Abstract
We report the proteomic and antivenomic characterization of Crotalus tigris venom. This venom exhibits the highest lethality for mice among rattlesnakes and the simplest toxin proteome reported to date. The venom proteome of C. tigris comprises 7–8 gene products from 6 toxin families: the presynaptic β-neurotoxic heterodimeric PLA2, Mojave toxin, and two serine proteinases comprise, respectively, 66% and 27% of the C. tigris toxin arsenal, whereas a VEGF-like protein, a CRISP molecule, a medium-sized disintegrin, and 1–2 PIII-SVMPs, each represents 0.1–5% of the total venom proteome. This toxin profile really explains the systemic neuro- and myotoxic effects observed in envenomated animals. In addition, we found that venom lethality of C. tigris and other North American rattlesnake type II venoms correlates with the concentration of Mojave toxin A-subunit, supporting the view that the neurotoxic venom phenotype of crotalid type II venoms may be described as a single-allele adaptation. Our data suggest that the evolutionary trend towards neurotoxicity, which has been also reported for the South American rattlesnakes, may have resulted by paedomorphism. The ability of an experimental antivenom to effectively immunodeplete proteins from the type II venoms of C. tigris, C. horridus, C. oreganus helleri, C. scutulatus scutulatus, and S. catenatus catenatus, indicated the feasibility of generating a pan-American anti-Crotalus type II antivenom, suggested by the identification of shared evolutionary trends among South American and North American Crotalus.
Keywords: North American rattlesnake, Crotalus tigris, snake venomics, snake venom neurotoxicity, antivenomics
INTRODUCTION
Crotalus tigris, the tiger rattlesnake, is a ground-dwelling, medium-sized pitviper (the largest specimen on record measured 88.5 cm1). The monotypic C. tigris is found in isolated populations in rocky habitats or in mesquite grasslands at elevations of near sea level to about 1400 m, in southwestern United States (central, south-central, and extreme southeastern Arizona), extending southward into northwestern Mexico (Sonoran Desertscrub, Chihuahuan Desertscrub, Interior Chaparral, and Madrean Evergreen Woodland), and on Isla Tiburón in the Gulf of California1–3. 35–52 irregular, grey or brownish diffuse dorsal crossbands (“tiger bands”), and the pink to lavender tinge of its body distinguish the tiger rattlesnake from other species of rattlesnakes which occur within some areas of its range, C. atrox, C. cerastes, C. mitchellii, C. molossus, and C. scutulatus3. C. tigris has a very small triangular head relative to the size of the body and a large rattle, which can make a lot of sound, reaching a loudness of about 77 dB4 (http://www.californiaherps.com/noncal/southwest/swsnakes/pages/c.tigris.html).
Little is known about the natural history of the C. tigris. It is chiefly nocturnal during the hot summer months, diurnal and crepuscular in fall, and hibernates over the cold months of late fall and winter in rock crevices or animal burrows1–3. In spite of being a ground-dwelling inhabitant of the desert, its activity is not restricted to the ground. It swims readily and also has been found in bushes 60 cm above the floor1. The tiger rattlesnake ambushes much of its prey but also active forages small rodents and lizards2,5, juveniles relying heavily on lizards and adults depending more on rodents. In addition, these small rattlesnakes have been known to eat fairly large prey, including kangaroo rats, packrats, and even spiny lizards6. This is based upon its venom’s high lethality, rated the highest of all rattlesnake venoms (LD50 value for mice is 0.07 mg/kg intraperitoneal, 0.056 mg/kg intravenous, and 0.21 mg/kg subcutaneous)7–9.
Approximately 7,000–8,000 reptile bites are reported to the American Association of Poison Control Centers (AAPCC) each year10,11. Most bites result from the eastern diamondback rattlesnake (C. adamanteus), the western diamondback rattlesnake (C. atrox), the prairie and Pacific rattlesnakes (C. viridis), the timber rattlesnake (C. horridus), and the pygmy rattlesnake (S. miliarius), when a snake is handled or abused. The eastern and western diamondback rattlesnakes account for the most fatalities. Human bites by C. tigris are infrequent, and literature available on bites by this snake is scarce. The several recorded human envenomations by tiger rattlers produced little local pain, swelling, or other reaction following the bite, and despite the toxicity of its venom no significant systemic symptoms12,13. The comparatively low venom yield (6.4–11 mg dried venom) and short 4.0–4.6 mm fangs1,2 of C. tigris possibly prevent severe envenoming in adult humans. However, the clinical picture could be very more serious if the person bitten was a child or a slight build individual. The early therapeutic use of antivenom is important if significant envenomation is suspected. The purpose of this report was to characterize the venom toxin proteome of the tiger rattlesnake, establish composition-toxicity correlations, and investigate the immunoreactivity profile of an experimental and a commercial antivenom.
EXPERIMENTAL SECTION
Venoms and antivenoms
The venoms of adult C. tigris (Tiger rattlesnake), C. horridus (Timber rattlesnake), C. scutulatus scutulatus type A (Mohave rattlesnake), and C. oreganus helleri (Southern Pacific rattlesnake) were extracted from specimens kept in captivity in the serpentarium of the National Natural Toxins Research Center (Kingsville, TX, http://ntrc.tamuk.edu) by biting on a parafilm-wrapped container.
The anti-crotalic antivenom used for in vivo neutralization assays study was produced at Instituto Butantan (But, São Paulo, Brazil, http://www.butantan.gov.br) by hyperimmunization of horses with a pool of equal amounts of C. d. terrificus and C. d. collilineatus venoms collected in Southeastern and Midwestern Brazil, in the states of São Paulo, Mato Grosso and Minas Gerais (Marisa Maria Teixeira da Rocha, Instituto Butantan, personal communication). The antivenom comprise purified F(ab′)2 fragments generated by pepsic digestion of ammonium sulphate-precipitated IgGs. F(ab′)2 concentration was determined spectrophotometrically using an extinction coefficient (ε) of 1.4 for a 1mg/ml concentration at 280 nm using a 1cm light pathlength cuvette14.
An experimental antiserum was raised in rabbits by subcutaneous injections of sublethal amounts of a mixture of venoms from C. d. terrificus, C. simus, and C. lepidus lepidus. First injection comprised 100 μg venom in 100 μl of PBS (20 mM phosphate, 135 mM NaCl, pH 7.3) emulsified with an equal volume of Freund’s complete adjuvant. Booster injections comprising increasing amounts (200–500 μg) of immunogen emulsified in Freund’s incomplete adjuvant were administered every 2 weeks for a period of 2–3 month (depending on the titer of the antiserum determined by a standard ELISA procedure). Terminal cardiac blood collection, done by intracardiac puncture performed under general anesthesia, was approved by the IBV’s Ethics Commission. The IgG fraction was purified by ammonium sulphate precipitation followed by affinity chromatography on Sepharose-Protein A (Agarose Bead Technologies, Tampa, FL, USA) following the manufacturer’s instructions.
Venomics
Venom proteins were separated by reverse-phase HPLC using a Teknokroma Europa C18 (0.4 cm × 25 cm, 5 μm particle size, 300 Å pore size) column as described15. Protein detection was at 215 nm and peaks were collected manually and dried in a Speed-Vac (Savant). The relative abundances (% of the total venom proteins) of the different protein families in the venoms were estimated from the relation of the sum of the areas of the reverse-phase chromatographic peaks containing proteins from the same family to the total area of venom protein peaks. In a strict sense, and according to the Lambert-Beer law, the calculated relative amounts correspond to the “% of total peptide bonds in the sample”, which is a good estimate of the % by weight (g/100g) of a particular venom component. The relative contributions of different proteins eluting in the same chromatographic fraction were estimated by densitometry after SDS-PAGE separation.
