Abstract
We employ non-radioactive in situ hybridization techniques, which combine good tissue morphology preservation with high sensitivity of transcript detection, to map gene expression in the regenerating digestive tube of the sea cucumber Holothuria glaberrima. We investigated localization of transcripts of Wnt9, TCTP, Bmp1/Tll, the genes that have been previously known to be implicated in embryogenesis and cancer. The choice was determined by our long-term goal of trying to understand how the developmental regulatory pathways known to be involved in tumor development can be activated in post-traumatic regeneration without leading to malignant growth. The gene expression data combined with the available morphological information highlight the gut mesothelium (the outer layer of the digestive tube) as a highly dynamic tissue, whose cells undergo remarkable changes in their phenotype and gene expression in response to injury. This reversible transition of the gut mesothelium from a complex specialized tissue to a simple epithelium composed of rapidly proliferating multipotent cells seems to depend on the expression of genes from multiple developmental/cancer-related pathways.
1. Introduction
Regeneration, although a distinct phenomenon, is known to rely in part on the mechanisms, that are normally involved in embryogenesis, but recapitulation of the developmental program is never absolute (Carlson, 2007). On the other hand, there are also clear similarities between regeneration and cancer (see, for instance, White and Zon, 2008). The fundamental question here is twofold. First, to what extent does regeneration depend on the mechanisms of normal development? Answering this question will help to understand whether and how re-deployment of developmental programs will help to design new therapeutic approaches to treat the poorly regenerating human body parts. The second issue is what makes regenerative processes stay under control for the benefit of the organism as a whole, without leading to runaway malignant growth. To answer these questions, one needs an extensive knowledge of the mechanisms that underlie regenerative responses.
Arguably, one of the most suitable models in which to seek the answer to the above questions is visceral regeneration in sea cucumbers (holothurians). These marine invertebrates are classified within the phylum Echinodermata, which, on the one hand, is closely related to chordates, and, on the other hand, many members of the phylum are among the best deuterostomian regenerators. Visceral regeneration in sea cucumbers is a naturally occurring phenomenon, which follows autotomy (evisceration) of internal organs in response to adverse stimuli (Byrne, 2001; Hyman, 1955; Wilkie, 2001). A wealth of information has been accumulated on the morphology of the digestive tube in uninjured and regenerating animals and on the cellular mechanisms of regeneration (García-Arrarás et al., 1998; Mashanov and Garcia-Arraras, 2011; Mashanov et al., 2005; Mashanov et al., 2010). Over the last decade, the molecular events involved in regeneration have also started to be uncovered (Mashanov and Garcia-Arraras, 2011; Mashanov et al., 2010; Ortiz-Pineda et al., 2009; Sun et al., 2011). Of particular interest in the context of the present study is the observation that the regenerating animals showed significant up-regulation of survivin and mortalin, two genes that are known not only to be involved in embryogenesis and normal stem cell maintenance, but also in cancer progression (Mashanov et al., 2010).
Here, we continue this line of research. A recent microarray study indicated Wnt9, TCTP and Bmp1/Tll as being significantly up-regulated in sea cucumber gut regeneration, as compared with non-injured animals (Ortiz-Pineda et al., 2009). These genes have been previously known to be implicated both in embryogenesis and cancer progression, and here we report their spatio-temporal expression patterns in the regenerating gut of the sea cucumber Holothuria glaberrima.
Wnt genes code for highly conserved secreted signalling proteins that are involved in regulation of diverse cell functions, including cell division, cell death, fate decision, cytoskeleton dynamics, cell adhesion, establishing cell polarity, etc (Croce and McClay, 2008; Nejak-Bowen and Monga, 2011; Taipale and Beachy, 2001; Wend et al., 2010). Wnt 9 is one of the least studied members of Wnt family. Most published studies have focused on the role of two paralogs, Wnt 9a and Wnt 9b, in vertebrate embryogenesis. For instance, Wnt 9a was found to be essential for normal liver morphogenesis (Matsumoto et al., 2008), and development of iris and corneal epithelium (Fokina and Frolova, 2006). Wnt9b plays an important role in craniofacial morphogenesis (Juriloff et al., 2006; Lan et al., 2006) and also in kidney development (Karner et al., 2011). However, there have been no reports on the role of Wnt9 subfamily members in regeneration.
Translationally controlled tumor protein (TCTP) is a regulator of cell division and cell growth, which is ubiquitously found in all eukaryotes. TCTP is up-regulated in highly proliferating normal tissues and in various cancers. Its expression levels also change significantly under various stress conditions, such as starvation, heat shock, proapoptotic/cytotoxic signals (Bommer and Thiele, 2004; Pollins et al., 2007; Zhu et al., 2008). TCTP is also an important developmental gene. In mice, knockout of TCTP leads to early embryonic lethality associated with defects in cell cycle progression and excessive cell death (Chen et al., 2007a). Its expression is also reported to be associated with the mesoderm development (notochord, presomitic mesoderm, nascent somites) in amphioxus (Chen et al., 2007b). Besides involvement in control of cell proliferation, known cellular functions of TCTP include prevention of apoptosis via inducing degradation of p53, a major tumour suppressor protein (Bommer and Thiele, 2004; Rho et al., 2011). TCTP has also been implicated in regeneration, however, all known reports are restricted to mammals (Jiménez et al., 2005; Pollins et al., 2007; Zhu et al., 2008). Since mammals, and human in particular, have very limited regenerative potential, we therefore, asked whether the role of TCTP in post-traumatic recovery is a more general phenomenon and whether it would be also involved in more robust regeneration responses, such as those that are seen in echinoderms.
Bone morphogenic protein1 (BMP1)/Tolloid (TLD)-like proteins constitute a subgroup of multidomain secreted zinc endopeptidases within the astacin (M12A) family. In mammals, there are four BMP1-like proteinases, including BMP1, Tolloid (TLD), Tolloid-like 1 (TLL1), and Tolloid-like 2 (TLL2). These peptidases play multiple roles in developmental morphogenetic processes. First, they are essential for the proper formation of the extracellular matrix via cleavage of the precursor molecules followed by release of the mature connective tissue proteins (Ge and Greenspan, 2006; Hopkins et al., 2007). The second function of BMP1-like metalloproteinases is activation of TGFβ-like growth factors by liberating them from latent complexes with their inhibitors (Ge and Greenspan, 2006; Hopkins et al., 2007). Among these morphogenetic factors are TGFβ-like BMPs, which are involved in a vast diversity of developmental processes (Bragdon et al., 2011), myostatin and CDF11, negative regulators of muscle growth and neurogenesis, respectively, and TGFβ-1, which controls cell differentiation, growth, and apoptosis (Ge and Greenspan, 2006). Significant up-regulation of Bmp1 transcription level was reported in human osteosarcoma cells, and can therefore contribute to cancer progression (Lee et al., 1997).
We show here that Wnt9, TCTP, and Bmp1/Tll transcripts are extensively expressed in sea cucumber gut regeneration and hypothesize that the products of these genes contribute to the enormous injury-triggered plasticity of adult tissues seen in echinoderms.
2. Results
2.1. Sequence analysis
Unlike vertebrates, who have two paralogs Wnt9a and Wnt9b, basal deuterostomes, including echinoderms, are characterized by the presence of a single Wnt9 in their genomes (Supplementary Fig. 1) (Croce and McClay, 2008; Croce et al., 2006). Wnt9 gene of H. glaberrima, encodes a 368 amino acid protein, which shares typical characteristics with its vertebrate orthologs (Cox et al., 2010; Fokina and Frolova, 2006; Katoh and Katoh, 2005; Qian et al., 2003) (Supplementary Fig. 1, 2). As predicted by SignalP3.0 (http://www.cbs.dtu.dk/services/SignalP), it has a 32 amino acid-long N-terminal secretory signal peptide. In eukaryotes, secretory signal peptides allow a protein to be translocated across the endoplasmic reticulum membrane. Once in the endoplasmic reticulum, the proteins are usually targeted for secretion to the outside of the cells, unless they have specific retention signals (Emanuelsson et al., 2007).
The predicted protein sequence of H. glaberrima TCTP is 175 amino acids long. It shows a high degree of conservation in the N-terminal 40 amino-acid long region (16 of 40 amino acids in this region are identical between the sea cucumber and human sequences), which is known to be necessary for the anti-apoptotic function of TCTP. Other conserved features include the TCTP1 signature and the C-terminal self-interaction domain (Yang et al., 2005) (Supplementary Fig. 3).
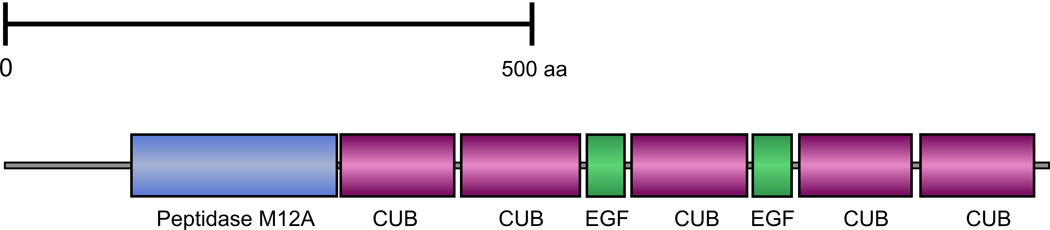
The sequence of the BMP1/TLL protein of H. glaberrima has not been reported before. We identified the homolog of Bmp1 among 5173 ESTs representing three cDNA libraries from the normal and regenerating digestive tube (Rojas-Cartagena et al., 2007) by BLAST search against the non-redundant NCBI protein database. The full length of the coding region was reconstructed by performing 5' and 3' RACE. Phylogenetic analysis of the sea cucumber BMP1/TLL shows that the sea cucumber protein clusters together with BMP1-like sequences of amphioxus and sea urchin, and that these three proteins form an outgroup relative to the BMP1, TLL1 and TLL2 of vertebrates (Supplementary Fig. 4). Therefore, we prefer to keep the name BMP1/TLL for the sea cucumber sequence. The deduced sea cucumber BMP1/TLD-like protein is 983 amino acids long. It shows exactly the same domain organization as full-length BMP1 of vertebrates (Mac Sweeney et al., 2008) and contains a highly conserved N-terminal metalloprotease domain, two calcium-binding EGF-like domains separated by a CUB domain and flanked on either side by pairs of CUB domains (Fig. 1). The protease domain contains all the residues known to be essential for catalytic activity (Angerer et al., 2006; Mac Sweeney et al., 2008): the three histidine residues that bind the zinc ion in the active center of the enzyme, as well as the conservative glutamic acid and tyrosine (Supplementary Fig. 5). The CUB domains are thought to be involved into protein-protein interactions, whereas the ability of the EGF-like domains to bind calcium ions has been hypothesized to affect the configuration of the BMP1/TLD-like proteinase (Hopkins et al., 2007).
Figure 1.
Domain organization of the predicted BMP1/TLL protein of the sea cucumber H. glaberrima.
