each time point gave highly consistent results. With a fold change of ≥ 1.5 at p ≤ 0.001, 2322 genes were regulated by IL-21, which included well-recognized STAT3-regulated genes such as Socs3 and Bcl3.
To identify which IL-21-regulated genes might be regulated by STAT3, we identified those “within reach” of a STAT3-binding site by ChIP-Seq analysis. Because transcriptional regulators can bind upstream, within, or downstream of target genes, and act from a distance, knowing binding-site locations does not necessarily indicate their importance for gene regulation. Nevertheless, genes with such sites were reasonable candidates. On the basis of the ChIP-Seq experiments, 811 of the 2322 IL-21-regulated genes (Table S4) were associated with 1682 STAT3-binding sites, or an average of approximately two STAT3 sites per IL-21-regulated gene. Most of these sites (1308 or 78%) also bound IRF4, and we observed a cooperative effect of IRF4 on STAT3 binding and vice versa: sites binding both STAT3 and IRF4 had on average twice as many tags for each protein as sites binding only STAT3 or IRF4.
We next examined the location of the STAT3 and IRF4 sites relative to the IL-21 regulated genes by using gene annotations defined in AceView (Thierry-Mieg and Thierry-Mieg, 2006), which includes all experimentally supported alternatively spliced transcripts. STAT3-binding sites lie inside 63% of the regulated genes, upstream of 41%, and downstream of 25% (Tables S5, S6, and S7, respectively); this sums to >100%, reflecting the frequent presence of several STAT3 sites in more than one location relative to a given gene. The sites are broadly distributed, but with a relative enrichment in the promoter area, with 14% of sites between 2 kb 5′ upstream and 1 kb downstream of the most 5′ TSS (Figure S7). The broad distribution is consistent with the ability of STATs to act at a distance, as for example is seen in the IL2RA gene (Kim et al., 2001). Interestingly, similar to the Prdm1 site, STAT3 sites located downstream of genes more often exhibit IRF4 cobinding (96%, 329 of 345 sites) than sites within (71%, 600 of 850 sites) or upstream (78%, 422 of 543 sites) of genes.
Strikingly, 310 of the 811 IL-21 sensitive genes near a STAT3-binding site were previously associated to a phenotype by MGI annotation (Table S4): 199 genes (25%) have particularly interesting phenotypes, given that the genetic defects affect the immune system (180 genes) or the hematopoietic system (145 genes) or lead to carcinogenesis (44 genes), as compared to only 5% of 33,774 genes tested on the Affymetrix chip. When we integrated in AceView the tissues of origin of the cDNA libraries reported in GenBank and dbEST, we found that 791 of the 811 IL-21-regulated genes (98%) with a STAT3 site nearby were expressed within lymphoid tissues versus 66% of genes on the Affymetrix chip.
STAT3 and IRF4 Broadly Cooperate to Mediate IL-21-Induced Gene Expression in CD4+ T Cells
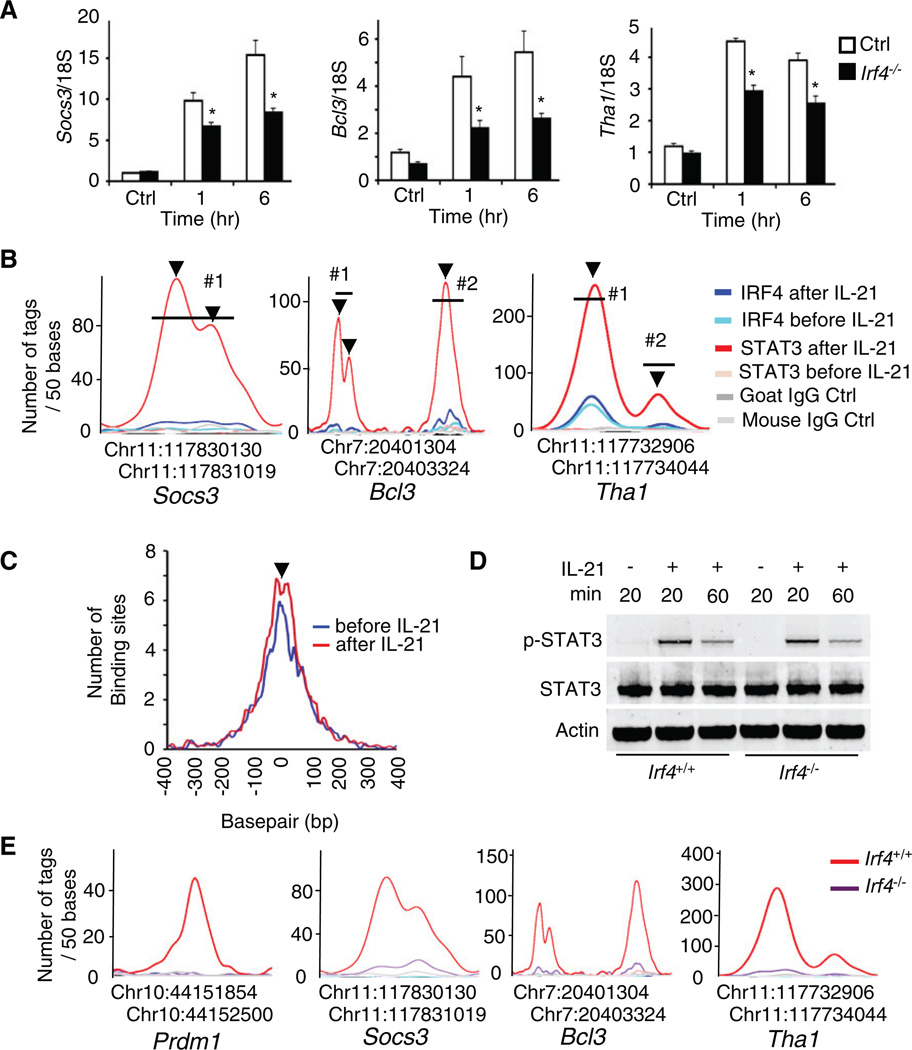
To extend our findings, we selected a number of genes induced >1.5-fold with colocalization of STAT3 and IRF4 binding (a list of these genes is in Tables S5–S7). As shown in Figure 6A (open bars), IL-21 significantly enhanced the expression of Socs3, Bcl3, and Tha1 in CD4+ T cells from WT littermate controls, but their induction was diminished in cells from Irf4−/− mice (solid bars) and Stat3−/− mice (data not shown). This is consistent with closely positioned STAT3- and IRF4-binding sites in these genes (Figures 6B) and establishes that STAT3 and IRF4 more broadly functionally cooperate (see Figure S9 for the sequences spanning the regions in the Socs3, Bcl3, and Tha1 genes that are depicted in Figure 6B).
