Abstract
Organs from nonheart-beating donors are attractive for use in cell therapy. Understanding the nature of molecular perturbations following reperfusion/reoxygenation will be highly significant for nonheart-beating donor cells. We studied nonheart-beating donor rats for global gene expression with Affymetrix microarrays, hepatic tissue integrity, viability of isolated hepatocytes, and engraftment and proliferation of transplanted cells in dipeptidyl peptidase IV-deficient rats. In nonheart-beating donors, liver tissue was morphologically intact for >24 hours with differential expression of 1, 95, or 372 genes, 4, 16 or 34 hours after death, respectively, compared with heart-beating donors. These differentially-expressed genes constituted prominent groupings in ontological pathways of oxidative phosphorylation, adherence junctions, glycolysis/gluconeogenesis, as well as other discrete pathways. We successfully isolated viable hepatocytes from nonheart-beating donors, especially up to 4 hours after death, although the hepatocyte yield and viability were inferior to hepatocytes from heart-beating donors, p<0.05. Similarly, although hepatocytes from nonheart-beating donors engrafted and proliferated after transplantation in recipient animals, this was inferior to hepatocytes from heart-beating donors, p<0.05. Gene expression profiling in hepatocytes isolated from nonheart-beating donors showed far greater perturbations compared with corresponding liver tissue, including representation of pathways in focal adhesion, actin cytoskeleton, extracellular matrix-receptor interactions, multiple ligand-receptor interactions, and signaling in insulin, calcium, wnt, Jak-Stat, or other cascades. Conclusion: Liver tissue remained intact over prolonged periods after death in nonheart-beating donors but extensive molecular perturbations following reperfusion/reoxygenation impaired viability of isolated hepatocytes from these donors. Insights into molecular changes in hepatocytes from nonheart-beating donors offers opportunities for improving donor cell viability, which will advance utility of nonheart-beating donor organs for cell therapy or other applications.
Keywords: Cell therapy, gene expression, hepatocytes, liver, nonheart-beating donor
Early experiences of liver cell therapy have been promising (1). However, worsening shortages of donor organs for isolating cells is a major hurdle. Therefore, alternative sources of donor cells will be highly significant, although current alternatives are unsatisfactory. For instance, availability of surgically-resected healthy liver specimens is sporadic, such specimens are generally small, and specimens adjacent to tumors may harbor malignant cells. Living-related organ donation solely for isolating cells poses risks to the donor and seems premature because efficacy of cell therapy needs to be proven by further work. Similarly, the ability to generate hepatocytes by differentiating embryonic, fetal or adult stem cells is in its infancy. Whether extrahepatic cells, i.e., hematopoietic, mesenchymal, or other cells, could replace liver tissue is controversial. By contrast, nonheart-beating (NHB) cadaveric donors offer a large and widely available resource. Studies of cadaveric fetal livers indicated that highly-viable hepatocytes could be isolated many hours after death and tissue anoxia (2). These fetal cells expanded extensively in culture and could be cryopreserved multiple times. This differed from adult tissues and mature hepatocytes, which are much less anoxia-tolerant. While resistance to anoxia is not an intrinsic biological property of mammalian cells, as some animal species do resist anoxia (3), this focused attention on molecular mechanisms in tissue viability, e.g., brain, after hypoxia or anoxia (4). On the other hand, molecular changes in liver after anoxia, including hepatocytes from NHB donors, have not been determined (5-7).
To understand the nature of molecular perturbations in NHB donor liver, including isolated hepatocytes from such livers in comparison with heart-beating (HB) donor liver, we studied Fischer 344 rats. The fate of transplanted cells was determined in dipeptidyl peptidase IV-deficient (DPPIV-) rats, where syngeneic DPPIV+ cells are readily identified (8). Preconditioning of recipient rats by retrorsine, a genotoxic alkaloid, plus two-thirds partial hepatectomy (PH) induces transplanted cell proliferation to permit analysis of liver repopulation (9). These systems provided insights into molecular changes in NHB donor livers, as well as the cell therapy potential of NHB donor livers.
Experimental Procedures
Animals
F334 rats were 10 to 12-weeks old (National Cancer Institute, Bethesda, MD). Syngeneic DPPIV- F334 rats were 6 to 8-weeks old and were from Special Animal Core of Marion Bessin Liver Research Center. Rats were housed in Institute for Animal Studies under 14/10 hour light/dark cycles with unrestricted food and water. The Animal Care and Use Committee at Albert Einstein College of Medicine approved animal protocols in conformity with NIH requirements.
For NHB donor rats, midline laparotomy was performed under anesthesia, 20-gauge cannula was placed in portal vein, and filled with 0.2 millilters of normal saline containing 200 units of heparin (NDC 63323-540-31; American Pharmaceutical Partners Inc., Schaumburg, IL). Immediately afterwards, 0.2 milliliter 15% potassium chloride (KCl) was injected into inferior vena cava. When cells were not to be isolated, KCl was injected into tail vein or heart for cardiac arrest. Cadavers were stored at 4°C. HB donor rats were controls.
Hepatocyte isolation
Chemicals were from Sigma Chemical Co. (St. Louis, MO). Collagenase was from Worthington Corp. (Lakewood, NJ). Hepatocytes were isolated by two-step collagenase perfusion (10), except glucose was substituted with 20 mM fructose (F0127, Sigma), according to Anundi et al (11).
Liver repopulation assay
At 6 and 8-weeks of age, DPPIV- rats received 30 mg/kg retrorsine intraperitoneally (i.p.), and 4 weeks later, PH was performed according to Higgins and Anderson method. Cells were transplanted 7d after PH. Liver repopulation analysis was done 3 weeks after cell transplantation.
Cell viability
Cells were manually counted in Neubaur chamber with exclusion of 0.2% trypan blue dye. For attachment to culture dishes, 3×104 cells/cm2 were plated in Roswell Park Memorial Institute (RPMI) 1640 medium with serum and antibiotics. For apoptosis, DNA was extracted (DNeasy Blood & Tissue Kit, Qiagen Inc., Valencia, CA), and resolved in 1% agarose gels with ethidium bromide.
Metabolic capacity of cells
Cells cultured overnight in dishes at 3×104 cells/cm2 were incubated with 5 mM ammonium chloride in RPMI 1640 medium for 2h at 37°C. To 100 μL aliquots of medium, we added 0.3 milliliter urease buffer reagent (U-3383, Sigma) for 20 min at room temperature. This was followed by 0.6 milliliter phenol nitroprusside (P-6994, Sigma), 0.6 milliliter alkaline hypochlorite (A-1727, Sigma), and 3.0 milliliter water. The mixture was incubated for 30 min at room temperature. Absorbance was read at 540 nm. Standard curves were made with 10 mg/ml urea (U-5128, Sigma). The data were corrected for the number of cells attached in dishes, which was measured manually in a Neubaur chamber, after medium was harvested.
