Abstract
Proteomics discovery of novel cancer serum biomarkers is hindered by the great complexity of serum, patient-to-patient variability, and triggering by the tumor of an acute-phase inflammatory reaction. This host response alters many serum protein levels in cancer patients, but these changes have low specificity as they can be triggered by diverse causes. We addressed these hurdles by utilizing a xenograft mouse model coupled with an in-depth 4-D protein profiling method to identify human proteins in the mouse serum. This strategy ensures identified putative biomarkers are shed by the tumor, and detection of low-abundance proteins shed by the tumor is enhanced because the mouse blood volume is more than a thousand times smaller than that of a human. Using TOV-112D ovarian tumors, more than 200 human proteins were identified in the mouse serum, including novel candidate biomarkers and proteins previously reported to be elevated in either ovarian tumors or the blood of ovarian cancer patients. Subsequent quantitation of selected putative biomarkers in human sera using label-free multiple reaction monitoring (MRM) mass spectrometry (MS) showed that chloride intracellular channel 1, the mature form of cathepsin D, and peroxiredoxin 6 were elevated significantly in sera from ovarian carcinoma patients.
Keywords: Biomarker discovery, ovarian cancer, cancer biomarkers, xenograft mouse model, proteomics, multidimensional protein profiling
Introduction
Ovarian cancer is the fifth-leading cause of cancer-related death in women in the United States, and is the most lethal of all gynecological malignancies.1 In 2010, an estimated 21,880 women were diagnosed with ovarian cancer, and 13,850 deaths occurred in the United States alone.1 The most common and deadly form of ovarian cancer is epithelial ovarian cancer, which further can be divided into four major histopathological groups: serous, endometrioid, mucinous, and clear cell tumors.2,3 The high mortality rate of ovarian cancer is due largely to the lack of effective screening strategies for early detection. When ovarian cancer is diagnosed at an early stage (stages I or II), treatment is highly effective, with a five-year survival rate of up to 90%, whereas the five-year survival rate for patients with advanced disease (stages III and IV) is reduced to 30% or less.4, 5 Unfortunately, most ovarian cancers are not diagnosed until after the cancer has spread, primarily because earlier-stage diseases are asymptomatic and the ovaries are buried deep within the body.
Current screening methods for ovarian cancer typically use a combination of pelvic examination, transvaginal ultrasonography, and serum CA125, but these methods are not effective in detecting early-stage ovarian cancer.6–8 In addition, CA125 is recognized as a poor protein biomarker for early detection due to its high false positive rate and poor sensitivity and specificity.9, 10 Other promising biomarkers have been reported,11, 12 but a recently completed study comparing many of these protein biomarkers showed that none of them performed better than CA125 as a biomarker for ovarian cancer.13 A few groups also have used panels of biomarkers and obtained better sensitivity and specificity than CA125 alone when used in diagnostic samples.14–17 However, a recent study found that available biomarker panels did not outperform CA125 when used in prediagnostic samples.18 Therefore, better biomarkers that could diagnose early-stage ovarian cancer with high sensitivity and specificity are needed. Furthermore, it is unlikely that any single protein will have adequate specificity and sensitivity for early diagnosis of most solid-tumor cancers. Instead, multiple novel biomarkers must be identified and analyzed in combination to identify biomarker panels that can outperform the use of CA125 alone.
Proteomics technology offers a conceptually attractive platform for cancer biomarker discovery.19 Human blood, in the form of plasma or serum, is one of the most valuable specimens for protein biomarker discovery because it is routinely collected, collection is minimally invasive, and it contains thousands of proteins, including those secreted or shed into the blood by tumors.20 However, systematic discovery of serological biomarkers directly from human serum using proteomics has proven extremely challenging due to the extremely wide concentration range of blood proteins that span more than 10 orders of magnitude. In addition, the most tumor-specific proteins are very likely to primarily be shed by the tumor and will be very low abundant in blood, as exemplified by well-known cancer biomarkers such as PSA and CEA, which are present in serum in the low ng/mL to pg/mL range.20, 21 Most cancers and other diseases also elicit a wide range of host response mechanisms, producing many acute-phase or inflammation-related proteins. It is unlikely that most such relatively general host responses will have sufficient specificity and sensitivity for cancer detection in at-risk populations, although selected inflammation-related biomarkers could contribute to panels of biomarkers that include proteins specifically shed by the tumor. Regardless, it is clear that these common, acute-phase-related changes in serum proteins hamper discovery of tumor-specific proteins when directly profiling sera in human populations. Finally, individual protein levels in blood are highly variable in the human population due to extensive genetic, physiological, and environmental variations, requiring analysis of many patient and control samples before statistically significant, disease-related differences can be identified.
The dynamic range and complexity of the blood proteome can partially be addressed by major protein depletion and multidimensional sample prefractionation. We and others have shown that multidimensional sample prefractionation prior to mass spectrometry analysis greatly enhances proteome coverage and allows detection of low-abundance proteins, at least down to the low ng/mL range.22–27 To overcome the genetic, physiological, and environmental variability associated with analyzing human samples, many less complex experimental models, including cancer cell lines in culture,28, 29 cancer tissue specimens,30, 31 ascites fluid,32, 33 secretomes,34, 35 and mouse models,36–38 have been used in ovarian cancer biomarker discovery. Each model has its benefits, but most strategies, except for the use of mouse models, are not able to determine if the discovered biomarkers are actually shed into blood. In ovarian cancer, the use of both genetically engineered and xenograft mouse models to facilitate serum biomarker discovery has been described.36–38 Even though subject-to-subject heterogeneity is considerably reduced with the use of genetically engineered mouse models, these models still produce many host-response protein changes that can be difficult to distinguish from more tumor-specific protein changes.37
The use of xenograft mouse models has several advantages over other models. First, proteins shed by human tumors into mouse blood can be unambiguously distinguished from less-specific host responses by exploiting species differences in peptide sequences identified by liquid chromatography-tandem mass spectrometry (LC-MS/MS). Second, the blood volume of a mouse is approximately 5,000 times less than an adult human. Therefore, proteins shed by similar-sized small tumors in a mouse and an adult human are likely to be at least 1,000 times more concentrated in a xenograft mouse as compared to the same size tumor in a human. Third, the minimal biological heterogeneity of the xenograft mouse model means that only a small number of samples need to be profiled in order to make inferences about putative biomarkers.
While the use of xenograft mouse models potentially can improve detection of novel cancer biomarkers, mouse serum is still a very complex proteome and requires multidimensional sample prefractionation for sufficient depth of analysis. For example, in a prior xenograft mouse study using two-dimensional gel electrophoresis and without any sample prefractionation, only acute-phase proteins were identified successfully.39 In the case of ovarian cancer, two different studies using a xenograft model with human SKOV-3 serous ovarian cancer cells have been described. In one study, mouse sera were trypsin-digested and analyzed directly by LC-MS/MS, resulting in identification of 13 human proteins.38 The other study focused on the low-molecular weight serum proteome/peptidome of the xenograft model and reported the identification of five human proteins.36 While both prior xenograft ovarian cancer studies successfully identified a few candidate biomarkers (14-3-3 zeta and S100A6), we reasoned that the combination of a xenograft mouse model with more extensive fractionation of serum proteins could identify much larger numbers of novel human candidate biomarkers. The most difficult-to-detect proteins are expected to be lower abundance and, therefore, may be more tumor specific.
In this study, we established a xenograft mouse model using the ovarian endometrioid TOV-112D cell line and analyzed the serum proteome using a 4-D protein profiling strategy. We demonstrated that it is possible to detect many tumor-derived human proteins, including low-abundance human proteins that are present at < 100 ng/mL in normal human serum. In a proof-of-concept validation analysis, we quantified the levels of three high-priority candidate biomarkers in serum from ovarian patients, as well as normal controls and patients with benign disease, using label-free MRM-MS.
Materials and Methods
Cell culture
The human epithelial ovarian cancer cell line TOV-112D was obtained from the American Type Culture Collection (ATCC, Manassas, VA). The TOV-112D cells were maintained in a 37° C incubator with a 5% CO2−95% air atmosphere in a 1:1 mixture of MCDB 105 media and Medium 199 (Sigma-Aldrich, St. Louis, MO) supplemented with 15% fetal calf serum, as described previously.40
Ovarian cancer growth in vivo and mouse serum collection
Approximately 3 million TOV-112D cells in 100 µl PBS were injected subcutaneously into the flanks of nine severe combined immunodeficient (SCID) female mice. Tumor volumes were monitored by caliper measurements. When tumors were approximately 1 cm in length, blood was collected from mice by cardiac puncture under anesthesia, animals were immediately euthanized, and the tumor at the injection site and other internal organs were collected. This study protocol was approved by the Wistar Institute’s Institutional Animal Care and Use Committee (IACUC).
The collected blood was allowed to clot at room temperature and followed by centrifugation for 15 min at 4° C to collect the serum. Individual aliquots of serum from each mouse were then snap-frozen and stored at −80° C. Serum subsequently was thawed briefly and pooled based on assessment of tumor burden and extent of tumor necrosis. The total protein concentrations of pooled serum samples were measured using a BCA Protein Assay (Pierce Chemical, Rockford, IL), after which the pooled serum was re-aliquoted, snap-frozen, and stored at −80° C until future use. Tumor necrosis was assessed by microscopic inspection of hematoxylin and eosin (H&E) stained paraffin-embedded sections (5 µm), and other organs were macroscopically and microscopically examined for evidence of tumor metastasis.
Immunoaffinity removal of major mouse serum proteins
The pooled mouse serum was depleted using a 4.6 × 100 mm MARS Mouse-3 HPLC column (Agilent Technologies, Wilmington, DE). A total of 225 µL pooled serum was diluted five-fold with the manufacturer’s equilibration buffer, filtered through a 0.22 µm microcentrifuge filter, and briefly stored on ice. This sample subsequently was applied to the antibody column in five serial injections of 200–250 µL per depletion. The flow-through fractions containing unbound proteins were collected and pooled. The immunodepletion equilibration buffer was removed by buffer exchange into 10 mM sodium phosphate, pH 7.0, and the sample was concentrated to the initial serum volume using a 5K molecular weight cutoff (MWCO) spin concentrator. Bound proteins were eluted with the manufacturer’s elution buffer, neutralized with 1 M NaOH, pooled, aliquoted, and stored at −20° C for possible future analysis. Protein concentrations from the unbound and bound fractions were estimated using standard and reducing-reagent-compatible BCA assays, respectively.
MicroSol-IEF fractionation
Depleted and concentrated mouse serum (2.6 mg) was reduced with 20 mM DTT for 30 min and alkylated with 50 mM N,N-dimethylacrylamide (DMA) for 30 min at room temperature in 550 µL of buffer (final volume) containing 8 M urea, 20 mM Tris-HCL, pH 8.5. Alkylation was quenched with 1% DTT, and serum was diluted to 670 µL (final volume) in a sample buffer consisting of 8 M urea, 2 M thiourea, 4% CHAPS, 1% DTT, 1% pH 3–7 ZOOM focusing buffer, and 1% pH 7–12 ZOOM focusing buffer. Serum was fractionated by MicroSol-IEF, as previously described23, 41 using a ZOOM-IEF fractionator (Invitrogen, Carlsbad, CA), into five small-volume (550–650 µL) pools where the separation chambers were defined by IPG gel membranes having pH values of 3.0, 4.6, 5.4, 6.2, and 7.0, respectively. After focusing, samples were removed and each chamber was rinsed with a small volume of sample buffer, which was combined with the solution removed from that chamber. IPG gel membranes were extracted twice for 1 h each with 100 µL of 1% SDS, 20mM Tris, 1% 2-mercaptoethanol. To maximize protein loads for SDS-PAGE, MicroSol-IEF fractions were precipitated overnight with nine volumes of 200 proof ethanol, prechilled to −20° C. Ethanol supernatants were carefully removed and protein pellets were re-suspended in 50% ethanol, centrifuged, and pellets were frozen and stored at −20° C until further use. Membrane extracts were concentrated to approximately 50 µL with 5K MWCO spin concentrators.
SDS-PAGE/In-gel trypsin digestion of mouse serum
Frozen protein pellets from ethanol precipitation of MicroSol IEF fractions were thawed briefly and resuspended in SDS gel sample buffer. For fractions 2–4, aliquots derived from 15 µL of original serum per lane were loaded into 10-well 12% NuPAGE mini-gels (Invitrogen) and separated using MES running buffer until the tracking dye had migrated 4 cm. For fractions 1 and 5 and for membrane extractions, the equivalent of 37 µL and 80 µL of original serum, respectively, was loaded and separated for 1 cm. Gels were stained with Colloidal Blue (Invitrogen), and each lane subsequently was sliced into either 40 (fractions 2–4) or 10 (fractions 1, 5, and membranes) uniform 1 mm slices using a disposable gel cutter (The Gel Company, San Francisco, CA). For fractions 2–4, two adjacent slices in a single lane were combined in a digestion well. Slices from duplicate lanes of fractions 1, 5, and membrane extractions were combined and all samples were digested overnight using 0.02 µg/µL modified trypsin (Promega, Madison, WI). A total of 140 digests were performed from the five IEF fractions and six membrane extracts.