2.2. Introduction to the model
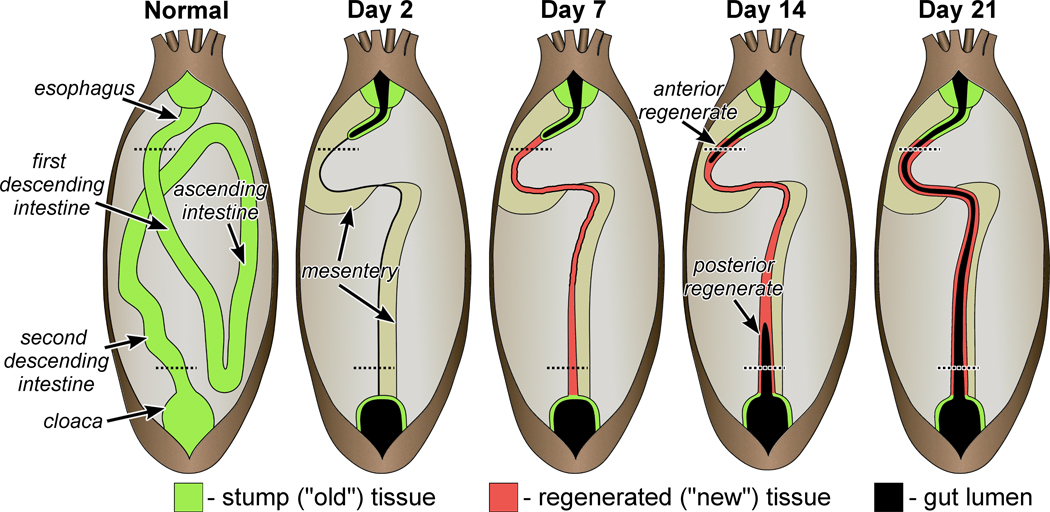
It is necessary here to give the reader a brief background on visceral regeneration in sea cucumbers. Organization of the normal and regenerating digestive tube of the sea cucumber H. glaberrima is schematically shown on Fig. 2. In this species, evisceration involves detachment of the entire intestine from the supporting mesentery, as well as from the esophagus and cloaca. The autotomized viscera are discarded through the anus. The early response to injury involves wound healing at the posterior end of the esophageal stump and at the anterior end of the cloaca. Regeneration per se begins with the formation of a solid rudiment in the free edge of the mesentery, which later serves to guide and enclose the developing lumen that forms as two outgrowth from both the anterior and posterior stumps. For the detailed description of the anatomy of the normal and regenerating digestive tube and for a review of the cellular mechanisms involved, the reader is referred to Hyman (1955), Garcia-Arraras and Greenberg (2001), and Mashanov and Garcia-Arraras (2011).
Figure 2.
Anatomy of the regenerating digestive tube in non-eviscerated individuals of the sea cucumber H. glaberrima and at different time points of regeneration. The dashed lines correspond to the position of the cross sections shown on Figures 3 and 4, for the anterior and posterior regenerates, respectively
2.3. Expression patterns in the normal gut
Wnt9
The Wnt9 transcripts are mostly undetectable by in situ hybridization in the tissues of the non-regenerating digestive tube (Fig. 5A).
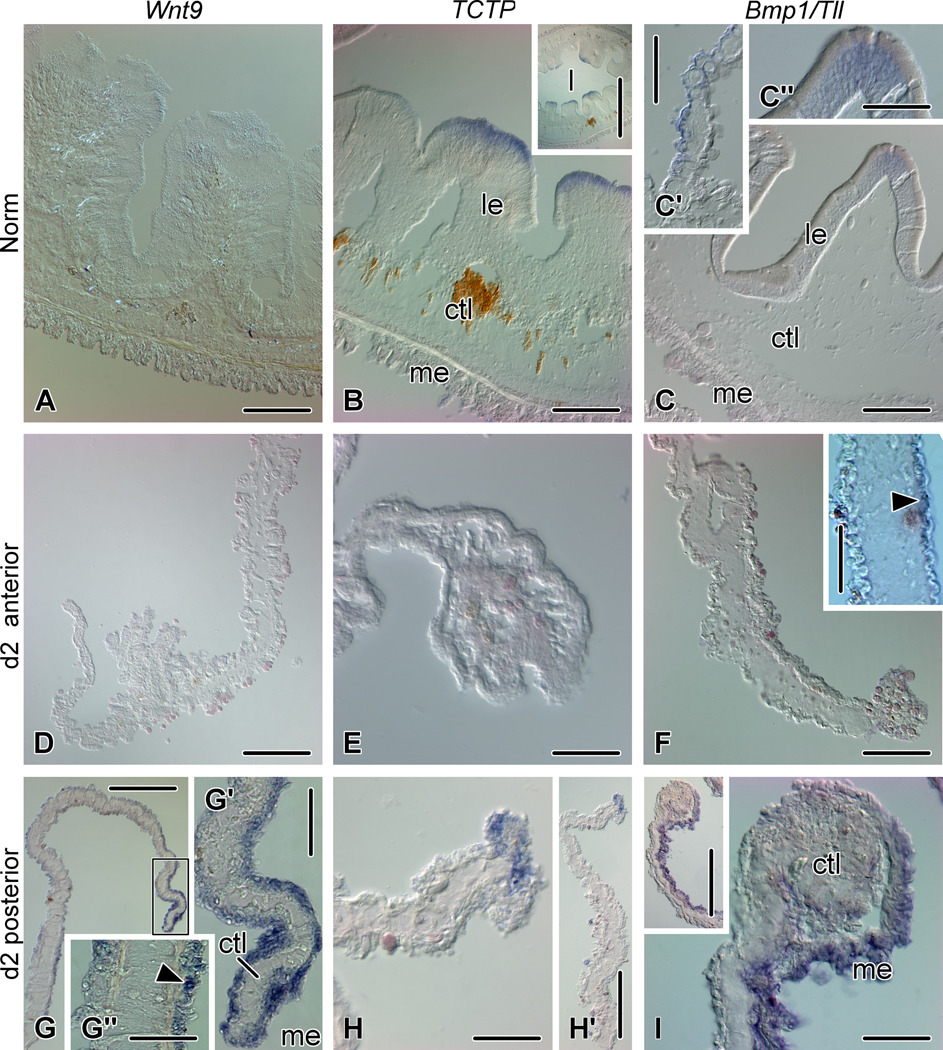
Figure 5.
Expression of Wnt9, TCTP, and Bmp1/Tll in the non-eviscerated gut (A - C'') and at early stages of regeneration (day 2) (D – I). (A) No Wnt9 in situ hybridization signal is seen in the tissues of the normal gut (esophagus). (B) TCTP expression in the apices of luminal epithelial folds in the esophagus of non-eviscerated animals. The inset shows a low-magnification view of the esophagus. (C) Bmp1/Tll expression in the normal gut (second descending intestine). (C') shows expression in the mesothelium of the gut mesentery and (C'') is a higher magnification of the apical part of a fold of the luminal epithelium; note moderate Bmp1/Tll expression in the perinuclear cytoplasm of the epithelial cells. (D) and (E) Absence of Wnt9 and TCTP in situ hybridization signal in the anterior regenerate on day 2. (F) Bmp1/Tll transcripts are mostly absent from the early anterior regenerate on day 2 after evisceration, however, weak asymmetrical expression (arrowhead) is seen in the mesothelium of the mesentery (inset). (G) Wnt9 expression in the posterior regenerate on day 2. (G') High magnification view of the free margin of the mesentery (boxed area on G). (G'') Asymmetrical Wnt9 expression in the proximal mesentery. Arrowhead shows a positively stained cell in the apical region of the mesothelium. (H) Expression of TCTP in mesothelial cells at the free edge of the posterior regenerate on day 2. (H') Lower magnification view of the posterior regenerate on day2. (I) Bmp1/Tll expression in the posterior regenerate on day 2. The inset shows a low magnification view of the regenerate. ctl – connective tissue layer; l – gut lumen; le – luminal epithelium; me – mesothelium. Scale bars: A – C', F - 100 µm; D, G, H', I inset - 200 µm; C'', F inset, G', G'', E, H, I - 50 µm; B inset - 500 µm.
TCTP
Many (although not all) of the folds of the luminal epithelium show moderate in situ hybridization signal for TCTP. The signal is restricted to apices and is not detected in the crypts (Fig. 5B).
Bmp1/Tll
Very weak expression is occasionally seen in the mesothelium of the gut mesentery and also in the luminal epithelium of the second descending intestine. Similarly to the TCTP expression, Bmp1/Tll transcripts in the luminal epithelium are restricted to the apices of the folds (Fig. 5C – C").
2.4. Expression patterns: day 2 after evisceration
Wnt9
Wnt9 transcripts are completely absent from the anterior regenerate (Fig. 3A, 5D). In the posterior regenerate, moderate to strong hybridization signal is widely distributed throughout the mesothelium of the free distant edge of the mesentery (Fig. 4A, 5G, G'). Wnt9 expression is seen in this region of the posterior regenerate even before the mesenterial thickening starts to develop. Moderate to intense hybridization signal is also often observed in more proximal regions of the mesentery (i.e. in the regions, which are positioned further from the free distal edge and closer to the body wall) (Fig. 4A, 5G''). This expression is often asymmetrical with stronger reaction on one side of the mesentery than on the other. In these proximal regions of the mesentery, the normal organization of the mesothelium is still evident, and the hybridization signal is clearly restricted to the apical half of the epithelial sheet, which is known to be occupied by cell bodies of peritoneal cells (Fig. 5G''). A similar asymmetrical expression pattern is seen in the mesentery attached to the cloacal stump (Supplementary Fig. 6A).
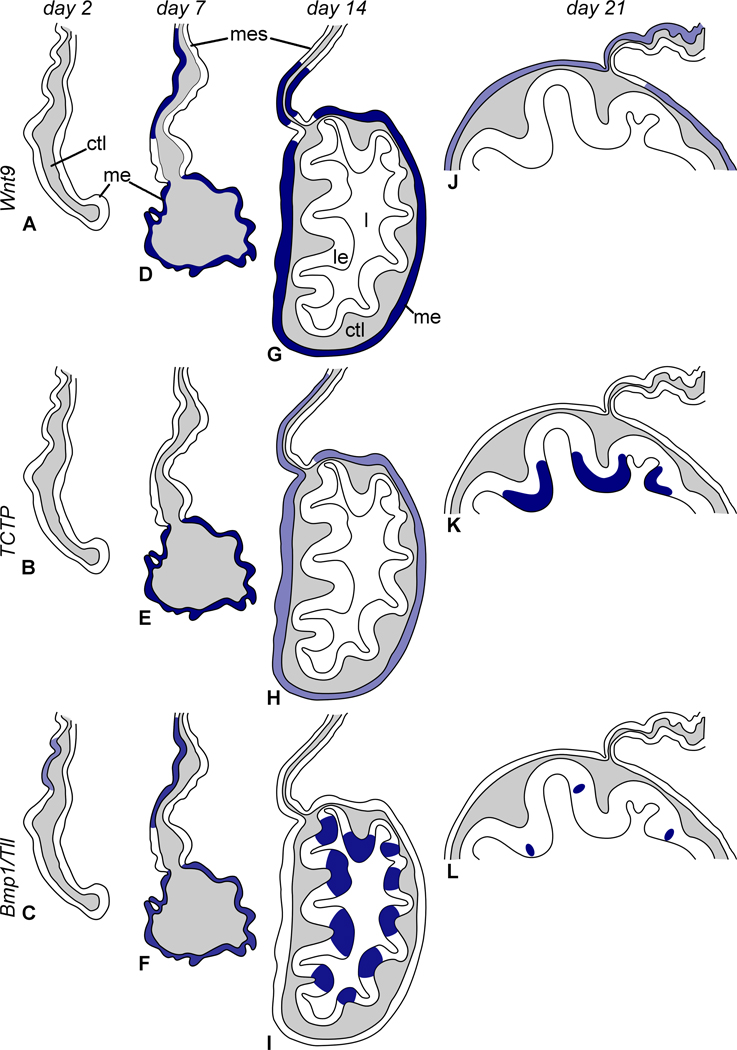
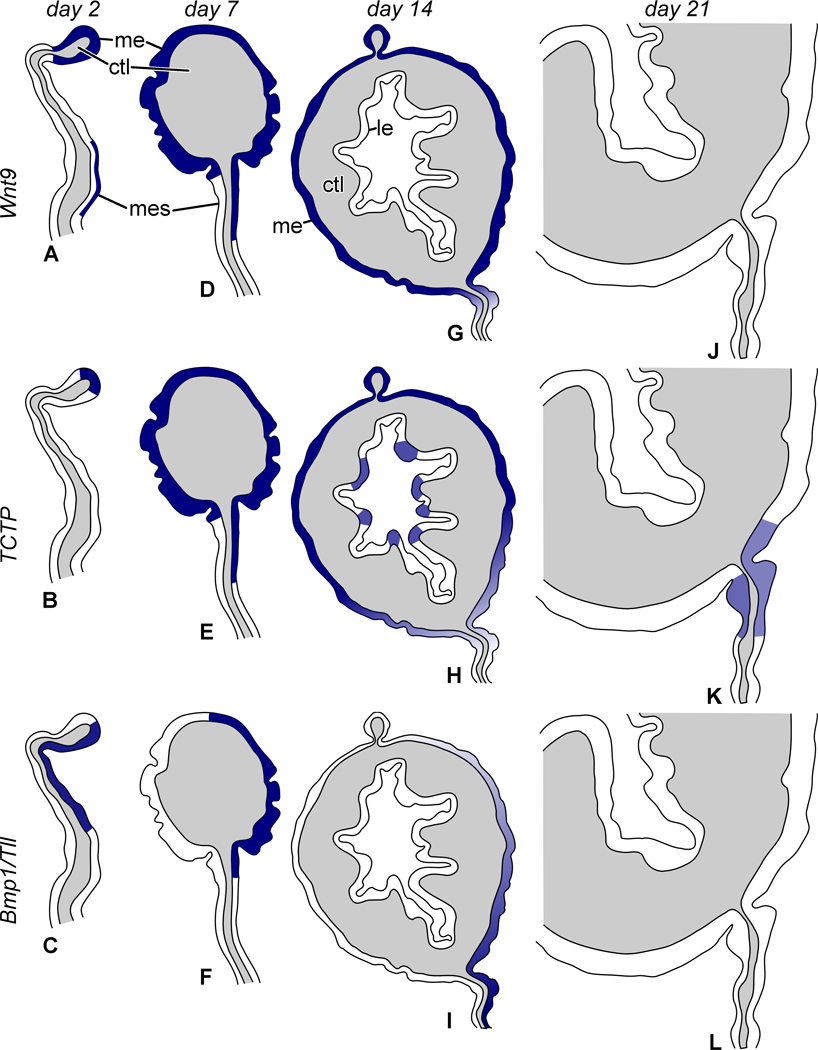
Figure 3.