Cell transplantation
For cell engraftment analysis, 1×107 cells in 0.5 ml serum-free RPMI 1640 were injected as a bolus into spleen isolated by subcostal laparotomy. In rats conditioned with retrorsine and PH, 0.5×107 cells were injected in 0.25 milliliter RPMI 1640 medium via spleen.
Tissue studies
To examine tissue integrity under light microscopy, liver sections from HB and NHB donors were stained by hematoxylin and eosin. For apoptosis, terminal deoxynucleotidyl transferase biotin-dUTP nick end-labeling (TUNEL) was performed (ApopTag peroxidase in situ apoptosis kit; Millipore, Billerica, MA). In negative controls, terminal deoxynucleotidyl transferase was omitted. Color was developed with diaminobenzidine (DAB) (K3467; Dako North America Inc., Carpinteria, CA).
Global gene expression
Total cellular RNA was extracted (RNEasy Mini Kit, Qiagen Inc., Valencia, CA), and hybridized with Rat Genome 230 2.0 arrays (Affymetrix Inc., Santa Clara, CA), according to manufacturer. Tissue samples were from HB donors immediately after death (n=3), and from NHB donors 4, 16 and 30h after death (n=3 each, total 12 arrays). Freshly isolated hepatocytes were from HB donors or NHB donors 4 and 16h after death (n=3 each, total 9 arrays).
For data analysis, microarray background was corrected at probe levels by robust multi-array normalization followed by quintiles method for normalizing perfect-match probe intensities. Normalized raw probe signal values were log2 transformed (12). Ratios of intergroup and intragroup sums of squares (BW ratio) were computed per probe (13). Datasets were segregated for analysis with p-values ≤0.01, while controlling false discovery rates to <1% and fold-differences in mean expression of ≥|2.0|. Gene lists of interest were obtained from venn diagrams (SAS software, SAS Institute Inc., Cary, NC). Genes were annotated and grouped by Database for Annotation, Visualization and Integrated Discovery (DAVID) (14,15). Gene pathways were mapped according to Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.genome.jp/kegg/pathway.html).
Cell engraftment and liver repopulation analysis
Liver samples were frozen to −80°C in methylbutane. Cryosections of 5 μm were fixed in cold chloroform-acetone for DPPIV staining, as previously described (8). For morphometry, 3 sections per liver lobe were studied per rat with 100 consecutive fields centered on portal areas under ×100 magnification. For liver repopulation, multiple microphotographs were obtained under ×40 magnification by Spot RT digital camera (Diagnostic Instrument Inc., Sterling Heights, MI), and transplanted cell areas were measured by ImageJ (National Cancer Institute, Bethesda, MD).
Experimental design
For tissue integrity, NHB donor livers were studied 15 min, and 2, 4, 6, 8, 10, 16, 24, 30, and 40h after death compared with HB donors (n=3-4 ea, total 40). Samples were collected for global gene expression analysis (n=3 ea). Hepatocytes were isolated for cell viability studies (n=3-6 each). Gene expression was profiled in isolated hepatocytes (n=3 per condition). We determined engraftment capacity of hepatocytes from HB and NHB donors (n=3-4 per condition). For cell engraftment, hepatocytes from each donor were transplanted into DPPIV- rats (n=3-4 each, total 50), and animals were killed 7d later. For liver repopulation, cells were transplanted from NHB (n=4) or HB donors (n=3) into DPPIV- rats preconditioned with retrorsine and PH (n=3-6 per donor, total 40). These animals were killed after 3 weeks.
Statistical analysis
Data are shown as means±SD. Analyses included t-tests, Mann Whitney tests or ANOVA with Dunn's test by SigmaStat 3.1 software (SysStat Software, Point Richmond, CA). P <.05 was considered significant.
Results
Integrity of NHB liver tissue
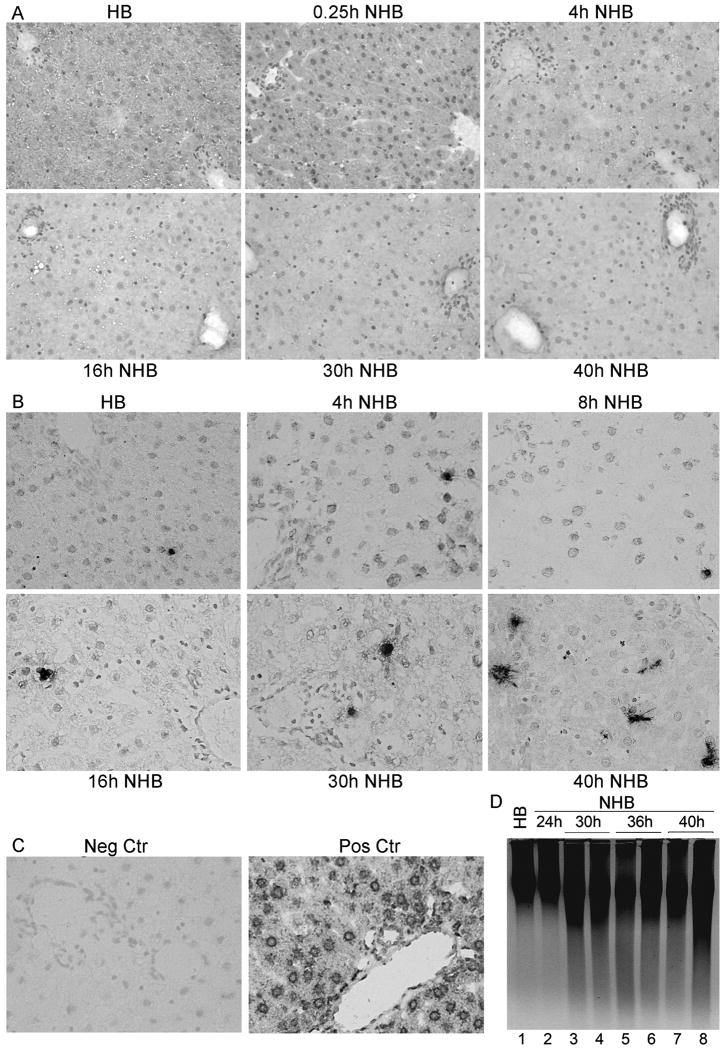
Liver was morphologically intact despite several hours after death in NHB donors, including after 15 min, and 2, 4, 6, 8, 10, 16, 24, 30, or 40h (Fig. 1A). Hepatic necrosis or inflammatory infiltrates were absent. Hepatocytes and bile duct cells appeared unremarkable. This was similar to hepatic morphology in HB donors. TUNEL showed limited apoptosis (Fig. 1B, 1C). Only 0-1 apoptotic cells were found per section under ×200 magnification, up to 24h after death, with slightly more apoptosis after 30h and 40h in NHB donor livers, although still only 2-3 or 6-8 TUNEL+ cells were found per section, respectively. DNA laddering confirmed limited apoptosis in NHB donor livers (Fig. 1D).