LC-MS/MS
Tryptic digests were analyzed on an LTQ-FT hybrid mass spectrometer (Thermo Electron, San Jose, CA) coupled with a NanoLC pump (Eksigent Technologies, Livermore, CA) and autosampler. Tryptic peptides were separated as described previously22 by RP-HPLC on a PicoFrit column (75 µm i.d., 15 nm tip opening; New Objective, Woburn, MA), packed with 8 cm of Hypersil C18 1.9-µm resin (Thermo Electron). Eluted peptides were analyzed by the mass spectrometer set to repetitively scan m/z from 400 to 1600 followed by data-dependant MS/MS scans on the six most abundant ions with dynamic exclusion enabled.
Data analyses
Peptides from each LC-MS/MS run were interpreted from MS/MS spectra using SEQUEST in Bioworks 3.2 (Thermo Electron). DTA files were created and searched against a combined mouse and human database generated from Uniprot (5/16/06), National Center for Biotechnology Information non-redundant (2/05/06), and International Protein Index (version 3.17) databases. This composite database also contained the reversed sequences of each entry appended to the beginning of the forward database. The database was indexed with the following parameters: monoisotopic mass range of 750 to 3500, length of 4 to 100, partial tryptic cleavages with a maximum of two internal missed cleavage sites, static modification of Cys by dimethylacrylamide (+99.0684 Da) and dynamic modification of Met to methionine sulfoxide (+15.9946 Da). The DTA files were searched with a 2.5 Da peptide mass tolerance. Other search parameters were identical to those used for database indexing.
Outputs from all SEQUEST searches were combined, filtered using in-house scripts, and grouped into non-redundant proteins using DTASelect version 1.9.42 An in-house script was used to correct the wrong peptide m/z assignments to the C13 peaks. Peptides were filtered using mass accuracy ≤ 8 ppm, Sf ≥ 0.4 and requiring full tryptic specificity for all identified peptides. This filtering scheme resulted in 1.4% peptide false positives calculated as the number of unique reversed sequence hits/number of unique forward sequence hits. Keratin identifications were removed from the datasets as probable contaminations. Additional Java scripts were developed to compress the non-redundant protein identifications reported by DTASelect into the smallest sets of unique proteins. Peptide counts were derived after collapsing different forms (charge states and modifications) of the same peptide into a single hit. Further reduction was applied by allowing a peptide to be assigned only once to the protein that had the highest sequence coverage.
Custom software also was developed to separate mouse and human proteins based on their sequence identifiers. Putative human peptides were then searched using BLAST against a mouse-only database from Uniprot (11/2007) to remove any putative human sequences that exactly matched mouse sequences.
Human serum collection
Sera from patients with benign ovarian tumors (n=9), and from late-stage ovarian cancer patients (stages III, n=15; or IV, n=3) were collected at the University of Turin, Turin, Italy, at the time of diagnosis. Control serum samples (n=6) were collected from healthy, post-menopausal female donors at the Wistar Institute, Philadelphia, PA. All specimens were processed in compliance with institutional review board and Health Insurance Portability and Accountability Act (HIPAA) requirements.
Processing of human serum samples for MRM analysis
Control and patient serum samples were analyzed either individually or as pools, as follows. Samples were depleted of 20 abundant serum proteins using a ProteoPrep20 Immunodepletion Column (Sigma-Aldrich). Typically, 30–50 µL of serum was depleted, concentrated, and prepared for SDS-PAGE, as previously described.43 SDS-PAGE conditions for human serum were identical to those described above for the analysis of mouse serum, with the exception that the equivalent of 10 µL of original serum were loaded into three adjacent lanes and separated for 4 cm. Gels were sliced and digested, as previously described.43
Label-free multiple reaction monitoring
MRM experiments were performed on a 4000 QTRAP hybrid triple quadrupole/linear ion trap mass spectrometer (AB Sciex, Foster City, CA) interfaced with a NanoACQUITY UPLC system (Waters, Milford, MA). Eight µL of tryptic digests were injected using the partial loop injection mode onto a UPLC Symmetry trap column (180 µm i.d. × 2 cm packed with 5 µm C18 resin; Waters) and then separated by RP-HPLC on a PicoFrit column (75-µm i.d., 15-µm tip opening) packed in-house with 25 cm of Magic C18 3-µm reversed-phase resin (Michrom Bioresources, Auburn, CA). Chromatography was performed with Solvent A (Milli-Q [Millipore, Billerica, MA] water with 0.1% formic acid) and Solvent B (acetonitrile with 0.1% formic acid). Peptides were eluted at 200 nL/min for 3–28% B over 42 min, 28–50% B over 26 min, 50–80% B over 5 min, 80% B for 4.5 min before returning to 3% B over 0.5 min. To minimize sample carryover, a fast blank gradient was run between each sample. An identical reference sample was run at the beginning of each set of samples and was used to normalize variation in MRM signals caused by changes in performance of the HPLC, reverse-phase column or mass spectrometer.
MRM data were acquired with a spray voltage of 2,500 V, curtain gas of 20 p.s.i., nebulizer gas of 10 p.s.i., interface heater temperature of 150 °C, and a pause time of 5 ms. Multiple MRM transitions were monitored using unit resolution in both Q1 and Q3 quadrupoles to maximize specificity. Each MRM transition had a minimum dwell time of 15 s. Data analyses were performed using MultiQuant version 1.1 software (AB Sciex). The most abundant transition for each peptide was used for quantitation unless interference from the matrix was observed. In these cases, another transition free of interference was chosen for quantitation.
Statistical analyses
Serum levels of candidate biomarkers were compared across patient groups using an unpaired, two-tailed Student’s t-test. Welch’s correction was applied to the t-test when the variances between the two sets were significantly different. Statistical significance was determined if the P-value of the test was less than 0.05. Calculations, scatter plots, and receiver operator characteristic (ROC) curves were generated using the GraphPad Prism 5 (GraphPad, San Diego, CA). Optimal cut-points were obtained by identifying a threshold for each biomarker that resulted in maximum sensitivity and specificity when used to classify serum as tumor or control. Both sensitivity and specificity for each decision rule defined by biomarker-specific optimal cut-point were computed, as well as the positive and negative predictive values. The odds ratio between the group classification and the result from each decision rule from the logistic regression was used as a measure of their association. Multivariate models were fit using logistic regression analysis.
Results and Discussion
Overview of the ovarian cancer biomarker discovery and verification/validation strategies
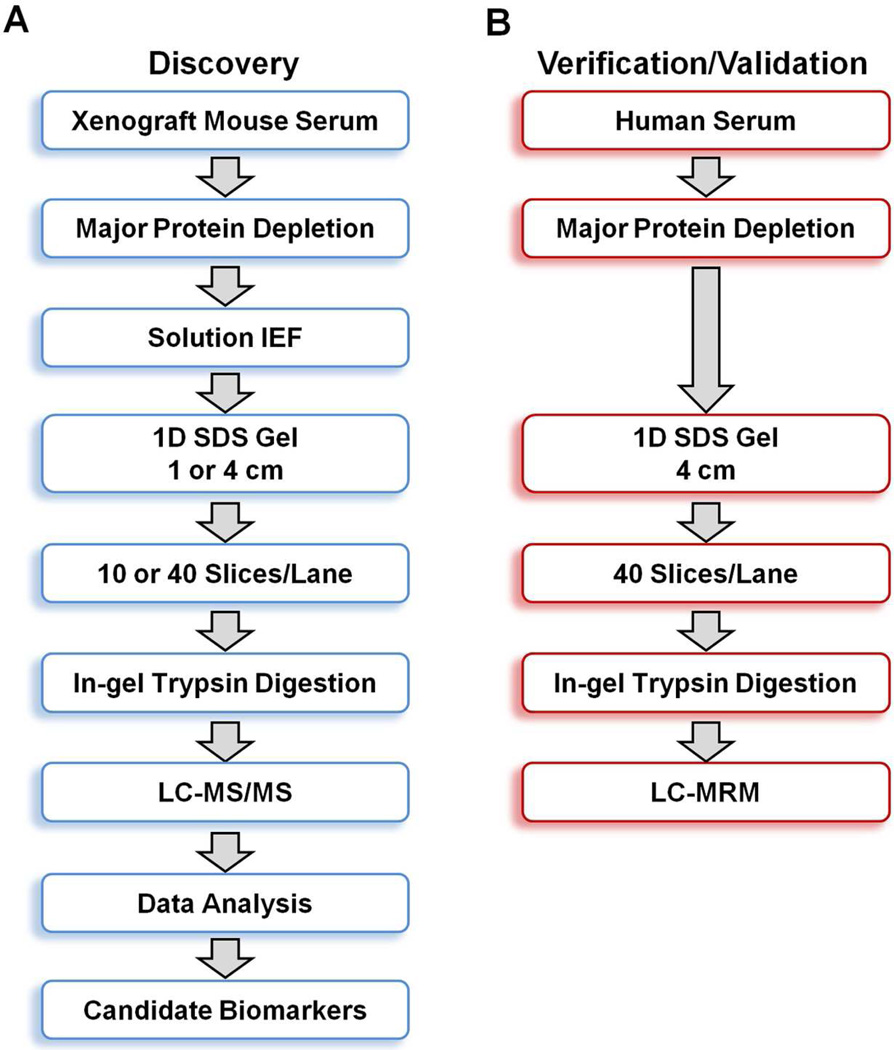
The general experimental workflow we used for discovery and verification of candidate ovarian cancer protein biomarkers is shown in Figure 1. For discovery of candidate human biomarkers, serum proteins obtained from SCID mice harboring human ovarian cancer tumors were subjected to a 4-D separation consisting of three sequential, and substantially orthogonal, protein separations, i.e., major protein depletion, solution IEF, and 1-D SDS-PAGE, followed by online, reversed-phase LC peptide separation prior to MS/MS analysis. We previously developed this 4-D protein profiling method for comprehensive analysis of human serum and plasma proteomes, which resulted in the most comprehensive coverage of a serum sample in the pilot phase of the Human Proteome Organization Plasma Proteome Project (HUPO PPP).22, 44 That study also demonstrated that 14 of the 20 proteins known at that time to be in the 1–100 ng/mL range in normal human serum could be detected. While this method has the capacity to detect many low-abundance proteins, the extensive fractionations and large number of associated LC-MS/MS runs require several weeks to analyze a single proteome, making it impractical to analyze large numbers of samples. However, depth of proteome analysis is much more important than throughput for discovery of candidate cancer biomarkers in the xenograft mouse cancer model, which makes this 4-D method ideally suited for this application.
Figure 1.
Overview of the experimental workflow for ovarian cancer biomarker discovery and verification/validation. (A) Sera from xenograft SCID mice harboring TOV-112D ovarian cancer cells were subjected to four-dimensional separation prior to LC-MS/MS analysis on an LTQ-FT mass spectrometer. (B) Control and cancer patient sera were subjected to three-dimensional separation prior to quantitative LC-MRM analysis on a 4000 QTRAP mass spectrometer.
Verification and initial laboratory-scale validation of candidate biomarkers in human serum represent an important second hurdle in biomarker identification, because appropriate sandwich ELISA or other immunoassays rarely are available for novel candidate biomarkers. We therefore developed an independent, multiplexed, targeted mass spectrometry verification strategy utilizing label-free MRM analysis for this purpose (Figure 1B). Previous MRM studies have shown that multidimensional separations are essential to quantitate plasma proteins in the low ng/mL range unless targeted enrichment is used.45, 46 We also recently demonstrated that this label-free GeLC-MRM workflow is highly reproducible and is capable of quantitating proteins in serum down to approximately 200 pg/mL.47 The throughput of this method is much lower than sandwich ELISA assays or their equivalent. Hence, MRM methods as used herein are not directly applicable as a clinical test. However, such MRM assays are a useful tool for initial screening of candidate biomarkers in modest-sized patient cohorts for biomarkers where validated immunoassays have not yet been established. Proteins that can distinguish between patients and controls will be carried forward for further testing, which is likely to ultimately require the costly, time-consuming task of developing sandwich ELISA assays or related immunoassays. We have recently applied this strategy to verify ADAM12, a disintegrin and metalloprotease domain-containing protein, as a novel biomarker for the diagnosis of ectopic pregnancy.48, 49 Another important advantage of the GeLC-MRM method is that SDS gel fractionation can resolve different molecular weight-forms of targeted proteins and consequently permit separate quantitation of each form of the targeted proteins. Similarly, it can distinguish between closely related protein isoforms with high confidence by targeting isoform-specific peptide sequences. Such specificity is often defined only for a few of the best-characterized immunoassays. Therefore, the label-free GeLC-MRM workflow enables rapid, sensitive, and economical initial screening of large numbers of candidate biomarkers prior to setting up stable-isotope dilution MRM assays or immunoassays for the most promising candidate biomarkers.