Diagram summarizing organization of the anterior gut rudiment and expression patterns of Wnt9, TCTP, and Bmp1/Tll. (A, D, G, J) Distribution of Wnt9 transcripts on day 2, 7, 14, and 21, respectively. (B, E, H, K) Expression pattern of TCTP on day 2, 7, 14 and 21, respectively. (C, F, I, L) Expression pattern of Bmp1/Tll on day 2, 7, 14 and 21, respectively. ctl – connective tissue layer; l – gut lumen; le – luminal epithelium; me – mesothelium; mes – mesentery. Not to scale.
Figure 4.
Diagram of organization of the posterior gut rudiment showing expression patterns of Wnt9, TCTP, and Bmp1/Tll. (A, D, G, J) Expression of Wnt9 on day 2, 7, 14, and 21, respectively. (B, E, H, K) Expression pattern of TCTP on day 2, 7, 14, and 21, respectively. (C, F, I, L) Expression pattern of Bmp1/Tll on day 2, 7, 14, and 21, respectively. ctl – connective tissue layer; le – luminal epithelium; me – mesothelium; mes – mesentery. Not to scale.
TCTP
No in situ hybridization signal is seen in the early anterior regenerate (Fig. 3B, 5E). However, as in the normal digestive tube, moderate expression is present in the apices of the folds of the luminal epithelium in the esophageal stump (Supplementary Fig. 6B). In the posterior rudiment, there is moderate to strong expression of TCTP, which is restricted to mesothelial cells at the free edge of the mesentery (Fig. 4B, 5H, H').
Bmp1/Tll
On day 2, Bmp1/Tll transcripts are mostly absent from the anterior regenerate with the exception of weak asymmetric expression in the mesothelium of the middle region of the mesentery (located approximately half-way between the body wall and the free margin) (Fig. 3C, 5F). In the posterior rudiment, Bmp1/Tll shows moderate to intense asymmetrical expression in the mesothelium of the distal region of mesentery where the connective tissue thickening starts to form (Fig. 4C, 5I).
2.5. Expression patterns: day 7 after evisceration
Wnt9
By day 7 after evisceration, both the anterior and posterior regenerates develop a conspicuous solid connective tissue thickening in the free edge of the mesentery (Fig. 3D, 4D, 6A, B). At this stage, Wnt9 transcripts are present in both rudiments and show similar distribution patterns. Intense in situ hybridization signal is seen throughout the mesothelium that covers the rudiment, as well as in the mesothelium of the mesentery, where expression is often asymmetrical (Fig. 3D, 4D, 6A, B). Wnt9 transcripts are also expressed in the stump ('old') tissues, i.e., in the mesothelium of the cloaca (as in non-eviscerated animals) (Supplementary Fig. 6C), and both in the mesothelium and luminal epithelium of the esophagus (Supplementary Fig. 6D, D').
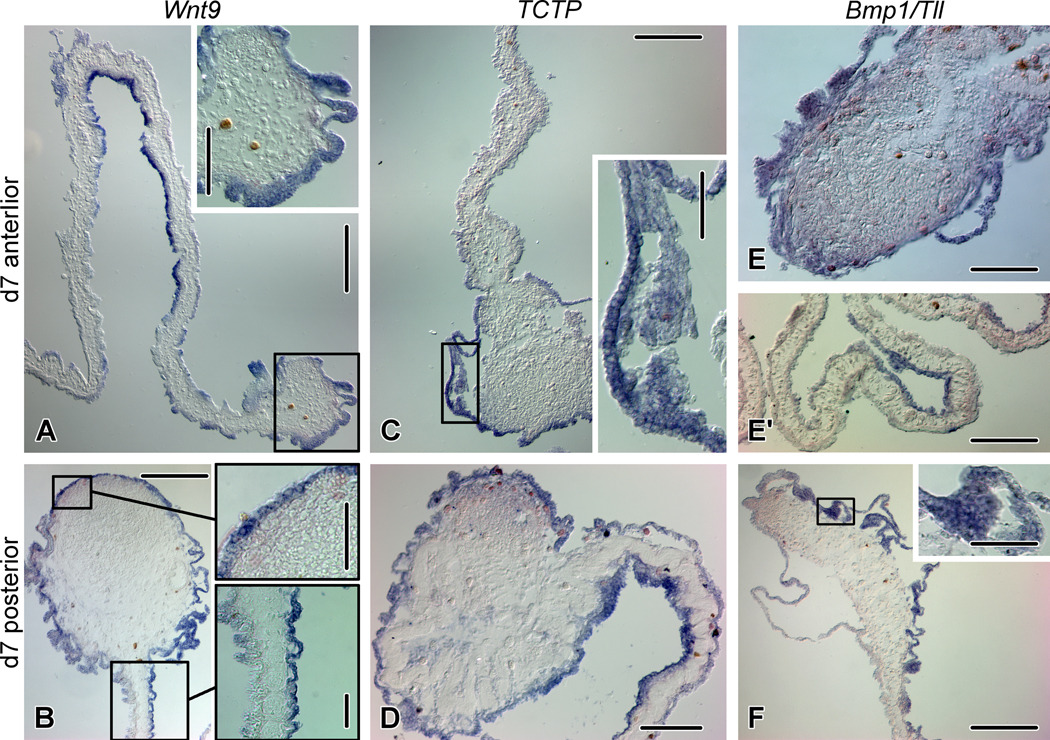
Figure 6.
Expression of Wnt9, TCTP, and Bmp1/Tll on day 7 of gut regeneration. (A) Wnt9 expression in the anterior rudiment. The inset shows a high magnification view of the rudiment (boxed area on the main image). (B) Wnt9 expression in the posterior rudiment. The insets correspond to the boxed areas on the main image and show the mesothelium of the rudiment and asymmetrical Wnt9 expression in the mesothelium of the mesentery. (C) TCTP expression in the anterior gut regenerate. The inset shows the mesothelium of the rudiment (boxed are on the main image). (D) TCTP expression in the posterior gut regenerate. (E) Moderate Bmp1/Tll expression in the anterior regenerate. (E') Asymmetrical expression of Bmp1/Tll in the anterior mesentery. (F) Asymmetrical expression of Bmp1/Tll in the mesothelium of the posterior regenerate. The inset shows a high magnification view of the mesothelium (boxed are on the main image). Scale bars: A, B, C, F – 200 µm; A inset, D, E – 100 µm; B inset, C inset, E', F inset – 50 µm.
TCTP
On day 7, TCTP is strongly expressed all over the mesothelium in both the anterior and posterior regenerates (Fig. 3E, 4E, 6C, D). In the posterior rudiment, moderate to strong asymmetric expression of TCTP transcripts is also seen in the mesothelium of the mesentery, which supports the regenerate (Fig. 4E, 6D). As in the non-eviscerated animals and at earlier stages of regeneration, TCTP expression on day 7 is also seen in the esophageal stump in the apical region of the luminal epithelium (Supplementary Fig. 6E).
Bmp1/Tll
Bmp1/Tll shows moderate expression throughout the mesothelium of the anterior gut regenerate (Fig. 3F, 6E). Asymmetrical expression at moderate to high level is also seen in the anterior supporting mesentery (Fig. 3F, 6E'). In the posterior regenerate, the transcripts are presentat high levels only in the mesothelium on one side of the rudiment, while the opposite side shows weak or no expression (Fig. 4F, 6F).
2.6. Expression patterns: day 14 after evisceration
Wnt 9
By day 12 – 14 after evisceration, the luminal epithelium of the stumps starts to invade the connective tissue thickening of both the anterior and posterior regenerates (Fig. 2), which, therefore, develop the typical trilaminar organization of their wall composed of the mesothelium, the connective tissue layer, and the luminal (digestive) epithelium (Fig. 3G, 4G, 7A, B). In both the anterior and posterior regenerates, Wnt9 is extensively expressed in the mesothelium, but not in the luminal epithelium of the newly developed gut segments. Moderate expression is also seen in mesothelium of the distal mesentery (Fig. 3G, 4G, 7A, B). Wnt9 transcripts almost completely disappear from the 'old' stump tissue. However, positively stained cells are occasionally seen in the luminal epithelium of the cloaca (Supplementary Fig. 6F).