Figure 1. Integrity of liver in NHB donors.
(A) Hematoxylin and eosin-stained sections from HB and NHB donors after death, as indicated, showing tissues were intact without necrosis or autolysis. (B) TUNEL+ cells were infrequent in HB and NHB donor livers. (C) Negative control liver without TUNEL and DNAse-treated control liver with extensive TUNEL. (D) DNA laddering showing little hepatic apoptosis over time. Orig. Mag., A and B, ×400; B, methylgreen counterstain.
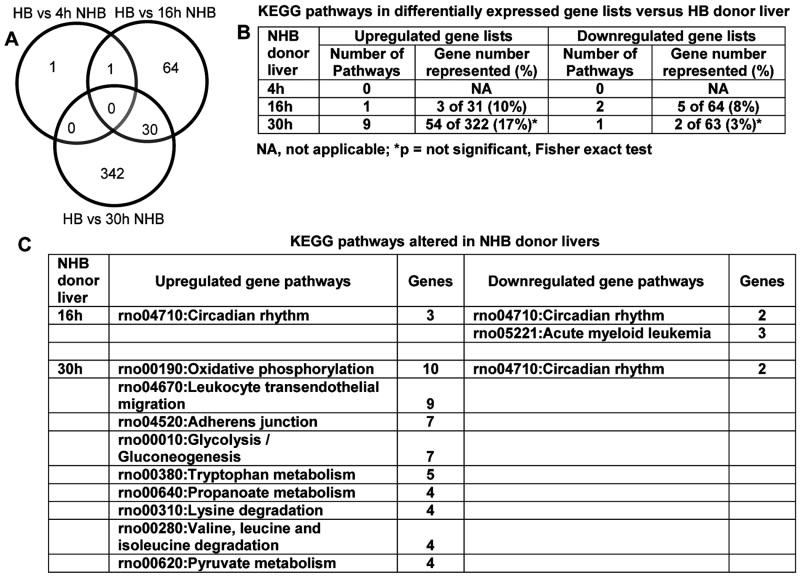
These restricted morphological changes in NHB liver were reflected by gene expression profiles (Fig. 2A). Remarkably, only one gene was differentially expressed in NHB livers 4h after death: downregulation of lipid synthesis regulator, stearoyl-coenzyme A desaturase 2. By contrast, gene expression changed more in NHB donor livers 16h and 30h after death, with differential expression, either up or down versus HB livers, of 95 and 372 genes, respectively. These genes were clustered in relatively few curated KEGG pathways (Fig. 2B). Further study indicated perturbations in discrete pathways, including oxidative phosphorylation, leukocyte migration, cell integrity (adherens junctions), intermediary metabolism, or circadian rhythm (Fig. 2C).
Figure 2. Gene expression profiles in HB and NHB donor livers.
(A) Differentially expressed genes in NHB livers 4, 16 and 30h after death versus HB livers. Only one gene was differentially expressed in NHB liver 4h after death. After 16h, 64 genes were differentially expressed, including the downregulated gene after 4h, and after 30h, 342 genes were differentially expressed, including 30 of 64 genes (47%) differentially expressed 16h after death in NHB livers. (B) KEGG pathways in differentially expressed gene lists, including numbers of genes in pathways. Total numbers of genes in (B) differed from (A) as the former included one or more transcripts per gene. (C) Differentially regulated pathways in NHB livers 16 or 30h after death.
Functional gene groups showed similar perturbations in NHB donor livers 16h and 30h after death (Table 1). Therefore, tissue changes in NHB donor livers after death were gradual, since 12h elapsed from differential expression of 1 gene after 4h versus 95 genes after 16h, and another 14h elapsed for differential expression of 372 genes after 30h. However, differentially-expressed gene lists in NHB donors did not include genes in apoptosis or cell death pathways, which was in agreement with tissues showing limited apoptosis.
Table 1. Representation of major functionally annotated groups in differentially expressed gene lists in NHB donor liver versus HB donor liver.
| Gene ontology groups | NHB donor liver 16h | NHB donor liver 30h | ||
|---|---|---|---|---|
| Number Upregulated genes (%) | Number Downregulated genes (%) | Number Upregulated genes (%) | Number Downregulated genes (%) | |
| Cellular process | 195 (61%) | |||
| Metabolic process | 27(43%) | 136 (48%) | ||
| Cellular metabolic process | 14 (45%) | 26 (41%) | 143 (44%) | |
| Primary metabolic process | 14 (45%) | 136 (42%) | ||
| Transport | 10 (32%) | 59 (18%) | 11 (17%) | |
| Cellular component organization and biogenesis | 10 (32%) | 55 (17%) | ||
| Localization | 10 (32%) | 68 (21%) | ||
| Biological regulation | 10 (32%) | 21 (33%) | 17 (27%) | |
| Regulation of biological process | 9 (29%) | 19 (30%) | 15 (24%) | |
| Developmental process | 8 (26%) | 13 (21%) | 12 (19%) | |
| Macromolecule metabolic process | 9 (29%) | 23 (37%) | 103 (32%) | |
| Establishment of localization | 10 (32%) | 61 (19%) | 11 (17%) | |
| Protein metabolic process | 9 (29%) | 65 (20%) | ||
| Cellular protein metabolic process | 64 (20%) | |||
| Regulation of cellular process | 9 (29%) | 16 (25%) | ||
| Regulation of cellular metabolic process | 13 (21%) | |||
| Biosynthetic process | 6 (19%) | 46 (14%) | ||
| Response to stress | 5 (16%) | 7 (11%) | 26 (8%) | 8 (13%) |
| Negative regulation of biological process | 5 (16%) | |||
| Immune system process | 5 (16%) | |||
| Secretion | 4 (13%) | |||
| Cell death | 4 (13%) | 23 (7%) | ||
| Leukocyte activation | (10%) | |||
| Multicellular organismal development | (10%) | 11 (17%) | ||
| Biopolymer metabolic process | 17 (27%) | |||
| Regulation of metabolic process | 14 (22%) | |||
| Nucleobase, nucleoside, nucleotide and nucleic acid metabolic process | 13 (21%) | |||
| Transcription, DNA-dependent | 11 (17%) | |||
| Transcription | 11 (17%) | |||
| Regulation of transcription, DNA-dependent | 11 (17%) | |||
| RNA biosynthetic process | 11 (17%) | |||
| Regulation of gene expression | 11 (17%) | |||
| RNA metabolic process | 11 (17%) | |||
| Regulation of transcription | 11 (17%) | |||
| Intracellular signaling cascade | 10 (16%) | 31 (10%) | ||
Mapping of differentially expressed genes along functional pathways, including mitochondrial oxidative phosphorylation, transendothelial leukocyte migration, adherence junctions, and glycolysis/gluconeogenesis was consistent with depletion of energy, need for glucose production, cell-cell interaction-type events, e.g., leukocyte recruitment, and cytoskeletal alterations, in NHB donor livers after death (Supplementary Figs. 1-4).