A potential concern with our discovery and initial validation workflows is the use of immunoaffinity protein depletion to facilitate in-depth proteome analysis. Major proteins such as albumin can act as carrier proteins that, when removed, could lead to loss of interacting non-targeted proteins.50 In fact, using Cibachron blue dye affinity chromatography, the ‘molecular sponge’ property of albumin has been exploited to identify peptide biomarker candidates for ovarian cancer.51 However, the degree of non-targeted protein losses appeared to depend on the method used to remove major proteins. Less specific methods, such as Cibachron blue dye, result in greater losses compared to the more specific immunodepletion methods commonly used currently.52 Recent studies using the MARS and IgY-14 columns showed that immunodepletion of major proteins was highly reproducible, and less than 40 non-targeted proteins were detected in the bound fraction.53, 54 In addition, these non-targeted bound proteins were mostly captured at a low level in a reproducible manner. Major protein immunodepletion also resulted in a 25% increase in identified proteins, including some low-abundance (<10 ng/ml) plasma proteins, and enriched nontargeted plasma proteins by ~ 4-fold compared to undepleted plasma.54 While the identified low-abundance proteins only accounted for ~6% of total identified proteins, it is unclear whether alternative untargeted proteomic strategies are able to greatly outperform major protein immunodepletion in detecting low-abundance proteins. Therefore, despite its limitations, major protein immunodepletion is an effective method for reducing plasma or serum proteome complexity to enhance protein detection.
Xenograft mouse model of human ovarian endometrioid cancer
To identify candidate serum biomarkers using the xenograft mouse model, TOV-112D ovarian endometrioid tumor cells were injected subcutaneously. This cell line was originally derived from a patient with a malignant ovarian tumor that had never been exposed to radiation or chemotherapy.55 This cell line was chosen because it has a fast growth rate and has been demonstrated to form tumors readily in immune-compromised mice. More importantly, the in vitro growth characteristics of the cell line mimic the aggressive clinical behavior of the cancer.55 In addition, previous proteomics biomarker discovery studies used human SKOV-3 serous ovarian cancer cells.36, 38 It is important to note that ovarian cancer is a heterogeneous disease with four major subtypes: serous, clear cell, endometrioid, and mucinous that develop differently, have distinct underlying molecular events during oncogenesis, respond differently to chemotherapy, and have differences in gene expression.2, 3 Therefore, we expect proteomics biomarker discovery using different ovarian cancer cell subtypes will lead to identification of sets of biomarkers where some proteins are common to all or multiple cancer subtypes, while other proteins may be specific to a single subtype.
Four-dimensional protein profiling of xenograft mouse serum proteome
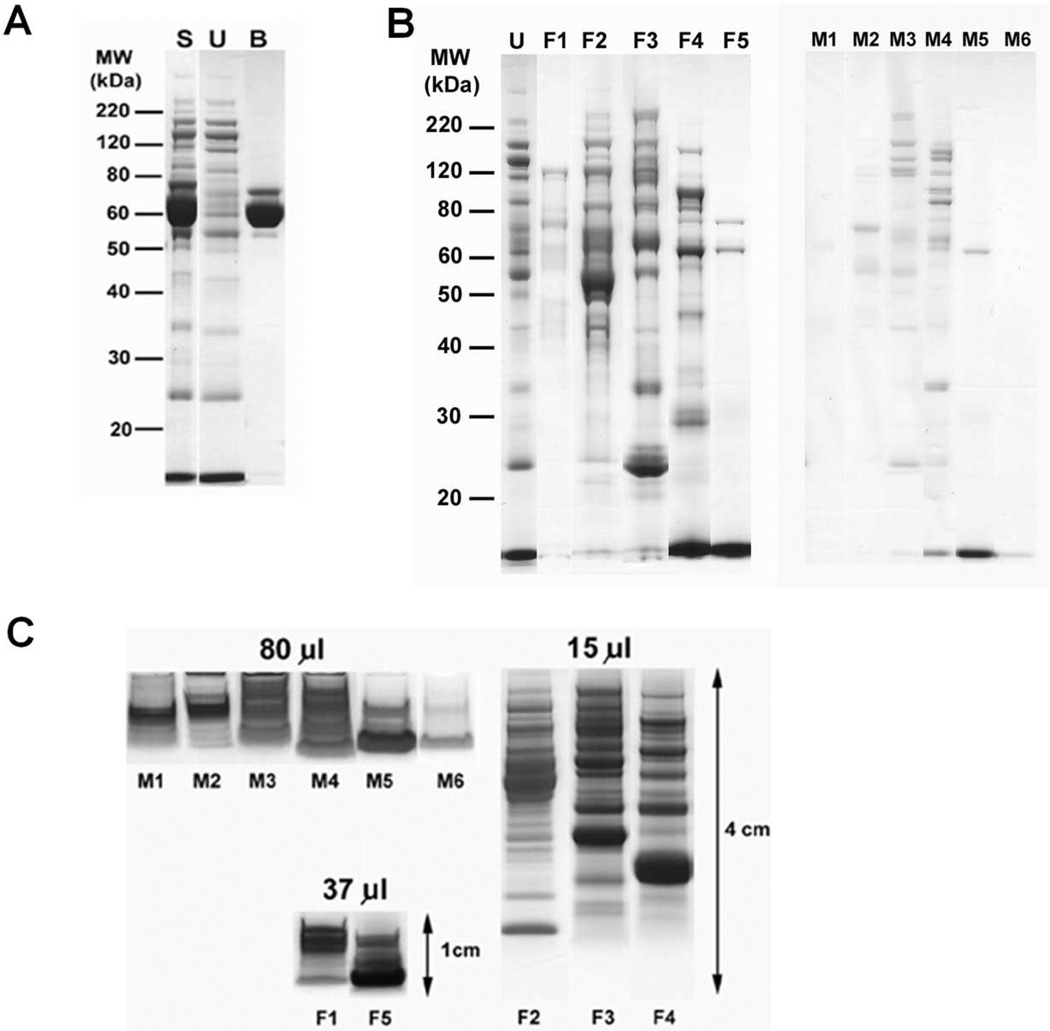
The mouse serum was first subjected to depletion of three major proteins from a total of 225 µl (10.2 mg) serum using a MARS Mouse-3 HPLC column. Following depletion, 3.1 mg of total unbound proteins were recovered. SDS-PAGE analysis of the unbound and bound fractions showed good depletion of albumin (69 kDa) and transferrin (77 kDa), as expected (Figure 2A). The third protein expected to be depleted by this column, IgG, was not apparent in this experiment because SCID mice have very low levels of immunoglobulins. Following major protein depletion, the unbound proteins were separated into five pI fractions using MicroSol-IEF, and proteins with pIs identical to the pH of the MicroSol-IEF separator membranes were extracted (Figure 2B). Although the total amount of protein trapped in the MicroSol-IEF membrane partitions was very low, these fractions were included in the proteome analysis to increase its comprehensiveness. The third fractionation step utilized 1-D SDS-PAGE, and to enhance detection of low-abundance protein, MicroSol IEF fractions and membrane extracts were concentrated and the largest possible protein loads that avoided excessive band distortion were applied onto the gels. Furthermore, because trypsin digestion of large gel volumes containing low protein amounts can be inefficient with disproportionally high adsorptive losses, the length of the electrophoretic separation was adjusted based upon sample complexity and the number of fractions desired. Hence, the most complex fractions (F2 to F4) were separated for 4 cm and cut into 20 × 2-mm slices, while remaining, less complex fractions were separated for 1 cm and cut into10 × 1-mm slices, with corresponding slices from duplicate lanes combined for trypsin digestion (Figure 2C). This yielded a total of 140 samples for trypsin digestion and LC-MS/MS analysis.
Figure 2.
Four-dimensional protein profiling of human ovarian TOV-112D xenograft mouse serum proteome. (A) SDS-PAGE of unfractionated serum (S), unbound fraction (U) of the MARS Mouse 3 LC-Column, and the bound fraction (B) containing the abundant serum proteins. (B) SDS-PAGE of MicroSol-IEF fractions (F1 to F5) of depleted mouse serum, and proteins extracted from MicroSol-IEF membrane partitions (M1 to M6). (C) Separation of MicroSol-IEF samples for in-gel trypsin digestion and LC-MS/MS analysis. Membrane (M1–M6) and solution fractions (F1–F5) were separated as indicated. Volumes indicate the original serum volumes from which the loaded fractions were derived.
Ovarian cancer xenograft mouse serum proteome
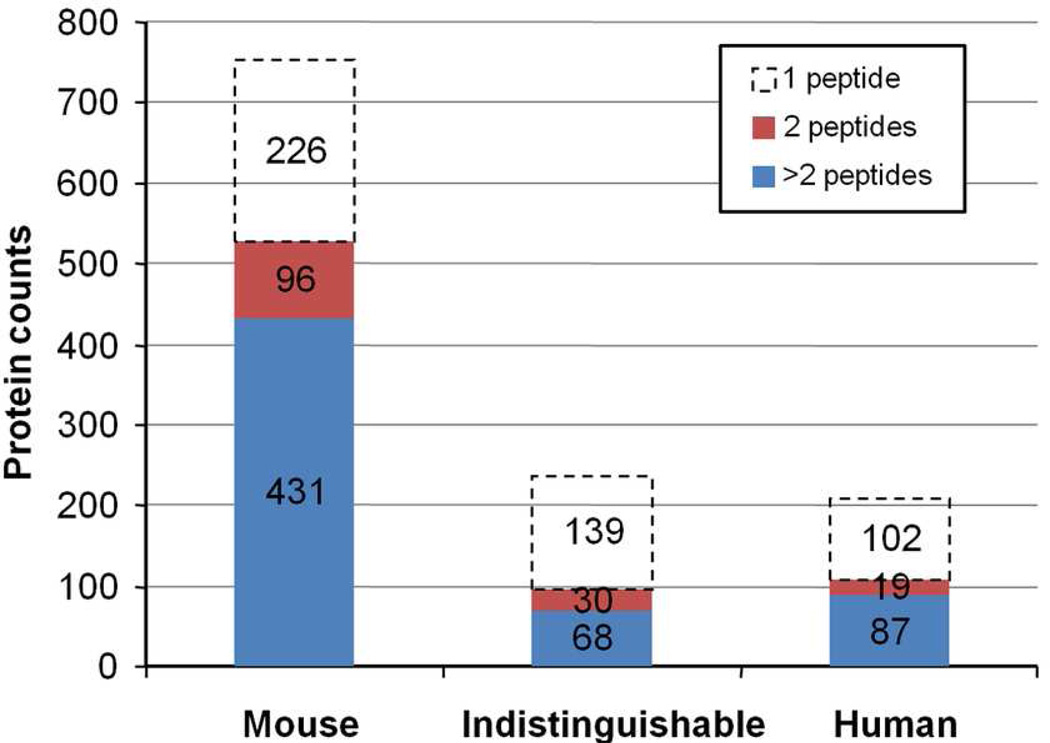
From a total of 140 LC-MS/MS runs performed on a LTQ-FT mass spectrometer, 1.2 million MS/MS spectra were acquired and searched against a mouse and human composite database. After stringent data filtering and removal of redundant entries and common contaminants, a total of 1,198 unique proteins were identified from 6,014 unique peptides (Figure 3). The peptide false discovery rate estimated from the number of hits against the reversed entries in the composite database was 1.4%. As expected, the majority of identified proteins (753) were mouse proteins, as they contained peptide sequences unique to the mouse database. Based on the database search results, 222 proteins were initially identified as human proteins because they contained at least one apparent human-specific peptide. To confirm the species assignment, all peptides for these putative human proteins were searched against a mouse database using BLAST, and the results were used to remove proteins where all peptides were identical to mouse sequences or contained only isobaric differences (Ile/Leu). After the BLAST analysis, a total of 573 unique human-specific peptides remained and they defined 208 high-confidence human proteins identified by at least one human-specific peptide. The apparent molecular weights of these human proteins ranged from 10 to 435 kDa.
Figure 3.
Non-redundant proteins identified in the ovarian TOV-112D xenograft mouse serum proteome. The bar chart shows the number of unique mouse proteins (Mouse), proteins where all identified peptides were common to homologous mouse and human proteins (Indistinguishable), and unique human proteins (Human). The distribution of proteins identified by one, two, and greater than two unique peptides are shown.
