Abstract
The specification of temporal identity within single progenitor lineages is essential to generate functional neuronal diversity in Drosophila and mammals. In Drosophila, four transcription factors are sequentially expressed in neural progenitors (neuroblasts) and each regulates the temporal identity of the progeny produced during its expression window. The first temporal identity is established by the Ikaros-family zinc finger transcription factor Hunchback (Hb). Hb is detected in young (newly-formed) neuroblasts for about an hour and is maintained in the early-born neurons produced during this interval. Hb is necessary and sufficient to specify early-born neuronal or glial identity in multiple neuroblast lineages. The timing of hb expression in neuroblasts is regulated at the transcriptional level. Here we identify the cis-regulatory elements that confer proper hb expression in “young” neuroblasts and early-born neurons. We show that the neuroblast element contains clusters of predicted binding sites for the Seven-up transcription factor, which is known to limit hb neuroblast expression. We identify highly conserved sequences in the neuronal element that are good candidates for maintaining Hb transcription in neurons. Our results provide the necessary foundation for identifying trans-acting factors that establish the Hb early temporal expression domain.
Introduction
Temporal patterning is particularly important for central nervous system (CNS) development, due to the vast number of cell types that need to be produced by relatively few progenitors. In the mammalian CNS, individual neural progenitors give rise to an ordered series of cell types in the cerebral cortex, retina, and spinal cord (reviewed in Pearson and Doe, 2004). In Drosophila, embryonic neuroblasts (NBs) undergo a series of asymmetric cell divisions to generate smaller progeny called ganglion mother cells (GMCs) that typically produce a pair of neurons (Doe, 2008). We and others have identified four transcription factors – Hunchback (Hb), Krüppel (Kr), Pdm, Castor (Cas) – that are sequentially expressed in embryonic NBs and maintained in the GMC and neuronal progeny born during the expression window (Brody and Odenwald, 2000; Isshiki et al., 2001; Kambadur et al., 1998). The early factor Hb is necessary and sufficient to specify the fate of early-born neurons in multiple NB lineages (Cleary and Doe, 2006; Grosskortenhaus et al., 2005; Isshiki et al., 2001; Kanai et al., 2005; Novotny et al., 2002; Pearson and Doe, 2003; Tran and Doe, 2008).
Genetic experiments using Gal4 lines to drive UAS-hb expression beyond its normal window of expression in NBs leads to overproduction of early-born neuronal fates and a delay or loss of the later-born neuronal fates in all tested NB lineages (Cleary and Doe, 2006; Isshiki et al., 2001; Kanai et al., 2005; Mettler et al., 2006; Novotny et al., 2002; Pearson and Doe, 2003). Thus, it is critical to downregulate hb expression to generate neuronal diversity. A study of hb neuroblast expression patterns using (a) intron probes to detect active transcription, (b) coding sequence probes to detect cytoplasmic mature mRNA, and (c) antibody probes to detect protein revealed a strong conrguence among all three probes, indicating that the timing of hb neuroblast expression is primarily regulated at the transcriptional levels (Grosskortenhaus et al., 2005). Furthermore, experiments using in vitro cultured NBs show that lineage-extrinsic cues are not required for hb downregulation (Grosskortenhaus et al., 2005); however, blocking cytokinesis will prevent hb downregulation (Grosskortenhaus et al., 2005). These cellular studies suggest that repression of hb requires either the segregation of regulatory factors out of the NB during asymmetric cell division, or “feedback” signaling from the new-born GMC to the NB. In addition, the orphan steroid hormone receptor Seven-up (Svp) is required for timely repression of hb expression (Kanai et al., 2005; Mettler et al., 2006), as is the Pipsqueak domain nuclear protein Distal antenna (Dan) (Kohwi et al., 2011). Recently it has been shown that the transcriptional repressor Prospero is required to keep Svp levels low (Mettler et al., 2006); Prospero is normally segregated out of the NB into the GMC (Spana and Doe, 1995), which may explain why NBs that do not undergo cytokinesis maintain hb expression: they have nuclear Prospero which prevents accumulation of Svp and thus allows persistent hb expression. This attractive model remains to be tested.
To understand the temporal regulation of hb expression in NBs, we have identified the cis-regulatory DNA that confers proper temporal expression of hb in NBs, and identified additional cis-regulatory elements that promote hb expression in early-born neurons and repress hb expression in late-born neurons. In addition, we have found an unexpected cis-regulatory element that drives expression only in late-born neurons, where hb is normally never detected.