Hepatocytes from NHB donor livers showed extensive perturbations
The yield of hepatocytes from HB donor livers was 300±92 × 106 with viability of 83±2%. HB hepatocytes attached in dishes with 60-80% efficiency. Cells showed characteristic slightly-rounded and then flattened morphology over several hours.
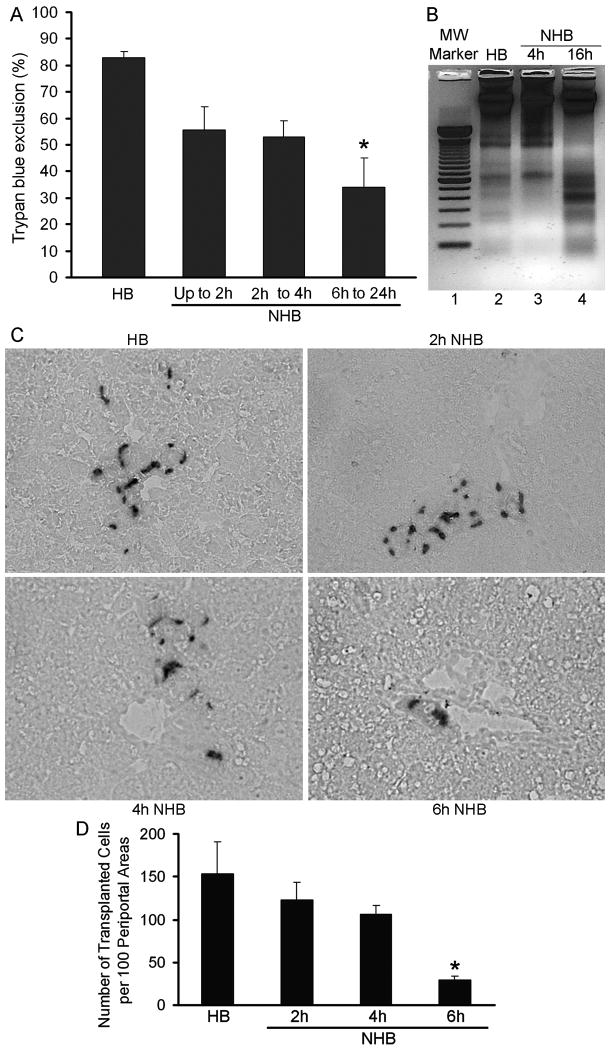
Hepatocyte yield from NHB donor livers was lower at various times after death: 15 min to 1h, 150±24 × 106 cells; 2 to 4h, 114±50 × 106 cells; and 6 to 24h, 56±25 × 106 cells, p<0.05, ANOVA with Dunn's test. Cell viability was also lower, particularly beyond 4h after death: 15 min to 1h, 56±9%; 2 to 4h, 53±6%; and 6 to 24h, 34±11%; p<0.05, ANOVA with Dunn's test (Fig. 3A). Only <20% hepatocytes from NHB donors isolated 15 min to 2h after death attached in dishes. These cells remained rounded subsequently. Hepatocytes from NHB donors 6h or longer after death did not attach or survive in dishes. However, the functional integrity was maintained in viable cells isolated 1h after death from NHB donors or from HB donor livers by assays of ureagenesis. Cells in both groups were metabolically active. NHB donor cells produced 280±56 μg urea and HB donor cells 312±82 μg urea per milliliter medium per 1×106 attached cells, respectively, p=n.s. This indicated generally equivalent function in those cells that attached and survived after overnight culture. Although intact tissues had not shown significant apoptosis, we found onset of apoptosis with DNA laddering in hepatocytes from NHB donors, especially cells isolated 6h or longer after death (Fig. 3B). When hepatocytes from HB livers were transplanted into DPPIV- rats, cells engrafted in liver, as was expected (Fig. 3C). Cells from NHB donor livers 2, 4 or 6h after death, also engrafted. However, cells from NHB donor livers beyond 6h after death engrafted in liver extremely rarely. Therefore, we restricted analysis of cell engraftment to hepatocytes from NHB donors 2, 4 or 6h after death. In recipients of HB donor hepatocytes, we found 154±37 transplanted cells per 100 periportal areas after 7d (Fig. 3D). By contrast, cell transplantation from NHB donors 2, 4 or 6h after death produced 123±20, 107±10, and 29±5 hepatocytes per 100 periportal areas, which was 20%, 30% and 82% less, respectively, p<0.05, ANOVA, Dunn's method,
Figure 3. Viability of hepatocytes isolated from HB and NHB donor livers.
(A) Trypan blue dye exclusion in hepatocytes from HB and NHB donors. (B) DNA ladder analysis. Lane 1, molecular weight marker of 100 base pairs; lane 2, HB hepatocytes; lanes 3 and 4, NHB hepatocytes 4 and 16h after death. (C) Cell engraftment after 7d in DPPIV- rats with transplanted cells in periportal areas of liver. Representative findings are from recipients of HB or NHB hepatocytes isolated 2, 4 or 6h after death. Original magnification, ×400; methylgreen counterstain. (D) Morphometric analysis of cell engraftment. Asterisks in A and D, p<0.05 versus HB hepatocytes, ANOVA with Dunn's test (n=3-5 donors ea with 3-4 recipients of cells per donor).
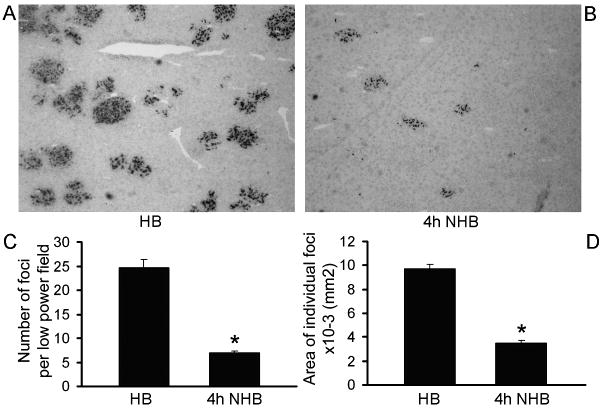
We examined liver repopulation ability of NHB donor cells in retrorsine-PH-conditioned DPPIV- rats. Hepatocytes from both HB and NHB donors engrafted in the liver of preconditioned rats. HB donor cells formed readily visible clusters of proliferating transplanted cells (Fig. 4A). Fewer transplanted cell clusters were observed in recipients of NHB cells, including cells from donors 1, 4 or 6h after death (Fig. 4B). Morphometric analysis showed 25±2 transplanted cell foci per low power field (LPF) in recipients of HB donor cells (Fig. 4C). Fewere transplanted cell foci were found in recipients of NHB hepatocytes 4h after death, 7±0.4 per LPF, which was 3.6-fold less than HB cells, p<0.001, t-test. The size of transplanted cell foci was 9.7±0.4×10-3 mm2 in recipients of HB cells (Fig. 4D). By contrast, the size of transplanted cell foci was 3.5±0.2×10-3 mm2 after transplantation of cells from NHB donors 4h after death, which was 2.8-fold smaller, p<0.001, Mann-Whitney rank sum test. In view of significant differences in engraftment and proliferation of NHB donor hepatocytes, we further studied molecular perturbations.