HPLC fractions were analyzed by SDS-PAGE (using 15% polyacrylamide gels) and N-terminal sequencing (using a Procise instrument, Applied Biosystems, Foster City, CA, USA). Amino acid sequence similarity searches were performed against the available databanks using the BLAST program16 implemented in the WU-BLAST2 search engine at http://www.bork.embl-heidelberg.de. In some cases, this information is sufficient to assign a venom toxin to a protein family represented in the databases. Molecular mass determination was performed by MALDI-TOF mass spectrometry (using an Applied Biosystems Voyager-DE Pro™ instrument) and electrospray ionization (ESI) mass spectrometry (using an Applied Biosystems QTrap™ 2000 mass spectrometer). Protein bands of interest were excised from Coomassie Brilliant Blue-stained SDS-PAGE gels and subjected to automated reduction, alkylation, and in-gel digestion with sequencing grade porcine pancreatic trypsin (Promega). The tryptic peptide mixtures were dried in a SpeedVac and dissolved in 5 ml of 50% ACN and 0.1% TFA. 0.85 ml of digest were spotted onto a MALDI-TOF sample holder, mixed with an equal volume of a 1:10 (v/v) dilution of a saturated solution of α-cyano-4-hydroxycinnamic acid (Sigma) in 50% ACN containing 0.1% TFA, dried, and analyzed with an Applied Biosystems Voyager-DE Pro MALDI-TOF mass spectrometer, operated in delayed extraction and reflectror modes. For peptide ion sequencing, the protein digest mixtures were loaded in nanospray capilars (http://www.proxeon.com) and submitted to electrospray ionization mass spectrometric analysis using an Applied Biosystem’s QTrap 2000 mass spectrometer. Enhanced Multiply Charged mode was run at 250 amu/s across the entire mass range to determine the charge state of the ions. Monoisotopic doubly- or triply-charged precursor ions were selected (within a window of ± 0.5 m/z) and sequenced by CID-MS/MS using the Enhanced Product Ion mode with Q0 trapping option; Q1 was operated at unit resolution, the Q1-to-Q2 collision energy was set to 30 (for m/z ≤ 700) or 35 eV (for m/z > 700), the Q3 entry barrier was 8 V, the LIT (linear ion trap) Q3 fill time was 250 ms, and the scan rate in Q3 was 1000 amu/s. CID spectra were interpreted manually (i.e. de novo sequenced) or using MASCOT as a seach engine, either through its public-available website (http://www.matrixscience.com), or using a licensed version (2.0) of the MASCOT program. Searches were done against the default non-redundant datababes or a private database containing 1763 viperid protein sequences deposited in the SwissProt/TrEMBL database (http://www.uniprot.org/) plus the previously assigned peptide ion sequences from snake venomics projects carried out in our laboratories15,17. MS/MS mass tolerance was set to ± 0.6 Da. Carbamidomethyl cysteine and oxidation of methionine were fixed and variable modifications, respectively.
Neutralization of venom lethality
To assess the ability of the anticrotalic antivenom produced at Instituto Butantan to neutralize the lethal activity of C. tigris venom, five mice received an i.p. injection of a venom challenge dose of 5 μg (~ 4 LD50 for mice of 16–18 g body weight) in 250 μl of phosphate-buffered saline (0.12 M NaCl, 0.04 M sodium phosphate, pH 7.2). An intraperitoneal (i.p.) median lethal dose (LD50) of 0.07 μg/g body weight was assumed9. Another group of five mice received an identical injection of venom that had been preincubated with the antivenom, at a ratio of 4 μl antivenom/μg venom, for 30 min at room temperature. Deaths were scored after a period of 48 hr.
Antivenomics
For antivenomics17, 1 mg of crude C. tigris venom in 300 μl of 0.2 M phosphate, pH 7.0, was incubated overnight at room temperature and with gentle stirring with 10 mg of rabbit IgG antibodies affinity-purified from the antiserum raised against a mixture of venoms from C. d. terrificus, C. simus, and C. lepidus lepidus. IgG-antigen immunocomplexes were pulled down with Agarose-Protein-A (ABT) beads capable of retaining 25 mg of IgG molecules. After centrifugation at 13,000 rpm for 3 min in an Eppendorf centrifuge, the supernatant containing the non-bound venom proteins was submitted to reverse-phase separation as described above. Control samples were subjected to the same procedure except that (i) pre-immune rabbit serum IgGs were employed, or (ii) antivenom IgGs were not included in the reaction mixture.
RESULTS AND DISCUSSION
The minimalist venom proteome of C. tigris suggests that neurotoxicity represents a paedomorphic trend in Neartic type II venoms
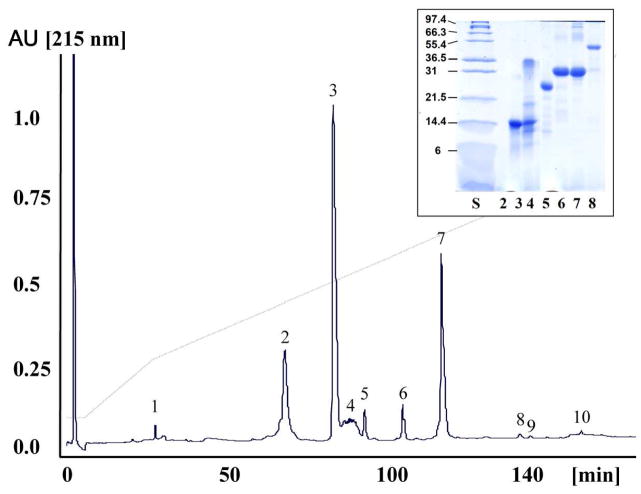
Rattlesnake venoms belong to one of two distinct phenotypes, which broadly correspond to type I (high levels of SVMPs and low toxicity, LD50 >1 mg/g mouse body weight) and type II (low metalloproteinase activity and high toxicity, LD50 <1 mg/g mouse body weight) defined by Mackessy18,19. The high toxicity of type II venoms and the characteristic systemic neuro- and myotoxic effects observed in envenomations appear to be directly related to the high concentration of the presynaptic β-neurotoxic heterodimeric PLA2 molecules in these venoms. The venom proteome of C. tigris appears to be composed by only 7–8 gene products from 6 different toxin families (Table 1, Figs. 1 and 2). In particular, the low metalloproteinase content, the high concentration of Mohave toxin subunits (66% of the total venom proteins) (Table 2), and its high toxicity, LD50 0.05 (i.v)-0.07 (i.p.) mg/g mouse body weight, which is the highest known for any rattlesnake venom7–9, place C. tigris venom into the type II class defined by Mackessy18,19. This is by far the simplest viperid snake venom toxin proteome reported to date. Hence, most snake venoms of family Viperidae (vipers and pitvipers) analyzed by state-of-the-art proteomic tecniques comprise several tens to some hundreds of molecules.17,20–26 Cerberus rynchops venom represents another very low complexity proteome. It appears to contain a total of five major proteins, one isoform each of metalloproteinase, CRISP and C-type lectin and two major isoforms of ryncolins.27 C. rynchops (dog-faced water snake) belongs to Homalopsidae of Colubroidea (rear-fanged snakes). The pharmacological profile of C. rynchops venom remains elusive, but given the central role that diet has played in the adaptive radiation of snakes28–30, venom may represent a key trophic adaptative trait.31 In the frame of this view, the relative composition of C. tigris venom (Table 2) suggests that the pharmacological relevance of its toxins may vary widely: the two subunits of Mojave toxin and two serine proteinases comprise, respectively, 66% and 27% of the C. tigris toxin arsenal, whereas a VEGF-like protein, a CRISP molecule, a medium-sized disintegrin, and 1–2 PIII-SVMPs, each represents 0.1–5% of the total venom proteome (Table 2).
TABLE 1.