1. Results and discussion
1.1 Generation of hb enhancer-reporter constructs
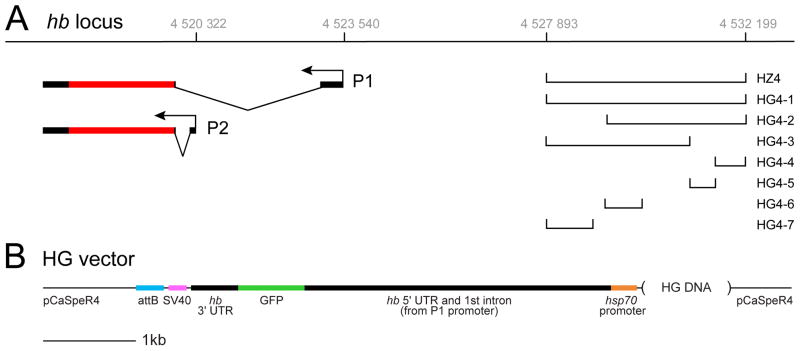
During previous studies of the hb cis-regulatory region controlling early blastoderm expression (Margolis et al., 1994; Margolis et al., 1995), we discovered an ~4kb element that conferred expression in neuroblasts and GMCs but not neurons (HZ4 element; Fig. 1A). Here, we extend our analysis to subdivide this 4kb element using a series of hb-green fluorescent protein (hb- GFP) constructs containing (from 5′ to 3′) hb cis-regulatory DNA, the hb P2 5′ UTR including the first intron and the P1 promoter, a basal hsp70 promoter, GFP coding sequence, the hb 3′ UTR, the SV40 transcription termination sequence, and an attB sequence (see Experimental procedures for details). We then used the PhiC recombinase system to insert all transgenes into the same genomic site on chromosome 2 (Figure 1A,B; see Experimental procedures). This eliminates variability in transgene expression due to chromosomal position effects can be a problem when inserting transgenes randomly in the genome. We used EvoPrinter software (Odenwald et al., 2005; Yavatkar et al., 2008) to identify blocks of evolutionarily-conserved sequence within the largest construct (HG4-1; Fig. 2), and then designed smaller constructs centered around these blocks of conservation (HG4-2 through HG4-7) as shown in Figs. 1 and 2. We originally designed the constructs using a more restrictive EvoPrint showing sequences conserved among all twelve Drosophila species, but present the EvoPrint of sequences conserved among 11/12 species. This resulted in a small block of conservation between HG4-6 and HG4-7 not being included in these smaller constructs. This region of DNA could contain regulatory information that might help sum HG4-6 and HG4-7 to match the larger HG4-3 construct.
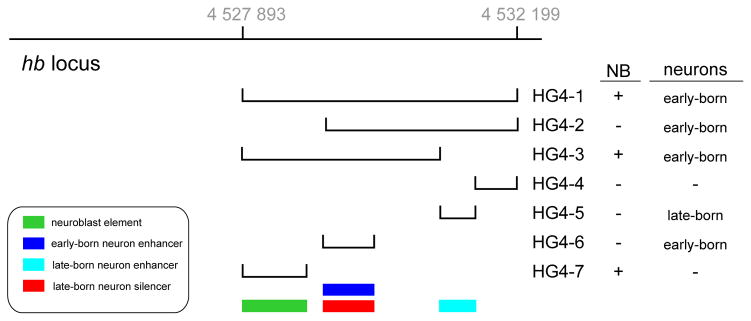
Fig. 1.
Location of enhancer-reporter constructs at the hunchback (hb) locus. The two hb transcripts produced from the P1 and P2 promoters are shown (black, untranslated regions; red, coding region). Grey numbers at top are the Flybase coordinates for the region. Lines shown at right indicate the DNA elements used in the reporter constructs (see methods for construct details). Each of these DNA elements lie within the first intron of CG8112, whose promoter start site is 4526704 on the strand opposite that of hb.
Fig. 2.
Evoprint of the cis-regulatory DNA used in this study. Bold capital letters represent conservation in either all 12 or 11/12 Drosophila species for which genomic data exists. The Flybase coordinates for the first nucleotide of each row is shown to the right, and the approximate positions of the constructs used is shown to the left (see methods for exact location of these constructs). Transcription factor predicted binding sites within the conserved blocks of DNA were determined using the TESS website (http://www.cbil.upenn.edu/cgi-bin/tess). Blue, Seven-up (Svp) binding sites; gray, Hb binding sites; yellow, Pou domain binding sites, including Nubbin/Pdm2; blue underlines, homeodomain core motifs (ATTA/TAAT) sites.