Figure 4. Proliferation of transplanted hepatocytes from HB and NHB donors.
(A and B) DPPIV+ transplanted cells in liver from HB and NHB donors 4h after death. Original magnification ×400; methylgreen counterstain. (C and D) Morphometric quantitation of transplanted cell foci (C), and area occupied by individual transplanted cell foci (n=3-4 recipients from each of 3 donors). NHB hepatocytes produced fewer transplanted cell foci and area occupied by these transplanted cells was smaller. Asterisks, p<0.05 versus HB donor hepatocytes, Mann-Whitney rank sum tests.
Gene expression changed profoundly in NHB hepatocytes
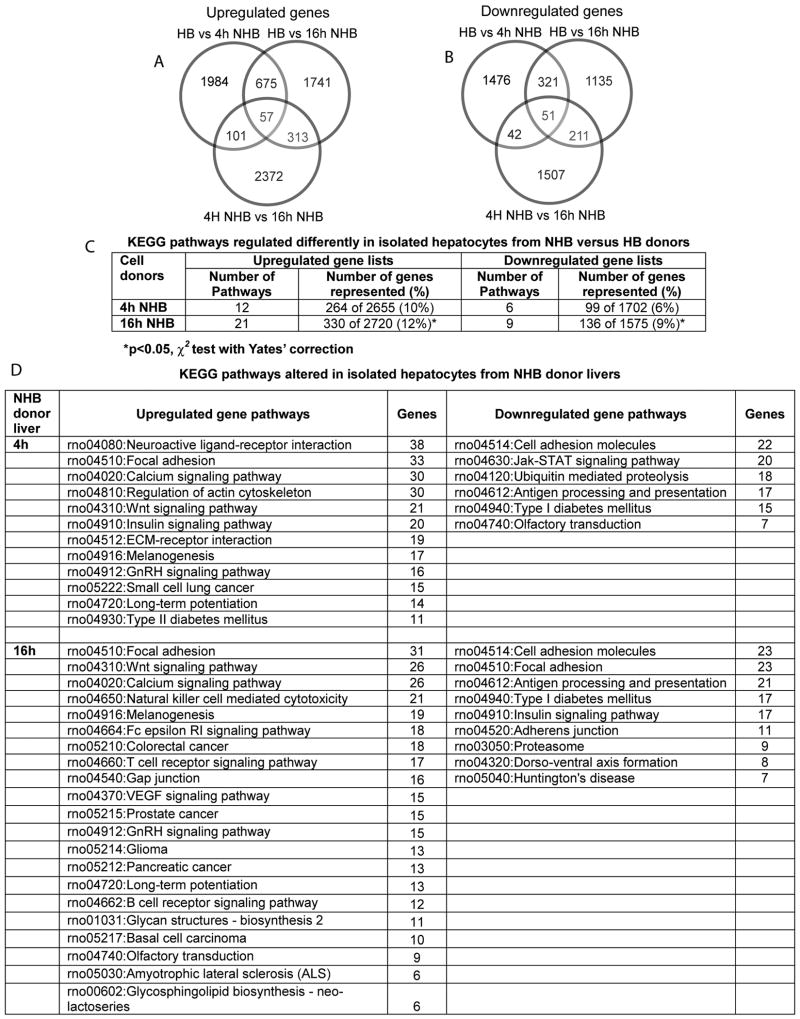
First, we compared gene expression profiles in HB donor hepatocytes and hepatocytes isolated from NHB donors 4h and 16h after death (Fig. 5). This showed extensive perturbations in NHB donor cells with differential expression of 1,000-2,000 transcripts (Fig. 5A, 5B). More KEGG pathways were represented in upregulated genes in NHB cells. Also, more KEGG pathways were represented with increasing time after death, i.e., 16h versus 4h, p<0.05 (Fig. 5C). Therefore, to further identify gene expression differences, we categorized functionally-active pathways in upregulated and downregulated gene lists from NHB donor cells 4h and 16h after death (Fig. 5D).
Figure 5. Gene expression profiles in hepatocytes from NHB versus HB donors.
(A and B) Up- and downregulated genes in NHB hepatocytes 4h and 16h after death compared with HB hepatocytes. Gene lists were larger in cells compared with tissues even 4h after death (see Fig. 2). This difference was more pronounced in cells NHB cells 16h after death. (C) KEGG pathways represented in differentially expressed genes from NHB hepatocytes. The numbers of these pathways and genes in individual pathways rose in NHB hepatocytes, as time-after-death increased. (D) Pathways with largest numbers of differentially expressed genes in NHB hepatocytes 4h or 16h after death.
The data established commonalities in gene expression pathways in NHB donor cells 4h and 16h after death (Table 2). Remarkably, apoptotic pathways were again not represented in NHB donor cells. In view of similarities in gene expression changes and because inferior cell engraftment and proliferation was obvious in cells from NHB donors 4h after death, we restricted analysis of molecular pathways to that time. We focused particularly on pathways in focal adhesion, cell adhesion, actin cytoskeleton, and extracellular matrix (ECM)-receptor interactions, because these should have affected cell attachment, and cell engraftment and proliferation. This identified a number of candidate pathway genes expressed differentially that could have profoundly affected these processes (Supplementary Figs. 5-7).