Assignment of the reverse-phase fractions from the venoms of Crotalus tigris, isolated as in Fig. 1, to protein families by N-terminal Edman sequencing, mass spectrometry, and collision-induced fragmentation by nESI-MS/MS of selected peptide ions from in-gel digested protein bands separated by SDS-PAGE (insert in Fig. 1). In MS/MS-derived sequences, X = Ile or Leu; Z, pyrrolidone carboxylic acid; B, Lys or Gln. Unless other stated, for MS/MS analyses, cysteine residues were carbamidomethylated; Molecular masses of native proteins were determined by electrospray-ionization (± 0.02%) or MALDI-TOF (± 0.2%) mass spectrometry. Apparent molecular masses were determined by SDS-PAGE of reduced samples. “de novo”, peptide sequence determined by manual interpretation of MS/MS spectra that did not produce any hit by MASCOT search. Peptide sequences from the MS/MS-based assigned proteins matching ions present in the tryptic peptide mass fingerprint spectra are labeled with asterisks.
| HPLC Fraction | N-terminal sequence | Molecular mass | Peptide ion | MS/MS-derived sequence | Mowse score | Protein family | |
|---|---|---|---|---|---|---|---|
| m/z | z | ||||||
| 1 | AGEECDCGSPANP | 7320 Da | Disintegrin | ||||
| 2 | SPEXCQG SYGCYCG |
9 kDa | Mojave toxin acid chain [P18998] | ||||
| 3 | HLLQFNKMIKFETRK | 14187 Da | 649.8 | 2 | YGYMFYPDSR | 75 | Mojave toxin basic chain [P62023] |
| 871.8 | 2 | GTWCEEQICECDR | 75 | ||||
| 2158.9 | 1 | NAIPFYAFYGCYCGWGGR* | |||||
| 1978.0 | 1 | SLSTYKYGYMFYPDSR* | |||||
| 4 | ND | 36 kDa | 692.3 | 2 | ZAMPFMEVYER | de novo | svVEGF |
| 5 | SVDFDSESPRKPEIQ | 24 kDa | 569.6 | 2 | SVDFDSESPR | 36 | CRISP |
| 769.3 | 2 | MEWYPEAAANAER | 68 | ||||
| 6 | VVGGDECNINEHR | 31 kDa | 749.8 | 2 | VVGGDECDINEHR | 60 | Serine proteinase |
| 657.4 | 2 | XGNWGSXTPXTR | de novo | ||||
| 552.6 | 2 | TXCAGXVQGGK | 88 | ||||
| 7,10 | VIGGDECNINEHRFL | 31 kDa | 756.8 | 2 | VIGGDECNINEHR | 42 | Serine proteinase [~ ABY65929] |
| 1321.8 | 1 | HILIYVGVHDR * | |||||
| 1770.9 | 1 | ILCAGVLEGGIDTCHR* | |||||
| 8,9 | ND | 46 kDa | 578.3 | 3 | MYDXVNVXTPXYHR | 42 | PIII-SVMP (~ Q9DGB9) |
| 1155.6 | 1 | EGNHYGYCR* | |||||
| 1283.8 | 1 | EGNHYGYCRK* | |||||
| 604.6 | 3 | QGAQCAEGLCCDQCR | 35 | ||||
Fig. 1. Characterization of the venom proteome of Crotalus tigris.
Reverse-phase HPLC separation of the venom proteins of C. tigris. Insert, SDS-PAGE of the reverse-phase HPLC separated venom proteins run under reduced conditions. Molecular mass markers (in kDa) are indicated at the side of each gel. Protein bands were excised and characterized by mass fingerprinting and CID-MS/MS of selected doubly- or triply-charged peptide ions (Table 1).
Fig. 2. Overall protein composition of the venoms of C. tigris.
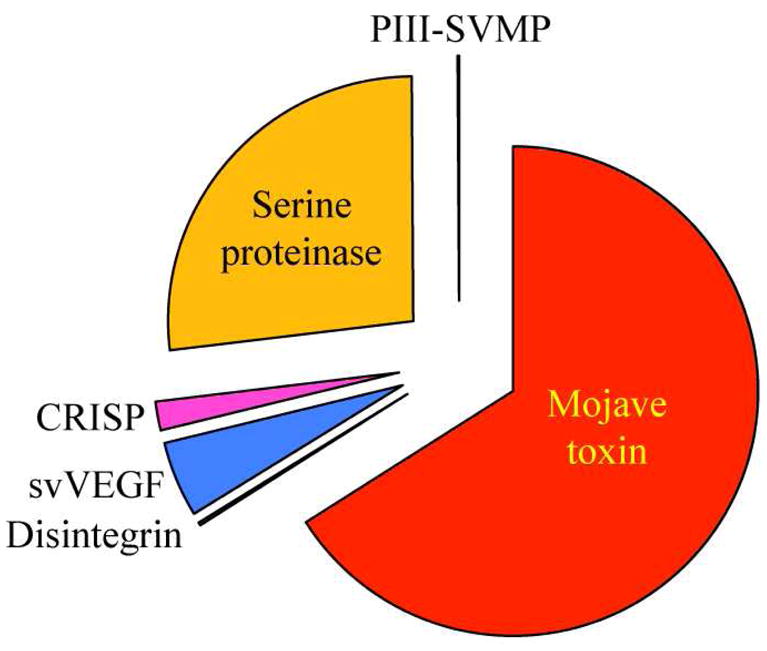
Relative occurrence of proteins from different toxin families in the venoms of adult C. tigris. PIII-SVMP, snake venom Zn2+-metalloproteinase (SVMPs) of class III; CRISP, cysteine-rich secretory protein; svVEGF, snake venom vascular endothelial growth factor. For details of the individual proteins characterized consult Table 1. The percentages of the different toxin families in the venoms are listed in Table 2.
TABLE 2.
Overview of the relative occurrence of proteins (in percentage of the total HPLC-separated proteins of the different families) in the venom of Crotalus tigris.
| Protein family | % of total venom proteins |
|---|---|
| Mojave toxin | 66.2 |
| • acid chain | 22.1 |
| • basic chain | 44.1 |
| Serine proteinase | 26.8 |
| svVEGF | 5.0 |
| CRISP | 1.9 |
| Disintegrin | 0.2 |
| PIII-SVMP | 0.1 |
The toxin profile of C. tigris venom may explain the effects observed in envenomated animals8,32. Mice injected s.c. with C. tigris venom characteristically showed circling movements, ataxia, and flaccid paralysis. Local subcutaneous hemorrhage was not observed except with doses about ten times the LD50%. In addition, although the venom exhibits low, but significant protease activity, it does not seem to cause any hemolytic activity. These systemic neuro- and myotoxic effects appear to be directly related to the concentration of the presynaptic β-neurotoxic heterodimeric PLA2 molecules, Mojave toxin (in Neartic rattlesnakes)33, crotoxin (in Central and South American rattlesnake venoms)34–36, and sistruxin (in Sistrurus catenatus catenatus and S. c. tergeminus venoms)37,38. The occurrence of high concentration of a toxin immunologically related to Mojave toxin in C. tigris venom had been reported by Weinstein et al.39, and subsequently the presence of Mojave toxin subunits A and B in C. tigris was verified using DNA analysis of blood and toxin specific immunological analysis of venom40. Hawgood41 compiled a review of pathophysiological effects of Mojave toxin: Castinolia et al.42 demonstrated inhibition of neuromuscular transmission, and Ho and Lee43 and Gopalakrishnakone et al.44 found the site of action to be presynaptic and also described the toxin as myonecrotic and capable of causing pulmonary hemorrhage. Mice injected with the isolated toxin at a dosage of about the LD50 level of crude venom displayed ataxia and a short period of hyperexcitability, which were followed by prostration and tachypnea. Extensive respiratory distress occured rapidly and led to death within 15 min postinjection39. This pattern of pharmacological activities resembles the symptoms observed in envenomings by South American rattlesnakes (Crotalus durissus sp.), characterized by severe systemic effects associated with neurotoxicity and systemic myotoxicity45,46.