1.2 Identification of hb cis-regulatory DNA conferring proper temporal expression in NBs
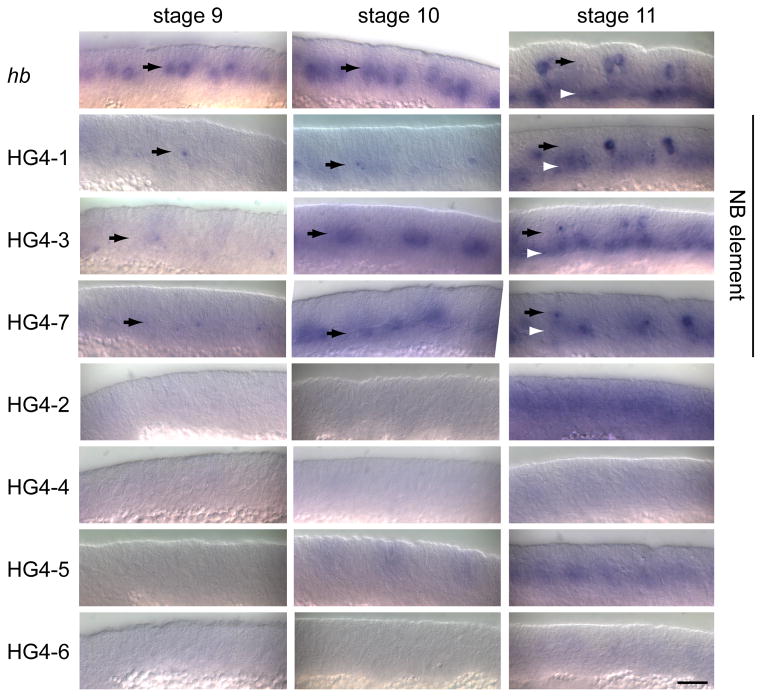
We performed RNA in situ hybridization with an antisense GFP probe to whole mount embryos expressing each of the hb-GFP reporter transgenes (Fig. 3). The endogenous hb gene (detected with a hb-specific probe) shows maximal expression in NBs during stages 9 and 10 (Fig 3, NB layer marked with black arrows). Importantly, expression in the neuroblast layer is lost by stage 11 (Fig. 3, black arrows), although there is clear expression in early-born GMCs at this stage (Fig. 3, white arrowheads). Using a GFP probe, we found that the HG4-1, HG4-3, and HG4-7 transgenes are all expressed in a pattern similar to endogenous hb: expression in NBs at stage 9 and stage 10, followed by early-born GMC expression at stage 11). Although these transgenes may have been expressed at lower levels than endogenous hb (or the probes were less effective), it is important to note that all three transgenes showed the proper temporal pattern. The remaining HG transgenes (HG4-2, HG4-4, HG4-5, HG4-6) did not show NB expression (Fig. 3).
Fig. 3.
Expression of the endogenous hunchback gene (hb) and enhancer-reporter transgenes (HG series) in neuroblasts and GMCs. Drosophila embryonic NBs first delaminate at early stage 9 and express hb at the time of their delamination; they maintain hb for about an hour until stage 10. In addition, some NBs first delaminate at stage 10 or stage 11, and they also typically express hb for about an hour after delaminating, so that there are scattered Hb+ NBs persisting into stage 11. Nevertheless, the bulk of hb expression occurs at stage 9 and 10. RNA in situ hybridization shown for the indicated genotypes. Black arrows indicate the neuroblast layer in stage 9–11 embryos; white arrowheads indicate the GMC layer in the stage 11 embryos. Only lines HG4-1, HG4-3, and HG4-7 show neuroblast expression (“neuroblast element” label, to right). Scale bar, 20 μm.
We do not show ventral views of the transgene RNA patterns in figure 3 because the expression of endogenous hb, and the transgenes, is transient in neuroblasts (at the time they form), and neuroblasts form at different times from the earliest at stage early 9 (e.g. 7–1) to the latest at late stage 11 (e.g. 7–3). Thus, the RNA in situ’s will never show the full complement of neuroblasts at a single time point. Instead, to determine if the three HG transgenes with proper temporal expression also had proper spatial expression within the NB population, we stained for GFP and the neuroblast marker Deadpan (Dpn) and imaged the field of NBs (Fig. 4). Because GFP has a relatively long half-life, this experiment allowed us to “sum up” the expression of the HG transgenes during stages 9 and 10 to determine if they were properly expressed in the majority of NBs in each segment. We stained for GFP, the neuroblast marker Deadpan, and the positional marker Engrailed (not shown; to identify segment boundaries and embryo staging). We found that the HG4-1, HG4-3, and HG4-7 transgenes each expressed in >90% of NBs per segment (Fig. 4), similar to endogenous Hb protein (Isshiki et al., 2001). We conclude that the nested DNA elements contained in HG4-1, HG4-3, and HG4-7 all possess the cis-regulatory DNA necessary for proper temporal expression in newly-formed NBs; this maps the element to the smallest transgene DNA, HG4-7, which contains 961 base pairs. We conclude that a 961 nucleotide cis-regulatory DNA element is sufficient to generate the hb temporal expression pattern within embryonic NBs.
Fig. 4.