Table 2. Representation of major functionally annotated groups in differentially expressed gene lists in NHB donor hepatocytes versus HB donor hepatocytes.
| Gene ontology groups | Cells from NHB donor liver 4h | Cells from NHB donor liver 16h | ||
|---|---|---|---|---|
| Number Upregulated genes (%) | Number Downregulated genes (%) | Number Upregulated genes (%) | Number Downregulated genes (%) | |
| Cellular process | 984 (37%) | 732 (43%) | 745 (47%) | |
| Primary metabolic process | 480 (28%) | 595 (22%) | 511 (32%) | |
| Cellular metabolic process | 485 (29%) | 595 (22%) | 508 (32%) | |
| Macromolecule metabolic process | 410 (24%) | 512 (19%) | 450 (29%) | |
| Biological regulation | 455 (17%) | 361 (21%) | 458 (17%) | 346 (22%) |
| Regulation of biological process | 394 (15%) | 322 (19%) | 390 (14%) | 308 (20%) |
| Biopolymer metabolic process | 366 (14%) | 320 (19%) | 380 (14%) | 332 (21%) |
| Regulation of cellular process | 345 (13%) | 287 (17%) | 351 (13%) | 276 (17%) |
| Developmental process | 323 (12%) | 252 (15%) | 316 (12%) | 236 (15%) |
| Protein metabolic process | 240 (15%) | |||
| Cellular macromolecule metabolic process | 227 (14%) | |||
| Cellular protein metabolic process | 221 (14%) | |||
| Cellular localization | 314 (12%) | 208 (12%) | 326 (12%) | 204 (13%) |
| Cellular component organization and biogenesis | 279 (11%) | 210 (12%) | 281 (10%) | 208 (13%) |
| Establishment of localization | 271 (10%) | 175 (10%) | 294 (11%) | 175 (11%) |
| Transport | 263 (10%) | 172 (10%) | 282 (10%) | 169 (11%) |
| Nucleobase, nucleoside, nucleotide and nucleic acid metabolic process | 258 (10%) | 243 (14%) | 265 (10%) | 251 (16%) |
| Gene expression | 234 (9%) | 204 (12%) | 248 (9%) | 214 (14%) |
| Regulation of metabolic process | 211 (8%) | 180 (11%) | 176 (11%) | |
| Multicellular organismal development | 240 (9%) | 179 (11%) | 229 (8%) | 156 (10%) |
| Establishment of localization | 271 (10%) | 175 (10%) | 294 (11%) | 175 (11%) |
| RNA metabolic process | 188 (7%) | 172 (10%) | 201 (7.4%) | 182 (12%) |
| Anatomical structure development | 232 (9%) | 168 (10%) | 238 (9%) | 155 (10%) |
| Regulation of cellular metabolic process | 193 (7%) | 168 (10%) | 198 (7%) | 170 (11%) |
| Regulation of gene expression | 186 (7%) | 162 (10%) | 178 (6.5%) | 162 (10%) |
| Transcription | 176 (6.6%) | 160 (9.4%) | 174 (6.4%) | 158 (10%) |
| Others | ||||
| Intracellular signaling cascade | 142 (5%) | 121 (7%) | 161 (6%) | 123 (8%) |
| Response to stress | 105 (4%) | 102 (6%) | 116 (4%) | 104 (7%) |
| Cell adhesion | 103 (4%) | 84 (3%) | ||
| Cell-cell signaling | 90 (3%) | 77 (3%) | 56 (4%) | |
| Potassium ion transport | 30 (1%) | 23 (1%) | ||
| Induction of programmed cell death | 31 (2%) | 26 (1%) | 27 (2%) | |
| Wnt receptor signaling pathway | 16 (0.6%) | |||
| Calcium mediated signaling | 10 (0.6%) | 10 (0.4%) | 9 (0.6%) | |
Multiple cell signaling pathways were perturbed. Sublists of genes regulating calcium signaling, along with mapping of those genes in calcium signaling pathways, indicated changes in multiple calcium-dependent intracellular processes (Supplementary Fig. 8). Major changes were obvious in insulin signaling (Supplementary Fig. 9), which was consistent with energy needs. We found wnt signaling was perturbed (Supplementary Fig. 10). Among downregulated genes were included Jak-Stat signaling (Supplementary Fig. 11), which is involved in cell injury. Surprisingly, genes involved in neuroactive ligand-receptor interactions constituted the single largest pathway in NHB donor cells (Supplementary Table 1). The pathway representing ubiquitin-mediated proteolysis indicated further anoxia-dependent perturbations (Supplementary Table 2).
Discussion
This study advanced insights in the potential of NHB donors for cells. We found no hepatic autolysis, necrosis or significant apoptosis in NHB livers over many hours, whereas molecular perturbations were due to hepatic reperfusion/reoxygenation. This identification of molecular changes in NHB livers offers opportunities for mechanistic interventions to preserve cell viability. As cells isolated from NHB donors did possess appropriate metabolic activity, i.e., ureagenesis, and also because hepatocytes from NHB donors did engraft and proliferate to some extent, selected cell therapy applications should be feasible, e.g., where liver repopulation is unnecessary (16).
Hyperkalemia caused rapid death and avoided confounding issues, e.g., with hepatic venous outflow obstruction following thoracotomy, phrenotomy, exsanguination, or cardiac clamping (17-19). This was confirmed by essentially no gene expression changes at early times. The consequences of anoxia were apparent later with gene expression changes, including mitrochondrial inner-membrane electron transport and coupled oxidative phosphorylation, although genes in complexes I (NADH-coenzyme Q reductase, Ndufs5) II (succinate ubiquinone oxidoreductase) and IV (cytochrome c oxidase) were expressed at higher levels 30h after death (Supplementary Fig. 1). Such relative resistance to anoxia was a good portent for isolating cells from NHB livers. Organ- and species-specific mechanisms in anoxia-tolerance should be relevant for maintenance of hepatic tissue integrity after death. For instance, in anoxia-resistant deep-sea turtles, NADH and cytochrome c oxidase transcripts accumulate rapidly and persist at high levels during anoxia (20). Some tissue changes identified by gene expression were typical of anoxia/reperfusion, e.g., gradients of chemotaxis, adherence, and transendothelial leukocyte migration, as observed in heart (21). Activation of adherence junction genes likely represented attempts to conserve tissue integrity. Many hepatic changes after death, i.e., expression of glycolytic genes, of lactate dehydrogenase A to generate pyruvate, of genes in amino acid metabolism, likely reflected efforts at metabolic homeostasis with declining energy stores.
Previously, NHB donor livers were typically evaluated within 40 to 60 min after death (5). In one study, cell isolation 8 to 12 hours after anoxia produced viability <15% (6). By contrast, in a mouse study, viable cells were isolated even 24 hours after death, although the cell viability assay was liver repopulation in FAH-/- mice, where even few viable cells can proliferate extensively (7). We found viable cells were best isolated from NHB livers up to 4 hours after death. Damage in isolated cells was clearly due to reperfusion/reoxygenation, implying roles for energy depletion and pro-oxidant stress. Among structural changes of greatest concern were focal adhesion, cell adhesion, actin cytoskeleton, extracellular matrix component (ECM)-receptor interactions, and adherence junctions. This was not previously known. However, gene expression profiling of post-mortem muscle showed ECM-receptor interactions or calcium signaling were deleterious, decreasing water-holding capacity of myocytes (22). No doubt, actin cytoskeleton weakening should have increased damage after enzymatic liver digestion, and impaired cell adhesion was reflected by inferior attachment in dishes of NHB hepatocytes. These mechanisms were relevant because focal adhesions and ECM-receptor interactions are required for engraftment of transplanted hepatocytes (23).