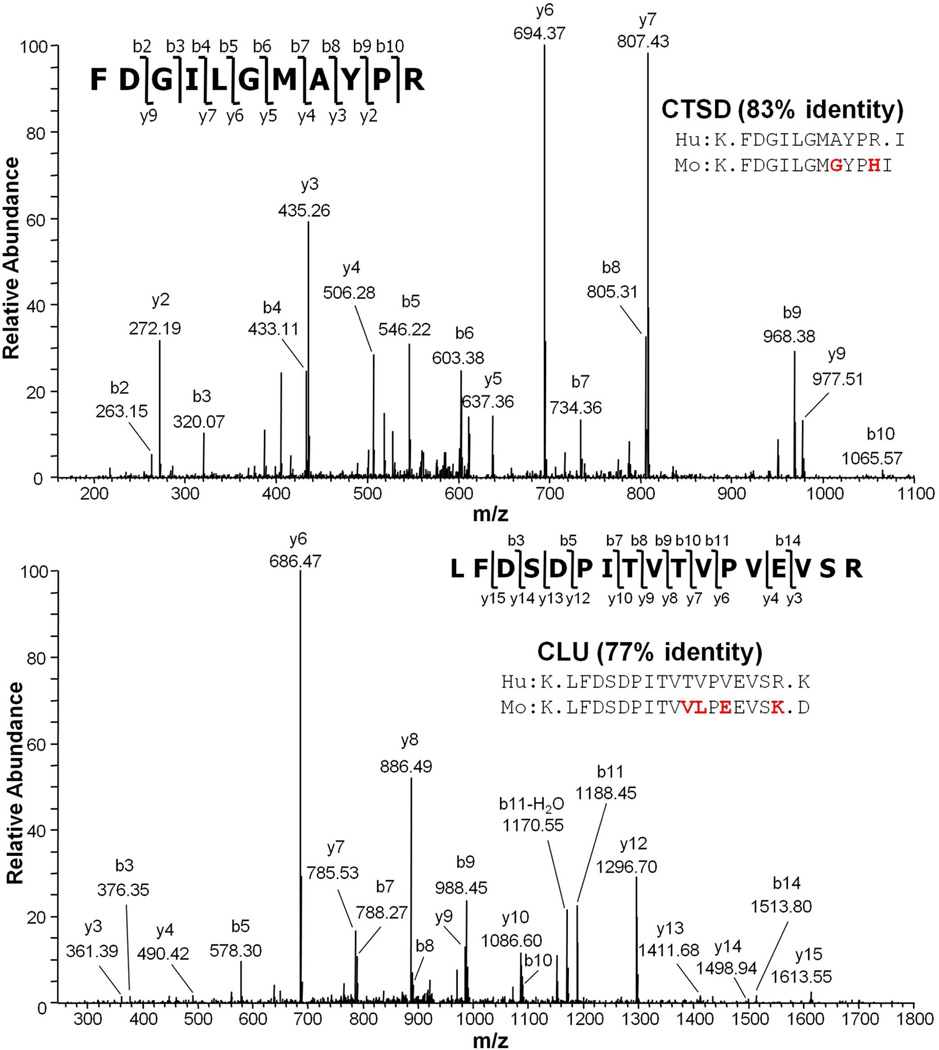
It is possible that some apparently human proteins were misidentified as human due to single nucleotide polymorphisms (SNPs) or deamidation of an asparagine to an aspartic acid. Therefore, such assignments were flagged as tentatively human. However, 87% of identified human-specific peptides differed from the homologous mouse sequences by more than a single nucleotide, indicating that this was a relatively minor concern. Two examples of MS/MS spectral assignments for human-specific peptides and the corresponding mouse sequences are shown in Figure 4. In both examples, the identified human-specific peptide sequences differ from the homologous mouse sequences by more than one residue, unambiguously indicating that the identified proteins had to be secreted by the human ovarian tumors into the mouse blood. In addition to the mouse or human-specific proteins, 237 proteins were identified by peptides common to both mouse and human sequences, and are therefore species indistinguishable at this stage.
Figure 4.
MS/MS spectra of two human-specific peptides identified from the ovarian TOV-112D xenograft mouse serum. Sequence alignments of human peptides (Hu) with their corresponding mouse peptides (Mo) are shown, and the protein sequence homology between the two species is indicated. CTSD, cathepsin D. CLU, clusterin.
These results demonstrate the feasibility of identifying large numbers of human-specific proteins from xenograft mouse models of solid tumor cancers when an in-depth analysis of the serum proteome is performed using a 4-D protein profiling strategy. Of course, as in any proteomics studies, human-specific proteins identified by only a single peptide will have a somewhat higher probability of being a false positive assignment despite the low overall peptide false discovery rate used here. Despite the somewhat higher uncertainty, these single-peptide proteins are not disregarded at this stage of analysis, because the most specific cancer biomarkers are likely to be very low-abundance proteins that are only shed by the tumor. Proteins identified by single peptides are likely to be among the lowest abundance proteins in these datasets because there is a rough correlation between protein abundance and the number of peptides identified. Even if all single-peptide proteins were disregarded, 106 human proteins were identified by at least two peptides.
Previous serum proteomics studies of ovarian cancer xenograft mouse models only identified a few human-specific proteins, presumably due at least in part to lower levels of sample fractionation used in those studies.36, 38 In the study that analyzed the low-molecular weight serum proteome using LC-MS/MS analysis, only 400 peptides were identified, and these peptides corresponded to a total combination of 300 human and mouse proteins. By using MS/MS spectral counts, five human-specific proteins were identified at a statistically significant higher level in the cancer versus control xenograft model.36 In contrast, a similar low-molecular weight serum proteome analysis of a lung tumor xenograft mouse model that incorporated an additional fractionation by strong cation exchange chromatography prior to LC-MS/MS analysis was able to identify more than a thousand proteins, although no effort was made to distinguish between human and mouse proteins in that study.56 Another ovarian cancer study directly analyzed the xenograft serum proteome by LC-MS/MS and identified 13 human proteins. However, most of the human species assignments were made by comparing the results with those obtained from a separately analyzed SKOV-3 cell line secretome.38 In fact, the candidate biomarker reported in that study (14-3-3 zeta) was identified by a single peptide that is indistinguishable from the corresponding mouse sequence. Proteomics profiling of xenograft mouse models of prostate, breast, and oral squamous cell carcinomas also has been reported.57–59 The number of human-specific proteins reported using a xenograft mouse model could be influenced by the specific cell line used, the number of cells injected, the site of cell injection (subcutaneously vs. orthotopically), total human tumor burden at blood collection, and other tumor properties such as extent of necrosis. However, the most critical factors that affect depth of analysis are likely to be the total human tumor burden at the time of blood collection and the extent of plasma fractionation prior to LC-MS/MS analysis. For example, the previous xenograft mouse studies utilized three or less dimensions of sample fractionation and resulted in identification of less than 20 human-specific proteins. In contrast to these earlier studies, the current study demonstrates that a more in-depth protein profiling strategy, such as the 4-D method, is crucial for successful identification of substantial numbers of human-specific proteins in xenograft mouse serum.
Human-specific proteins released by ovarian tumors
A group of interesting human proteins identified in this study are summarized in Table 1, and details of the peptide identifications are reported in Supplemental Table 1. Interestingly, a substantial number of common, relatively abundant serum proteins, such as ITIH4, APOA1, TTR, and TF are shed by the ovarian tumors. Some of these common plasma proteins were observed in human ovarian tumor specimens in prior reports, but it was not clear whether these proteins infiltrated the tissue from the blood or if the tumor produced these proteins. Some of these abundant proteins also have been associated with host-response or acute-phase reaction to biological insults and are primarily synthesized by the liver. However, the identification of multiple, human-specific peptides for these proteins unambiguously demonstrates that they were produced and shed into the blood by the ovarian tumors. Despite their unambiguous tumor origin, these proteins are unlikely to be useful for diagnosing or monitoring ovarian cancer in humans, because the contribution to blood levels from small tumors is likely to be swamped by shedding from other tissues and normal variations in the protein’s level in the normal population. Interestingly, three of these proteins, APOA1, TTR and a fragment of ITIH4 used in a multimarker panel, were reported to have higher diagnostic accuracy than CA125 for detection of early-stage ovarian cancer.14 However, a subsequent study found that the use of these proteins in biomarker panels did not outperform CA125 when used in prediagnostic samples.18 Nevertheless, the identification of these proteins that previously have been shown to be produced by ovarian cancer cells or tissues further demonstrates that the xenograft mouse model system is a realistic system for serum biomarker discovery.
Table 1.
Selected human proteins shed by ovarian TOV-112D tumors into SCID mouse blood
| UniProt | Name | Descriptive Name | Hu/Ina | Peptides Verifiedb |
MRM Assay |
Comments |
|---|---|---|---|---|---|---|
| Medium and High Abundance Serum Proteins | ||||||
| Q14624 | ITIH4 | Inter-alpha-trypsin inhibitor heavy chainH4 |
5/5 | n.d. | n.d. | Biomarker for ovarian cancer14 |
| P02647 | APOA1 | Apolipoprotein A–I | 10/0 | n.d. | n.d. | Biomarker for ovarian cancer14, 80 |
| P61769 | B2M | Beta-2-microglobulin | 1/0 | n.d. | n.d. | Overexpressed in many cancer cells, including ovarian81 |
| P02766 | TTR | Transthyretin | 4/0 | n.d. | n.d. | Biomarker for ovarian cancer14, 80 |
| P02787 | TF | Transferrin | 4/1 | n.d. | n.d. | Biomarker for ovarian cancer80 |
| P07195 | LDHB | L-lactate dehydrogenase B chain |
1/6 | n.d. | n.d. | Increased in tissue cytosol from patients with ovarian cancer82 |
| P23528 | CFL1 | Cofilin-1 | 1/7 | n.d. | n.d. | Identified in TOV-112D secretome35 |
| P05090 | APOD | Apolipoprotein D | 3/0 | n.d. | n.d. | Increased in ovarian cancer tumors83 |
| P10909 | CLU | Clusterin | 7/0 | 8 | yes | Higher level in serum of ovarian cancer patients35 |
| Candidate Biomarkers Evaluated | ||||||
| P07339 | CTSD | Cathepsin D | 3/1 | 8 | yes | Prognostic factor for ovarian cancer61 |
| O00299 | CLIC1 | Chloride intracellular channel protein1 |
1/4 | 7 | yes | Identified in TOV-112D secretome2 |
| P15121 | AKR1B1 | Aldose reductase | 1/3 | 1 | no | Increase in breast, ovarian cervical, and rectal cancer tissues63 |
| Q9NP08 | HMX1 | Homeobox protein HMX1 | 1/0 | 1 | no | |
| Q7Z4N2 | TRPM1 | Melastatin 1 | 1/0 | 0 | no | |
| O60888 | CUTA | Protein CutA | 1/0 | 0 | no | Identified in TOV-112D secretome35 |
| Q3SYB4 | SERPINB12 | SERPINB12 protein | 1/0 | 0 | no | |
| P30041 | PRDX6 | Peroxiredoxin-6 | 0/4 | 6 | yes | Identified in TOV-112D secretome35 |
Number of unique human / indistinguishable peptides. MS/MS spectra of proteins identified by a single human peptide are shown in Supplemental Figure 1.
Number of peptides successfully verified in targeted Orbitrap MS/MS analysis of pooled advanced ovarian cancer sera. n.d., not determined.
Some of the human proteins identified were previously reported to be possible serum biomarkers for ovarian cancer. For example, CTSD level has been proposed as a prognostic factor in a variety of cancers, including ovarian cancer.60, 61 Recently, it also was shown that quantitation of circulating autoantibody against CTSD can differentiate benign ovarian conditions from ovarian carcinoma.62 Similarly, CLU was shown to be present at a higher level in serum of late-stage ovarian cancer patients versus normal controls.35 A number of other proteins such as LDHB, CFL1, CLIC1, and AKR1B1 also were identified previously in ovarian cancer tissues or in conditioned media for ovarian cancer cell lines,35, 63 but they were not previously known to be shed into the blood.
Most of the human proteins identified have at least 70% sequence identity with their mouse counterpart, and some share ≥ 90% identity with the mouse protein. An example of a highly homologous protein is CLIC1, which shares 98% sequence identity between the two species. Despite this very high sequence homology, a human-specific peptide, LAALNPESNTAGLDIFAK, was identified that indicated the protein was shed by the human ovarian tumor (Figure 5). This demonstrates that very highly homologous human and mouse proteins can be distinguished using the xenograft mouse model and the 4-D serum protein profiling method.
Figure 5.
Sequence alignment of the human and mouse CLIC1 protein. The identified CLIC1 peptides are indicated. Tryptic sites (K or R) are indicated in bold.