Expression of the endogenous hunchback gene (hb) and enhancer-reporter transgenes (HG series) in stage 11 neuroblasts. Confocal imaging of embryos stained for GFP (top row) and the neuroblast marker Deadpan (Dpn, bottom row). Lines HG4-1, HG4-3, and HG4-7 show expression in most or all neuroblasts (“neuroblast element” label, at bottom). Two segments are shown, determined by Engrailed staining (not shown). Scale bar, 10 μm.
Interestingly, this region of DNA contains six predicted Svp binding sites (AGGTCA or its complement, with one mismatch allowed; Fig. 2, blue highlighting), including one pair spaced 10 nucleotides apart that is predicted to allow Svp homo- or hetero-dimer binding (Zelhof et al., 1995); this site is not detected in any other conserved sequence blocks within the 4kb HZ4 region. In addition to the predicted Svp binding sites, which may be important for limiting Hb expression, there are other blocks of conserved sequence within the HG4-7 DNA that may represent binding sites for direct or indirect Dan-mediated repression of hb transcription (Kohwi et al., 2011), or binding sites for currently unknown transcriptional activators of hb expression in NBs. Mutation of individual small blocks of conserved sequence would be necessary to test these hypotheses; sites found to be functionally important would be excellent candidates for biochemical purification and identification of trans-acting factors that regulate the timing of hb expression in neuroblasts.
1.3 Identification of hb cis-regulatory DNA conferring proper expression in early-born neurons
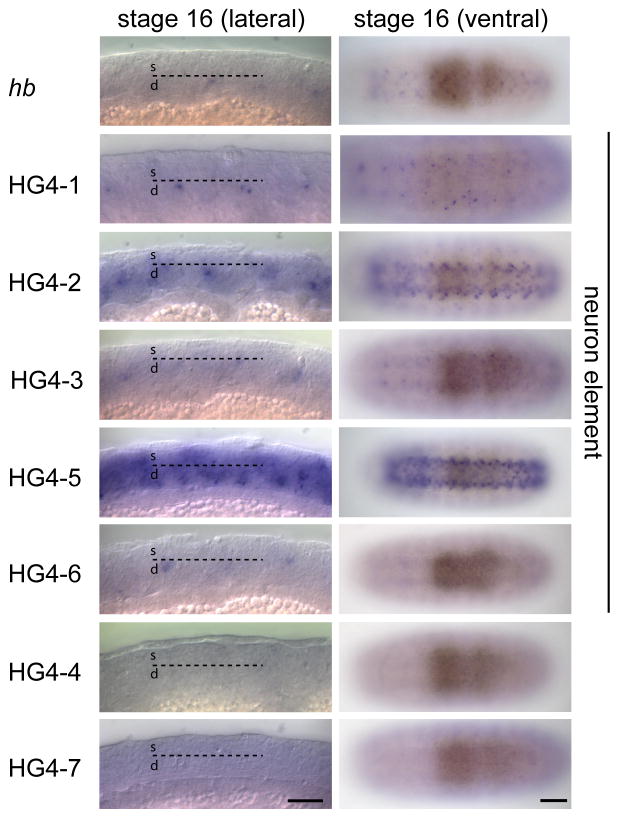
We next examined embryos at stage 16 to determine the expression pattern of each transgene in post-mitotic neurons. At this stage, early-born neurons are located in the deepest layer of the CNS, whereas later-born neurons occupy a more superficial position (Isshiki et al., 2001). The endogenous hb gene is weakly detected in deep layer neurons in lateral views (Fig. 5, top row, black arrows indicate deep layer of CNS). Using an antisense GFP probe, we found that the HG4-1, HG4-2, HG4- 3, and HG4-6 transgenes are all expressed in deep layer neurons (Fig. 5); in addition HG4-5 appears to be expressed in deep layer neurons and strongly expressed in superficial (late-born) neurons (Fig. 5, white arrowhead represents superficial layer of the CNS). The transgenes HG4-4 and HG4-7 show little or no RNA expression in the stage 16 CNS (Fig. 5). Lower magnification ventral views of stage 16 embryos show the relative level of RNA in the CNS generated by each transgene (Fig. 5, right column, bracket indicates CNS). The observation of neuronal expression for the HG4-1 construct was somewhat surprising, as the HZ4 transgene did not show neuronal expression (data not shown). This could be due to position effects acting on the HZ4 transgene (which was randomly inserted into the genome, as opposed to placed at same attP genomic site like all the HG transgenes), or due to the lack of regulatory sequences mapping to the hb 5′ or 3′ UTR (which were not present in the HZ4 construct).
Fig. 5.
Expression of the endogenous hunchback gene (hb) and enhancer-reporter transgenes (HG series) in neurons of stage 16 embryos, shown in lateral (left) or ventral (right) views. RNA in situ hybridization shown for the indicated genotypes. In the left column, deep neurons (d) are located below the dashed line and superficial neurons (s) are located above the dashed line; examples of deep neurons are shown by black arrows; examples of superficial neurons are shown by white arrowheads; brackets indicate the CNS (right column). Lines HG4-1, HG4-2, HG4-3, HG4-5 and HG4-6 show neuronal expression (“neuron element” label, to right). Scale bar, 20 μm (left column) and 50 μm (right column).