Little is known about intracellular signaling after hepatic anoxia. Since multiple signaling pathways were altered in NHB donor hepatocytes, these processes were likely important in cell damage. For instance, mapping of focal adhesion pathways identified changes in extracellular signal regulated kinase-1 (ERK1). ERK-1 was strongly induced by reactive oxygen species after tissue reoxygenation and contributed in disruption of cell adhesion, focal adhesion complexes and cytoskeleton disorganization (24). After chemical hypoxia, prevention of energy depletion or acidosis, and incorporation of glycine, was hepatoprotective, albeit without restoring actin cytoskeleton or focal adhesions (25, 26), implying further complexities.
Intracellular calcium ion fluxes following anoxia have been of significance for hepatotoxicity or toxicity in other cell types, e.g., neurons (27,28). Therefore, we were intrigued by activation of neuroactive ligand-receptor pathways in NHB hepatocytes, especially because engagement of excitatory receptors is harmful (29). Currently, no information is available about these potential anoxia-modulating mechanisms in liver, which offers new directions for hepatoprotection. Perturbation in insulin signaling pathways was another aspect of NHB cells, most likely due to glycolysis in the setting of energy depletion.
In NHB donor hepatocytes, changes in wnt signaling, which included canonical wnt as well as wnt/calcium pathways, was unexpected, since these pathways have not been associated with anoxia. Noncanonical wnt/calcium signaling should be of interest because it may serve roles in remodeling of actin cytoskeleton (30), which was prominent in NHB cells. Moreover, wnt/calcium signaling is associated with cell survival (31). The significance of several downregulated genes, e.g., Jak-Stat signaling pathway, was unclear. Certainly, in view of the pleiotropic roles of wnt and Jak-Stat signaling in cell proliferation, injury, etc., these genes may contribute in hepatocyte viability. The contributions of proteolytic pathways served by ubiquitination, which also stood out in our gene lists, have not been examined in hypoxic tissue injury (32).
The integrity of NHB donor livers after death should be encouraging for isolating viable hepatocytes from people, although in studies with cadaveric tissues from monkeys and resected specimens from humans, hepatocyte viability was limited (7). The molecular profile of hepatocytes from NHB donor livers offers frameworks to understand reasons for inferior viability of isolated cells, while suggesting potential directions to reverse perturbations in pathway-specific fashion. Besides mitigating cell damage due to oxidative stress, regulation of pathways in adherence junctions, cytoskeletal integrity, extracellular matrix interactions, cell signaling, etc., by various interventions, should be of particular interest. These studies will require detailed and careful prospective manipulations that could be guided by progressive reversal of molecular changes identified here in cells from NHB donors.
Supplementary Material
Acknowledgments
Ms. Chaoying Zhang provided technical assistance. The studies were published in abstract form (Enami Y, et al. Hepatology 2008;48:503A).
Funding: Partly supported by NIH grants R01 DK071111, R01 DK088561, P30 DK41296 and P30 CA13330.
Footnotes
No conflicts of interest exist.
Role of authors: YE, conceived the study, acquired, analyzed and interpreted data, and drafted the manuscript; BJ acquired, analyzed and interpreted data; SB acquired, analyzed and interpreted data; JL analyzed and annotated data; SG conceived and supervised the study, obtained funding, analyzed and interpreted data, and drafted and made critical conceptual contributions in the manuscript.
References
- 1.Fisher RA, Strom SC. Human hepatocyte transplantation: worldwide results. Transplantation. 2006;82:441–9. doi: 10.1097/01.tp.0000231689.44266.ac. [DOI] [PubMed] [Google Scholar]
- 2.Malhi H, Irani AN, Gagandeep S, Gupta S. Isolation of human progenitor liver epithelial cells with extensive replication capacity and differentiation into mature hepatocytes. J Cell Sci. 2002;115:2679–2688. doi: 10.1242/jcs.115.13.2679. [DOI] [PubMed] [Google Scholar]
- 3.Issartel J, Hervant F, de Fraipont M, Clobert J, Voituron Y. High anoxia tolerance in the subterranean salamander Proteus anguinus without oxidative stress nor activation of antioxidant defenses during reoxygenation. J Comp Physiol B. 2009;179:543–51. doi: 10.1007/s00360-008-0338-9. [DOI] [PubMed] [Google Scholar]
- 4.Muramatsu Y, Furuichi Y, Tojo N, Moriguchi A, Maemoto T, Nakada H, Hino M, Matsuoka N. Neuroprotective efficacy of FR901459, a novel derivative of cyclosporin A, in in vitro mitochondrial damage and in vivo transient cerebral ischemia models. Brain Res. 2007;1149:181–90. doi: 10.1016/j.brainres.2007.02.036. [DOI] [PubMed] [Google Scholar]
- 5.Hughes RD, Mitry RR, Dhawan A, Lehec SC, Girlanda R, Rela M, Heaton ND, Muiesan P. Isolation of hepatocytes from livers from non-heart-beating donors for cell transplantation. Liver Transpl. 2006;12:713–717. doi: 10.1002/lt.20732. [DOI] [PubMed] [Google Scholar]
- 6.Porretti L, Gatti S, Gramignoli R, Colombo F, Lopa R, Cattaneo A, Scalamogna M, Colombo G, Rossi G, Bonino F, Rebulla P, Prati D. Animal model for liver cell banking from non-heart beating donors after prolonged ischemia time. Dig Liver Dis. 2006;38:905–911. doi: 10.1016/j.dld.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 7.Erker L, Azuma H, Lee AY, Guo C, Orloff S, Eaton L, Benedetti E, Jensen B, Finegold M, Willenbring H, Grompe M. Therapeutic liver reconstitution with murine cells isolated long after death. Gastroenterology. 2010;139:1019–29. doi: 10.1053/j.gastro.2010.05.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enami Y, Bandi S, Kapoor S, Krohn N, Joseph B, Gupta S. Hepatic stellate cells promote hepatocyte engraftment in rat liver after prostaglandin-endoperoxide synthase inhibition. Gastroenterology. 2009;136:2356–64. doi: 10.1053/j.gastro.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laconi S, Pillai S, Porcu PP, Shafritz DA, Pani P, Laconi E. Massive liver replacement by transplanted hepatocytes in the absence of exogenous growth stimuli in rats treated with retrorsine. Am J Pathol. 2001;158:771–7. doi: 10.1016/s0002-9440(10)64019-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benten D, Kumaran V, Joseph B, Gupta S. Hepatocyte transplantation activates hepatic stellate cells with beneficial modulation of cell engraftment. Hepatology. 2005;42:1072–1081. doi: 10.1002/hep.20889. [DOI] [PubMed] [Google Scholar]