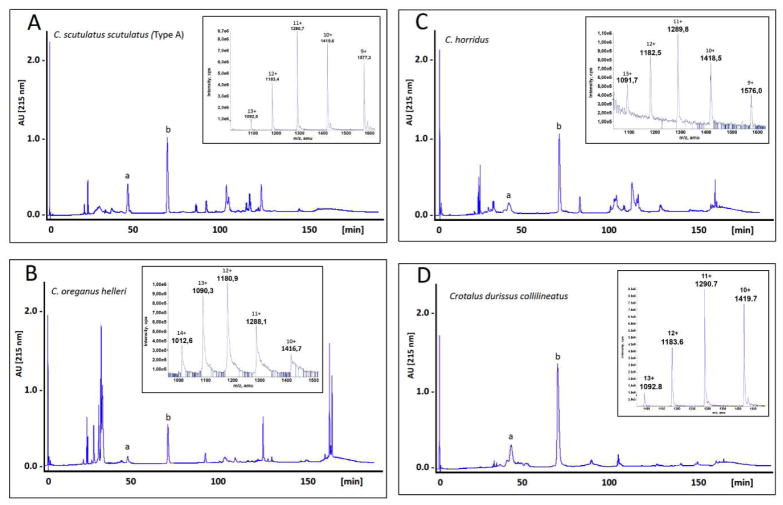
C. tigris also exhibits a toxin venom profile and lethal median toxicity (LD50) closely resembling those of neurotoxic South American Crotalus durissus subspecies (terrificus, cascavella, collilineatus)47,48 (compare Fig. 1 and panel D of Fig. 3; Table 3). Moreover, reverse-phase HPLC profiling of the venoms from C. scutulatus scutulatus (Css, Mohave rattlesnake), C. horridus (Ch, Timber rattlesnake), and the Southern Pacific rattlesnake, C. oreganus helleri (Coh), displayed in Figure 3 highlights the occurrence of Mojave toxin subunits in these New World rattlesnakes and shows the close resemblance between North American type II and neurotoxic South American crotalid venoms. Mojave toxin molecules isolated from these taxa exhibit identical N-terminal sequences (A-subunit, SSYGCYCGAGGQGWP + SPENCQGESQPC; B-subunit, HLLQFNKMIKFETRK) and very similar electrospray-ionization isotope-averaged molecular masses (A-subunits, 9 kDa; B-subunits: 14186 Da (Css), 14156 Da (Ch), 14177 Da (Coh); 14187 Da (Cdc)).
Fig. 3. Comparison of the toxin profiles of type II venoms.
The venom proteins of C. scutulatus scutulatus (type A) (panel A), C. oreganus helleri (B), C. horridus (C), and C. durissus cascavella (D) were separated by reverse-phase HPLC. Peaks containing the acidic and the basic subunits of neurotoxin Mojave toxin or crotoxin are labelled “a” and “b”, respectively. Insert, electrospray-ionization mass spectra of the neurotoxin B-subunits.
TABLE 3.
Correlation between intraperitoneal median lethal toxicity (LD50, in mg venom/g mouse) and relative abundance (in percentage of the total HPLC-separated proteins) of Mojave toxin or crotoxin subunits (Figs. 1 and 3) in the venom proteomes of crotalid species. %AB, estimated percentage of AB heterodimer.
| A-chain | B-chain | %AB | LD50 | |
|---|---|---|---|---|
| C. oreganus helleri | 2 | 9 | 22 | 1.518 |
| S. catenatus catenatus | 3.6 | 12.8 | 28 | 0.13–0.4330 |
| C. scutulatus scutulatus (A) | 4.5 | 19 | 24 | 0.22–0.4662 |
| C. oreganus concolor | 21 | 0.4618 | ||
| C. horridus | 7 | 22 | 32 | 0.22–1.063 |
| C. durissus collilineatus | 17 | 50 | 34 | 0.07–0.164 |
| C. durissus terrificus | 19 | 40.5 | 46 | 0.06–00964 |
| C. durissus cascavella | 20 | 52.5 | 39 | 0.07–0.164 |
| C. tigris | 21 | 45 | 46 | 0.05–0.077–9 |
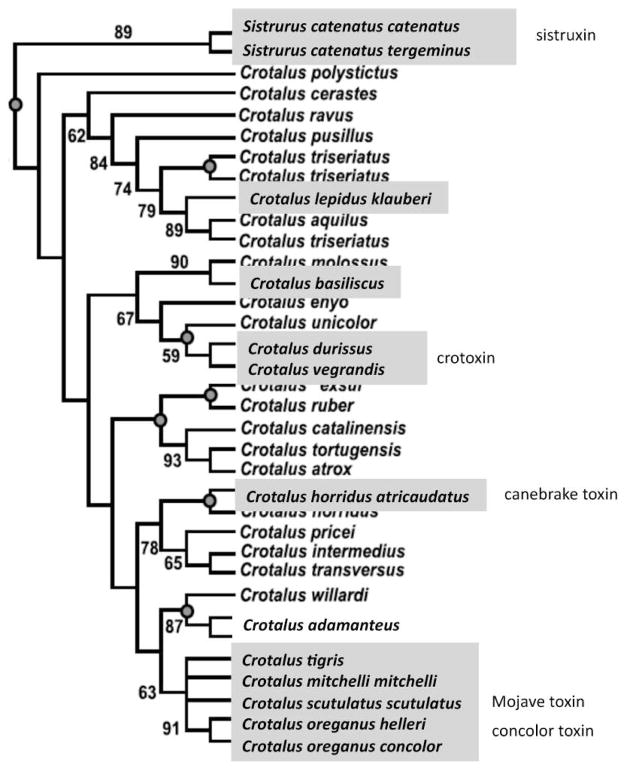
Rattlesnakes had its origin ~20 Mya in the Sierra Madre Occidental in the north-central Mexican Plateau49, and dispersed northward into North America and southward into South America1,3. Gain of neurotoxicity and lethality to rodents represents a paedomorphic trend that correlates with increased concentration of crotoxin along the axis of Crotalus radiation in South America47,48. The phylogeny and evolution of β-neurotoxic PLA2s present in the venoms of rattlesnakes has been investigated by Werman49. The distribution of the highly closely related Mojave toxin resembles a mosaic from Mexico northward50. The lack of phylogenetic clustering among rattlesnakes with neurotoxin PLA2 molecules in their venoms (Fig. 4) indicates that phylogeny may not be an important consideration in the evolution of rattlesnake type II venoms51. The distribution of Mojave toxin varies not only between species, but also between geographic populations within the same species51. Powell and Lieb52 have suggested that the extremely high neurotoxicity exhibited by North American rattlesnakes represents a transitory populational phenomenon associated with novel prey bases.
Fig. 4. Phylogenetic distribution of rattlesnake type II venoms.
Taxa confirmed for the presence of neurotoxic PLA2 complexes (sistruxin, crotoxin, canebrake toxin, Mojave toxin, concolor toxin) in their venoms are boxed in gray background. The cladogram, based on Castoe and Parkinson65, shows taxonomic relationships among rattlesnakes (Crotalus and Sistrurus) and highlights the lack of phylogenetic clustering among species with neurotoxin molecules in their venoms.
The evolution of rattlesnakes in the warm deserts of western North America has been investigated by evaluating mitochondrial DNA (mtDNA) sequences53. The most recent common ancestor for the rattlesnakes Sistrurus/Crotalus was estimated at 12.7 Mya (mid-Miocene), the subsequent divergence of S. miliarius anbd S. catenatus sp. appears to have occurred 10.2 Mya, and C. tigris seemingly diverged from C. mitchellii at the Late Miocene/Early Pliocene boundary (5.6 Ma)53. On the other hand, Wüster and co-workers54 have traced the dispersal of C. durissus in South America, revealing a classical pattern of stepwise colonization progressing from a northern center of origin in Mexico to northern South America and across the Amazon Basin. The biogeographical data suggested an ancient basal cladogenesis in the Central American C. simus clade dated back to the late Miocene/early Pliocene (6.4–6.7 Mya), and a relatively recent (1.7–1.1 Mya) basal South American dispersal across a central trans-Amazonian corridor during the middle Pleistocene (1.1–1.0 Mya)54,55. The timing of these cladogenetic events yielding taxa with type II venoms scattered through the phylogenetic tree of the rattlesnakes (Fig. 4) may reflect the convergence of an evolutionary trend towards neurotoxicity. Assuming that the evolutionary trend reported for the South American rattlesnakes47,48 holds true for the North American Crotalus species, the occurrence of Mojave toxin in the venom of adults specimens of certain populations within terminal clades of recently divergent North American taxa (Fig. 4) may also have resulted by paedomorphism. Furthermore, and taking for granted that experimental verification is required (e.g. through comparative proteomic analysis of neonate and adult venoms), the reported ontogenetic variation in the venom composition of North American rattlesnakes55,56, would support this hypothesis. Of note is that neonate, juvenile, and adult C. o. concolor venoms are essentially similar in composition, with respect to toxicity, amount of concolor toxin (PLA2), and low metalloproteinase activity55.