Xenograft mouse models of ovarian cancer are widely used in therapeutic and tumor biology studies.64, 65 Most xenografts are created by subcutaneous, intraperitoneal, or orthotopic intrabursal implantation of tumor cells/tissues. Some studies have shown that the site of implantation has no effect on the histopathological characteristics of the tumor66 or the ability to support early follicular growth of ovarian tissue.67 While secretion of some tumor proteins are likely to be affected by the implantation site, the similarities of histopathological characteristics suggest that many proteins shed by the tumor will be independent of tumor microenvironment. In support of the biological relevance of our xenograft mouse model, more than 50% of the human proteins identified in the current study were also identified in secretomes of primary human ovarian cancer tumor tissue in short-term organ culture (data not shown).
The production by the ovarian tumors and shedding into the mouse blood of proteins previously associated with ovarian cancer argue that this model effectively mimics normal protein shedding of ovarian cancer in humans. In addition, the xenograft mouse model allows detection of low-abundance serum proteins such as CTSD, which has a reported serum concentration of about 16 ng/mL.68, 69 Another important benefit of the use of the xenograft mouse model over cell lines, tumor tissues, and secretomes from cells or tissues is the ability to show that the identified proteins are actually shed into blood. Since the human proteins identified in this study were both unambiguously produced by the ovarian tumor and shed into the blood, they are likely to be shed into the bloodstream of ovarian cancer patients. Hence, they are candidate biomarkers worth testing in human patients. As noted above, the most cancer-specific biomarkers probably will be those proteins shed primarily by the tumor and not other organs such as the liver. Therefore, candidate biomarkers from the xenograft mouse model that either have not been reported previously in normal human serum or are known to be low abundance are considered high-priority candidates for screening in ovarian cancer patient sera.
Verification of selected candidate ovarian cancer biomarkers in human ovarian cancer patient sera
The potential utility of several high-priority candidate biomarkers for screening ovarian cancer patients was evaluated in a small patient cohort, as outlined in Figure 1 and described in Tang et al.47 In this proof-of-concept experiment, a subset of seven human proteins within the 20 to 55 kDa range was selected (CTSD, CLIC1, AKR1B1, HMX1, TRPM1, CUTA, and SERPINB12) for initial evaluation (Table 1). In addition, a higher abundant known ovarian-cancer-associated serum protein (CLU) was included as a positive control.
Although these human proteins were detected in the xenograft mouse serum, the ability to detect them in human serum using GeLC-MRM was unknown. Therefore, multiple peptides from each of these proteins initially were targeted by LC-MS/MS on an LTQ-Orbitrap XL mass spectrometer using a pool of abundant protein-depleted serum from nine late-stage ovarian cancer patients (Table 1). The targeted MS/MS analysis was able to identify multiple peptides for CLU, CTSD, and CLIC1. However, only a single peptide was identified for AKR1B1 and HMX1, and three proteins (TRPM1, CUTA, and SERPINB12) could not be identified. The inability to detect TRPM1, CUTA, and SERPINB12 might be due to either very low concentrations in human serum, as suggested by the identification of only a single peptide in the xenograft mouse system, or these single peptide proteins might be false positive identifications from the discovery experiment. Another possibility is that these proteins could be relatively specific to endometriod ovarian tumors, since the majority of the patient samples tested so far are from serous ovarian cancer patients (Supplemental Table 2). This possibility could be further tested by obtaining larger numbers of endometrioid subtype samples in future validation tests. Our minimum criteria for high-confidence GeLC-MRM quantitation required detection and quantitation of at least two peptides per protein and, therefore, AKR1B1 and HMX1 were not included in the MRM assays.
Preliminary validation of candidate biomarkers using GeLC-MRM quantitation
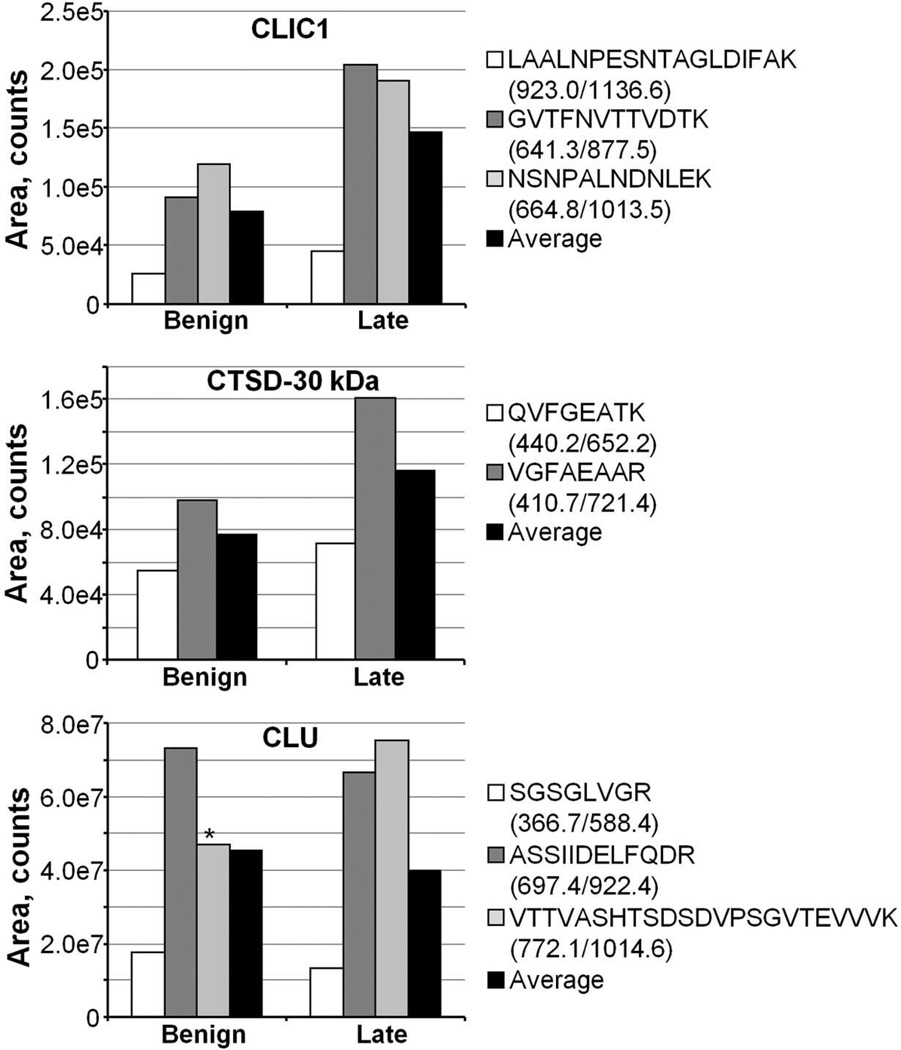
GeLC-MRM quantitation was performed initially on separate pools of nine serum samples from patients diagnosed with benign, and late- (stages III and IV) stage ovarian cancer. To assess the robustness of the MRM methods for the selected proteins and to obtain a preliminary indication of the predictive value of CLU, CTSD, and CLIC1, gel slices identified as containing the proteins in the xenograft mouse analysis, as well as adjacent fractions, were analyzed, and relative peptide amounts were summed across gel slices (Figure 6). As expected, typically all peptides from a given protein displayed similar trends across the three pooled serum samples. In the case of CLU, one peptide was disproportionately low in the benign samples. Examination of the raw data showed splitting of the peptide peak, apparently due to variations in spray in the triple quadrupole nanosource. Hence, this peptide was not used for protein quantitation in this dataset. It should also be noted that some serum proteins such as CTSD and CLU undergo proteolytic processing to yield mature forms of the protein. Although both the full-length and mature forms are detectable in serum if all fractions of the gel are analyzed, it is more efficient if analysis focuses on a discrete region of the gel to maximize throughput. In this study, we focused on the 20–55 kDa region, which included the mature forms of CTSD and CLU but not the full-length (unprocessed) forms of these proteins, which will be quantified in a future study that evaluates the higher molecular weight region of the gel.
Figure 6.
GeLC-MRM quantitation of CLIC1, CTSD-30 kDa, and CLU in pooled serum samples from patients with benign (n=9) and advanced (stage III, n= 6; stage IV, n=3) ovarian cancer. Asterisk indicates a CLU peptide affected by spray instability, and was not used in computing the Average value. The MRM transitions used for quantitation are indicated.
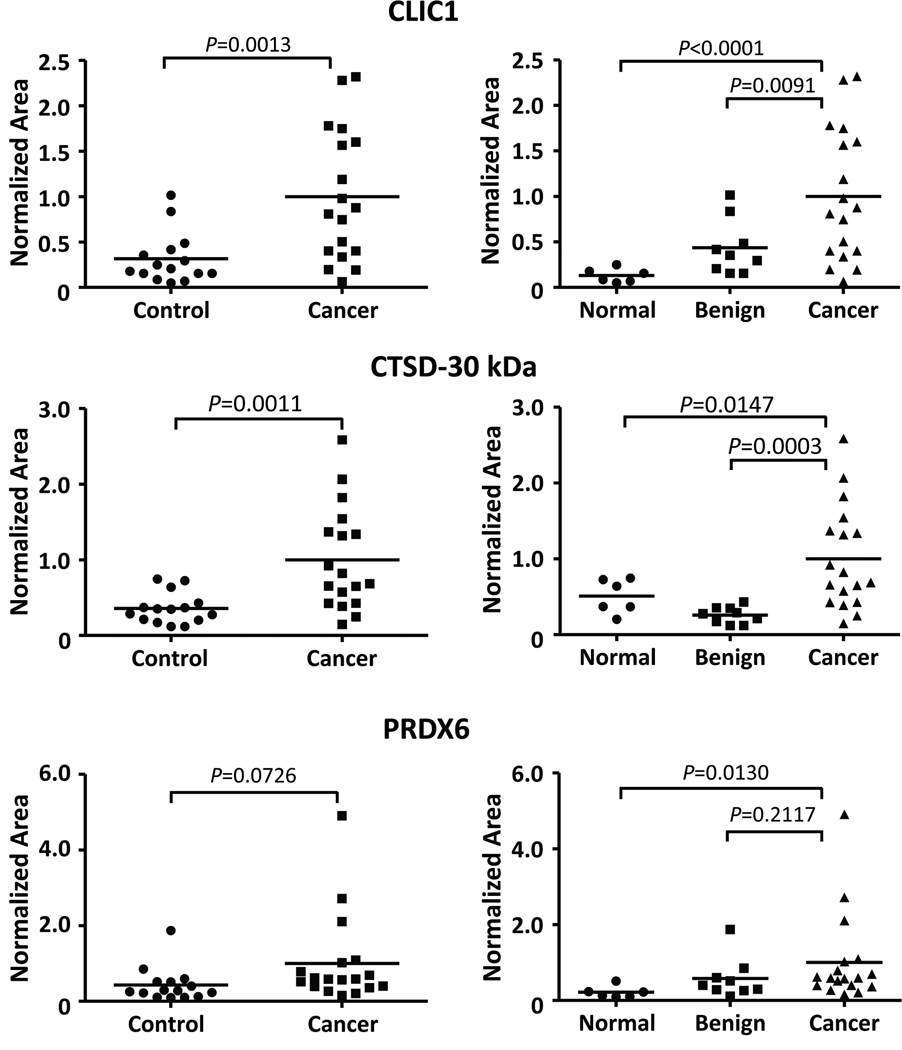
The results obtained from a preliminary analysis of the pooled samples showed that the CLIC1 and the CTSD mature forms (henceforth referred to as CTSD-30 kDa) exhibited the greatest difference between benign and late-stage ovarian cancer and therefore warranted further analysis. The levels of these proteins were measured in individual control serum samples (six normal and nine benign), and late-stage cancer samples (15 stage III and 3 stage IV). We did not continue to evaluate CLU because it is a known high-abundant plasma protein with reported concentration ranging from 58 to 150 µg/mL,35, 70 and its level is also elevated by many acute-phase stimuli such as inflammation, heat shock, and injury.71, 72 In addition to CLIC1 and CTSD-30 kDa, PRDX6 was included in subsequent analyses. PRDX6 is a 25 kDa bifunctional 1-Cys peroxiredoxin that has been hypothesized to promote cancer growth and invasiveness, with increased expression observed in some malignancies.73–75 Although human PRDX6 was not conclusively identified in this study, the mouse PRDX6 was identified with four peptides that are indistinguishable from human. In addition, PRDX6 was identified in a TOV-112D secretome study.35 Taken together, these results suggest PRDX6, which is in the 25 kDa region being assayed, could be a potential biomarker for ovarian cancer.