Figure 6. IRF4 Promotes STAT3 Binding In Vivo.
(A) CD4+ T cells from Irf4−/− and littermate control mice were preactivated and rested as in Figure 5A. mRNA for Socs3, Bcl3, and Tha1 determined by quantitative RT-PCR 1 and 6 hr after treatment with 100 ng/ml IL-21. Shown are means ± SEM of two independent experiments (total combined samples = 4). *p < 0.05 (compared with WT control mice).
(B) High-resolution mapping of STAT3 and IRF4-ChIP-Seq tags in the Socs3, Bcl3, and Tha1 genes.
(C) The distance in base pairs between the precise position of IRF4 binding, before (blue) or after (red) IL-21 treatment, and the precise position of STAT3 binding after IL-21 is plotted for all composite IRF4- and STAT3-binding sites with >30 tags.
(D) CD4+ T cells from Irf4+/+ and Irf4−/− mice were preactivated and stimulated with 100 ng/ml IL-21 as indicated, followed by immunoblotting with antibodies to phospho-STAT3, STAT3, or Actin. The representative result of two independent experiments is shown.
(E) High-resolution mapping of the STAT3 ChIP-Seq tags in the Prdm1, Socs3, Bcl3, and Tha1 genes in CD4+ T cells from Irf4−/− and littermate control mice.
We next partitioned the genes into those “within reach” of either a composite site binding both STAT3 and IRF4, of two independent sites with one binding STAT3 and the other IRF4, of a site binding STAT3 or IRF4 alone, or near no candidate site. Interestingly, genes whose expression was regulated by IL-21 (>1.5-fold change in the Affymetrix array) were much more frequently found in the vicinity of composite sites (Table S8). A chi-square analysis revealed that over the entire genome, the cooperative binding of STAT3 and IRF4 to the same sites correlates with a greater propensity to mediate IL-21-induced regulation of gene expression as compared to sites binding only STAT3 (p = 3 × 10−4). Moreover, a chromosomal map depicting the location of STAT3 and IRF4 sites throughout the genome reveals the high frequency of association of IRF4 with STAT3 and the rarity of apparently functional STAT3 sites that exist in the absence of IRF4 (Figure S6). Very close binding of STAT3 with IRF4 was evident in a genome-wide fashion (Figure 6C). The interesting “dip” in the peak (arrowhead in Figure 6C) that is evident after IL-21 stimulation suggests steric constraints when both proteins are present.
IRF4 Promotes STAT3 Binding
Because of the colocalization of STAT3 and IRF4, we investigated whether STAT3 binding is affected by IRF4 deficiency. We first examined STAT3 phosphorylation and found no differences in STAT3 phosphorylation in Irf4+/+ and Irf4−/− CD4+ T cells after IL-21 treatment (Figure 6D). We next used ChIP-Seq to map the STAT3-binding sites on the whole genome in Irf4−/− CD4+ T cells. As shown in Figure 6E, binding of STAT3 to the Prdm1, Socs3, Bcl3, and Tha1 genes was greatly diminished in the absence of IRF4. Moreover, only ~14% of the STAT3-binding sites previously identified (Table S2) were still present in the Irf4-deficient CD4+ T cells (Table S9), and even those sites were generally much more weakly seen.