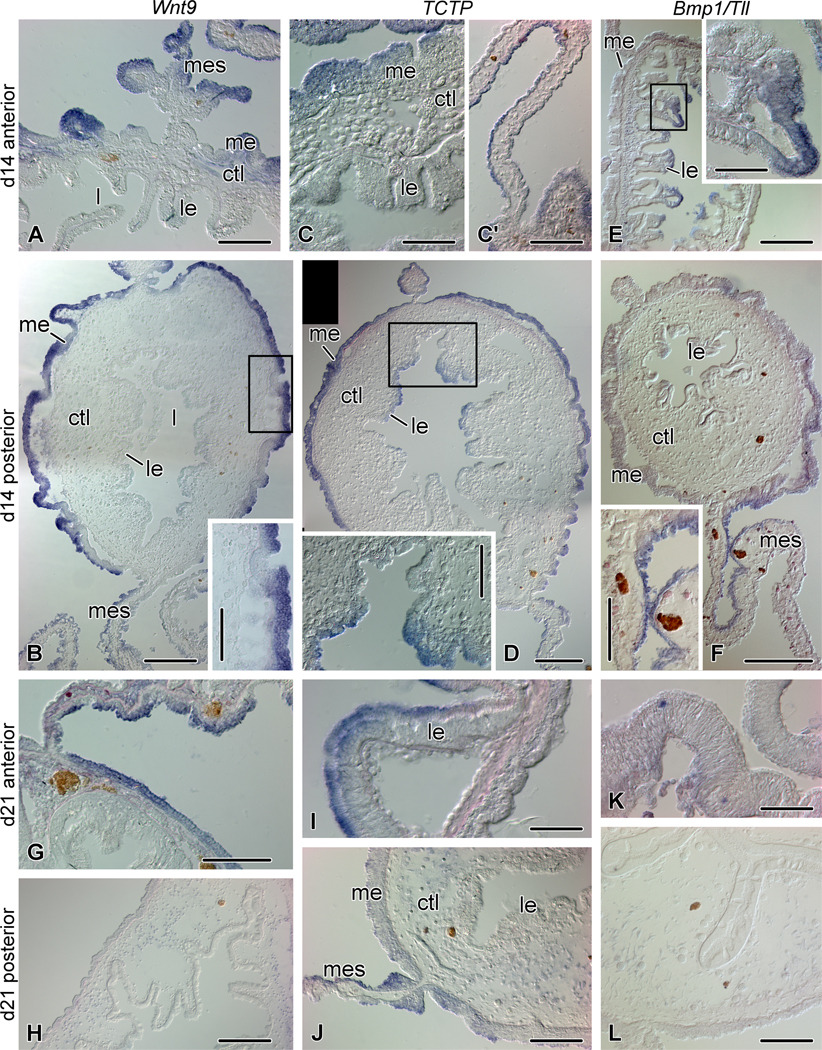
Figure 7.
Expression of Wnt9, TCTP, and Bmp1/Tll at advanced stages of regeneration: day 14 (A – F) and day 21 (G - L) after evisceration. (A) Wnt9 expression in the anterior rudiment on day 14. (B) Wnt9 expression in the posterior rudiment on day 14. The inset shows the mesothelium of the rudiment (boxed area on the main image). (C) Moderate TCTP expression in the anterior rudiment on day 14 after evisceration. (C') Asymmetrical TCTP expression in the supporting anterior mesentery on day 14. (D) Expression of TCTP in the posterior regenerate on day 14. Note medium expression in the cells at the apices of the folds of the luminal epithelium (inset, corresponding to the boxed region on the main image). (E) Bmp1/Tll expression in the anterior regenerate on day 14. The inset shows a fold of the luminal epithelium and corresponds to the boxed region on the main image. (F) Expression of Bmp1/Tll in the posterior gut regenerate on day 14. Note weak expression in the mesothelium of the regenerate and stronger asymmetrical hybridization signal in the supporting mesentery (inset). (G) Wnt9 expression in the mesothelium of the anterior part of the newly formed digestive tube on day 21. Note also asymmetrical expression in the supporting mesentery. (H) No Wnt9 expression is seen in the posterior part of the gut regenerate on day 21. (I) Expression of TCTP in the apices of the folds of the luminal epithelium in the anterior part of the regenerate on day 21. (J) Moderate expression of TCTP in the distal region of the mesentery at its attachment to the posterior part of the gut regenerate on day 21. (K) Bmp1/Tll transcripts detected in scattered cells in the luminal epithelium of the anterior part of the regenerate on day 21. (L) No Bmp1/Tll expression is seen in the posterior part of the newly formed digestive tube on day 21. ctl – connective tissue layer; l – gut lumen; le – luminal epithelium; me – mesothelium; mes - mesentery. Scale bars: A, B inset, C', D inset, F inset, G, J, L – 100 µm; B, D, E, F, H – 200 µm; C, E inset, I, K - 50 µm.
TCTP
In the anterior regenerate, weak to moderate TCTP expression is seen in the mesothelium (Fig. 3H, 7C). The distal part of the supporting mesentery also shows asymmetric expression of TCTP transcripts (Fig. 3H, 7C'). In the posterior regenerate, TCTP is widely expressed throughout the mesothelium of the rudiment at medium to high levels, but is almost completely absent from the supporting mesentery. The apices of the luminal epithelial folds also show medium in situ hybridization signal (Fig. 4H, 7D).
Bmp1/Tll
In the anterior regenerate, Bmp1/Tll transcripts disappear from the mesothelium of the rudiment, however, moderate to strong expression is seen in the cells at the apices of the folds of the digestive epithelium, which lines the newly created lumen of the gut regenerate (Fig. 3I, 7E). Bmp1/Tll expression in the posterior regenerate is highly asymmetrical and is mostly restricted to the distal region of the supporting mesentery and to the mesothelium of the adjacent regions of the rudiment proper. Expression in the rudiment is weaker than at the previous stage, whereas the distal region of the supporting mesentery (at its attachment to the gut regenerate) shows strong in situ hybridization signal (Fig. 4I, 7F).
2.7. Expression patterns: day 21 after evisceration
Wnt9
By day 21, the anterior and posterior regenerates meet and fuse together to form a continuous digestive tube (Fig. 2). At this stage moderate expression of Wnt9 is still seen in the mesothelium of the anterior segment of the newly formed intestine (Fig. 3J, 7G), but not in the posterior part of the regenerate (Fig. 4J, 7H). Asymmetrical expression is also seen in the distal region of the anterior supporting mesentery at its attachment to the intestine (Fig. 3J, 7G).
TCTP
On day 21 after evisceration, the expression pattern of TCTP in the newly regenerated gut is similar to what is seen in non-eviscerated animals. In the anterior region of the regenerate, the transcripts are restricted to the apices of the luminal epithelial folds (Fig. 3K, 7I). In the posterior part of the regenerate, TCTP transcripts are mostly absent, with an exception of weak expression in the restricted area of the distal mesentery at its attachment to the gut (Fig.4K, 7J).
Bmp1/Tll
Bmp1/Tll transcripts largely disappear from the tissues of newly regenerated tissues of the digestive tube by day 21 with the only exception of single scattered cells in the luminal epithelium of the anterior regions of the gut (Fig. 3L, 4L, 7K, L).
2.8. Reverse transcription polymerase chain reaction (RT-PCR)
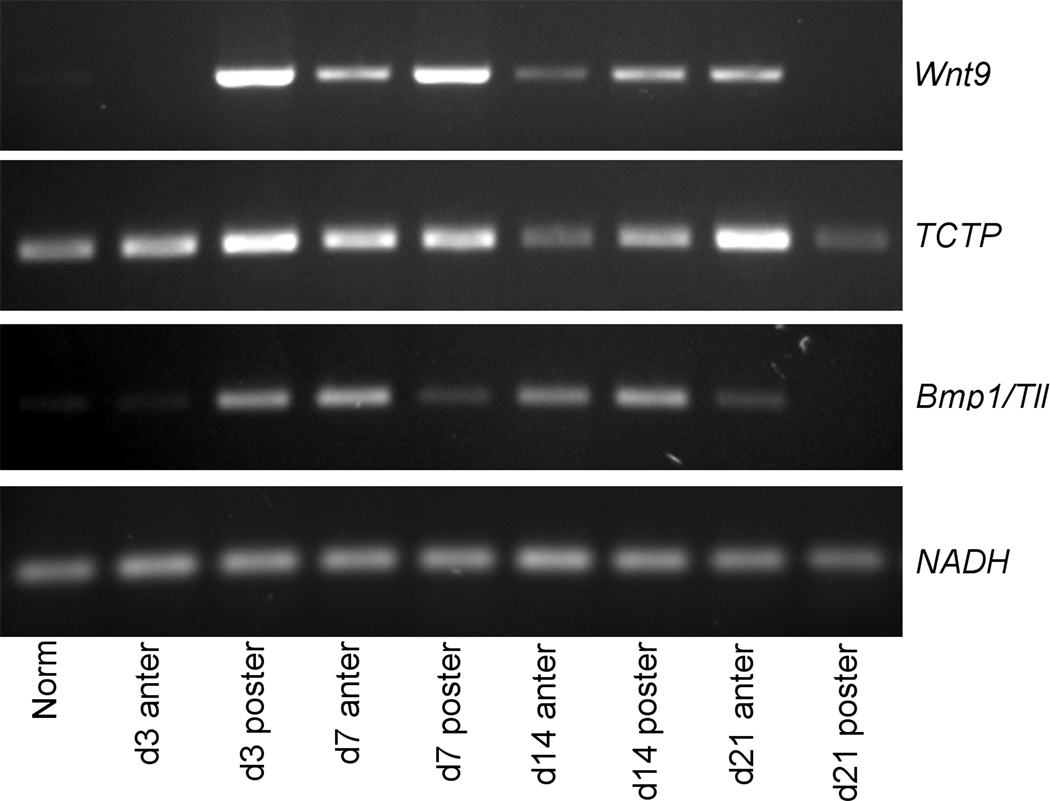
We validated our in situ results by performing end-point RT-PCR (Fig. 8). NADH dehydrogenaze subunit 5 was used as an internal standard to control for variations in RNA concentration. Obtaining precise quantification of gene expression was outside the scope of the present paper. Nevertheless, the absence or presence of the particular PCR product unambiguously correlated to the absence or presence of the in situ hybridization signal for the corresponding transcript in the normal and regenerating animals. Moreover, differences in the intensity of agarose gel bands representing a particular transcript in the normal and regenerating animals corresponded well with the spread and/or strength of the hybridization signal (Fig. 8). The specificity of our in situ hybridization probes is further confirmed by the distinct expression patterns and by the fact that hybridization with control probes produced no detectable staining.
Figure 8.
RT-PCR showing temporal expression of Wnt9, TCTP, and Bmp1/Tll at different time points of gut regeneration and in non-eviscerated animals. NADH dehydrogenase subunit 5 was used as an internal control. Total RNA for RT-PCR was extracted from pieces of the normal digestive tube or from the anterior and posterior regenerates plus 2–3 mm of the adjacent 'old' stump tissues.
3. Discussion
The present study showed up-regulation of Wnt9, TCTP, and Bmp1/Tll in visceral regeneration in the sea cucumber H. glaberrima. Involvement of these three genes in post-traumatic recovery is of particular interest, since they have been previously implicated in normal development and in cancer progression. On the other hand, their role in regeneration has not been studied extensively.
To our knowledge, the present study provides the first account of involvement of a member of Wnt9 subfamily in post-traumatic regeneration and also the first attempt to map the expression domains of Wnt9 in a species outside the vertebrate lineage. As pointed out above, the single Wnt9 found in modern basal deuterosomes is representative of an ancestral state that existed before the duplication in vertebrates (Croce and McClay, 2008). It has been shown that the resulting two paralogs, Wnt9a and Wnt9b, although being highly similar in their amino-acid sequence, show little overlap of their expression domains, and, therefore, control developmental processes in different regions of the body (Cox et al., 2010). The initial function of the ancestral form, before the duplication, is not known. In the digestive tube of non-autotomized individuals of the sea cucumber H. glaberrima, Wnt9 is quiescent, but its expression is triggered shortly after evisceration and is specifically restricted to the mesothelium of the regenerating digestive tube and to the mesothelium of the supporting mesentery. Unfortunately, the available data do not allow us to arrive at any definitive conclusion on the role of Wnt9 in sea cucumber visceral regeneration. The main reason for this is the complexity of Wnt signalling network. Depending on the cellular context, Wnt signalling can play different, and sometimes opposite, roles. Besides the Wnt/beta-catenin pathway there at least two other, 'non-canonical' pathways. In certain cases, activation of different pathways in the same organ leads to different consequences, and, moreover, different Wnt signalling pathways can antagonize one another (Croce and McClay, 2008; Topol et al., 2003; Weidinger and Moon, 2003). The regenerating sea cucumber H. glaberrima can express homologs of all the key protein components of the canonical and non-canonical Wnt signalling pathways (Mashanov et al., in preparation). Which of those pathways are activated during visceral regeneration and the functional consequences of this activation has yet to be studied.