Venom lethality in North American rattlesnake type II venoms correlates with concentration of Mojave toxin A-subunit
The relative abundances of the Mojave toxin (or crotoxin) A- and B-subunits in the proteomes of type II venoms were estimated from the areas of their reverse-phase chromatographic peaks (Table 3). In line with the fact that these neurotoxic PLA2s are composed of a non-toxic acidic (A-) 3-chain subunit, derived from the proteolytic cleavage of a single PLA2 precursor molecule, and a mildy toxic basic (B-), single-chain PLA2 subunit that associate noncovalently into dimers, increasing thereby the toxicity 10–30 times33–35, there is a clear correlation between heterodimeric Mojave toxin (or crotoxin) concentration and the reported venom LD50 (Table 3). In the venoms sampled in this work, the B-subunit was present in 2.1–4.5-fold excess with respect to the A-subunit, suggesting that translation into the venom of the acidic subunit is the limiting factor for confering enhanced venom neurotoxicity. In line with this view, the Mojave toxin B-subunit gene is widespread among Crotalus species, and its occurrence is independent of the A-subunit gene51–53. The neurotoxic venom phenotype may thus be described as a single-allele (A-subunit) adaptation51,52. This view is supported by previous reports showing that Mojave rattlesnakes (Crotalus scutulatus scutulatus) lacking the acidic subunit DNA sequence lack Mojave toxin in their venom57.
Antivenomics of C. tigris and other crotalid type II venoms
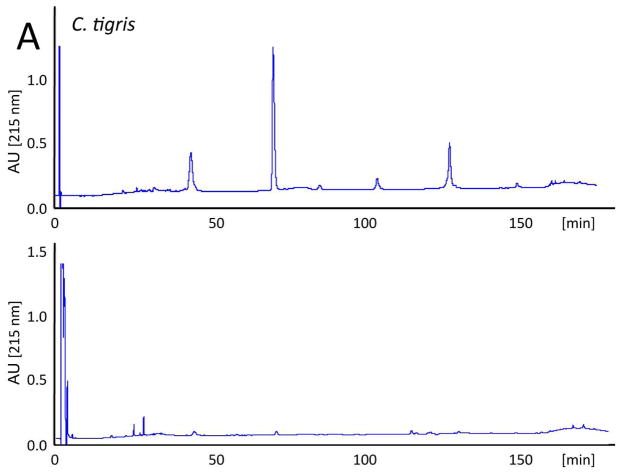
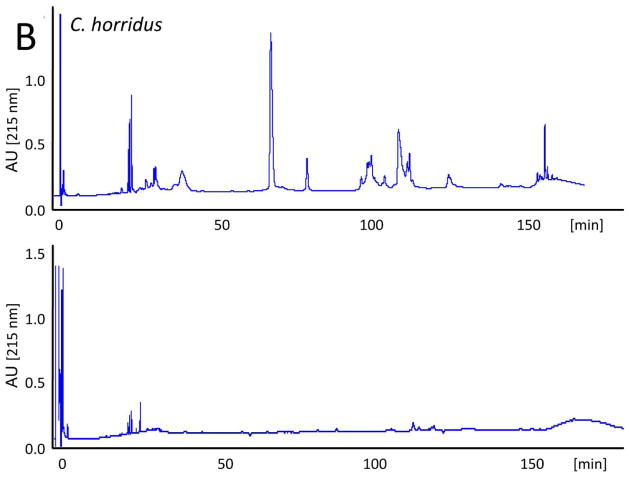
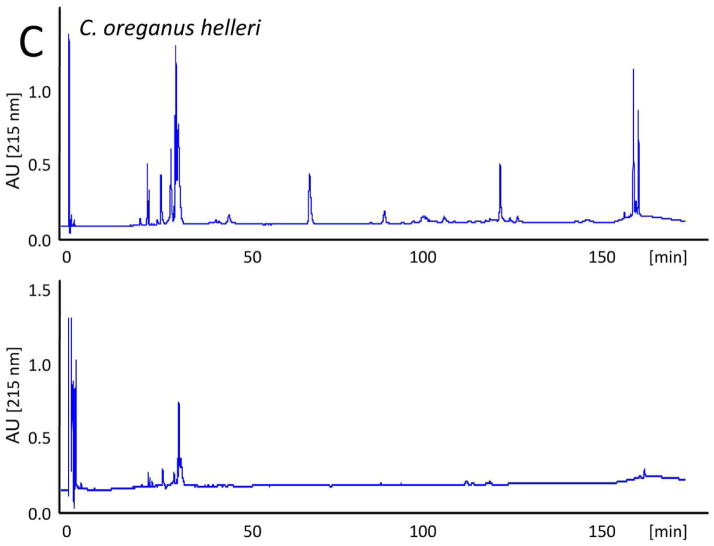
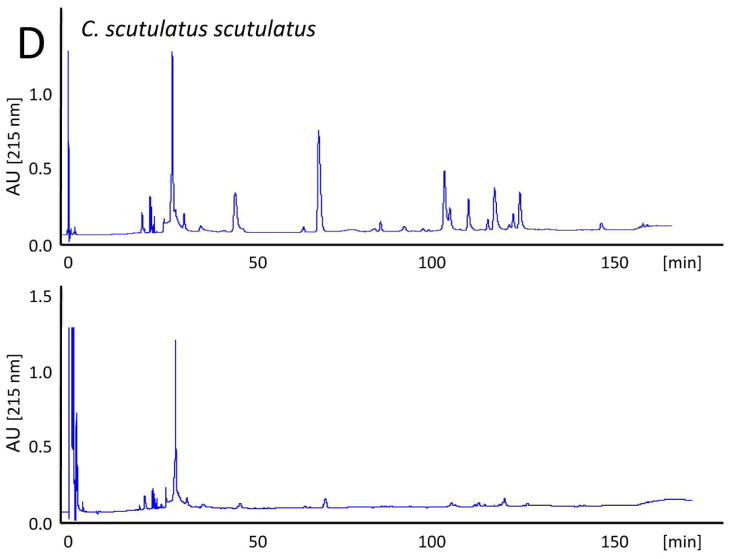
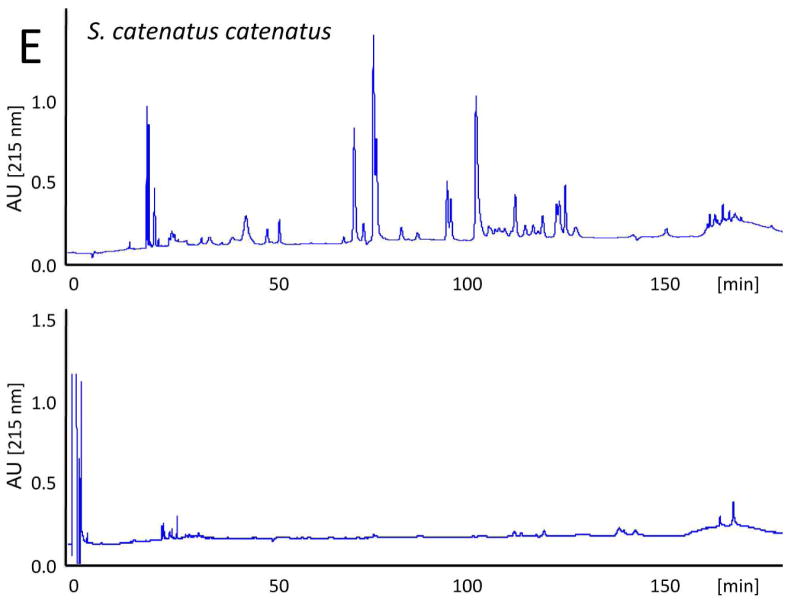
The evolutionary trend towards neurotoxicity observed in South American and North American rattlesnakes suggested the feasibility of generating a pan-American anti-crotalid type II venoms. To check this possibility, we first assessed the ability of the anticrotalic antivenom produced at Instituto Butantan against C. d. terrificus venom to neutralize the lethal activity of C. tigris venom. This antivenom showed a very high effectiveness in the neutralization of the lethal, myotoxic, and neurotoxic effects of the crotoxin-rich venoms of C. durissus subspecies and newborn C. simus58. All five mice receiving an intraperitoneal injection of ~ 4 LD50 of venom died within 16 h, whereas all mice that received the venom/antivenom mixture survived throughout the 48 h observation period, demonstrating that the Butantan anti-C. d. terrificus antivenom is also able to neutralize the lethal action of C. tigris venom. Next, we applied our antivenomics protocol17,59,60 to assess the ability of an experimental (C. simus, C. l. lepidus, C. d. terrificus) antivenom to immunodeplete proteins from the venoms of C. tigris, C. horridus, C. oreganus helleri, C. scutulatus scutulatus, and S. catenatus catenatus. The results, illustrated in Fig. 5, clearly showed that the trivalent antivenom was very effective targeting the toxins of these type II venoms.