- 11.Anundi I, King J, Owen DA, Schneider H, Lemasters JJ, Thurman RG. Fructose prevents hypoxic cell death in liver. Am J Physiol. 1987;253:G390–G396. doi: 10.1152/ajpgi.1987.253.3.G390. [DOI] [PubMed] [Google Scholar]
- 12.Inada M, Follenzi A, Cheng K, Surana M, Joseph B, Benten D, Bandi S, Qian H, Gupta S. Phenotype reversion in fetal human liver epithelial cells identifies the role of an intermediate meso-endodermal stage before hepatic maturation. J Cell Sci. 2008;121:1002–13. doi: 10.1242/jcs.019315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dudoit S, Fridlyand J, Speed TP. Comparison of discrimination methods for the classification of tumors using gene expression data. J Am Statist Assoc. 2002;97:77–87. [Google Scholar]
- 14.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 15.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nature Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 16.Bandi S, Joseph B, Berishvili E, Singhania R, Wu YM, Cheng K, Gupta S. Perturbations in Atm signaling pathways following drug-induced acute liver failure and their reversal during rescue of animals by cell therapy. Am J Pathol. 2011;178:161–74. doi: 10.1016/j.ajpath.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saad S, Minor T, Kötting M, Fu ZX, Hagn U, Paul A, Nagelschmidt M. Extension of ischemic tolerance of porcine livers by cold preservation including postconditioning with gaseous oxygen. Transplantation. 2001;71:498–502. doi: 10.1097/00007890-200102270-00003. [DOI] [PubMed] [Google Scholar]
- 18.Minor T, Olschewski P, Tolba RH, Akbar S, Kočálková M, Dombrowski F. Liver preservation with HTK: salutary effect of hypothermic aerobiosis by either gaseous oxygen or machine perfusion. Clin Transplant. 2002;16:206–211. doi: 10.1034/j.1399-0012.2002.01128.x. [DOI] [PubMed] [Google Scholar]
- 19.He XS, Ma Y, Ju WQ, Wu LW, Wu JL, Liang YJ, Hu RD, Chen GH, Huang JF. Dynamic microcirculatory changes in liver graft from non-heart-beating donor with warm ischemia injury in rat. Hepatobiliary Pancreat Dis Int. 2004;3:179–182. [PubMed] [Google Scholar]
- 20.Cai Q, Storey KB. Anoxia-induced gene expression in turtle heart. Upregulation of mitochondrial genes for NADH-ubiquinone oxidoreductase subunit 5 and cytochrome c oxidase subunit 1. Eur J Biochem. 1996;241:83–92. doi: 10.1111/j.1432-1033.1996.0083t.x. [DOI] [PubMed] [Google Scholar]
- 21.Rui T, Cepinskas G, Feng Q, Ho YS, Kvietys PR. Cardiac myocytes exposed to anoxia-reoxygenation promote neutrophil transendothelial migration. Am J Physiol Heart Circ Physiol. 2001;281:H440–7. doi: 10.1152/ajpheart.2001.281.1.H440. [DOI] [PubMed] [Google Scholar]
- 22.Ponsuksili S, Jonas E, Murani E, Phatsara C, Srikanchai T, Walz C, Schwerin M, Schellander K, Wimmers K. Trait correlated expression combined with expression QTL analysis reveals biological pathways and candidate genes affecting water holding capacity of muscle. BMC Genomics. 2008;9:367. doi: 10.1186/1471-2164-9-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumaran V, Joseph B, Benten D, Gupta S. Integrin and extracellular matrix interactions regulate engraftment of transplanted hepatocytes in the rat liver. Gastroenterology. 2005;129:1643–1653. doi: 10.1053/j.gastro.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 24.Sáenz-Morales D, Conde E, Escribese MM, García-Martos M, Alegre L, Blanco-Sánchez I, García-Bermejo ML. ERK1/2 mediates cytoskeleton and focal adhesion impairment in proximal epithelial cells after renal ischemia. Cell Physiol Biochem. 2009;23:285–94. doi: 10.1159/000218175. [DOI] [PubMed] [Google Scholar]
- 25.Marsh DC, Vreugdenhil PK, Mack VE, Belzer FO, Southard JH. Glycine protects hepatocytes from injury caused by anoxia, cold ischemia and mitochondrial inhibitors, but not injury caused by calcium ionophores or oxidative stress. Hepatology. 1993;17:91–8. [PubMed] [Google Scholar]
- 26.Nishimura Y, Romer LH, Lemasters JJ. Mitochondrial dysfunction and cytoskeletal disruption during chemical hypoxia to cultured rat hepatic sinusoidal endothelial cells: the pH paradox and cytoprotection by glucose, acidotic pH, and glycine. Hepatology. 1998;27:1039–49. doi: 10.1002/hep.510270420. [DOI] [PubMed] [Google Scholar]
- 27.Pastorino JG, Snyder JW, Hoek JB, Farber JL. Ca2+ depletion prevents anoxic death of hepatocytes by inhibiting mitochondrial permeability transition. Am J Physiol. 1995;268:C676–85. doi: 10.1152/ajpcell.1995.268.3.C676. [DOI] [PubMed] [Google Scholar]
- 28.Sirabella R, Secondo A, Pannaccione A, Scorziello A, Valsecchi V, Adornetto A, Bilo L, Di Renzo G, Annunziato L. Anoxia-induced NF-kappaB-dependent upregulation of NCX1 contributes to Ca2+ refilling into endoplasmic reticulum in cortical neurons. Stroke. 2009;40:922–9. doi: 10.1161/STROKEAHA.108.531962. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Su P, Liang P, Liu T, Liu X, Liu XY, Zhang B, Han T, Zhu YB, Yin DM, Li J, Zhou Z, Wang KW, Wang Y. The DREAM protein negatively regulates the NMDA receptor through interaction with the NR1 subunit. J Neurosci. 2010 Jun 2;30(22):7575–86. doi: 10.1523/JNEUROSCI.1312-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Q, Symes AJ, Kane CA, Freeman A, Nariculam J, Munson P, Thrasivoulou C, Masters JR, Ahmed A. A novel role for Wnt/Ca2+ signaling in actin cytoskeleton remodeling and cell motility in prostate cancer. PLoS One. 2010 May 4;5(5):e10456. doi: 10.1371/journal.pone.0010456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gregory MA, Phang TL, Neviani P, Alvarez-Calderon F, Eide CA, O'Hare T, Zaberezhnyy V, Williams RT, Druker BJ, Perrotti D, Degregori J. Wnt/Ca2+/NFAT signaling maintains survival of Ph+ leukemia cells upon inhibition of Bcr-Abl. Cancer Cell. 2010 Jul 13;18(1):74–87. doi: 10.1016/j.ccr.2010.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calzado MA, De La Vega L, Munoz E, Schmitz ML. From top to bottom: the two faces of HIPK2 for regulation of the hypoxic response. Cell Cycle. 2009;8:1659–64. doi: 10.4161/cc.8.11.8597. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.