IRF4 Is Critical for Tfh Cell Differentiation
Because STAT3 is a critical mediator for IL-21-induced follicular helper T (Tfh) cell differentiation (Nurieva et al., 2008; Vogelzang et al., 2008), and we show an IL-21-induced cooperation of STAT3 and IRF4, we immunized Irf4−/− mice with keyhole limpet hemocyanin (KLH) to determine the role of IRF4 in Tfh cell differentiation. The proportion of KLH-induced CXCR5+ICOS+CD4+ Tfh cells was markedly diminished in Irf4−/− mice, consistent with a role for IRF4 in Tfh cell differentiation (Figures S8A and S8B). As expected, the proportion of PNA+B220+ germinal center B cells was also low in Irf4−/− mice (Figures S8C and S8D).
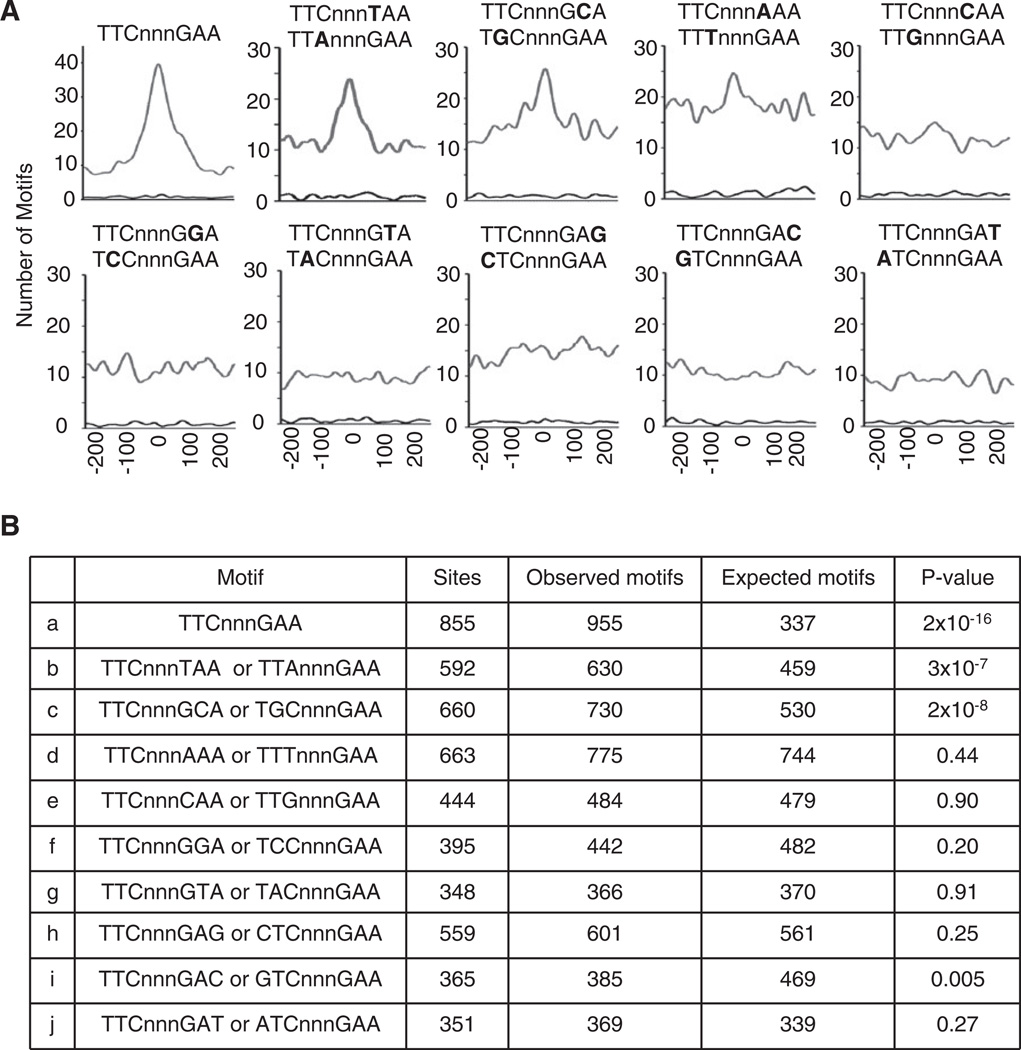
The TTCnnnTAA Motif that Binds STAT3 in the Prdm1 Gene Is Widely Used
In the Prdm1 IL-21 response element, STAT3 binds to a TTCnnnTAA variant GAS motif. We thus evaluated the utilization by STAT3 of the canonical TTCnnnGAA GAS motif versus all single-nucleotide variants in the 4397 STAT3-binding sites observed by ChIP-Seq. To evaluate the contribution of each motif to STAT3 binding, we measured its distance in base pairs relative to the maximum of the ChIP-Seq tag density and plotted the distribution of these distances over all STAT3-binding sites; if a motif binds STAT3, its position should coincide with the maximum of the tag density. The canonical TTCnnnGAA GAS motif was the most frequent and best centered (Figure 7A), and its peak is best approximated by a Gaussian centered at zero, with an accuracy of a single nucleotide. The TTCnnnTAA/TTAnnnGAA single-base variant found in the Prdm1 gene was the next best centered, followed by TTCnnnGCA/TGCnnnGAA, whereas other variants were less well centered. We compared the number of motifs observed in the central versus the marginal 200 bp of each distribution. Only the top three motifs (TTCnnnGAA, TTCnnnTAA/TTAnnnGAA, and TTCnnnGCA/TGCnnnGAA) were markedly enriched in the center of the binding sites, and we found no evidence for binding to the other variant motifs (Figure 7B). Thus, the canonical GAS motif and the TTCnnnTAA and TTCnnnGCA variant motifs are the ones broadly used for binding STAT3 in response to IL-21.