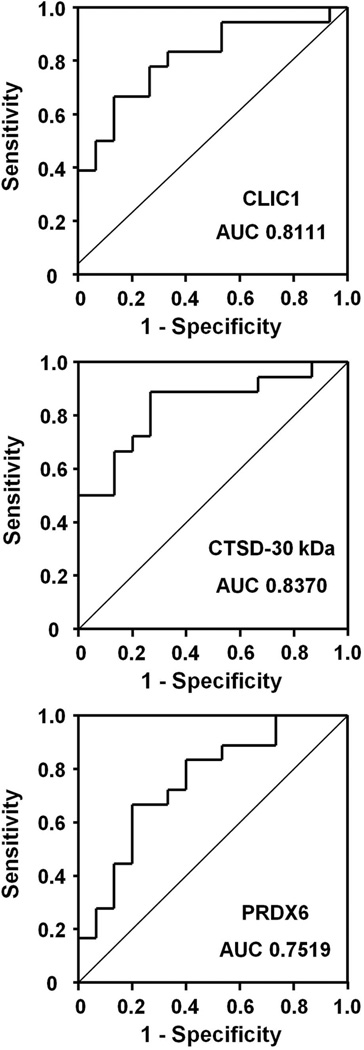
Label-free MRM quantitation of individual serum samples showed a significant difference (P<0.05, Student’s t-test) between the control (normal and benign) and cancer groups for CLIC1 and CTSD-30 kDa (Figure 7, left panels). The normal and benign samples also were compared separately to the cancer group, since the normal and benign samples were collected at two different sites and benign conditions often are more difficult to distinguish from cancer than healthy controls (Figure 7, right panels). PRDX6 showed a significant difference between normal controls and cancer but not between benign disease and cancer, whereas there were significant differences between cancer and either non-cancer group for the other two biomarkers. To further evaluate the potential diagnostic efficacy for each of the three proteins, receiver operating characteristic curve analyses were performed on the control and cancer groups (Figure 8). In agreement with the t-test, both CLIC1 and CTSD-30 kDa showed a larger area under the ROC (AUC) compared to PRDX6. The sensitivity and specificity, as well as the positive and negative predictive values for each biomarker at the optimal cut-point, are presented in Supplemental Table 3. A binary decision rule for CTSD1-30kDa, CLIC1, and PRDX6 was created using their optimal cut-point. Each binary variable was a significant predictor of tumor samples (p=0.001. 0.005, and 0.011, respectively). In the multivariate analysis, only CTSD1-30kDa and CLIC1 remained in the final model (p=0.009 and 0.052, respectively). The AUC for the predicted probability of a tumor sample from the multivariate model, including these two biomarkers, was 0.893.
Figure 7.
Scatter plots showing LC-MRM quantitation of CLIC1, CTSD-30 kDa, and PRDX6 in serum of ovarian cancer patients (Cancer: 15 stage III, 3 stage IV) and individuals without ovarian cancer (Control: 6 normal, 9 benign). Right panels, scatter plots of the same data comparing Normal and Benign groups separately with the Cancer samples. P-values were calculated using Student’s t-test. Horizontal bars in each dataset indicate the average serum level of the protein. MRM transitions used and quantitation values for all samples are listed in Supplemental Table 2.
Figure 8.
ROC curves of CLIC1, CTSD-30 kDa, and PRDX6. ROC curves were generated from Control (6 normal, 9 benign) and Cancer (15 stage III, 3 stage IV) datasets. The area under the ROC curve (AUC) is indicated for each candidate.
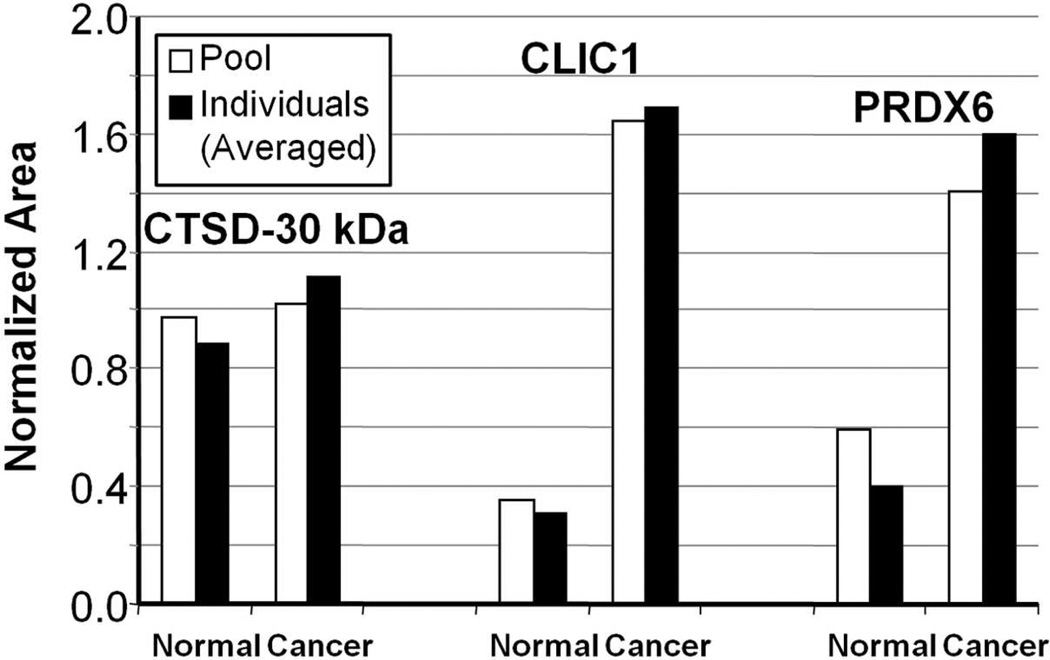
We previously demonstrated the reproducibility of the label-free GeLC-MRM workflow in quantitating biomarkers for ectopic pregnancy.47, 48 To evaluate the reproducibility of the entire label-free GeLC-MRM workflow for the ovarian cancer biomarkers, we prepared two separate serum pools of normal (n=6) and ovarian cancer (n=9) samples and subjected the two pooled samples to major protein depletion and GeLC-MRM quantitation, as shown in Figure 1B. MRM quantitation of CTSD-30 kDa, CLIC1, and PRDX6 in this new pool of samples then were compared to the averaged quantitation values of individual samples that made up the pooled samples. As shown in Figure 9, MRM quantitation of the normal and cancer samples are very similar for the pool samples and the average of individual samples, demonstrating the reproducibility of the entire label-free GeLC-MRM workflow. In this analysis, the level of CTSD did not appear to be significantly different between the two groups due to the inclusion of more low responders in the cancer group. This highlights the potential risks of using pooled serum samples to attempt to gauge the predictive value of a biomarker, and the importance of analyzing individual samples in biomarker validation. These results indicate that CTSD-30 kDa and CLIC1 are promising biomarkers of ovarian cancer, although the performance of these two markers for early detection remains to be determined as larger cohorts, as well as early-stage cancer samples and pre-diagnosis specimens, need to be tested in future studies. While PRDX6 is a less promising biomarker based upon the current data, further studies using a larger cohort may show efficacy of this biomarker, particularly as part of a multi-protein panel. To our knowledge, CTSD, CLIC1, and PRDX6 levels in serum of ovarian cancer patients have not been reported previously—although, a recent report did show that quantitation of circulating autoantibody against CTSD can differentiate between benign and other stages of ovarian carcinoma including stage I,62 which supports our argument that CTSD is a biomarker for ovarian cancer. Interestingly, CLIC1 recently was discovered as a novel plasma marker for nasopharyngeal carcinoma,76 although its role in ovarian cancer is still unclear.
Figure 9.
Comparison of GeLC-MRM quantitation of CTSD-30 kDa, CLIC1, and PRDX6 from pooled samples versus average of individual samples. White bars, GeLC-MRM quantitation obtained from a pool of Normal (n=6) and a pool of Cancer (n=9) serum samples. Black bars, GeLC-MRM quantitation obtained from the six individual Normal (average) and nine individual Cancer (average) serum samples.
Conclusions
New proteomic technologies or strategies are constantly being refined or developed to improve identification of novel cancer biomarkers. One interesting approach is the use of nanoparticle capture technology to enrich for the low-molecular weight peptidome that has been suggested to contain a rich source of cancer-specific diagnostic information.77, 78 Another strategy involves the use of combinatorial peptide libraries to enrich for low-abundance proteins in ovarian cancer ascites.79 Here, we demonstrate that analysis of serum from an ovarian cancer xenograft mouse model using a 4-D protein profiling method is capable of deep mining of the mouse plasma with resulting identification of more than two hundred human proteins that are unambiguously shed by human tumors into the blood. While the xenograft mouse model is not expected to fully recapitulate the in vivo human tumor microenvironment in patients, many of the proteins identified in this study have been previously associated with ovarian cancer, although many others were not previously known to be produced by the tumor and shed into the blood. Verification and small-scale validation of selected high-priority candidate biomarkers show that a substantial portion of the tested candidate biomarkers correlate with ovarian cancer in patient serum specimens.
Supplementary Material
Acknowledgements
This work was supported by National Institutes of Health grants CA131582 and CA120393 to D.W.S., as well as an institutional grant to the Wistar Institute (NCI Cancer Core Grant CA010815). We gratefully acknowledge Dr. Dionyssios Katsaros, University of Turin, Turin, Italy, for providing benign and ovarian cancer sera. We gratefully acknowledge the Wistar Institute Proteomics Core, the Histotechnology Core, and the Animal Facility for their assistance with the project.