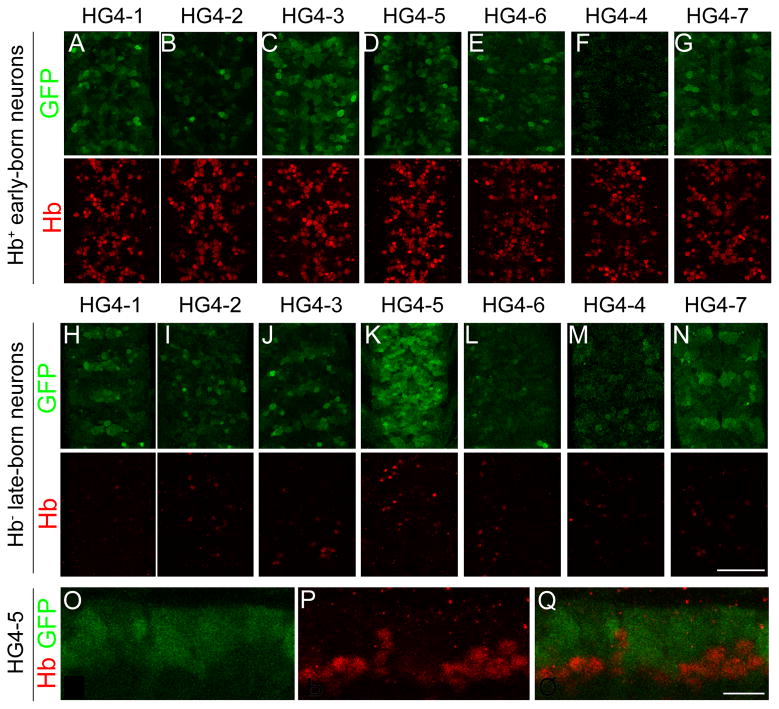
To determine the expression pattern of each transgene within early-born and late-born neurons of the CNS at higher resolution, we double stained each of the transgenic lines for GFP and Hb protein (a marker for deep-layer, early-born neurons). We found that HG4-1, HG4-2, HG4-3, and HG4-6 transgenes all showed GFP staining in deep layer neurons (Fig. 6A-G), although it was not a perfect 1-to-1 register with the Hb+ deep layer neurons. Some GFP+ neurons were in Hb-negative neurons, whereas some Hb+ neurons lacked GFP. For example, line HG4-6 had Hb+ neurons that were 19% strong GFP, 37% weak GFP, 44% no GFP (n=328 neurons from two embryos); the lack of GFP in some Hb+ neurons is most likely due to lack of full cis-regulatory information in the transgene, but we can’t rule out low levels of GFP being present. In addition, HG4-5 showed strong GFP staining in Hb-negative superficial neurons (Fig. 6K), whereas the other transgenes were weakly or not detected in superficial neurons (Fig. 6H-J, L-N); see next section. Unexpectedly, we noticed HG4-7 exhibited a clear GFP protein signal within the CNS (Fig. 6G, N) despite lack of detectable GFP RNA at this stage (Fig. 5). The GFP protein may be due to persistence of GFP from neuroblasts and GMCs into the neurons. We conclude that the smallest DNA element that drives early-born neuron expression is the 866 base pair HG4-6 element, and that all larger elements containing this fragment also show early-born neuron expression.
Fig. 6.
Expression of the endogenous hunchback gene (hb) and enhancer-reporter transgenes (HG series) in the stage 16 CNS. Confocal images of the CNS stained for HG reporter construct patterns (GFP, green) and endogenous Hb protein (red). (A–G) Ventral view of deep layer, early-born, Hb+ neurons. Lines HG4-1, HG4-2, HG4-3, HG4-5 and HG4-6 show expression in these Hb+ neurons. (H–N) Ventral view of superficial layer, late-born, Hb-negative neurons. Only line HG4-5 shows strong expression in these Hb-negative neurons; other lines show scattered expression (HG4-1, HG4- 2, HG4-3) or high background due to increasing the confocal gain to try and detect expression (HG4- 4, HG4-6, HG4-7). For A–N, two segments are shown, determined by Engrailed staining in the fourth confocal channel (not shown). (O–Q) Lateral view of a stage 16 CNS showing HG4-5 expression in superficial neurons (O, Q; green GFP staining) and almost no expression in Hb+ deep layer neurons (P, Q; red endogenous Hb staining). Scale bar is 50 μm in A–N, and 10 μm in O–Q.