Figure 7. Distance between the Maxima of STAT3 Binding and GAS or GAS-like Motifs.
(A) The presence of TTCnnnGAA GAS motifs and single-nucleotide variants in the 4397 STAT3-binding sites were analyzed genome-wide. The position of each motif relative to the maximum of STAT3-binding tags (within 250 bp on each side) was recorded and the distribution over all sites was plotted.
(B) Shown are the number of sites with the motif within 100 bp of maximal STAT3 binding, the number of observed and expected motifs, assuming a locally flat distribution, and p value of the observed overrepresentation of each motif in the center (Pearson χ2 test).
DISCUSSION
We previously demonstrated that IL-21 augments expression of BLIMP1, explaining the dramatic ability of IL-21 to drive terminal B cell differentiation to plasma cells. To explain the basis for IL-21-induced BLIMP1 expression, we analyzed the Prdm1 gene and identified an IL-21 response element ~4.7 kb 3′ of the polyadenylation signal. Importantly, we showed that both STAT3 and IRF4 bind to this element and are critical for IL-21-induced Prdm1 expression in B and T cells.
IL-21 is known to signal via STAT3 (Diehl et al., 2008; Zeng et al., 2007), but a role for IRF4 in IL-21 signaling was not clear. Irf4 is an immediate early gene induced in B and T cells, but it also is highly expressed in plasma cells (Klein et al., 2006; Sciammas et al., 2006). Although one group reported that IRF4 is not required for LPS-induced Prdm1 induction (Klein et al., 2006), another showed that Irf4−/− B cells have diminished LPS-induced Prdm1 expression and detected a putative LPS-induced IRF4-binding site in intron 5 (Sciammas et al., 2006). A study of multiple myeloma also found IRF4 binding to the PRDM1 gene (Shaffer et al., 2008). Although our ChIP-Seq data reveal IRF4-binding sites within intron 5 for all IRF4-binding sites in mouse CD4+ T cells, this region, which is included in reporter R6, exhibited little if any IL-21-induced Prdm1 expression. Instead, the IL-21 response element is 3′ of the gene, suggesting that IL-21 and LPS induce Prdm1 expression via at least partially distinct elements, with our data indicating a functional cooperation between STAT3 and IRF4.
Although IL-21, IL-6, and IL-10 all activate STAT3, their physiological actions differ, presumably as a result of when and where the cytokines are produced and their receptors are expressed. Additionally, they differ in other signaling pathways used and other STATs that are activated. For example, IL-21 also activates STAT1 and STAT5 as well as ERK and PI3-kinase-Akt (Zeng et al., 2007). In this study, we showed that IL-21 induced STAT3 phosphorylation more potently than did IL-6 or IL-10, and correspondingly, IL-21 was the most potent in inducing Socs3 expression in CD4+ T cells. Socs3 expression was also IRF4 dependent, suggesting that STAT3 and IRF4 cooperate for regulating a range of genes.
On the basis of ChIP-Seq analysis, after IL-21 stimulation, STAT3 binds close to IRF4, not only 3′ of the Prdm1 gene but also at thousands of sites throughout the genome. IRF4 was frequently bound to sites prior to IL-21 stimulation, suggesting possibly enhanced binding of STAT3 to IRF4-occupied sites. Anti-CD3 plus anti-CD28-mediated preactivation of T cells and anti-CD40 stimulation of B cells can induce Irf4 expression (Basso et al., 2004; Grumont and Gerondakis, 2000), and consistent with this, the ChIP-Seq data revealed broad IRF4 binding even before IL-21 treatment and substantial rearrangement of IRF4 binding after IL-21 treatment. Strikingly, ChIP-Seq analysis with Irf4−/− CD4+ T cells revealed that IL-21-induced STAT3 binding was dramatically diminished, including in the Prdm1, Socs3, Bcl3, and Tha1 genes. Thus, IRF4 is critical for optimal STAT3 binding after IL-21 treatment in T cells. Our data indicate that prebound IRF4 may guide STAT3 to specific binding sites and/or may enhance STAT3 binding. As noted, we were not able to establish a definitive STAT3-IRF4 physical interaction, although this cannot be excluded, and it is possible that additional factors might be required for such an interaction. Moreover, it is possible that the binding of STAT3 is dependent on the overall chromatin structure and that the presence of IRF4 alters this structure to allow and/or facilitate the binding of STAT3.