Footnotes
Supporting Information Available
Figure showing MS/MS spectra of all single-peptide proteins listed in Table 1 showing details of Sequest peptide identification of selected human proteins shed by ovarian TOV-112D tumors into SCID mouse; Table showing normalized MRM peak area values for CLIC1, CTSD, and PRDX6 peptides and their averages in all samples analyzed; Table showing the sensitivity, specificity, and positive and negative predicted values for tumor vs. normal and benign. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60(5):277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Bell DA. Origins and molecular pathology of ovarian cancer. Mod Pathol. 2005;18 Suppl 2:S19–s32. doi: 10.1038/modpathol.3800306. [DOI] [PubMed] [Google Scholar]
- 3.Kobel M, Kalloger SE, Boyd N, McKinney S, Mehl E, Palmer C, Leung S, Bowen NJ, Ionescu DN, Rajput A, Prentice LM, Miller D, Santos J, Swenerton K, Gilks CB, Huntsman D. Ovarian carcinoma subtypes are different diseases: implications for biomarker studies. PLoS Med. 2008;5(12):e232. doi: 10.1371/journal.pmed.0050232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cannistra SA. Cancer of the ovary. N Engl J Med. 2004;351(24):2519–2529. doi: 10.1056/NEJMra041842. [DOI] [PubMed] [Google Scholar]
- 5.Mutch DG. Surgical management of ovarian cancer. Semin Oncol. 2002;29 1 Suppl 1:3–8. doi: 10.1053/sonc.2002.31589. [DOI] [PubMed] [Google Scholar]
- 6.Grover SR, Quinn MA. Is there any value in bimanual pelvic examination as a screening test. Med J Aust. 1995;162(8):408–410. [PubMed] [Google Scholar]
- 7.Clarke-Pearson DL. Clinical practice. Screening for ovarian cancer. N Engl J Med. 2009;361(2):170–177. doi: 10.1056/NEJMcp0901926. [DOI] [PubMed] [Google Scholar]
- 8.Mutch DG. Ovarian cancer: to screen or not to screen. Obstet Gynecol. 2009;113(4):772–774. doi: 10.1097/AOG.0b013e31819ece24. [DOI] [PubMed] [Google Scholar]
- 9.Moss EL, Hollingworth J, Reynolds TM. The role of CA125 in clinical practice. J Clin Pathol. 2005;58(3):308–312. doi: 10.1136/jcp.2004.018077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nick AM, Sood AK. The ROC ‘n’ role of the multiplex assay for early detection of ovarian cancer. Nat Clin Pract Oncol. 2008;5(10):568–569. doi: 10.1038/ncponc1214. [DOI] [PubMed] [Google Scholar]
- 11.Terry KL, Sluss PM, Skates SJ, Mok SC, Ye B, Vitonis AF, Cramer DW. Blood and urine markers for ovarian cancer: a comprehensive review. Dis Markers. 2004;20(2):53–70. doi: 10.1155/2004/241982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dutta S, Wang FQ, Phalen A, Fishman DA. Biomarkers for ovarian cancer detection and therapy. Cancer Biol Ther. 2010;9(9):668–677. doi: 10.4161/cbt.9.9.11610. [DOI] [PubMed] [Google Scholar]
- 13.Cramer DW, Bast RC, Jr, Berg CD, Diamandis EP, Godwin AK, Hartge P, Lokshin AE, Lu KH, McIntosh MW, Mor G, Patriotis C, Pinsky PF, Thornquist MD, Scholler N, Skates SJ, Sluss PM, Srivastava S, Ward DC, Zhang Z, Zhu CS, Urban N. Ovarian cancer biomarker performance in prostate, lung, colorectal, and ovarian cancer screening trial specimens. Cancer Prev Res (Phila) 2011;4(3):365–74. doi: 10.1158/1940-6207.CAPR-10-0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Z, Bast RC, Jr, Yu Y, Li J, Sokoll LJ, Rai AJ, Rosenzweig JM, Cameron B, Wang YY, Meng XY, Berchuck A, Van Haaften-Day C, Hacker NF, de Bruijn HW, van der Zee AG, Jacobs IJ, Fung ET, Chan DW. Three biomarkers identified from serum proteomic analysis for the detection of early stage ovarian cancer. Cancer Res. 2004;64(16):5882–5890. doi: 10.1158/0008-5472.CAN-04-0746. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Z, Yu Y, Xu F, Berchuck A, van Haaften-Day C, Havrilesky LJ, de Bruijn HW, van der Zee AG, Woolas RP, Jacobs IJ, Skates S, Chan DW, Bast RC., Jr Combining multiple serum tumor markers improves detection of stage I epithelial ovarian cancer. Gynecol Oncol. 2007;107(3):526–531. doi: 10.1016/j.ygyno.2007.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorelik E, Landsittel DP, Marrangoni AM, Modugno F, Velikokhatnaya L, Winans MT, Bigbee WL, Herberman RB, Lokshin AE. Multiplexed immunobead-based cytokine profiling for early detection of ovarian cancer. Cancer Epidemiol Biomarkers Prev. 2005;14(4):981–987. doi: 10.1158/1055-9965.EPI-04-0404. [DOI] [PubMed] [Google Scholar]
- 17.Visintin I, Feng Z, Longton G, Ward DC, Alvero AB, Lai Y, Tenthorey J, Leiser A, Flores-Saaib R, Yu H, Azori M, Rutherford T, Schwartz PE, Mor G. Diagnostic markers for early detection of ovarian cancer. Clin Cancer Res. 2008;14(4):1065–1072. doi: 10.1158/1078-0432.CCR-07-1569. [DOI] [PubMed] [Google Scholar]
- 18.Zhu CS, Pinsky PF, Cramer DW, Ransohoff DF, Hartge P, Pfeiffer RM, Urban N, Mor G, Bast RC, Jr, Moore LE, Lokshin AE, McIntosh MW, Skates SJ, Vitonis A, Zhang Z, Ward DC, Symanowski JT, Lomakin A, Fung ET, Sluss PM, Scholler N, Lu KH, Marrangoni AM, Patriotis C, Srivastava S, Buys SS, Berg CD. A framework for evaluating biomarkers for early detection: validation of biomarker panels for ovarian cancer. Cancer Prev Res (Phila) 2011;4(3):375–383. doi: 10.1158/1940-6207.CAPR-10-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pitteri S, Hanash S. A systems approach to the proteomic identification of novel cancer biomarkers. Dis Markers. 2010;28(4):233–239. doi: 10.3233/DMA-2010-0696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1(11):845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 21.Rifai N, Gillette MA, Carr SA. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat Biotechnol. 2006;24(8):971–983. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 22.Tang HY, Ali-Khan N, Echan LA, Levenkova N, Rux JJ, Speicher DW. A novel four-dimensional strategy combining protein and peptide separation methods enables detection of low-abundance proteins in human plasma and serum proteomes. Proteomics. 2005;5(13):3329–3342. doi: 10.1002/pmic.200401275. [DOI] [PubMed] [Google Scholar]
- 23.Tang HY, Speicher DW. Complex proteome prefractionation using microscale solution isoelectrofocusing. Expert Rev Proteomics. 2005;2(3):295–306. doi: 10.1586/14789450.2.3.295. [DOI] [PubMed] [Google Scholar]
- 24.Jacobs JM, Adkins JN, Qian WJ, Liu T, Shen Y, Camp DG, 2nd, Smith RD. Utilizing human blood plasma for proteomic biomarker discovery. J Proteome Res. 2005;4(4):1073–1085. doi: 10.1021/pr0500657. [DOI] [PubMed] [Google Scholar]
- 25.Lee HJ, Lee EY, Kwon MS, Paik YK. Biomarker discovery from the plasma proteome using multidimensional fractionation proteomics. Curr Opin Chem Biol. 2006;10(1):42–49. doi: 10.1016/j.cbpa.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 26.Zeng Z, Hincapie M, Pitteri SJ, Hanash S, Schalkwijk J, Hogan JM, Wang H, Hancock WS. A Proteomics Platform Combining Depletion, Multi-lectin Affinity Chromatography (M-LAC), and Isoelectric Focusing to Study the Breast Cancer Proteome. Anal Chem. 2011;83(12):4845–4854. doi: 10.1021/ac2002802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Faca V, Pitteri SJ, Newcomb L, Glukhova V, Phanstiel D, Krasnoselsky A, Zhang Q, Struthers J, Wang H, Eng J, Fitzgibbon M, McIntosh M, Hanash S. Contribution of protein fractionation to depth of analysis of the serum and plasma proteomes. J Proteome Res. 2007;6(9):3558–3565. doi: 10.1021/pr070233q. [DOI] [PubMed] [Google Scholar]
- 28.Gagne JP, Gagne P, Hunter JM, Bonicalzi ME, Lemay JF, Kelly I, Le Page C, Provencher D, Mes-Masson AM, Droit A, Bourgais D, Poirier GG. Proteome profiling of human epithelial ovarian cancer cell line TOV-112D. Mol Cell Biochem. 2005;275(1–2):25–55. doi: 10.1007/s11010-005-7556-1. [DOI] [PubMed] [Google Scholar]
- 29.Dai L, Li C, Shedden KA, Misek DE, Lubman DM. Comparative proteomic study of two closely related ovarian endometrioid adenocarcinoma cell lines using cIEF fractionation and pathway analysis. Electrophoresis. 2009;30(7):1119–1131. doi: 10.1002/elps.200800505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu Y, Wu R, Sangha N, Yoo C, Cho KR, Shedden KA, Katabuchi H, Lubman DM. Classifications of ovarian cancer tissues by proteomic patterns. Proteomics. 2006;6(21):5846–5856. doi: 10.1002/pmic.200600165. [DOI] [PubMed] [Google Scholar]
- 31.Bengtsson S, Krogh M, Szigyarto CA, Uhlen M, Schedvins K, Silfversward C, Linder S, Auer G, Alaiya A, James P. Large-scale proteomics analysis of human ovarian cancer for biomarkers. J Proteome Res. 2007;6(4):1440–1450. doi: 10.1021/pr060593y. [DOI] [PubMed] [Google Scholar]
- 32.Gortzak-Uzan L, Ignatchenko A, Evangelou AI, Agochiya M, Brown KA, St Onge P, Kireeva I, Schmitt-Ulms G, Brown TJ, Murphy J, Rosen B, Shaw P, Jurisica I, Kislinger T. A proteome resource of ovarian cancer ascites: integrated proteomic and bioinformatic analyses to identify putative biomarkers. J Proteome Res. 2008;7(1):339–351. doi: 10.1021/pr0703223. [DOI] [PubMed] [Google Scholar]
- 33.Kuk C, Kulasingam V, Gunawardana CG, Smith CR, Batruch I, Diamandis EP. Mining the ovarian cancer ascites proteome for potential ovarian cancer biomarkers. Mol Cell Proteomics. 2009;8(4):661–669. doi: 10.1074/mcp.M800313-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Faca VM, Ventura AP, Fitzgibbon MP, Pereira-Faca SR, Pitteri SJ, Green AE, Ireton RC, Zhang Q, Wang H, O’Briant KC, Drescher CW, Schummer M, McIntosh MW, Knudsen BS, Hanash SM. Proteomic analysis of ovarian cancer cells reveals dynamic processes of protein secretion and shedding of extra-cellular domains. PLoS One. 2008;3(6):e2425. doi: 10.1371/journal.pone.0002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gunawardana CG, Kuk C, Smith CR, Batruch I, Soosaipillai A, Diamandis EP. Comprehensive analysis of conditioned media from ovarian cancer cell lines identifies novel candidate markers of epithelial ovarian cancer. J Proteome Res. 2009;8(10):4705–4713. doi: 10.1021/pr900411g. [DOI] [PubMed] [Google Scholar]
- 36.Wei BR, Hoover SB, Ross MM, Zhou W, Meani F, Edwards JB, Spehalski EI, Risinger JI, Alvord WG, Quinones OA, Belluco C, Martella L, Campagnutta E, Ravaggi A, Dai RM, Goldsmith PK, Woolard KD, Pecorelli S, Liotta LA, Petricoin EF, Simpson RM. Serum S100A6 concentration predicts peritoneal tumor burden in mice with epithelial ovarian cancer and is associated with advanced stage in patients. PLoS One. 2009;4(10):e7670. doi: 10.1371/journal.pone.0007670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pitteri SJ, JeBailey L, Faca VM, Thorpe JD, Silva MA, Ireton RC, Horton MB, Wang H, Pruitt LC, Zhang Q, Cheng KH, Urban N, Hanash SM, Dinulescu DM. Integrated proteomic analysis of human cancer cells and plasma from tumor bearing mice for ovarian cancer biomarker discovery. PLoS One. 2009;4(11):e7916. doi: 10.1371/journal.pone.0007916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He Y, Wu X, Liu X, Yan G, Xu C. LC-MS/MS analysis of ovarian cancer metastasis-related proteins using a nude mouse model: 14-3-3 zeta as a candidate biomarker. J Proteome Res. 2010;9(12):6180–6190. doi: 10.1021/pr100822v. [DOI] [PubMed] [Google Scholar]
- 39.Juan HF, Chen JH, Hsu WT, Huang SC, Chen ST, Yi-Chung Lin J, Chang YW, Chiang CY, Wen LL, Chan DC, Liu YC, Chen YJ. Identification of tumor-associated plasma biomarkers using proteomic techniques: from mouse to human. Proteomics. 2004;4(9):2766–2775. doi: 10.1002/pmic.200400785. [DOI] [PubMed] [Google Scholar]
- 40.Dafou D, Ramus SJ, Choi K, Grun B, Trott DA, Newbold RF, Jacobs IJ, Jones C, Gayther SA. Chromosomes 6 and 18 induce neoplastic suppression in epithelial ovarian cancer cells. Int J Cancer. 2009;124(5):1037–1044. doi: 10.1002/ijc.24058. [DOI] [PubMed] [Google Scholar]