Fig. 5. Immunodepletion of venom proteins by an experimental antivenom.
Reverse-phase separations of the venom proteins (upper chromatograms) from C. tigris (A), C. horridus (B), C. oreganus helleri (C), C. scutulatus scutulatus (type A) (D), and S. catenatus catenatus (E), and the toxins recovered after incubation of the crude venoms with the experimental trivalent antivenom against C. simus, C. l. lepidus, C. d. terrificus (lower chromatograms), followed by immunoprecipitation with Agarose-Protein A.
Concluding remarks and perspectives
The characterization of the venom of C. tigris and finding of largely conserved toxin profile in the venoms of other neurotoxic North American rattlesnakes provides a proteomic framework to interpret previous biochemical, immunochemical, and pharmacological investigations on crotalid type II venoms. Of particular relevance is the correlation between the translational level of Mojave toxin A-subunit and venom lethal activity. In addition, the crystal structure of crotoxin from C. d. terrificus, recently solved at 1.35 Å resolution, indicates that posttranslational cleavage of the acidic subunit precursor is a prerequisite for the assembly of the heterodimeric β-neurotoxin61. However, the identity of the protease responsible for the proteolytic processing of the acidic subunit precursor of neurotoxic PLA2 complexes, Mojave toxin and crotoxin, remains elusive. Whether any of the two serine proteinases present in C. tigris venom bears this activity deserves further investigation.
Our antivenomic results and the lack of phylogenetic clustering among rattlesnakes with neurotoxin PLA2 molecules in their venoms (a paedomorphic trend?) strongly indicate that proteomic-guided identification of evolutionary and immunological trends among venoms may aid replacing the traditional geographic- and phylogenetic-driven hypotheses for antivenom production strategies by a more rationale approach based on venom proteome phenotyping and immunological profile similarities. In this respect we predict that the neurotoxic venoms of C. lepidus klauberi and C. mitchelli mitchelli may exhibit the proteomic and evolutionary trends outlined here for type II Crotalus venoms. The identification of shared evolutionary trends among South American and North American Crotalus reported here may impact the choice of venoms for immunization to produce an effective pan-American anti-Crotalus antivenom.
Supplementary Material
Synopsis.
Crotalus tigris, the deadliest rattlesnake, possesses a minimalist venom.
Neurotoxicity of C. tigris venom correlates with its Mojave A-subunit content.
The trend towards neurotoxicity may represent a paedomorphic trait.
A trivalent antivenom efficiently immunodepletes type II venom toxins.
Evolutionary and immunological trends may aid generating an effective pan-American anti-Crotalus antivenom.
Acknowledgments
This work has been financed by grants BFU2010-17373 (from the Ministerio de Ciencia e Innovación, Madrid, Spain), CRUSA-CSIC (project 2009CR0021), and PROMETEO/2010/005 from the Generalitat Valenciana (Valencia, Spain), NIH/VIPER resource grant (#5 P40 RR018300-09), and Texas A&M University-Kingsville.
Footnotes
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Klauber LM. Rattlesnakes: Their Habitats, Life Histories, and Influence on Mankind. 2. University of California Press; Berkeley: 1997. [Google Scholar]
- 2.Ernst CH, Ernst EM. Snakes of the United States and Canada. Smithsonian Books; Washington and London: 2003. [Google Scholar]
- 3.Campbell JA, Lamar WW. The Venomous Reptiles of the Western Hemisphere. Vol. 2. Comstock Publishing Associates; Ithaca, NY: 2004. [Google Scholar]
- 4.Cook PM, Rowe MP, van Devender RW. Allometric scaling and interspecific differences in the rattling sounds of rattlesnakes. Herpetologica. 1994;50:358–368. [Google Scholar]
- 5.Armstrong BL, Murphy JB. Univ Kansas, Mus Nat Hist, Special Publication. Vol. 5. 1979. The natural history of Mexican rattlesnakes; pp. 1–88. [Google Scholar]
- 6.Sonoran Dessert Digital Library. ( http://www.desertmuseumdigitallibrary.org/public/detail.php?id=ASDM09389&sp=Crotalus%20tigris)
- 7.Brown JH. Toxicology and Pharmacology of Venoms from Poisonous Snakes. Charles C. Thomas; Springfield, Illinois: 1973. [Google Scholar]
- 8.Minton SA, Jr, Weinstein SA. Protease activity and lethal toxicity of venoms from some little known rattlesnakes. Toxicon. 1984;22:828–830. doi: 10.1016/0041-0101(84)90169-7. [DOI] [PubMed] [Google Scholar]
- 9.Weinstein SA, Smith LA. Preliminary fractionation of tiger rattlesnake, Crotalus tigris, venom. Toxicon. 1990;28:1447–1456. doi: 10.1016/0041-0101(90)90158-4. [DOI] [PubMed] [Google Scholar]
- 10.Watson WA, Litovitz T, Rodgers GC, Jr, Klein-Schwartz W, Reid N, Youniss J, Flanagan A, Wruk KM. 2004 Annual report of the American Association of Poison Control Centers Toxic Exposure Surveillance System. Am J Emerg Med. 2005;23:589–666. doi: 10.1016/j.ajem.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 11.Bronstein AC, Spyker DA, Cantilena LR, Jr, Green J, Rumack BH, Heard SE. 2006 Annual Report of the American Association of Poison Control Centers’ National Poison Data System (NPDS) Clin Toxicol (Phila) 2007;45:815–917. doi: 10.1080/15563650701754763. [DOI] [PubMed] [Google Scholar]
- 12.Ernst CH. Venomous reptiles of North America. Smithsonian Institution Press; Washington, D.C: 1992. pp. 1–236. [Google Scholar]
- 13.Norris R. Venom Poisoning in North American Reptiles. In: Campbell JA, Lamar WW, editors. The Venomous Reptiles of the Western Hemisphere. Comstock Publishing Associates; Ithaca and London: 2004. pp. 683–708. [Google Scholar]
- 14.Fasman DG. Practical Handbook of Biochemistry and Molecular Biology. CRC Press; Boston: 1992. [Google Scholar]
- 15.Calvete JJ, Juárez P, Sanz L. Snake venomics. Strategy and applications. J Mass Spectrom. 2007;42:1405–1414. doi: 10.1002/jms.1242. [DOI] [PubMed] [Google Scholar]
- 16.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Calvete JJ. Snake Venomics, Antivenomics, and Venom Phenotyping: The Ménage à Trois of proteomic tools aimed at understanding the biodiversity of venoms. In: Kini RM, Clemetson K, Markland F, McLane MA, Morita T, editors. Toxins and Hemostasis. Springer; Amsterdam: 2010. pp. 285–300. [Google Scholar]