IRF4 was previously shown to be critical for Th2 (Lohoff et al., 2002) and Th17 (Brüstle et al., 2007) cell differentiation. Consistent with IRF4 cooperatively acting with STAT3, which is known to be important for IL-21-induced Tfh cell differentiation (Nurieva et al., 2008; Vogelzang et al., 2008), our data indicate that IRF4 may also contribute to Tfh cell differentiation.
In summary, we have identified an IL-21 response element 3′ to the Prdm1 gene that binds STAT3 and IRF4, and we show a functional cooperation between these two proteins. ChIP-Seq genome wide analysis revealed that this effect is not restricted to the Prdm1 gene, establishing the broad importance of STAT3-IRF4 cooperation. We further show defective Tfh cell differentiation in Irf4−/− mice analogous to the reported defective Tfh cell differentiation in Stat3−/− mice. Whether there is broader cooperativity of other STAT proteins with IRF4 or of STAT3 with other IRF family members is an area for future investigation. Finally, we have elucidated the relative utilization of canonical versus noncanonical GAS motifs for STAT3 binding in vivo and demonstrated broad use of a variant motif that is critical in the Prdm1 IL-21 response element.
EXPERIMENTAL PROCEDURES
Mice and Cell Culture
Animal experiments used protocols approved by the NHLBI Animal Use and Care Committee and followed NIH guidelines. Details of mice and cell culture are provided in the Supplemental Experimental Procedures.
Preactivation of Cells and Cytokines
For preactivation of CD4+ T cells, cells were stimulated with 3 µg/ml plate-bound anti-CD3, 1 µg/ml soluble anti-CD28, and 50 U/ml IL-2 for 3 days and rested for 24 hr in fresh RPMI medium. Cytokines were used at the following concentrations: 100 U/ml IL-2, 50 ng/ml IL-4, 10 ng/ml IL-5, 10 ng/ml IL-6, 20 ng/ml IL-10, 100 ng/ml IL-21, or 50 ng/ml IFN-γ.
Antibodies
Antibodies to STAT3, STAT5A, and STAT5B were from Zymed (South San Francisco, CA), antibodies to STAT1, IRF4, IRF8, and PU.1 were from Santa Cruz (Santa Cruz, CA), and antibodies to CD3ε and CD28 were from PharMingen (San Diego, CA).
Quantitative Real-Time PCR Analysis
This was performed with standard methods (see Supplemental Experimental Procedures).
Immunoblotting and FACS
For immunoblotting (Kovanen et al., 2008), cell lysates were resolved on 10% Bis-Tris gels (NuPAGE) and immunoblotted with antibodies to phospho-STAT3 (Cell Signaling Technology, CA), STAT3 (PharMingen, CA), BLIMP1 (Novus,CO), and β-actin (Santa Cruz, CA). FACS was performed with standard methods.
Enhancer Constructs, Transient Transfections, and Luciferase Assays
The mouse −979 and +183 Prdm1 promoter fragment was cloned between the HindIII and XhoI polylinker sites in pGL3-Basic or pGL4-Basic luciferase reporter vectors (Promega, Madison, WI) (Supplemental Experimental Procedures). A total of 5 × 106 NFS201 cells were transfected with 5 µg of test plasmid and 40 ng of pRL-TK as a transfection efficiency control, with use of the DEAE-dextran method. Twenty-four hours later, cells were stimulated with medium or 50 ng/ml of mouse IL-21 (R&D Systems) for 18 hr. Splenic B cells, CD4+ T cells, or CD8+ T cells were isolated from 6- to 8-week-old C57BL/6 mice with anti-B220, anti-CD4, or anti-CD8-microbeads (Miltenyi). B cells were preactivated with 1 µg/ml anti-CD40 for 3 days, rested for 24 hr, and electroporated (Electropulser, Bio-Rad) with 20 µg of test plasmid and 10 µg of pRL-TK. Splenic CD4+ and CD8+ T cells were activated with plate-bound anti-CD3ε (3 µg/ml), soluble anti-CD28 (1 µg/ml), and IL-2 (50 U/ml) for 3 days, rested for 24 hr, and electroporated with 10 µg of test plasmid and 5 µg pRL-TK. B and T cells were rested, stimulated with medium or 100 ng/ml mouse IL-21 for 6 hr, and analyzed for luciferase activity with a luminometer (Victor2 1420 Multilabel Counter; PerkinElmer Life Sciences) and a Dual Luciferase Assay System Kit (Promega).