In spite of the lack of direct experimental evidence, some interesting speculations on the role of Wnt9 in gut regeneration can still be drawn by a synthesis of the known information on the cellular processes in Wnt9-expressing regenerating tissues of the sea cucumber and the data on the role of Wnt9 orthologs in other organisms. The strongest expression of Wnt9 is observed on days 7 – 14 in the anterior gut regenerate and on days 2 – 14 in the posterior regenerate. At these time points the mesothelium (the outer epithelial layer) of the regenerating gut undergoes drastic transformations. Under normal conditions (in non-eviscerated animals), the gut mesothelium shows complex organization being composed of a supporting framework of peritoneal cells, a basal musculature formed by myoepithelial cells, and an associated nervous plexus (Feral and Massin, 1982; Mashanov et al., 2004). Visceral autotomy involves detachment of the digestive tube from its supporting mesentery. It is the breakage zone along the free distal edge of the remaining mesentery that initiates the development of the early gut regenerate. The mesothelium of this region (and to a lesser degree of the rest of the mesentery) undergoes drastic de-differentiation and simplification in organization (Garcia-Arraras et al., unpublished; Mashanov and Garcia-Arraras, 2011; Mashanov et al., 2005), and this is exactly the region where Wnt9 starts to be expressed. The net result of this transformation is a simplified epithelium composed of irregularly shaped actively proliferating cells. The underlying basal lamina is either breached or completely degraded. Some of these cells detach from the epithelium and invade the underlying connective tissue of the developing gut rudiment as free-moving mesenchymal cells (Garcia-Arraras et al., unpublished).
Epithelial-mesenchymal transition and the reverse process, mesenchymal-epithelial transition, are critical for the normal embryogenesis, but are also implicated in cancer and wound healing. Wnt signalling pathways are known to be involved in the control of these changes in cell phenotype (Iwai et al., 2010; Micalizzi et al., 2010). In the mouse embryo, Wnt9b transcripts were detected in organs or tissues (lung, kidney) that undergo signalling interaction between epithelial and mesenchymal tissues and/or transformation between these tissue types (Fokina and Frolova, 2006; Karner et al., 2011; Qian et al., 2003). Therefore, one of the possible roles of Wnt9 in sea cucumber regeneration can be the control of transitions between the dedifferentiated mesothelial cells and the mesenchyma.
In the non-eviscerated animals, the hybridization signal for TCTP is restricted to the apices of the luminal epithelial folds of the anterior regions of the digestive tube. The functional role of this expression remains unknown. In the regenerating gut, TCTP is mostly absent from the digestive epithelium, with the exception of the discontinuous mosaic pattern in the luminal epithelium of the growing posterior rudiment. However, a very prominent expression is seen in the regenerating mesothelium. This expression pattern is very similar to distribution of survivin and mortalin transcripts described previously (Mashanov et al., 2010). This similarity may reflect synergetic action of these genes, since all three of them are known to be involved in apoptosis suppression and regulation of cell division (Altieri, 2008; Bommer and Thiele, 2004; Conte et al., 2009; Wadhwa et al., 2002). Moreover, the peak of expression of survivin, mortalin, and TCTP in the regenerating gut mesothelium coincides with deep transient dedifferentiation of this tissue. The dedifferentiated cells acquire an ability to undergo rapid cell divisions and eventually give rise to all differentiated cell types of the mesothelium, and, in some species, even contribute to regeneration of the luminal epithelium (Garcia-Arraras et al., 1998; Garcia-Arraras et al., unpublished; Mashanov et al., 2005). An interesting correlation here is that all three genes, besides their other functions, are known be involved in maintenance and regulation of stem cells. Mortalin, for instance, is constitutively expressed in planarian neoblasts and is necessary both for normal cell turnover and for regeneration (Conte et al., 2009). Survivin is involved in maintenace of stem cell and protects them from apoptosis (Li et al., 2010; Marconi et al., 2007). TCTP is implicated in regulation of pluripotency via control of transcription of oct4 and nanog (Koziol et al., 2007). Therefore, as was hypothesized earlier (Mashanov et al., 2010), it is plausible that the plasticity of the holothurian mesothelium, with its ability to undergo transient dedifferentiation, can be due to temporary activation of stem cell regulatory-related pathways.
Bmp1/Tll is markedly up-regulated on days 3 through 12 after evisceration. Of particular interest is the strong expression in the apices of the developing folds of the luminal epithelial in the anterior rudiment, and also the unilateral asymmetric expression in the mesothelium of the posterior regenerate. It is important to note here that Bmp-1/Tolloid was reported to be up-regulated in the anterior, but not in the posterior part of the digestive tube during amphibian intestinal remodelling in metamorphosis and this expression correlated with development of the adult intestinal epithelial structures (Shimizu et al., 2002). Left-right asymmetry in gene expression is known to be involved in gut morphogenesis in vertebrate embryos (Danesh et al., 2009; Kurpios et al., 2008). This expression induces asymmetrical changes in cell adhesion and extracellular matrix composition, eventually leading to gut tilting and looping (Kurpios et al., 2008). We therefore hypothesize that in the regenerating digestive tube of H. glaberrima, Bmp1/Tll may be involved in morphogenetic movements leading to folding of the luminal epithelium and gut looping. BMP1-like proteinases are known to influence the key morphogenetic processes such as cell migration, differentiation, cell division, and cell death either via control of the composition and mechanical properties of the extracellular matrix or via activation of TGFβ-like growth factors (Ge and Greenspan, 2006; Hopkins et al., 2007). The exact mechanism of Bmp1/Tll action in sea cucumber gut regeneration still remains to be studied. However, possible involvement of Bmp1/Tll in the control over extracellular matrix properties seems quite plausible, since the intestinal regenerate in holothurians is known to undergo large-scale remodelling of its connective tissue (Quiñones et al., 2002).
Spatio-temporal expression pattern of five genes has been studied in detail so far in sea cucumber gut regeneration. Besides the three genes, described in the present study, involvement of two other genes, survivin and mortalin, has been investigated earlier (Mashanov et al., 2010). Although all those genes have certain peculiarities in their expression patterns, a general pattern still can be seen. Some of these genes, such as Bmp1/Tll, TCTP, and survivin are expressed in the luminal epithelium of the growing gut rudiments, but all the five genes show consistent upregulation in the mesothelium of the regenerating intestine starting on days 2 – 7 and persisting until at least day 14. As mentioned above, it is during this time period that the gut mesothelium undergoes drastic dedifferentiation, which leads to transient simplification in tissue architecture, increase in cell motility, and, most importantly, to a burst in cell proliferation and broad developmental plasticity (Mashanov and Garcia-Arraras, 2011). Therefore, genes from multiple developmental/cancer-related pathways, seem to contribute to the reversible transition of the gut mesothelium from a complex specialized tissue to a pool of rapidly proliferating multipotent cells.
4. Experimental procedures
4.1. Animal collection, maintenance, and evisceration
Adult individuals (body length 8.5 – 11 cm) of the sea cucumber Holothura glaberrima Selenka, 1867 (Echinodermata: Holothuroidea) were collected from the intertidal zone of the Atlantic coast of Puerto Rico. Evisceration was induced by injecting a few milliliters of 0.35 M KCl into the coelomic cavity. Regenerating animals were kept in aerated indoor seawater tanks.
4.2. Sequence analysis
The sequences of H. glaberrima Wnt9 and TCTP were retrieved from the NCBI database (GenBank ID: GQ243223.1 and GU191016.1, respectively). The homolog of Bmp1 was identified among 5173 EST sequences of three cDNA libraries representing the digestive tube of non-eviscerated and regenerating individuals of H. glaberrima (Rojas-Cartagena et al., 2007) by BLAST search against the non-redundant protein database of the NCBI. Full-length cDNA sequence was obtained by performing 5' and 3' RACE using SMARTer RACE cDNA Amplification kit (Clontech). The resulting sequence of H. glaberrima Bmp1/Tll was deposited into the GenBank under accession number JN602083. Conserved domains were identified by searching against Pfam database (Finn et al., 2010). The N-terminal secretory signal peptide of Wnt9 was identified by the online program SignalP3.0 (http://www.cbs.dtu.dk/services/SignalP). The sea cucumber sequences were further analysed by performing multiple alignment of the predicted protein sequences with the corresponding orthologs from other animals using ClustalX, version 2.0.12 (Larkin et al., 2007). The alignments were analysed and annotated using Jalview 2.6.1 (Waterhouse et al., 2009). The phylogenetic analysis was performed using MEGA 5 software by neighbour-joining method (Tamura et al., 2011).
4.3. In situ hybridization
At least three animals were used to study the expression pattern of each gene per time point. In situ hybridization was largely performed according to Holland et al. (1996) with some modifications as described in Mashanov et al. (2010). Briefly, DIG-labeled probes for in situ hybridization were synthesized from PCR-generated DNA templates. The primers used for template synthesis are listed in the Supplementary Table 1. The non-eviscerated and regenerating individuals of H. glaberrima were anaesthetized in 0.2% chlorobutanol (1,1,1-trichloro-2-methyl-2-propanol hydrate) (Sigma) in seawater for 10–15 min at room temperature. The pieces of the normal digestive tube or the anterior and posterior regenerates were excised and fixed overnight at 4°C in a mixture of 4% paraformaldehyde and 0.1% glutaraldehyde in 0.01 M PBS (pH 7.4, 1030 mOsm). The tissue samples were subject to whole-mount in situ hybridization staining in 96-well cell culture plates. Prehybridization steps included treatment with proteinase K and acetylation. The riboprobes were diluted in hybridization buffer to a final concentration of about 400 ng/ml and the hybridization step was carried out at 60°C overnight. After stringency washes, the samples were equilibrated in Blocking Solution (Roche) and incubated in alkaline-phosphatase-conjugated anti-DIG antibodies (Roche) (1:2000) overnight at 4°C. After washing off the unbound antibodies, the color reaction was developed in NBT/BCIP solution in the dark. The samples were then postfixed in 4% paraformaldehyde and 0.1% glutaraldehyde in PBS, cryoprotected in buffered sucrose, and cryoembedded in in OCT medium (Takara). Serial cryosections were cut with a Leica CM1850 cryostat, mounted in buffered gelatine/glycerol and then analyzed and photographed with a Nikon Eclipse 600 microscope equipped with DIC optics and a SPOT RT3 digital camera (Diagnostic Instruments, Inc.).
All micrographs in the present paper represent transverse sections, which were cut orthogonal to the main axis of the organs. All figures are orientated with the ventral side of the animal to the bottom.
4.4. RT-PCR
In order to validate in situ hybridization results, we performed RT-PCR to verify the presence or absence of the transcripts in the normal and regenerating tissues. Total RNA was extracted from pieces of the normal digestive tube or from the anterior and posterior regenerates (which were dissected out together with 2 – 3 mm of the adjacent 'old' stump tissues) using TRI reagent (Sigma). First-strand cDNA was produced from 1 µg of the total RNA with random hexamer primers and ImPromt-II reverse transcriptase (Promega). Amplification was carried out with the same gene-specific primers that were used to construct the DIG-labelled riboprobes for in situ hybridization (Supplementary Table 1). NADH dehydrogenase subunit 5 was used as the internal control. PCR products were analyzed by agarose gel electrophoresis.