It is currently unknown how hb expression is maintained in the GMCs and neurons born from Hb+ young neuroblasts. We can rule out simple positive autoregulation because misexpression of Hb in neurons does not activate hb transcription (Grosskortenhaus et al., 2005). The presence of several blocks of highly conserved sequence within the 866 base pair HG4-6 element would be excellent starting points for identifying the transcriptional activator(s) required to maintain Hb expression in early-born neurons.
1.4 Identification of a cis-regulatory DNA element conferring expression in late-born neurons
The HG4-5 showed strong reporter expression in Hb-negative superficial neurons (Fig. 6K), which was surprising because Hb is not detected in these neurons at any embryonic stage (Isshiki et al., 2001; Kambadur et al., 1998). None of the other transgenes were strongly expressed in these neurons (Fig. 6H-J, L-N), although we detected weak “stripy” GFP reporter expression in the superficial layer neurons in line HG4-7 (Fig. 6N). To test whether HG4-5 was preferentially expressed in late-born neurons, we assayed lateral confocal sections through the stage 16 CNS double stained for GFP and Hb protein. We observed a virtually complementary pattern of GFP and Hb expression (Fig. 6O-Q). This shows that the DNA contained in the HG4-5 transgene contains an enhancer that drives expression in late-born neurons. Interestingly, the HG4-2 construct contains the HG4-5 fragment but does not have similar strong expression in late-born neurons (compare Fig. 6I with 6K); this shows that HG4-2 must contain a late-born neuron silencer element. We do not know whether the late-born neuron enhancer found in HG4-5 is normally silenced by adjacent regulatory elements during wild type development, or if it drives expression of a flanking gene such as CG8112, whose promoter is closer to the HG4-5 element than the hb promoter (CG8112 is transcribed in the opposite direction as hb, and its predicted promoter lies between the HG4-5 element and the hb promoters; i.e. the HG4-5 element is in the first intron of CG8112). Nevertheless, the serendipitous identification of a late-neuron enhancer will be useful as a tool (e.g. making a Gal4 line with late-born neuron expression); future experiments will be necessary to determine its relevance to hb or CG8112 transcription.
HG4-5 is only 451 base pairs long, and contains just two blocks of conserved sequence. Interestingly, there are eight predicted Hb binding sites within these small blocks of conserved sequence, raising the possibility that Hb acts as a transcriptional auto-repressor to prevent the function of this enhancer fragment within early-born neurons (e.g. to keep the adjacent CG8112 gene off in early-born neurons). In this case, we might expect the remaining small region of conserved sequence to contain binding site(s) for a pan-neuronal transcriptional activator.
2. Experimental procedures
2.1 Enhancer-reporter transgenic lines and other stocks
The HZ4 construct was made as described previously (Margolis et al., 1994; Margolis et al., 1995); briefly, it contains the DNA from Flybase position 4527893 (XbaI site) to 4532199 (PstI site) – coordinates shown in Fig. 1A – attached to a basal hsp70 promoter directly upstream of the lacZ coding sequences. Unlike the HG constructs described below, there is no hb 5′ or 3′ UTR sequences in the HZ4 construct; furthermore HZ4 was randomly inserted into the genome. The HG-1 contstruct contains precisely the same DNA as the HZ4 construct, but driving a different reporter (GFP instead of lacZ). All HG contructs are summarized in Fig. 1A (different cis-regulatory fragments) and Fig. 1B (core construct that is common to all HG transgenes). To make the core HG construct, hb 5′UTR was amplified from a BAC clone, BACR01F13 using forward primer 5′CAGTTGTGCTCCGAGTCC and reverse primer 5′CTCGCCCTTGCTCACCATCTTGGCGGCTCTAGACGG, which contained the most 5′ region of the GFP coding sequence. The hb 3′UTR was amplified from BACR01F13 using forward primer 5′GCATGGACGAGCTGTACAAGTAAGTTCCCCATCACCATCACCTTG (which contained the most 3′ region of the GFP coding sequence) and reverse primer 5′TATATTGAATAATTGGATTTATTTGATTTGATTTCGTTC. We inserted the GFP coding sequence between the hb 5′ UTR and 3′ UTR utilizing forward primer 5′CCGTCTAGAGCCGCCAAGATGGTGAGCAAGGGCGAG, which contained hb 3′ 5′UTR sequence and reverse primer 5′CAAGGTGATGGTGATGGGGAACTTACTTGTACAGCTCGTCCATGC, which contained hb 5′ 3′UTR sequence. This core hb-gfp gene was cloned into SpeI-BamHI site of a modified pCaSpeR4 vector by inserting a basal heatshock promoter (hsp70 TATA box) at NotI-SpeI site, SV40 termination signal at XbaI-HpaI site and attB site at XhoI site to make the core HG vector shown in Fig. 1B.