Electrophoresis Mobility Shift Assays and Chromatin Immunoprecipitation Assays
These were done by standard methods (see Supplemental Experimental Procedures).
ChIP-Seq Analysis
CD4+ T cells were prepared and ChIP-Seq data were generated as described previously (Schones et al., 2008). A matched IgG control was associated to each experiment. Chromatin from 107 cells (~200 ng of DNA) was sonicated into 200–500 bp fragments, DNA ends were repaired with polynucleotide kinase and Klenow enzyme and treated with Taq polymerase for generation of a protruding 3′ “A” nucleotide for adaptor ligation. After ligation of Solexa adaptors, DNA was amplified with the adaptor primers for 17 cycles and fragments of ~220 bp (mononucleosome + adaptors) were isolated from agarose gels and used for cluster generation and sequencing on a Solexa 1G and Illumina GA2 Genome Analyzers. The sequence tags will be available from the NCBI Trace/Short Read Archive repository. To align sequence tags, analyze their clustering, identify binding motifs, and display results, we developed new code in C language as extensions of the AceView project (Thierry-Mieg and Thierry-Mieg, 2006). Each Solexa 25-mer tag was mapped on the mouse reference genome (version 37), allowing up to two mismatches (single-base insertion, deletion, or substitution), and retained only if it had a single best position genome-wide. Each tag was shifted downstream by half the effective length of the sonicated sequenced fragments, measured as the maximum of the genome-wide correlation function between hits to the sense and antisense strands of the chromosomes, and represented by a Gaussian with an area of 1 and σ of 100 bp, which dampens the sampling fluctuations and introduces a controlled level of fuzziness and “smoothness.” Summing these elementary Gaussians yields a smoothed tag density. Regions where the density locally exceeds a given threshold are analyzed. The number of tags was measured after subtracting the local tags from the matched IgG experiment. If the tag number exceeded a second threshold, it was scored as a peak. Regions with an IgG peak were deemed nonspecific and eliminated. The peak height indicates the protein binding strength. The position of the peak maximum (i.e., the maximum of the probability of binding the protein) is expected to coincide with the binding motif. The width covered by the peak (typically 200–600 bp) is the region protected by the protein or protein complex. Often, in large peaks, one can distinguish subpeaks, which probably correspond to the binding of multiple proteins. When multiple proteins bind to overlapping sequences, their peaks are clustered to generate a broader composite binding site. See Supplemental Experimental Procedures and Figures S4, S5, and S6 for details.
RNA Preparation and Microarray Analysis
Total RNA was prepared with RNeasy (QIAGEN, Valencia, CA). Probes were prepared with Affymetrix protocols and hybridized to mouse MOE 430 2.0 arrays (Affymetrix, Santa Clara, CA). The excellent reproducibility across replicas is shown in Figure S2.
Supplementary Material
ACKNOWLEDGMENTS
We thank H.C. Morse III for NFS201 and NFS202 cells, D.E. Schones for valuable discussions and assistance with handling the Illumina raw ChIP-Seq data and for processing ChIP-Seq data, K. Cui and T.-Y. Roh for help with Solexa sequencing, the NHLBI Affymetrix Core and N. Raghavachari, X. Xu, and D. Liu for help with generating, analyzing, and handling Affymetrix data, and J.-X. Lin, H. Young, K. Zhao, and K.-T. Jeang for valuable discussions and/or critical comments. This work was supported by the Division of Intramural Research, National Heart, Lung, and Blood Institute, and by the National Center for Biotechnology Information, National Library of Medicine. W.J.L. is an inventor on patents and patent applications related to IL-21.
Footnotes
ACCESSION NUMBERS
The microarray primary CEL files and Illumina raw Chip-Seq data are available in the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/gds) under the accession number GSE19198.
SUPPLEMENTAL DATA
Supplemental Data include Supplemental Experimental Procedures, nine figures, and eleven tables and can be found with this article online at http://www.cell.com/immunity/supplemental/S1074-7613(09)00511-1.