Highlights.
-
>
Wnt9, TCTP, and Bmp1/Tll are up-regulated in sea cucumber gut regeneration
-
>
All three genes are markedly up-regulated in the mesothelium of the regenerate
-
>
TCTP and Bmp1/Tll are expressed in the luminal epithelium of the regenerate
-
>
Changes in gene expression correlated with mesothelial dedifferentation
Supplementary Material
Phylogenetic position of H. glaberrima Wnt9 (indicated by an arrowhead) in relation to other members of Wnt9 subfamily (A). The sea cucumber Wnt9 is most similar to the sea urchin Wnt9 protein. The two vertebrate paralogs, Wnt9a and Wnt9b, form a well supported monophyletic clade with two clusters, to which the single Wnt9 of basal chordates is related as an outgroup. The tree was rooted using sequences of other invertebrate Wnt subfamiles (B). The phylogenetic analysis was based on nucleotide sequences of the coding regions of the genes which were aligned with ClustalW and then processed using the Neighbor-Joining method implemented in MEGA 5 software (Tamura et al., 2011). Support of the nodes was inferred by bootstraping method (2000 replicates), and the numbers represent the percentage of replicate trees in which the taxa clustered together in the bootsrap test. The following sequences were used for constructing the phylogenetic tree: Anolis carolinensis Wnt9a (RefSeq ID: XM_003229241.1); A. carolinensis Wnt9b (RefSeq ID: XM_003222531.1); Branchiostoma floridae Wnt1 (GenBank ID: AF061974); B. floridae Wnt4 (GenBank ID: AF061973.1); B. floridae Wnt14/Wnt9 (RefSeq ID: XM_002598581.1); Danio rerio Wnt9a (GenBank ID: FJ231750.1); D. rerio Wnt9b (RefSeq ID: NM_001137660); Gallus gallus Wnt9a (GenBank ID: AY753293.1); G. gallus Wnt9b (RefSeq ID: XM_001234393); Holothuria glaberrima Wnt9 (GenBank ID: GQ243223.1); Homo sapiens Wnt9a (RefSeq ID: NM_003395); H. sapiens Wnt9b (RefSeq ID: NM_003396); Mus musculus Wnt9a (RefSeq ID: NM_139298); M. musculus Wnt9b (RefSeq ID: NM_011719); Nematostella vectensis Wnt2 (GenBank ID: AY725201); N. vectensis Wnt4 (GenBank ID: AY687348.1); Paracentrotus lividus Wnt5 (GenBank ID: HM449806.1); Saccoglossus kowalevskii Wnt2 (RefSeq ID: NM_001164983); S. kowalevskii Wnt4 (RefSeq ID: XM_002737213); S. kowalevskii Wnt9 (RefSeq ID: NM_001168207); Strogylocentrotus purpuratus Wnt3 (RefSeq ID: XM_785502); S. purpuratus Wnt5 (RefSeq ID: XM_774853); S. purpuratus Wnt9 (RefSeq ID: XM_775730); S. purpuratus Wnt10 (RefSeq ID: XM_776471.1); Xenopus tropicalis Wnt9b (RefSeq ID: NM_001103083.1).
Multiple sequence alignment of Wnt9 subfamily members from H. glaberrima and other deuterostomes. Blue shading is used to show the degree of conservation in the column. The dotted red line indicates the cleavage site of the secretory signal peptide. The latter showed little conservation among the species, therefore it is only partially displayed on this figure. Red asterisks indicate 24 conserved cystein residues shared by all Wnt9 homologs of deuterostomes. The following sequences were used for the alignment: Branchiostoma floridae Wnt14 (Wnt9) (RefSeq ID: XP_002598627); Danio rerio Wnt9a (GenBank ID: ACI95254); D. rerio Wnt9b (RefSeq ID: NP_001131132); Gallus gallus Wnt9a (GenBank ID: AAW81995); G. gallus Wnt9b (RefSeq ID: XP_001234394); Holothuria glaberrima Wnt9 (GenBank ID: ACS74870); Homo sapiens Wnt9a (RefSeq ID: NP_003386); H. sapiens Wnt9b (RefSeq ID: NP_003387.1); Saccoglossus kowalevskii Wnt9 (RefSeq ID: NP_001161679.1).
Multiple sequence alignment of TCTP protein sequences showing conserved motifs. Residue conservation is shown in shade of blue. The following sequences were included in the alignment: Apostichopus japonicus TCTP (GenBank ID: ABC87996.1); Branchiostoma belcheri TCTP (Swiss-Prot ID: Q95VY2.1); Danio rerio TCTP (Swiss-Prot ID: Q9DGK4.1); Drosophila melanogaster TCTP (Swiss-Prot ID: Q9VGS2.1); Gallus gallus TCTP (Swiss-Prot ID: P43347.1); Holothuria glaberrima TCTP (GenBank ID: ACZ73830.1); Homo sapiens TCTP (Swiss-Prot ID: P13693.1); Lumbricus rubellus TCTP (Swiss-Prot ID: O18477.1); Rattus norvegicus TCTP (Swiss-Prot ID: P63029.1); Xenopus tropicalis TCTP (Swiss-Prot ID: Q66JC5.1).
Phylogenetic analysis showing the relationship of Bmp1/Tll of H. glaberrima (indicated by an arrowhead) with BMP1-like proteinases of other deuterostomes. The phylogenetic tree was constructed by the neighbor-joining algorithm in MEGA 5.0 (Tamura et al., 2011). The bootstrap test was performed with 2000 replicates. The tree was rooted using human CUB and Sushi multiple domains 2 protein (CSMD2). The database accession numbers of the sequences used to construct the tree are as follows: Branchiostoma floridae BMP1 (GenBank ID: AY986775.1); Danio rerio TLL1 (RefSeq ID: NM_131010.1); Holothuria glaberrima BMP1/TLL (GenBank ID: JN602083); Homo sapiens BMP1 (RefSeq ID: NM_006129.4); H. sapiens TLL1 (RefSeq ID: NM_012464.4); H. sapiens TLL2 (RefSeq ID: NM_012465.3); H. sapiens CSMD2 (RefSeq ID: NM_052896); Mus musculus BMP1 (RefSeq ID: NM_009755.3); M. musculus TLL1 (RefSeq ID: NM_009390.2); M. musculus TLL2 (RefSeq ID: NM_011904.3); Strogylocentrotus purpuratus BMP1 (RefSeq ID: NM_214563.1); Xenopus laevis BMP1 (RefSeq ID: NM_001090275.1); Xenopus laevis TLL1 (RefSeq ID: NM_001090425.1); Xenopus laevis TLL2 (RefSeq ID: NM_001090908.1).
Multiple sequence alignment of the astacin protease domain (red box) of H. glaberrima BMP1/TLL and BMP1 and Tolloid-like (TLL) proteins of other deuterostomes. Asterisks show three histidine residues that anchor the catalytic zinc ion in the active center of the protease. Filled and empty circles indicate the position of the conserved tyrosine and water-bound glutamic acid, respectively. The following sequences were used in the alignment: Branchiostoma floridae BMP1 (GenBank ID: AAX84844.1); Danio rerio BMP1a (RefSeq ID: NP_001035126.1); D. rerio BMP1b (RefSeq ID: NP_001034901.1); D. rerio TLL1 (RefSeq ID: NP_571085.1); Gallus gallus TLL1 (Swiss-Prot ID: Q9DER7.1); Holothuria glaberrima BMP1/TLL (GenBank ID: JN602083); Homo sapiens BMP1 (RefSeq ID: NP_006120.1); H. sapiens TLL1 (RefSeq ID: NP_036596.3); H. sapiens TLL2 (RefSeq ID: NP_036597.1); Mus musculus BMP1 (RefSeq ID: NP_033885.2); M. musculus TLL1 (Swiss-Prot ID: Q62381.1); M. musculus TLL2 (RefSeq ID: NP_036034.1); Strogylocentrotus purpuratus BMP1 (RefSeq ID: NP_999728.1); Xenopus laevis BMP1 (RefSeq ID: NP_001083744.1); X. laevis TLL2 (RefSeq ID: NP_001084377.1).
Expression of Wnt9, TCTP, and Bmp1/Tll in the stumps ('old' tissue) adjacent to the regenerates. (A) Asymmetrical expression of Wnt9 (arrowheads) in the mesentery attached to the cloacal stump on day 2. (B) Expression of TCTP in the apices of the luminal epithelium in the esophageal stump on day 2. (C) Wnt9 expression (arrowheads) in the mesothelium of the cloacal stump and in the supporting mesentery on day 7. (D) Wnt9 expression in both the mesothelium and luminal epithelium of the esophageal stump as well as in the distal region of the supporting mesentery on day 7. (D') Higher magnification of the gut wall of the esophageal stump (boxed region on D). (E) TCTP expression in the apical region of the luminal epithelium of the esophageal stump on day 7. (F) Scattered cells showing moderate expression of Wnt9 are occasionally seen in the luminal epithelium of the cloaca on day 14. ctl – connective tissue layer; me – mesothelium; mes – mesentery; le – luminal epithelium. Scale bars: A, C, D, - 200 µm; B, D', F - 50 µm; E – 100 µm.