- 41.Zuo X, Echan L, Hembach P, Tang HY, Speicher KD, Santoli D, Speicher DW. Towards global analysis of mammalian proteomes using sample prefractionation prior to narrow pH range two-dimensional gels and using one-dimensional gels for insoluble and large proteins. Electrophoresis. 2001;22(9):1603–1615. doi: 10.1002/1522-2683(200105)22:9<1603::AID-ELPS1603>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 42.Tabb DL, McDonald WH, Yates JR., 3rd DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res. 2002;1(1):21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beer LA, Tang HY, Barnhart KT, Speicher DW. Plasma Biomarker Discovery Using 3D Protein Profiling Coupled with Label-Free Quantitation. Methods Mol Biol. 2011;728:3–27. doi: 10.1007/978-1-61779-068-3_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Omenn GS, States DJ, Adamski M, Blackwell TW, Menon R, Hermjakob H, Apweiler R, Haab BB, Simpson RJ, Eddes JS, Kapp EA, Moritz RL, Chan DW, Rai AJ, Admon A, Aebersold R, Eng J, Hancock WS, Hefta SA, Meyer H, Paik YK, Yoo JS, Ping P, Pounds J, Adkins J, Qian X, Wang R, Wasinger V, Wu CY, Zhao X, Zeng R, Archakov A, Tsugita A, Beer I, Pandey A, Pisano M, Andrews P, Tammen H, Speicher DW, Hanash SM. Overview of the HUPO Plasma Proteome Project: results from the pilot phase with 35 collaborating laboratories and multiple analytical groups, generating a core dataset of 3020 proteins and a publicly-available database. Proteomics. 2005;5(13):3226–3245. doi: 10.1002/pmic.200500358. [DOI] [PubMed] [Google Scholar]
- 45.Anderson L, Hunter CL. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol Cell Proteomics. 2006;5(4):573–588. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- 46.Keshishian H, Addona T, Burgess M, Kuhn E, Carr SA. Quantitative, multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Mol Cell Proteomics. 2007;6(12):2212–2229. doi: 10.1074/mcp.M700354-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang HY, Beer LA, Barnhart KT, Speicher DW. Rapid Verification of Candidate Serological Biomarkers Using Gel-based, Label-free, Multiple Reaction Monitoring. J Proteome Res. 2011;10(9):4005–4017. doi: 10.1021/pr2002098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beer LA, Tang HY, Sriswasdi S, Barnhart KT, Speicher DW. Systematic discovery of ectopic pregnancy serum biomarkers using 3-D protein profiling coupled with label-free quantitation. J Proteome Res. 2011;10(3):1126–1138. doi: 10.1021/pr1008866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rausch ME, Beer L, Sammel MD, Takacs P, Chung K, Shaunik A, Speicher D, Barnhart KT. A disintegrin and metalloprotease protein-12 as a novel marker for the diagnosis of ectopic pregnancy. Fertil Steril. 2011;95(4):1373–1378. doi: 10.1016/j.fertnstert.2010.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou M, Lucas DA, Chan KC, Issaq HJ, Petricoin EF, 3rd, Liotta LA, Veenstra TD, Conrads TP. An investigation into the human serum “interactome”. Electrophoresis. 2004;25(9):1289–1298. doi: 10.1002/elps.200405866. [DOI] [PubMed] [Google Scholar]
- 51.Lopez MF, Mikulskis A, Kuzdzal S, Golenko E, Petricoin EF, 3rd, Liotta LA, Patton WF, Whiteley GR, Rosenblatt K, Gurnani P, Nandi A, Neill S, Cullen S, O’Gorman M, Sarracino D, Lynch C, Johnson A, McKenzie W, Fishman D. A novel, high-throughput workflow for discovery and identification of serum carrier protein-bound peptide biomarker candidates in ovarian cancer samples. Clin Chem. 2007;53(6):1067–1074. doi: 10.1373/clinchem.2006.080721. [DOI] [PubMed] [Google Scholar]
- 52.Zolotarjova N, Martosella J, Nicol G, Bailey J, Boyes BE, Barrett WC. Differences among techniques for high-abundant protein depletion. Proteomics. 2005;5(13):3304–3313. doi: 10.1002/pmic.200402021. [DOI] [PubMed] [Google Scholar]
- 53.Liu T, Qian WJ, Mottaz HM, Gritsenko MA, Norbeck AD, Moore RJ, Purvine SO, Camp DG, 2nd, Smith RD. Evaluation of multiprotein immunoaffinity subtraction for plasma proteomics and candidate biomarker discovery using mass spectrometry. Mol Cell Proteomics. 2006;5(11):2167–2174. doi: 10.1074/mcp.T600039-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tu C, Rudnick PA, Martinez MY, Cheek KL, Stein SE, Slebos RJ, Liebler DC. Depletion of abundant plasma proteins and limitations of plasma proteomics. J Proteome Res. 2010;9(10):4982–4991. doi: 10.1021/pr100646w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Provencher DM, Lounis H, Champoux L, Tetrault M, Manderson EN, Wang JC, Eydoux P, Savoie R, Tonin PN, Mes-Masson AM. Characterization of four novel epithelial ovarian cancer cell lines. In Vitro Cell Dev Biol Anim. 2000;36(6):357–361. doi: 10.1290/1071-2690(2000)036<0357:COFNEO>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 56.Hood BL, Lucas DA, Kim G, Chan KC, Blonder J, Issaq HJ, Veenstra TD, Conrads TP, Pollet I, Karsan A. Quantitative analysis of the low molecular weight serum proteome using 18O stable isotope labeling in a lung tumor xenograft mouse model. J Am Soc Mass Spectrom. 2005;16(8):1221–1230. doi: 10.1016/j.jasms.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 57.Bijian K, Mlynarek AM, Balys RL, Jie S, Xu Y, Hier MP, Black MJ, Di Falco MR, LaBoissiere S, Alaoui-Jamali MA. Serum proteomic approach for the identification of serum biomarkers contributed by oral squamous cell carcinoma and host tissue microenvironment. J Proteome Res. 2009;8(5):2173–2185. doi: 10.1021/pr800979e. [DOI] [PubMed] [Google Scholar]
- 58.Orazine CI, Hincapie M, Hancock WS, Hattersley M, Hanke JH. A proteomic analysis of the plasma glycoproteins of a MCF-7 mouse xenograft: a model system for the detection of tumor markers. J Proteome Res. 2008;7(4):1542–1554. doi: 10.1021/pr7008516. [DOI] [PubMed] [Google Scholar]
- 59.van den Bemd GJ, Krijgsveld J, Luider TM, van Rijswijk AL, Demmers JA, Jenster G. Mass spectrometric identification of human prostate cancer-derived proteins in serum of xenograft-bearing mice. Mol Cell Proteomics. 2006;5(10):1830–1839. doi: 10.1074/mcp.M500371-MCP200. [DOI] [PubMed] [Google Scholar]
- 60.Benes P, Vetvicka V, Fusek M. Cathepsin D--many functions of one aspartic protease. Crit Rev Oncol Hematol. 2008;68(1):12–28. doi: 10.1016/j.critrevonc.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Losch A, Schindl M, Kohlberger P, Lahodny J, Breitenecker G, Horvat R, Birner P. Cathepsin D in ovarian cancer: prognostic value and correlation with p53 expression and microvessel density. Gynecol Oncol. 2004;92(2):545–552. doi: 10.1016/j.ygyno.2003.11.016. [DOI] [PubMed] [Google Scholar]
- 62.Taylor DD, Gercel-Taylor C, Parker LP. Patient-derived tumor-reactive antibodies as diagnostic markers for ovarian cancer. Gynecol Oncol. 2009;115(1):112–120. doi: 10.1016/j.ygyno.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 63.Saraswat M, Mrudula T, Kumar PU, Suneetha A, Rao Rao TS, Srinivasulu M, Reddy B. Overexpression of aldose reductase in human cancer tissues. Med Sci Monit. 2006;12(12):CR525–CR529. [PubMed] [Google Scholar]
- 64.Connolly DC, Hensley HH. Xenograft and Transgenic Mouse Models of Epithelial Ovarian Cancer and Non Invasive Imaging Modalities to Monitor Ovarian Tumor Growth In situ -Applications in Evaluating Novel Therapeutic Agents. Curr Protoc Pharmacol. 2009;45 14 12 1–14 12 26. [PMC free article] [PubMed] [Google Scholar]
- 65.Shaw TJ, Senterman MK, Dawson K, Crane CA, Vanderhyden BC. Characterization of intraperitoneal, orthotopic, and metastatic xenograft models of human ovarian cancer. Mol Ther. 2004;10(6):1032–1042. doi: 10.1016/j.ymthe.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 66.Massazza G, Tomasoni A, Lucchini V, Allavena P, Erba E, Colombo N, Mantovani A, D’Incalci M, Mangioni C, Giavazzi R. Intraperitoneal and subcutaneous xenografts of human ovarian carcinoma in nude mice and their potential in experimental therapy. Int J Cancer. 1989;44(3):494–500. doi: 10.1002/ijc.2910440320. [DOI] [PubMed] [Google Scholar]
- 67.Dath C, Van Eyck AS, Dolmans MM, Romeu L, Delle Vigne L, Donnez J, Van Langendonckt A. Xenotransplantation of human ovarian tissue to nude mice: comparison between four grafting sites. Hum Reprod. 2010;25(7):1734–1743. doi: 10.1093/humrep/deq131. [DOI] [PubMed] [Google Scholar]
- 68.Merseburger AS, Hennenlotter J, Stenzl A, Beger G, Rinnab L, Kuczyk MA, Kuefer R. Cathepsin D serum levels are not a valid serum marker in renal cell carcinoma. Urol Int. 2007;79(1):41–43. doi: 10.1159/000102912. [DOI] [PubMed] [Google Scholar]
- 69.Fukuda ME, Iwadate Y, Machida T, Hiwasa T, Nimura Y, Nagai Y, Takiguchi M, Tanzawa H, Yamaura A, Seki N. Cathepsin D is a potential serum marker for poor prognosis in glioma patients. Cancer Res. 2005;65(12):5190–5194. doi: 10.1158/0008-5472.CAN-04-4134. [DOI] [PubMed] [Google Scholar]
- 70.Hortin GL, Sviridov D, Anderson NL. High-abundance polypeptides of the human plasma proteome comprising the top 4 logs of polypeptide abundance. Clin Chem. 2008;54(10):1608–1616. doi: 10.1373/clinchem.2008.108175. [DOI] [PubMed] [Google Scholar]
- 71.Falgarone G, Chiocchia G. Chapter 8: Clusterin A multifacet protein at the crossroad of inflammation and autoimmunity. Adv Cancer Res. 2009;104:139–170. doi: 10.1016/S0065-230X(09)04008-1. [DOI] [PubMed] [Google Scholar]
- 72.Trougakos IP, Gonos ES. Chapter 9: Oxidative stress in malignant progression: The role of Clusterin, a sensitive cellular biosensor of free radicals. Adv Cancer Res. 2009;104:171–210. doi: 10.1016/S0065-230X(09)04009-3. [DOI] [PubMed] [Google Scholar]
- 73.Manevich Y, Fisher AB. Peroxiredoxin 6, a 1-Cys peroxiredoxin, functions in antioxidant defense and lung phospholipid metabolism. Free Radic Biol Med. 2005;38(11):1422–1432. doi: 10.1016/j.freeradbiomed.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 74.Lee SB, Ho JN, Yoon SH, Kang GY, Hwang SG, Um HD. Peroxiredoxin 6 promotes lung cancer cell invasion by inducing urokinase-type plasminogen activator via p38 kinase, phosphoinositide 3-kinase, and Akt. Mol Cells. 2009;28(6):583–588. doi: 10.1007/s10059-009-0152-6. [DOI] [PubMed] [Google Scholar]
- 75.Ho JN, Lee SB, Lee SS, Yoon SH, Kang GY, Hwang SG, Um HD. Phospholipase A2 activity of peroxiredoxin 6 promotes invasion and metastasis of lung cancer cells. Mol Cancer Ther. 2010;9(4):825–832. doi: 10.1158/1535-7163.MCT-09-0904. [DOI] [PubMed] [Google Scholar]
- 76.Chang YH, Wu CC, Chang KP, Yu JS, Chang YC, Liao PC. Cell secretome analysis using hollow fiber culture system leads to the discovery of CLIC1 protein as a novel plasma marker for nasopharyngeal carcinoma. J Proteome Res. 2009;8(12):5465–5474. doi: 10.1021/pr900454e. [DOI] [PubMed] [Google Scholar]
- 77.Petricoin EF, Belluco C, Araujo RP, Liotta LA. The blood peptidome: a higher dimension of information content for cancer biomarker discovery. Nat Rev Cancer. 2006;6(12):961–967. doi: 10.1038/nrc2011. [DOI] [PubMed] [Google Scholar]
- 78.Fredolini C, Meani F, Luchini A, Zhou W, Russo P, Ross M, Patanarut A, Tamburro D, Gambara G, Ornstein D, Odicino F, Ragnoli M, Ravaggi A, Novelli F, Collura D, D’Urso L, Muto G, Belluco C, Pecorelli S, Liotta L, Petricoin EF., 3rd Investigation of the ovarian and prostate cancer peptidome for candidate early detection markers using a novel nanoparticle biomarker capture technology. AAPS J. 2010;12(4):504–518. doi: 10.1208/s12248-010-9211-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Drabovich AP, Diamandis EP. Combinatorial peptide libraries facilitate development of multiple reaction monitoring assays for low-abundance proteins. J Proteome Res. 2010;9(3):1236–1245. doi: 10.1021/pr900729g. [DOI] [PubMed] [Google Scholar]
- 80.Kozak KR, Su F, Whitelegge JP, Faull K, Reddy S, Farias-Eisner R. Characterization of serum biomarkers for detection of early stage ovarian cancer. Proteomics. 2005;5(17):4589–4596. doi: 10.1002/pmic.200500093. [DOI] [PubMed] [Google Scholar]
- 81.Scheffer GL, de Jong MC, Monks A, Flens MJ, Hose CD, Izquierdo MA, Shoemaker RH, Scheper RJ. Increased expression of beta 2-microglobulin in multidrug-resistant tumour cells. Br J Cancer. 2002;86(12):1943–1950. doi: 10.1038/sj.bjc.6600354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Simaga S, Osmak M, Babic D, Sprem M, Vukelic B, Abramic M. Quantitative biochemical analysis of lactate dehydrogenase in human ovarian tissues: correlation with tumor grade. Int J Gynecol Cancer. 2005;15(3):438–444. doi: 10.1111/j.1525-1438.2005.14410.x. [DOI] [PubMed] [Google Scholar]
- 83.Vazquez J, Gonzalez L, Merino A, Vizoso F. Expression and clinical significance of apolipoprotein D in epithelial ovarian carcinomas. Gynecol Oncol. 2000;76(3):340–347. doi: 10.1006/gyno.1999.5678. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.