Using EvoPrinterHD (Odenwald et al., 2005; Yavatkar et al., 2008), we found 4 regions evolutionally conserved in Drosophila melanogaster plus all 11 other Drosophila species, which was used to design fragments; a slightly more relaxed EvoPrint of D. melanogaster and 10/11 other species is shown (Fig. 2). Enhancer fragments were amplified from BACR01F13 using primers described in Supplemental Table 1 and cloned into KpnI-NotI site of HG vector. All transgenes were inserted into the same attP1 docking site on chromosome 2 by GenetiVision (Houston, TX). yellow white was used as wild type.
2.2 RNA in situ hybridization to embryos
Embryo whole-mount in situ RNA hybridization was carried out as previously published (Tautz and Pfeifle, 1989). The hb RNA probe was generated from BACR01F13 using forward primer 5′GTTCCCCATCACCATCACCTTG and reverse primer 5′CGTTCGATTCGAATTCGCTTTC with a T7 promoter sequence. For the GFP RNA probe, we used forward primer 5′CTTCTTCAAGTCCGCCATGC and reverse primer 5′AACTCCAGCAGGACCATGTG with a T7 promoter sequence. DIG Labeling Kit (Roche Diagnostics, Indianapolis, IN) was used for transcription and digoxygenin-labeling. Images were photographed by Zeiss AxioCam HRc camera on an Axioplan microscope DIC/Nomarski optics.
2.4 Immunofluorescence and DIC
Antibody staining was performed according to standard methods (Kohwi et al., 2011). Primary antibodies, dilutions and sources were: chicken anti-GFP 1:500 (Aves Labs, Inc., Tigard, OR USA); rabbit anti-Hb 1:200 (Tran and Doe, 2008); mouse anti-Even-skipped (3C10-c) and mouse anti-Engrailed (4D9) (Developmental Studies Hybridoma Bank, University of Iowa, IA, USA); rat anti-Dpn (Doe lab). Donkey anti-chicken Dylight 488- and donkey anti-mouse Dylight 649- conjugated secondary antibodies were from Jackson ImmunoResearch (West Grove, PA USA). Goat anti-rabbit Alexa Fluor 555- and Goat anti-rat Alexa Fluor 555-conjugated secondary antibodies were from Invitrogen (Eugene, OR USA). Confocal image stacks were collected using Zeiss LSM 700 confocal microscope, processed using ImageJ (NIH), and assembled into figures using Illustrator and Photoshop (Adobe Systems Inc., Mountain View, CA USA).
2.4 Transcription factor binding site identification
Transcription Element Search System software (TESS; http://www.cbil.upenn.edu/cgi-bin/tess) was used to identify predicted transcription factor binding sites within the conserved evoprinted regions shown in Figure 2. The Svp sites were annotated as CF1/USP sites, but these are equivalent to Svp sites (Zelhof et al., 1995). We only indicate high confidence sites; we excluded low information content sites under 5 base pairs. Where there were overlapping sites for a single factor, the site with the highest TESS score was used.
3. Conclusions
We conclude that the pattern of hb expression in the CNS can be recapitulated in large part by the cis-regulatory DNA contained in the ~4kb HG4-1 construct. Within this stretch of DNA are contained smaller elements: a 961 bp element that confers proper hb expression in newly-formed NBs (HG4-7), a 866 bp element that confers proper hb expression in early-born neurons (HG4-6), and an unexpected 451 bp element that confers expression in hb -negative late-born neurons (HG4-5); summarized in Fig. 7. The NB element contains predicted binding sites for a known trans-acting factor that regulates hb NB expression (Svp) and several other conserved regions that may represent transcriptional activator binding sites. The results of our study will provide a foundation for future functional analysis of this temporal cis-regulatory DNA, and the identification of trans-acting factors that control the precise timing of hb expression in the CNS.
Fig. 7.
Summary of the cis-regulatory elements controlling hunchback (hb) CNS expression. Grey numbers at top are the Flybase coordinates for the region. Lines shown at right indicate the DNA elements used in the reporter constructs. Color code for the four different regulatory functions discovered in this work are shown in the box, lower left.
Supplementary Material
Highlights.