- 18.Mackessy SP. Venom Composition in Rattlesnakes: Trends and Biological Significance. In: Hayes WK, Beaman KR, Cardwell MD, Bush SP, editors. The Biology of Rattlesnakes. Loma Linda University Press; Loma Linda, California: 2008. pp. 495–510. [Google Scholar]
- 19.Mackessy SP. Evolutionary trends in venom composition in the Western Rattlesnakes (Crotalus viridis sensu lato): Toxicity vs. tenderizers. Toxicon. 2010;55:1463–1474. doi: 10.1016/j.toxicon.2010.02.028. [DOI] [PubMed] [Google Scholar]
- 20.Fox JW, Ma L, Nelson K, Sherman NE, Derrano SMT. Comparison of indirect and direct approaches using ion-trap and Fourier transform ion cyclotron resonance mass spectrometry for exploring viperid venom proteomes. Toxicon. 2006;47:700–714. doi: 10.1016/j.toxicon.2006.01.022. [DOI] [PubMed] [Google Scholar]
- 21.Fox JW, Serrano SMT. Exploring snake venom proteomes: multifaceted analyses for complex toxin mixtures. Proteomics. 2008;8:909–920. doi: 10.1002/pmic.200700777. [DOI] [PubMed] [Google Scholar]
- 22.Georgieva D, Arni RK, Betzel C. Proteome analysis of snake venom toxins: pharmacological insights. Expert Rev Proteomics. 2008;5:787–797. doi: 10.1586/14789450.5.6.787. [DOI] [PubMed] [Google Scholar]
- 23.Calvete JJ, Fasoli E, Sanz L, Boschetti E, Righetti PG. Exploring the venom proteome of the western diamondback rattlesnake,Crotalus atrox, via snake venomics and combinatorial peptide ligand library approaches. J Proteome Res. 2009;8:3055–3067. doi: 10.1021/pr900249q. [DOI] [PubMed] [Google Scholar]
- 24.Fasoli E, Sanz L, Wagstaff S, Harrison RA, Righetti PG, Calvete JJ. Exploring the venom proteome of the African puff adder, Bitis arietans, using a combinatorial peptide ligand library approach at different pHs. J Proteomics. 2010;73:932–942. doi: 10.1016/j.jprot.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 25.Durban J, Juárez P, Angulo Y, Lomonte B, Flores-Diaz M, Alape-Girón A, Sasa M, Sanz L, Gutiérrez JM, Dopazo J, Conesa A, Calvete JJ. Profiling the venom gland transcriptomes of Costa Rican snakes by 454 pyrosequencing. BMC Genomics. 2011;12:259. doi: 10.1186/1471-2164-12-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rokyta DR, Wray KP, Lemmon AR, Lemmon EM, Caudle SB. A high-throughput venom-gland transcriptome for the Eastern Diamondback Rattlesnake (Crotalus adamanteus) and evidence for pervasive positive selection across toxin classes. Toxicon. 2011;57:657–671. doi: 10.1016/j.toxicon.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 27.OmPraba G, Chapeaurouge A, Doley R, Devi KR, Padmanaban P, Venkatraman C, Velmurugan D, Lin Q, Kini RM. Identification of a novel family of snake venom proteins veficolins from Cerberus rynchops using a venom gland transcriptomics and proteomics approach. J Proteome Res. 2010;9:1882–1893. doi: 10.1021/pr901044x. [DOI] [PubMed] [Google Scholar]
- 28.Greene HW. Dietary correlates of the origin and radiation of snakes. Am Zool. 1983;23:431–441. [Google Scholar]
- 29.Li M, Fry BG, Kini RM. Eggs-only diet: its implications for the toxin profile changes and ecology of the Marbled sea snake (Aipysurus eydouxii) J Mol Evol. 2005;60:81–89. doi: 10.1007/s00239-004-0138-0. [DOI] [PubMed] [Google Scholar]
- 30.Gibbs HL, Mackessy SP. Functional basis of a molecular adaptation: prey-specific toxic effects of venom from Sistrurus rattlesnakes. Toxicon. 2008;53:672–679. doi: 10.1016/j.toxicon.2009.01.034. [DOI] [PubMed] [Google Scholar]
- 31.Daltry JC, Wüster W, Thorpe RS. Diet and snake venom evolution. Nature. 1996;379:537–540. doi: 10.1038/379537a0. [DOI] [PubMed] [Google Scholar]
- 32.Githens TS, Wolff NO. The Polyvalency of crotalidic antivenins. J Immunol. 1939;37:33–51. [Google Scholar]
- 33.John TR, Smith LA, Kaiser II. Genomic sequences encoding the acidic and basic subunits of Mojave toxin: unusually high sequence identity of non-coding regions. Gene. 1994;139:229–234. doi: 10.1016/0378-1119(94)90761-7. [DOI] [PubMed] [Google Scholar]
- 34.Bon C, Bouchier C, Choumet V, Faure G, Jiang MS, Lambezat MP, Radvanyi F, Saliou B. Crotoxin, half-century of investigations on a phospholipase A2 neurotoxin. Acta Physiol Pharmacol Latinoam. 1989;39:439–448. [PubMed] [Google Scholar]
- 35.Bon C. Multicomponent neurotoxic phospholipases A2. In: Kini RM, editor. Venom Phospholipase A2 Enzymes: Structure, Function and Mechanism. Wiley; Chichester: 1997. pp. 269–285. [Google Scholar]
- 36.Sampaio SC, Hyslop S, Fontes MR, Prado-Franceschi J, Zambelli VO, Magro AJ, Brigatte P, Gutierrez VP, Cury Y. Crotoxin: novel activities for a classic beta-neurotoxin. Toxicon. 2010;55:1045–1060. doi: 10.1016/j.toxicon.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 37.Chen YH, Wang YM, Hseu MJ, Tsai IH. Molecular evolution and structure-function relationships of crotoxin-like and asparagine-6-containing phospholipases A2 in pit viper venoms. Biochem J. 2004;381:25–34. doi: 10.1042/BJ20040125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanz L, Gibbs HL, Mackessy SP, Calvete JJ. Venom proteomes of closely related Sistrurus rattlesnakes with divergent diets. J Proteome Res. 2006;5:2098–2112. doi: 10.1021/pr0602500. [DOI] [PubMed] [Google Scholar]
- 39.Weinstein SA, Minton SA, Wilde CE. The distribution among ophidian venoms of a toxin isolated from the venom of the Mojave rattlesnake (Crotalus scutulatus scutulatus) Toxicon. 1985:825–844. doi: 10.1016/0041-0101(85)90014-5. [DOI] [PubMed] [Google Scholar]
- 40.Powell RL, Lieb CS, Rael ED. Identification of a neurotoxic venom component in the Tiger rattlesnake, Crotalus tigris. J Herpetol. 2004;38:149–152. [Google Scholar]
- 41.Hawgood BJ. Physiological and pharmacological effects of rattlesnake venoms. In: Tu AT, editor. Rattlesnake venoms Their actions and treatment. New York: Marcel Dekker; 1982. pp. 121–162. [Google Scholar]
- 42.Castilonia RR, Pattabhiraman TR, Russell FE, González H. Electrophysiological studies on aprotein fraction (K′) from Mojave rattlesnake (Crotalus scutulatus scutulatus) venom. Toxicon. 1981;9:473–479. doi: 10.1016/0041-0101(81)90005-2. [DOI] [PubMed] [Google Scholar]