Acknowledgements
The authors thank Mr. Rey Rosa for his help with animal collection and maintenance. This research was funded by NIH (grant number: 1SC1GM084770-01), NSF (grant number: IOS-0842870), and the University of Puerto Rico.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altieri DC. New wirings in the survivin networks. Oncogene. 2008;27:6276–6284. doi: 10.1038/onc.2008.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angerer L, Hussain S, Wei Z, Livingston BT. Sea urchin metalloproteases: a genomic survey of the BMP-1/tolloid-like, MMP and ADAM families. Dev. Biol. 2006;300:267–281. doi: 10.1016/j.ydbio.2006.07.046. [DOI] [PubMed] [Google Scholar]
- Bommer U, Thiele B. The translationally controlled tumour protein (TCTP) Int. J. Biochem. Cell Biol. 2004;36:379–385. doi: 10.1016/s1357-2725(03)00213-9. [DOI] [PubMed] [Google Scholar]
- Bragdon B, Moseychuk O, Saldanha S, King D, Julian J, Nohe A. Bone morphogenetic proteins: a critical review. Cell. Signal. 2011;23:609–620. doi: 10.1016/j.cellsig.2010.10.003. [DOI] [PubMed] [Google Scholar]
- Byrne M. The morphology of autotomy structures in the sea cucumber Eupentacta quinquesemita before and during evisceration. J. Exp. Biol. 2001;204:849–863. doi: 10.1242/jeb.204.5.849. [DOI] [PubMed] [Google Scholar]
- Carlson BM. Principles of Regenerative Biology. London: Elsevier; 2007. [Google Scholar]
- Chen SH, Wu P, Chou C, Yan Y, Liu H, Weng S, Yang-Yen H. A knockout mouse approach reveals that TCTP functions as an essential factor for cell proliferation and survival in a tissue- or cell type-specific manner. Mol. Biol. Cell. 2007a;18:2525–2532. doi: 10.1091/mbc.E07-02-0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Zhang H, Yang H, Huang X, Zhang X, Zhang P. The expression of AmphiTCTP, a TCTP orthologous gene in amphioxus related to the development of notochord and somites. Comp. Biochem. Physiol. B, Biochem. Mol. Biol. 2007b;147:460–465. doi: 10.1016/j.cbpb.2007.02.012. [DOI] [PubMed] [Google Scholar]
- Conte M, Deri P, Isolani ME, Mannini L, Batistoni R. A mortalin-like gene is crucial for planarian stem cell viability. Dev. Biol. 2009;334:109–118. doi: 10.1016/j.ydbio.2009.07.010. [DOI] [PubMed] [Google Scholar]
- Cox AA, Jezewski PA, Fang P, Payne-Ferreira TL. Zebrafish Wnt9a,9b paralog comparisons suggest ancestral roles for Wnt9 in neural, oral-pharyngeal ectoderm and mesendoderm. Gene Expr. Patterns. 2010;10:251–258. doi: 10.1016/j.gep.2010.05.005. [DOI] [PubMed] [Google Scholar]
- Croce JC, McClay DR. Evolution of the Wnt pathways. Methods Mol. Biol. 2008;469:3–18. doi: 10.1007/978-1-60327-469-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce JC, Wu S, Byrum C, Xu R, Duloquin L, Wikramanayake AH, Gache C, McClay DR. A genome-wide survey of the evolutionarily conserved Wnt pathways in the sea urchin Strongylocentrotus purpuratus. Dev. Biol. 2006;300:121–131. doi: 10.1016/j.ydbio.2006.08.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danesh SM, Villasenor A, Chong D, Soukup C, Cleaver O. BMP and BMP receptor expression during murine organogenesis. Gene Expr. Patterns. 2009;9:255–265. doi: 10.1016/j.gep.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- Feral J, Massin C. In: Digestive system: Holothuroidea. Jangoux M, Lawrence J, editors. Balkema, Rotterdam: Echinoderm Nutrition; 1982. pp. 192–212. [Google Scholar]
- Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, Holm L, Sonnhammer ELL, Eddy SR, Bateman A. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fokina VM, Frolova EI. Expression patterns of Wnt genes during development of an anterior part of the chicken eye. Dev. Dyn. 2006;235:496–505. doi: 10.1002/dvdy.20621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Arrarás JE, Greenberg MJ. Visceral regeneration in holothurians. Microsc. Res. Tech. 2001;55:438–451. doi: 10.1002/jemt.1189. [DOI] [PubMed] [Google Scholar]
- García-Arrarás JE, Estrada-Rodgers L, Santiago R, Torres II, Díaz-Miranda L, Torres-Avillán I. Cellular mechanisms of intestine regeneration in the sea cucumber, Holothuria glaberrima Selenka (Holothuroidea:Echinodermata) J. Exp. Zool. 1998;281:288–304. doi: 10.1002/(sici)1097-010x(19980701)281:4<288::aid-jez5>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- Ge G, Greenspan DS. Developmental roles of the BMP1/TLD metalloproteinases. Birth Defects Res. C Embryo Today. 2006;78:47–68. doi: 10.1002/bdrc.20060. [DOI] [PubMed] [Google Scholar]
- Holland L, Holland P, Holland N. Whole mount in situ hybridization applicable to Amphioxus and other small larvae. In: Ferraris J, Palumbi S, editors. Molecular Zoology: Advances, Strategies and Protocols. New York: Wiley-Liss; 1996. pp. 476–483. [Google Scholar]
- Hopkins DR, Keles S, Greenspan DS. The bone morphogenetic protein 1/Tolloid-like metalloproteinases. Matrix Biol. 2007;26:508–523. doi: 10.1016/j.matbio.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman L. The Celomate Bilateria. New York: McGraw-Hill Book Co. Inc.; 1955. The Invertebrates. IV. Echinodermata. [Google Scholar]
- Iwai S, Yonekawa A, Harada C, Hamada M, Katagiri W, Nakazawa M, Yura Y. Involvement of the Wnt-β-catenin pathway in invasion and migration of oral squamous carcinoma cells. Int. J. Oncol. 2010;37:1095–1103. doi: 10.3892/ijo_00000761. [DOI] [PubMed] [Google Scholar]
- Jiménez CR, Stam FJ, Li KW, Gouwenberg Y, Hornshaw MP, De Winter F, Verhaagen J, Smit AB. Proteomics of the injured rat sciatic nerve reveals protein expression dynamics during regeneration. Mol. Cell Proteomics. 2005;4:120–132. doi: 10.1074/mcp.M400076-MCP200. [DOI] [PubMed] [Google Scholar]
- Juriloff DM, Harris MJ, McMahon AP, Carroll TJ, Lidral AC. Wnt9b is the mutated gene involved in multifactorial nonsyndromic cleft lip with or without cleft palate in A/WySn mice, as confirmed by a genetic complementation test. Birth Defects Res. Part A Clin. Mol. Teratol. 2006;76:574–579. doi: 10.1002/bdra.20302. [DOI] [PubMed] [Google Scholar]
- Karner CM, Das A, Ma Z, Self M, Chen C, Lum L, Oliver G, Carroll TJ. Canonical Wnt9b signaling balances progenitor cell expansion and differentiation during kidney development. Development. 2011;138:1247–1257. doi: 10.1242/dev.057646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh Y, Katoh M. Comparative genomics on Wnt9a orthologs. Oncol. Rep. 2005;13:989–992. [PubMed] [Google Scholar]
- Koziol MJ, Garrett N, Gurdon JB. Tpt1 activates transcription of oct4 and nanog in transplanted somatic nuclei. Curr. Biol. 2007;17:801–807. doi: 10.1016/j.cub.2007.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurpios NA, Ibañes M, Davis NM, Lui W, Katz T, Martin JF, Izpisúa Belmonte JC, Tabin CJ. The direction of gut looping is established by changes in the extracellular matrix and in cell:cell adhesion. Proc. Natl. Acad. Sci. U.S.A. 2008;105:8499–8506. doi: 10.1073/pnas.0803578105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Y, Ryan RC, Zhang Z, Bullard SA, Bush JO, Maltby KM, Lidral AC, Jiang R. Expression of Wnt9b and activation of canonical Wnt signaling during midfacial morphogenesis in mice. Dev. Dyn. 2006;235:1448–1454. doi: 10.1002/dvdy.20723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lee S, Solow-Cordero DE, Kessler E, Takahara K, Greenspan DS. Transforming growth factor-beta regulation of bone morphogenetic protein-1/procollagen C-proteinase and related proteins in fibrogenic cells and keratinocytes. J. Biol. Chem. 1997;272:19059–19066. doi: 10.1074/jbc.272.30.19059. [DOI] [PubMed] [Google Scholar]
- Li F, Cheng Q, Ling X, Stablewski A, Tang L, Foster BA, Johnson CS, Rustum YM, Porter CW. Generation of a novel transgenic mouse model for bioluminescent monitoring of survivin gene activity in vivo at various pathophysiological processes. Survivin expression overlaps with stem cell markers. Am. J. Pathol. 2010;176:1629–1638. doi: 10.2353/ajpath.2010.090414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mac Sweeney A, Gil-Parrado S, Vinzenz D, Bernardi A, Hein A, Bodendorf U, Erbel P, Logel C, Gerhartz B. Structural basis for the substrate specificity of bone morphogenetic protein 1/tolloid-like metalloproteases. Mol. Biol. 2008;384:228–239. doi: 10.1016/j.jmb.2008.09.029. [DOI] [PubMed] [Google Scholar]
- Marconi A, Dallaglio K, Lotti R, Vaschieri C, Truzzi F, Fantini F, Pincelli C. Survivin identifies keratinocyte stem cells and is downregulated by anti-beta1 integrin during anoikis. Stem Cells. 2007;25:149–155. doi: 10.1634/stemcells.2006-0165. [DOI] [PubMed] [Google Scholar]
- Mashanov V, Frolova L, Dolmatov I. Structure of the digestive tube in the holothurian Eupentacta fraudatrix (Holothuroidea:Dendrochirota) Russian Journal of Marine Biology. 2004;30:314–322. [Google Scholar]
- Mashanov VS, Garcia-Arraras JE. Gut regeneration in holothurians: a snapshot of recent developments. Biol. Bull. 2011;221 doi: 10.1086/BBLv221n1p93. [DOI] [PubMed] [Google Scholar]
- Mashanov VS, Dolmatov IY, Heinzeller T. Transdifferentiation in holothurian gut regeneration. Biol. Bull. 2005;209:184–193. doi: 10.2307/3593108. [DOI] [PubMed] [Google Scholar]
- Mashanov VS, Zueva OR, Rojas-Catagena C, Garcia-Arraras JE. Visceral regeneration in a sea cucumber involves extensive expression of survivin and mortalin homologs in the mesothelium. BMC Dev. Biol. 2010;10:117. doi: 10.1186/1471-213X-10-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K, Miki R, Nakayama M, Tatsumi N, Yokouchi Y. Wnt9a secreted from the walls of hepatic sinusoids is essential for morphogenesis, proliferation, and glycogen accumulation of chick hepatic epithelium. Dev. Biol. 2008;319:234–247. doi: 10.1016/j.ydbio.2008.04.021. [DOI] [PubMed] [Google Scholar]
- Micalizzi DS, Farabaugh SM, Ford HL. Epithelial-mesenchymal transition in cancer: parallels between normal development and tumor progression. J Mammary Gland Biol Neoplasia. 2010;15:117–134. doi: 10.1007/s10911-010-9178-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nejak-Bowen KN, Monga SPS. Beta-catenin signaling, liver regeneration and hepatocellular cancer: sorting the good from the bad. Semin. Cancer Biol. 2011;21:44–58. doi: 10.1016/j.semcancer.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Pineda PA, Ramírez-Gómez F, Pérez-Ortiz J, González-Díaz S, Santiago-De Jesús F, Hernández-Pasos J, Del Valle-Avila C, Rojas-Cartagena C, Suárez-Castillo EC, Tossas K, Méndez-Merced AT, Roig-López JL, Ortiz-Zuazaga H, García-Arrarás JE. Gene expression profiling of intestinal regeneration in the sea cucumber. BMC Genomics. 2009;10:262. doi: 10.1186/1471-2164-10-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollins AC, Friedman DB, Nanney LB. Proteomic investigation of human burn wounds by 2D-difference gel electrophoresis and mass spectrometry. J. Surg. Res. 2007;142:143–152. doi: 10.1016/j.jss.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian J, Jiang Z, Li M, Heaphy P, Liu Y, Shackleford GM. Mouse Wnt9b transforming activity, tissue-specific expression, and evolution. Genomics. 2003;81:34–46. doi: 10.1016/s0888-7543(02)00012-5. [DOI] [PubMed] [Google Scholar]