REFERENCES
- Basso K, Klein U, Niu H, Stolovitzky GA, Tu Y, Califano A, Cattoretti G, Dalla-Favera R. Tracking CD40 signaling during germinal center development. Blood. 2004;104:4088–4096. doi: 10.1182/blood-2003-12-4291. [DOI] [PubMed] [Google Scholar]
- Brüstle A, Heink S, Huber M, Rosenplänter C, Stadelmann C, Yu P, Arpaia E, Mak TW, Kamradt T, Lohoff M. The development of inflammatory T(H)-17 cells requires interferon-regulatory factor 4. Nat. Immunol. 2007;8:958–966. doi: 10.1038/ni1500. [DOI] [PubMed] [Google Scholar]
- Diehl SA, Schmidlin H, Nagasawa M, van Haren SD, Kwakkenbos MJ, Yasuda E, Beaumont T, Scheeren FA, Spits H. STAT3-mediated up-regulation of BLIMP1 Is coordinated with BCL6 down-regulation control human plasma cell differentiation. J Immunol. 2008;180:4805–4815. doi: 10.4049/jimmunol.180.7.4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenbeis CF, Singh H, Storb U. Pip, a novel IRF family member, is a lymphoid-specific, PU.1-dependent transcriptional activator. Genes Dev. 1995;9:1377–1387. doi: 10.1101/gad.9.11.1377. [DOI] [PubMed] [Google Scholar]
- Gabriele L, Ozato K. The role of the interferon regulatory factor (IRF) family in dendritic cell development and function. Cytokine Growth Factor Rev. 2007;18:503–510. doi: 10.1016/j.cytogfr.2007.06.008. [DOI] [PubMed] [Google Scholar]
- Grumont RJ, Gerondakis S. Rel induces interferon regulatory factor 4 (IRF-4) expression in lymphocytes: Modulation of interferon-regulated gene expression by rel/nuclear factor kappaB. J. Exp. Med. 2000;191:1281–1292. doi: 10.1084/jem.191.8.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Jiang M, Anthony A, Pernis AB. Lineage-specific modulation of interleukin 4 signaling by interferon regulatory factor 4. J. Exp. Med. 1999;190:1837–1848. doi: 10.1084/jem.190.12.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber M, Brüstle A, Reinhard K, Guralnik A, Walter G, Mahiny A, von Löw E, Lohoff M. IRF4 is essential for IL-21-mediated induction, amplification, and stabilization of the Th17 phenotype. Proc. Natl. Acad. Sci. USA. 2008;105:20846–20851. doi: 10.1073/pnas.0809077106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallies A, Hawkins ED, Belz GT, Metcalf D, Hommel M, Corcoran LM, Hodgkin PD, Nutt SL. Transcriptional repressor Blimp-1 essential for T cell homeostasis and self-tolerance. Nat. Immunol. 2006;7:466–474. doi: 10.1038/ni1321. [DOI] [PubMed] [Google Scholar]
- Kim HP, Kelly J, Leonard WJ. The basis for IL-2-induced IL-2 receptor alpha chain gene regulation: Importance of two widely separated IL-2 response elements. Immunity. 2001;15:159–172. doi: 10.1016/s1074-7613(01)00167-4. [DOI] [PubMed] [Google Scholar]
- Klein U, Casola S, Cattoretti G, Shen Q, Lia M, Mo T, Ludwig T, Rajewsky K, Dalla-Favera R. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat. Immunol. 2006;7:773–782. doi: 10.1038/ni1357. [DOI] [PubMed] [Google Scholar]
- Kovanen PE, Bernard J, Al-Shami A, Liu C, Bollenbacher-Reilley J, Young L, Pise-Masison C, Spolski R, Leonard WJ. T-cell development and function are modulated by dual specificity phosphatase DUSP5. J. Biol. Chem. 2008;283:17362–17369. doi: 10.1074/jbc.M709887200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard WJ. Cytokines and immunodeficiency diseases. Nat. Rev. Immunol. 2001;1:200–208. doi: 10.1038/35105066. [DOI] [PubMed] [Google Scholar]
- Levy DE, Darnell JE., Jr Stats: Transcriptional control and biological impact. Nat. Rev. Mol. Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- Lohoff M, Mittrücker HW, Prechtl S, Bischof S, Sommer F, Kock S, Ferrick DA, Duncan GS, Gessner A, Mak TW. Dysregulated T helper cell differentiation in the absence of interferon regulatory factor 4. Proc. Natl. Acad. Sci. USA. 2002;99:11808–11812. doi: 10.1073/pnas.182425099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins G, Calame K. Regulation and functions of Blimp-1 in T and B lymphocytes. Annu. Rev. Immunol. 2008;26:133–169. doi: 10.1146/annurev.immunol.26.021607.090241. [DOI] [PubMed] [Google Scholar]
- Martins GA, Cimmino L, Shapiro-Shelef M, Szabolcs M, Herron A, Magnusdottir E, Calame K. Transcriptional repressor Blimp-1 regulates T cell homeostasis and function. Nat. Immunol. 2006;7:457–465. doi: 10.1038/ni1320. [DOI] [PubMed] [Google Scholar]