- 43.Ho CL, Lee CY. Presynaptic actions of Mojave toxin isolated from Mojave rattlesnake (Crotalus scutulatus) venom. Toxicon. 1981;19:889–892. doi: 10.1016/0041-0101(81)90086-6. [DOI] [PubMed] [Google Scholar]
- 44.Gopalakrishinakone P, Hawgood BJ, Holbrooke SE, Marsh NA, Santana De Sa S, Tu AT. Sites of action of Mojave toxin isolated from the venom of the Mojave rattlesnake. Br J Pharmacol. 1980;69:421–431. doi: 10.1111/j.1476-5381.1980.tb07031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Warrell DA. Snakebites in Central and South America: Epidemiology, Clinical Features, and Clinical Management. In: Campbell JA, Lamar WW, editors. The Venomous Reptiles of the Western Hemisphere. Comstock Publishing Associates; Ithaca and London: 2004. pp. 709–761. [Google Scholar]
- 46.Fan HW, Cardoso JL. Clinical toxicology of snake bites South America. In: Meier J, White J, editors. Handbook of Clinical Toxicology of Animal Venoms and Poisons. CRC Press; Florida: 1995. pp. 667–688. [Google Scholar]
- 47.Calvete JJ, Sanz L, Cid P, de la Torre P, Flores-Díaz M, Dos Santos MC, Borges A, Bremo A, Angulo Y, Lomonte B, Alape-Girón A, Gutiérrez JM. Snake venomics of the Central American rattlesnake Crotalus simus and the South American Crotalus durissus complex points to neurotoxicity as an adaptive paedomorphic trend along Crotalus dispersal in South America. J Proteome Res. 2010;9:528–544. doi: 10.1021/pr9008749. [DOI] [PubMed] [Google Scholar]
- 48.Boldrini-França J, Corrêa-Netto C, Silva MM, Rodrigues RS, De La Torre P, Pérez A, Soares AM, Zingali RB, Nogueira RA, Rodrigues VM, Sanz L, Calvete JJ. Snake venomics and antivenomics of Crotalus durissus subspecies from Brazil: assessment of geographic variation and its implication on snakebite management. J Proteomics. 2010;73:1758–1776. doi: 10.1016/j.jprot.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 49.Place AJ, Abramson CI. A quantitative analysis of the ancestral area of rattlesnakes. J Herpetol. 2004;38:152–156. [Google Scholar]
- 50.Werman SD. Phylogeny and the Evolution of β-Neurotoxic Phospholipases A2 (PLA2) in the Venoms of Rattlesnakes, Crotalus and Sistrurus (Serpentes: Viperidae) In: Hayes WK, Beaman KR, Cardwell MD, Bush SP, editors. The Biology of Rattlesnakes. Loma Linda University Press; Loma Linda, California: 2008. pp. 511–536. [Google Scholar]
- 51.Powell RL, Lieb CS, Rael ED. Geographic distribution of Mojave toxin and Mojave toxin subunits among selected Crotalus species. In: Hayes WK, Beaman KR, Cardwell MD, Bush SP, editors. The Biology of Rattlesnakes. Loma Linda University Press; Loma Linda, California: 2008. pp. 537–550. [Google Scholar]
- 52.Powell RL, Lieb CS. Perspective on Venom Evolution in Crotalus. In: Hayes WK, Beaman KR, Cardwell MD, Bush SP, editors. The Biology of Rattlesnakes. Loma Linda University Press; Loma Linda, California: 2008. pp. 551–556. [Google Scholar]
- 53.Douglas ME, Douglas MR, Schuett GW, Porras LW. Evolution of rattlesnakes (Viperidae; Crotalus) in the warm deserts of western North America shaped by Neogene vicariance and Quaternary climate change. Mol Ecol. 2006;15:3353–3374. doi: 10.1111/j.1365-294X.2006.03007.x. [DOI] [PubMed] [Google Scholar]
- 54.Wüster W, Ferguson JE, Quijada-Mascareñas JA, Pook CE, Salomão MG, Thorpe RS. Tracing an invasion: landbridges, refugia and the phylogeography of the Neotropical rattlesnake (Serpentes: Viperidae:Crotalus durissus. Mol Ecol. 2005;14:1095–1108. doi: 10.1111/j.1365-294X.2005.02471.x. [DOI] [PubMed] [Google Scholar]
- 55.Mackessy SP, Williams K, Ashton KG. Ontogenetic variation in venom composition and diet of Crotalus oreganus concolor: A case of venom paedomorphosis? Copeia. 2003;2003:769–782. [Google Scholar]
- 56.Mackessy SP. Venom ontogeny in the Pacific Rattlesnakes Crotalus viridis helleri and C. v. oreganus. Copeia. 1988;1988:92–101. [Google Scholar]
- 57.Wooldridge BJ, Pineda G, Banuelas-Ornelas JJ, Dagda RK, Gasanov SE, Rael ED, Lieb CS. Mojave rattlesnakes (Crotalus scutulatus scutulatus) lacking the acidic subunit DNA sequence lack Mojave toxin in their venom. Comp Biochem Physiol B. 2001;130:169–179. doi: 10.1016/s1096-4959(01)00422-5. [DOI] [PubMed] [Google Scholar]
- 58.Gutiérrez JM, Dos Santos MC, Furtado MF, Rojas G. Biochemical and pharmacological similarities between the venoms of newborn Crotalus durissus durissus and adult Crotalus durissus terrificus rattlesnakes. Toxicon. 1991;29:1273–1277. doi: 10.1016/0041-0101(91)90201-2. [DOI] [PubMed] [Google Scholar]
- 59.Calvete JJ, Sanz L, Angulo Y, Lomonte B, Gutiérrez JM. Venoms, venomics, antivenomics. FEBS Lett. 2009;583:1736–1743. doi: 10.1016/j.febslet.2009.03.029. [DOI] [PubMed] [Google Scholar]
- 60.Calvete JJ. Antivenomics and venom phenotyping: A marriage of convenience to address the performance and range of clinical use of antivenoms. Toxicon. 2010;56:1284–1291. doi: 10.1016/j.toxicon.2009.12.015. [DOI] [PubMed] [Google Scholar]
- 61.Faure G, Xu H, Saul FA. Crystal structure of crotoxin reveals key residues involved in the stability and toxicity of this potent heterodimeric β-neurotoxin. J Mol Biol. 2011;412:176–191. doi: 10.1016/j.jmb.2011.07.027. [DOI] [PubMed] [Google Scholar]
- 62.Glenn JL, Straight RC, Wolfe MC, Hardy DL. Geographical variation in Crotalus scutulatus scutulatus (Mojave rattlesnake) venom properties. Toxicon. 1983;21:119–130. doi: 10.1016/0041-0101(83)90055-7. [DOI] [PubMed] [Google Scholar]
- 63.Glenn JL, Straight RC, Wolt TB. Regional variation in the presence of canebrake toxin in Crotalus horridus venom. Comp Biochem Physiol Pharmacol Toxicol Endocrinol. 1994;107:337–346. doi: 10.1016/1367-8280(94)90059-0. [DOI] [PubMed] [Google Scholar]
- 64.Santoro ML, Sousa-e-Silva MCC, Gonçalves LRC, Almeida-Santos SM, Cardoso DF, Laporta-Ferreira IL, Saiki M, Peres CA, Sano-Martins IS. Comparison of the biological activities in venoms from three subspecies of South-American rattlesnake Crotalus durissus terrificus, C. durissus cascavella and C. durissus collilineatus. Comp Biochem Physiol. 1999;122C:61–73. doi: 10.1016/s0742-8413(98)10079-8. [DOI] [PubMed] [Google Scholar]
- 65.Castoe TA, Parkinson CL. Bayesian mixed models and the phylogeny of pitvipers (Viperidae: Serpentes) Mol Phylogenet Evol. 2006;39:91–110. doi: 10.1016/j.ympev.2005.12.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.