- Quiñones JL, Rosa R, Ruiz DL, García-Arrarás JE. Extracellular matrix remodeling and metalloproteinase involvement during intestine regeneration in the sea cucumber Holothuria glaberrima. Dev. Biol. 2002;250:181–197. doi: 10.1006/dbio.2002.0778. [DOI] [PubMed] [Google Scholar]
- Rho SB, Lee JH, Park MS, Byun H, Kang S, Seo S, Kim J, Park S. Anti-apoptotic protein TCTP controls the stability of the tumor suppressor p53. FEBS Lett. 2011;585:29–35. doi: 10.1016/j.febslet.2010.11.014. [DOI] [PubMed] [Google Scholar]
- Rojas-Cartagena C, Ortíz-Pineda P, Ramírez-Gómez F, Suárez-Castillo EC, Matos-Cruz V, Rodríguez C, Ortíz-Zuazaga H, García-Arrarás JE. Distinct profiles of expressed sequence tags during intestinal regeneration in the sea cucumber Holothuria glaberrima. Physiol. Genomics. 2007;31:203–215. doi: 10.1152/physiolgenomics.00228.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu K, Ishizuya-Oka A, Amano T, Yoshizato K, Ueda S. Isolation of connective-tissue-specific genes involved in Xenopus intestinal remodeling: thyroid hormone up-regulates Tolloid/BMP-1 expression. Dev. Genes Evol. 2002;212:357–364. doi: 10.1007/s00427-002-0250-3. [DOI] [PubMed] [Google Scholar]
- Sun L, Chen M, Yang H, Wang T, Liu B, Shu C, Gardiner DM. Large scale gene expression profiling during intestine and body wall regeneration in the sea cucumber Apostichopus japonicus. Comp. Biochem. Physiol. Part D Genomics Proteomics. 2011;6:195–205. doi: 10.1016/j.cbd.2011.03.002. [DOI] [PubMed] [Google Scholar]
- Taipale J, Beachy PA. The Hedgehog and Wnt signalling pathways in cancer. Nature. 2001;411:349–354. doi: 10.1038/35077219. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011 doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topol L, Jiang X, Choi H, Garrett-Beal L, Carolan PJ, Yang Y. Wnt-5a inhibits the canonical Wnt pathway by promoting GSK-3-independent beta-catenin degradation. Cell Biol. 2003;162:899–908. doi: 10.1083/jcb.200303158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadhwa R, Yaguchi T, Hasan MK, Mitsui Y, Reddel RR, Kaul SC. Hsp70 family member, mot-2/mthsp70/GRP75, binds to the cytoplasmic sequestration domain of the p53 protein. Exp. Cell Res. 2002;274:246–253. doi: 10.1006/excr.2002.5468. [DOI] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2 - a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidinger G, Moon RT. When Wnts antagonize Wnts. Cell Biol. 2003;162:753–755. doi: 10.1083/jcb.200307181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wend P, Holland JD, Ziebold U, Birchmeier W. Wnt signaling in stem and cancer stem cells. Semin. Cell Dev. Biol. 2010;21:855–863. doi: 10.1016/j.semcdb.2010.09.004. [DOI] [PubMed] [Google Scholar]
- White RM, Zon LI. Melanocytes in development, regeneration, and cancer. Cell Stem Cell. 2008;3:242–252. doi: 10.1016/j.stem.2008.08.005. [DOI] [PubMed] [Google Scholar]
- Wilkie IC. Autotomy as a prelude to regeneration in echinoderms. Microsc. Res. Tech. 2001;55:369–396. doi: 10.1002/jemt.1185. [DOI] [PubMed] [Google Scholar]
- Yang Y, Yang F, Xiong Z, Yan Y, Wang X, Nishino M, Mirkovic D, Nguyen J, Wang H, Yang X. An N-terminal region of translationally controlled tumor protein is required for its antiapoptotic activity. Oncogene. 2005;24:4778–4788. doi: 10.1038/sj.onc.1208666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Cheng H, Han N, Liu D, Zhu W, Fan B, Duan F. Messenger RNA expression of translationally controlled tumor protein (TCTP) in liver regeneration and cancer. Anticancer Res. 2008;28:1575–1580. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic position of H. glaberrima Wnt9 (indicated by an arrowhead) in relation to other members of Wnt9 subfamily (A). The sea cucumber Wnt9 is most similar to the sea urchin Wnt9 protein. The two vertebrate paralogs, Wnt9a and Wnt9b, form a well supported monophyletic clade with two clusters, to which the single Wnt9 of basal chordates is related as an outgroup. The tree was rooted using sequences of other invertebrate Wnt subfamiles (B). The phylogenetic analysis was based on nucleotide sequences of the coding regions of the genes which were aligned with ClustalW and then processed using the Neighbor-Joining method implemented in MEGA 5 software (Tamura et al., 2011). Support of the nodes was inferred by bootstraping method (2000 replicates), and the numbers represent the percentage of replicate trees in which the taxa clustered together in the bootsrap test. The following sequences were used for constructing the phylogenetic tree: Anolis carolinensis Wnt9a (RefSeq ID: XM_003229241.1); A. carolinensis Wnt9b (RefSeq ID: XM_003222531.1); Branchiostoma floridae Wnt1 (GenBank ID: AF061974); B. floridae Wnt4 (GenBank ID: AF061973.1); B. floridae Wnt14/Wnt9 (RefSeq ID: XM_002598581.1); Danio rerio Wnt9a (GenBank ID: FJ231750.1); D. rerio Wnt9b (RefSeq ID: NM_001137660); Gallus gallus Wnt9a (GenBank ID: AY753293.1); G. gallus Wnt9b (RefSeq ID: XM_001234393); Holothuria glaberrima Wnt9 (GenBank ID: GQ243223.1); Homo sapiens Wnt9a (RefSeq ID: NM_003395); H. sapiens Wnt9b (RefSeq ID: NM_003396); Mus musculus Wnt9a (RefSeq ID: NM_139298); M. musculus Wnt9b (RefSeq ID: NM_011719); Nematostella vectensis Wnt2 (GenBank ID: AY725201); N. vectensis Wnt4 (GenBank ID: AY687348.1); Paracentrotus lividus Wnt5 (GenBank ID: HM449806.1); Saccoglossus kowalevskii Wnt2 (RefSeq ID: NM_001164983); S. kowalevskii Wnt4 (RefSeq ID: XM_002737213); S. kowalevskii Wnt9 (RefSeq ID: NM_001168207); Strogylocentrotus purpuratus Wnt3 (RefSeq ID: XM_785502); S. purpuratus Wnt5 (RefSeq ID: XM_774853); S. purpuratus Wnt9 (RefSeq ID: XM_775730); S. purpuratus Wnt10 (RefSeq ID: XM_776471.1); Xenopus tropicalis Wnt9b (RefSeq ID: NM_001103083.1).
Multiple sequence alignment of Wnt9 subfamily members from H. glaberrima and other deuterostomes. Blue shading is used to show the degree of conservation in the column. The dotted red line indicates the cleavage site of the secretory signal peptide. The latter showed little conservation among the species, therefore it is only partially displayed on this figure. Red asterisks indicate 24 conserved cystein residues shared by all Wnt9 homologs of deuterostomes. The following sequences were used for the alignment: Branchiostoma floridae Wnt14 (Wnt9) (RefSeq ID: XP_002598627); Danio rerio Wnt9a (GenBank ID: ACI95254); D. rerio Wnt9b (RefSeq ID: NP_001131132); Gallus gallus Wnt9a (GenBank ID: AAW81995); G. gallus Wnt9b (RefSeq ID: XP_001234394); Holothuria glaberrima Wnt9 (GenBank ID: ACS74870); Homo sapiens Wnt9a (RefSeq ID: NP_003386); H. sapiens Wnt9b (RefSeq ID: NP_003387.1); Saccoglossus kowalevskii Wnt9 (RefSeq ID: NP_001161679.1).
Multiple sequence alignment of TCTP protein sequences showing conserved motifs. Residue conservation is shown in shade of blue. The following sequences were included in the alignment: Apostichopus japonicus TCTP (GenBank ID: ABC87996.1); Branchiostoma belcheri TCTP (Swiss-Prot ID: Q95VY2.1); Danio rerio TCTP (Swiss-Prot ID: Q9DGK4.1); Drosophila melanogaster TCTP (Swiss-Prot ID: Q9VGS2.1); Gallus gallus TCTP (Swiss-Prot ID: P43347.1); Holothuria glaberrima TCTP (GenBank ID: ACZ73830.1); Homo sapiens TCTP (Swiss-Prot ID: P13693.1); Lumbricus rubellus TCTP (Swiss-Prot ID: O18477.1); Rattus norvegicus TCTP (Swiss-Prot ID: P63029.1); Xenopus tropicalis TCTP (Swiss-Prot ID: Q66JC5.1).
Phylogenetic analysis showing the relationship of Bmp1/Tll of H. glaberrima (indicated by an arrowhead) with BMP1-like proteinases of other deuterostomes. The phylogenetic tree was constructed by the neighbor-joining algorithm in MEGA 5.0 (Tamura et al., 2011). The bootstrap test was performed with 2000 replicates. The tree was rooted using human CUB and Sushi multiple domains 2 protein (CSMD2). The database accession numbers of the sequences used to construct the tree are as follows: Branchiostoma floridae BMP1 (GenBank ID: AY986775.1); Danio rerio TLL1 (RefSeq ID: NM_131010.1); Holothuria glaberrima BMP1/TLL (GenBank ID: JN602083); Homo sapiens BMP1 (RefSeq ID: NM_006129.4); H. sapiens TLL1 (RefSeq ID: NM_012464.4); H. sapiens TLL2 (RefSeq ID: NM_012465.3); H. sapiens CSMD2 (RefSeq ID: NM_052896); Mus musculus BMP1 (RefSeq ID: NM_009755.3); M. musculus TLL1 (RefSeq ID: NM_009390.2); M. musculus TLL2 (RefSeq ID: NM_011904.3); Strogylocentrotus purpuratus BMP1 (RefSeq ID: NM_214563.1); Xenopus laevis BMP1 (RefSeq ID: NM_001090275.1); Xenopus laevis TLL1 (RefSeq ID: NM_001090425.1); Xenopus laevis TLL2 (RefSeq ID: NM_001090908.1).
Multiple sequence alignment of the astacin protease domain (red box) of H. glaberrima BMP1/TLL and BMP1 and Tolloid-like (TLL) proteins of other deuterostomes. Asterisks show three histidine residues that anchor the catalytic zinc ion in the active center of the protease. Filled and empty circles indicate the position of the conserved tyrosine and water-bound glutamic acid, respectively. The following sequences were used in the alignment: Branchiostoma floridae BMP1 (GenBank ID: AAX84844.1); Danio rerio BMP1a (RefSeq ID: NP_001035126.1); D. rerio BMP1b (RefSeq ID: NP_001034901.1); D. rerio TLL1 (RefSeq ID: NP_571085.1); Gallus gallus TLL1 (Swiss-Prot ID: Q9DER7.1); Holothuria glaberrima BMP1/TLL (GenBank ID: JN602083); Homo sapiens BMP1 (RefSeq ID: NP_006120.1); H. sapiens TLL1 (RefSeq ID: NP_036596.3); H. sapiens TLL2 (RefSeq ID: NP_036597.1); Mus musculus BMP1 (RefSeq ID: NP_033885.2); M. musculus TLL1 (Swiss-Prot ID: Q62381.1); M. musculus TLL2 (RefSeq ID: NP_036034.1); Strogylocentrotus purpuratus BMP1 (RefSeq ID: NP_999728.1); Xenopus laevis BMP1 (RefSeq ID: NP_001083744.1); X. laevis TLL2 (RefSeq ID: NP_001084377.1).
Expression of Wnt9, TCTP, and Bmp1/Tll in the stumps ('old' tissue) adjacent to the regenerates. (A) Asymmetrical expression of Wnt9 (arrowheads) in the mesentery attached to the cloacal stump on day 2. (B) Expression of TCTP in the apices of the luminal epithelium in the esophageal stump on day 2. (C) Wnt9 expression (arrowheads) in the mesothelium of the cloacal stump and in the supporting mesentery on day 7. (D) Wnt9 expression in both the mesothelium and luminal epithelium of the esophageal stump as well as in the distal region of the supporting mesentery on day 7. (D') Higher magnification of the gut wall of the esophageal stump (boxed region on D). (E) TCTP expression in the apical region of the luminal epithelium of the esophageal stump on day 7. (F) Scattered cells showing moderate expression of Wnt9 are occasionally seen in the luminal epithelium of the cloaca on day 14. ctl – connective tissue layer; me – mesothelium; mes – mesentery; le – luminal epithelium. Scale bars: A, C, D, - 200 µm; B, D', F - 50 µm; E – 100 µm.