- Noguchi M, Yi H, Rosenblatt HM, Filipovich AH, Adelstein S, Modi WS, McBride OW, Leonard WJ. Interleukin-2 receptor gamma chain mutation results in X-linked severe combined immunodeficiency in humans. Cell. 1993;73:147–157. doi: 10.1016/0092-8674(93)90167-o. [DOI] [PubMed] [Google Scholar]
- Nurieva RI, Chung Y, Hwang D, Yang XO, Kang HS, Ma L, Wang YH, Watowich SS, Jetten AM, Tian Q, Dong C. Generation of T follicular helper cells is mediated by interleukin-21 but independent of T helper 1, 2, or 17 cell lineages. Immunity. 2008;29:138–149. doi: 10.1016/j.immuni.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohinata Y, Payer B, O’Carroll D, Ancelin K, Ono Y, Sano M, Barton SC, Obukhanych T, Nussenzweig M, Tarakhovsky A, et al. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature. 2005;436:207–213. doi: 10.1038/nature03813. [DOI] [PubMed] [Google Scholar]
- Ozaki K, Kikly K, Michalovich D, Young PR, Leonard WJ. Cloning of a type I cytokine receptor most related to the IL-2 receptor beta chain. Proc. Natl. Acad. Sci. USA. 2000;97:11439–11444. doi: 10.1073/pnas.200360997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki K, Spolski R, Feng CG, Qi CF, Cheng J, Sher A, Morse HC, 3rd, Liu C, Schwartzberg PL, Leonard WJ. A critical role for IL-21 in regulating immunoglobulin production. Science. 2002;298:1630–1634. doi: 10.1126/science.1077002. [DOI] [PubMed] [Google Scholar]
- Ozaki K, Spolski R, Ettinger R, Kim HP, Wang G, Qi CF, Hwu P, Shaffer DJ, Akilesh S, Roopenian DC, et al. Regulation of B cell differentiation and plasma cell generation by IL-21, a novel inducer of Blimp-1 and Bcl-6. J. Immunol. 2004;173:5361–5371. doi: 10.4049/jimmunol.173.9.5361. [DOI] [PubMed] [Google Scholar]
- Parrish-Novak J, Dillon SR, Nelson A, Hammond A, Sprecher C, Gross JA, Johnston J, Madden K, Xu W, West J, et al. Interleukin 21 and its receptor are involved in NK cell expansion and regulation of lymphocyte function. Nature. 2000;408:57–63. doi: 10.1038/35040504. [DOI] [PubMed] [Google Scholar]
- Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciammas R, Shaffer AL, Schatz JH, Zhao H, Staudt LM, Singh H. Graded expression of interferon regulatory factor-4 coordinates isotype switching with plasma cell differentiation. Immunity. 2006;25:225–236. doi: 10.1016/j.immuni.2006.07.009. [DOI] [PubMed] [Google Scholar]
- Shaffer AL, Yu X, He Y, Boldrick J, Chan EP, Staudt LM. BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity. 2000;13:199–212. doi: 10.1016/s1074-7613(00)00020-0. [DOI] [PubMed] [Google Scholar]
- Shaffer AL, Emre NC, Lamy L, Ngo VN, Wright G, Xiao W, Powell J, Dave S, Yu X, Zhao H, et al. IRF4 addiction in multiple myeloma. Nature. 2008;454:226–231. doi: 10.1038/nature07064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spolski R, Leonard WJ. Interleukin-21: Basic biology and implications for cancer and autoimmunity. Annu. Rev. Immunol. 2008;26:57–79. doi: 10.1146/annurev.immunol.26.021607.090316. [DOI] [PubMed] [Google Scholar]
- Tamura T, Yanai H, Savitsky D, Taniguchi T. The IRF family transcription factors in immunity and oncogenesis. Annu. Rev. Immunol. 2008;26:535–584. doi: 10.1146/annurev.immunol.26.021607.090400. [DOI] [PubMed] [Google Scholar]
- Thierry-Mieg D, Thierry-Mieg J. AceView: A comprehensive cDNA-supported gene and transcripts annotation. Genome Biol. 2006;7 Suppl 1:S12.1–S12.14. doi: 10.1186/gb-2006-7-s1-s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner CA, Jr, Mack DH, Davis MM. Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell. 1994;77:297–306. doi: 10.1016/0092-8674(94)90321-2. [DOI] [PubMed] [Google Scholar]
- Vogelzang A, McGuire HM, Yu D, Sprent J, Mackay CR, King C. A fundamental role for interleukin-21 in the generation of T follicular helper cells. Immunity. 2008;29:127–137. doi: 10.1016/j.immuni.2008.06.001. [DOI] [PubMed] [Google Scholar]
- Zeng R, Spolski R, Finkelstein SE, Oh S, Kovanen PE, Hinrichs CS, Pise-Masison CA, Radonovich MF, Brady JN, Restifo NP, et al. Synergy of IL-21 and IL-15 in regulating CD8+ T cell expansion and function. J. Exp. Med. 2005;201:139–148. doi: 10.1084/jem.20041057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng R, Spolski R, Casas E, Zhu W, Levy DE, Leonard WJ. The molecular basis of IL-21-mediated proliferation. Blood. 2007;109:4135–4142. doi: 10.1182/blood-2006-10-054973. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.