Temporal identity within progenitor lineages is important but understudied
Hunchback transcription factor specifies first-born temporal identity in Drosophila and mammalian CNS
Precise temporal regulation of hunchback transcription is essential for CNS development
We have identified cis-regulatory DNA controlling hunchback temporal expression
Acknowledgments
We thank Minoree Kohwi for comments on the manuscript. This work was funded by an NIH grant GM41100 to J.W.P. and NIH grant HD27056 to C.Q.D., who is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Brody T, Odenwald WF. Programmed transformations in neuroblast gene expression during Drosophila CNS lineage development. Dev Biol. 2000;226:34–44. doi: 10.1006/dbio.2000.9829. [DOI] [PubMed] [Google Scholar]
- Cleary MD, Doe CQ. Regulation of neuroblast competence: multiple temporal identity factors specify distinct neuronal fates within a single early competence window. Genes Dev. 2006;20:429–434. doi: 10.1101/gad.1382206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe CQ. Neural stem cells: balancing self-renewal with differentiation. Development. 2008;135:1575–1587. doi: 10.1242/dev.014977. [DOI] [PubMed] [Google Scholar]
- Grosskortenhaus R, Pearson BJ, Marusich A, Doe CQ. Regulation of temporal identity transitions in Drosophila neuroblasts. Dev Cell. 2005;8:193–202. doi: 10.1016/j.devcel.2004.11.019. [DOI] [PubMed] [Google Scholar]
- Isshiki T, Pearson B, Holbrook S, Doe CQ. Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell. 2001;106:511–521. doi: 10.1016/s0092-8674(01)00465-2. [DOI] [PubMed] [Google Scholar]
- Kambadur R, Koizumi K, Stivers C, Nagle J, Poole SJ, Odenwald WF. Regulation of POU genes by castor and hunchback establishes layered compartments in the Drosophila CNS. Genes Dev. 1998;12:246–260. doi: 10.1101/gad.12.2.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai MI, Okabe M, Hiromi Y. seven-up Controls switching of transcription factors that specify temporal identities of Drosophila neuroblasts. Developmental cell. 2005;8:203–213. doi: 10.1016/j.devcel.2004.12.014. [DOI] [PubMed] [Google Scholar]
- Kohwi M, Hiebert LS, Doe CQ. The pipsqueak-domain proteins Distal antenna and Distal antenna-related restrict Hunchback neuroblast expression and early-born neuronal identity. Development. 2011;138:1727–1735. doi: 10.1242/dev.061499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis JS, Borowsky M, Shim CW, Posakony JW. A small region surrounding the distal promoter of the hunchback gene directs maternal expression. Developmental biology. 1994;163:381–388. doi: 10.1006/dbio.1994.1156. [DOI] [PubMed] [Google Scholar]
- Margolis JS, Borowsky ML, Steingrimsson E, Shim CW, Lengyel JA, Posakony JW. Posterior stripe expression of hunchback is driven from two promoters by a common enhancer element. Development. 1995;121:3067–3077. doi: 10.1242/dev.121.9.3067. [DOI] [PubMed] [Google Scholar]
- Mettler U, Vogler G, Urban J. Timing of identity: spatiotemporal regulation of hunchback in neuroblast lineages of Drosophila by Seven-up and Prospero. Development. 2006;133:429–437. doi: 10.1242/dev.02229. [DOI] [PubMed] [Google Scholar]
- Novotny T, Eiselt R, Urban J. Hunchback is required for the specification of the early sublineage of neuroblast 7-3 in the Drosophila central nervous system. Development. 2002;129:1027–1036. doi: 10.1242/dev.129.4.1027. [DOI] [PubMed] [Google Scholar]
- Odenwald WF, Rasband W, Kuzin A, Brody T. EVOPRINTER, a multigenomic comparative tool for rapid identification of functionally important DNA. Proc Natl Acad Sci U S A. 2005;102:14700–14705. doi: 10.1073/pnas.0506915102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson BJ, Doe CQ. Regulation of neuroblast competence in Drosophila. Nature. 2003;425:624–628. doi: 10.1038/nature01910. [DOI] [PubMed] [Google Scholar]
- Pearson BJ, Doe CQ. Specification of temporal identity in the developing nervous system. Annu Rev Cell Dev Biol. 2004;20:619–647. doi: 10.1146/annurev.cellbio.19.111301.115142. [DOI] [PubMed] [Google Scholar]
- Spana EP, Doe CQ. The prospero transcription factor is asymmetrically localized to the cell cortex during neuroblast mitosis in Drosophila. Development. 1995;121:3187–3195. doi: 10.1242/dev.121.10.3187. [DOI] [PubMed] [Google Scholar]
- Tautz D, Pfeifle C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- Tran KD, Doe CQ. Pdm and Castor close successive temporal identity windows in the NB3-1 lineage. Development. 2008;135:3491–3499. doi: 10.1242/dev.024349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran KD, Miller MR, Doe CQ. Recombineering Hunchback identifies two conserved domains required to maintain neuroblast competence and specify early-born neuronal identity. Development. 2010;137:1421–1430. doi: 10.1242/dev.048678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yavatkar AS, Lin Y, Ross J, Fann Y, Brody T, Odenwald WF. Rapid detection and curation of conserved DNA via enhanced-BLAT and EvoPrinterHD analysis. BMC Genomics. 2008;9:106. doi: 10.1186/1471-2164-9-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelhof AC, Yao TP, Chen JD, Evans RM, McKeown M. Seven-up inhibits ultraspiracle-based signaling pathways in vitro and in vivo. Molecular and cellular biology. 1995;15:6736–6745. doi: 10.1128/mcb.15.12.6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.